Abstract
The arylacetonitrilase from Pseudomonas fluorescens EBC191 differs from previously studied arylacetonitrilases by its low enantiospecificity during the turnover of mandelonitrile and by the large amounts of amides that are formed in the course of this reaction. In the sequence of the nitrilase from P. fluorescens, a cysteine residue (Cys163) is present in direct neighborhood (toward the amino terminus) to the catalytic active cysteine residue, which is rather unique among bacterial nitrilases. Therefore, this cysteine residue was exchanged in the nitrilase from P. fluorescens EBC191 for various amino acid residues which are present in other nitrilases at the homologous position. The influence of these mutations on the reaction specificity and enantiospecificity was analyzed with (R,S)-mandelonitrile and (R,S)-2-phenylpropionitrile as substrates. The mutants obtained demonstrated significant differences in their amide-forming capacities. The exchange of Cys163 for asparagine or glutamine residues resulted in significantly increased amounts of amides formed. In contrast, a substitution for alanine or serine residues decreased the amounts of amides formed. The newly discovered mutation was combined with previously identified mutations which also resulted in increased amide formation. Thus, variants which possessed in addition to the mutation Cys163Asn also a deletion at the C terminus of the enzyme and/or the modification Ala165Arg were constructed. These constructs demonstrated increased amide formation capacity in comparison to the mutants carrying only single mutations. The recombinant plasmids that encoded enzyme variants which formed large amounts of mandeloamide or that formed almost stoichiometric amounts of mandelic acid from mandelonitrile were used to transform Escherichia coli strains that expressed a plant-derived (S)-hydroxynitrile lyase. The whole-cell biocatalysts obtained in this way converted benzaldehyde plus cyanide either to (S)-mandeloamide or (S)-mandelic acid with high yields and enantiopurities.
Nitrilases (EC 3.5.5.1) are hydrolytic enzymes found in many bacteria, fungi, and plants which convert nitriles to the corresponding carboxylic acids and ammonia. They are members of the CN hydrolase (or nitrilase) superfamily of enzymes, which also encompasses other enzymes which attack C-N bonds, such as aliphatic amidases, carbamoylases, and N-acyltransferases (1). Nitrilases possess a catalytic triad which is composed of a cysteine, a glutamate, and a lysine residue and form during the catalytic cycle a covalent adduct between the cysteine residue and the carbon atom of the nitrile group (11, 12, 29). Nitriles are important intermediates in chemical industry, and several processes which utilize the chemo-, regio-, or enantioselectivity of nitrilases for the production of commercially interesting products have been investigated (13, 16, 17, 18, 22, 26, 27, 33, 34). There is also growing biotechnological interest in nitrilases because they form (as other members of the so-called nitrilase superfamily) spiral quaternary structures which can be studied by electron microscopy and which might be useful as templates in nanotechnology (30, 31).
We are currently investigating a nitrilase from Pseudomonas fluorescens EBC191 which converts various substituted phenylacetonitriles [e.g., 2-phenylpropionitrile (2-PPN), mandelonitrile (2-hydroxyphenylacetonitrile), or phenylglycinonitrile (2-aminophenylacetonitrile)] and also aliphatic 2-acetoxynitriles with moderate enantioselectivities into the corresponding α-substituted carboxylic acids. This enzyme forms with certain nitriles also significant amounts of the corresponding amides as side products (3, 5, 8, 15, 19, 24). The enzyme has recently been studied intensively in order to analyze the molecular basis for the substrate specificity, reaction specificity, and enantiospecificity of nitrilases (9, 10). In the course of these investigations, the effects of various carboxy-terminal mutations and mutations in close proximity to the catalytic active cysteine residue were analyzed. These experiments demonstrated that deletions of 47 to 67 amino acids (aa) from the carboxy terminus of the nitrilase resulted in variant forms that demonstrated increased amide formation and an increased formation of the (R)-acids (9). In addition, it was demonstrated that the size of the amino acid residue in direct proximity to the catalytic active cysteine residue (toward the C terminus) is determinative of the enantioselectivity of acid formation. Thus, it was found that only enzyme variants with large amino acid residues at this position showed a high degree of enantioselectivity for the formation of (R)-mandelic acid from racemic mandelonitrile (10). In the present study, we investigated a set of enzyme variants that carried mutations located in the amino-terminal part of the enzyme (in relation to the catalytic active cysteine residue). Thus, several mutations that resulted in changes in the enantioselectivity of the reactions and increased formation of amides were identified.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Plasmid pIK9 was used for the construction of all enzyme variants. The plasmid encodes a His-tagged variant of the nitrilase from P. fluorescens EBC191 (8). The (S)-hydroxynitrile lyase from cassava (Manihot esculenta HNL [MeHNL]) was expressed from plasmid pJOE5361.1, which is compatible with pIK9 and contained the MeHNL gene under the control of the same rhamnose-inducible promoter as that used in pIK9 (28). The plasmid preparations and cloning experiments were carried out with Escherichia coli JM109 (32). Escherichia coli strains were usually grown at 30°C in 2×YT liquid medium or on LB agar plates (25) supplemented with 100 μg/ml ampicillin (plus 20 μg/ml chloramphenicol, if appropriate).
Construction of nitrilase mutants.
The amino acid exchanges at position 163 of the nitrilase from P. fluorescens EBC191 were introduced using splicing by overlap extension (SOE) as previously described (6, 8, 10). The “general flanking 5′ and 3′ primers” S3182_2 and S2949_2-rev were used in combination with the “specific primers” C163A-fwd, C163A-rev, C163S-fwd, C163S-rev, C163Q-fwd, C163Q-rev, C163N-fwd, and C163N-rev. The same strategy was also used for the introduction of site-specific mutations at positions 110, 111, 113, 114, 116, and 117 of the nitrilase. (For the sequences of the oligonucleotides used, see the supplemental material).
The enzyme variants with two or more different mutations were produced from pCK105 (encoding a C-terminally deleted nitrilase variant) (8) by using the QuikChange site-directed mutagenesis kit from Stratagene. The oligonucleotides (Eurofins MWG Operon, Ebersberg, Germany) for the mutations were deduced by using the QuikChange primer design program from Stratagene. Cells of E. coli XL-1 Blue (Stratagene) were transformed with the mutated plasmids and positive clones selected on LB agar plates with ampicillin (100 μg/ml). The generated mutants were verified by sequencing of the mutated genes.
DNA preparation, DNA manipulation, transformation, PCR, DNA sequencing, database searches and sequence alignments were performed as described previously (9).
Construction of recombinant E. coli strains that simultaneously express nitrilase variants and the (S)-oxynitrilase from cassava.
E. coli JM109(pJOE5361.1) which encoded the (S)-oxynitrilase from cassava was transformed with mutated forms of plasmid pIK9 which coded for the relevant nitrilase variants (28). The E. coli strains carrying the two types of plasmids were selected on LB-agar plus 100 μg/ml ampicillin and 20 μg/ml chloramphenicol.
Enzyme assays.
The nitrilase activities in the recombinant strains were induced by the addition of rhamnose (0.2% [wt/vol]) to the dYT growth medium (8). The cells were harvested by centrifugation in the late exponential or early stationary growth phase and subsequently suspended in Tris-HCl buffer (pH 7.5) or Na-citrate buffer (pH 5) before the substrates were added.
The nitrilase activity of resting cells was routinely determined in reaction mixtures (1 ml each) containing 50 μmol Tris-HCl buffer (pH 7.5) or 100 μmol Na-citrate buffer (pH 5), 10 μmol nitrile and an appropriate amount of cells. All nitrile stock solutions (200 mM each) were prepared in methanol. The reaction mixtures were incubated at 30°C in a thermo mixer at 1,100 rpm. After different time intervals, samples (90 μl each) were taken and the reactions were stopped by the addition of 1 M HCl (10 μl). The samples were centrifuged at 14,000 × g for 10 min and the supernatants were analyzed using high-pressure liquid chromatography (HPLC).
SDS-PAGE and protein determination were performed as previously described (9).
Analytical methods.
Mandelonitrile and 2-phenylpropionitrile and their corresponding amides and acids were analyzed by HPLC. The achiral analysis was performed using Lichrospher RP8 or Lichrospher RP18 columns (Trentec Analysentechnik, Rutesheim, Germany) and methanol (40 to 50% [vol/vol]) and H3PO4 (0.3% [vol/vol]) in H2O as mobile phases (9).
The enantiomers of 2-phenylpropionic acid (2-PPA), mandelic acid, and the corresponding amides were separated on a Chiral-HSA column (ChromTech AB, Hägersten, Sweden). The mobile phases consisted of sodium phosphate buffer (10 mM; pH 6.0) plus 0.5% (vol/vol) 2-propanol or sodium phosphate buffer (100 mM; pH 7.0) containing 4.5% (vol/vol) acetonitrile.
Chemicals.
The sources of all chemicals have been described before (9, 28).
RESULTS
Construction of mutant enzymes carrying different amino acid exchanges at position 163.
The previously performed sequence comparisons among different nitrilases (9) demonstrated pronounced differences in the amino acid residue which is located directly adjacent (toward the N terminus) to the indispensable catalytic active cysteine residue. Cysteine residues were found at this position in P. fluorescens EBC191 and Alcaligenes faecalis JM3; asparagine residues in Rhodococcus rhodochrous NCIMB11216, R. rhodochrous K22, and Klebsiella ozaenae; glutamine residues in Bacillus sp. strain OxB-1 and Comamonas testosteroni; and an alanine residue in Synechocystis sp. strain PCC6803 (9). Therefore, it was decided that this residue in the nitrilase from P. fluorescens EBC191 would be modified, because it was expected that modifications of this residue could have a pronounced influence on the substrate specificity of the enzyme. The Cys163Ser, Cys163Ala, Cys163Gln, and Cys163Asn mutants were constructed. The initial analysis of the mutants obtained from the attempt to generate the Cys163Asn variant demonstrated two different ratios of acid/amide formed by individual clones. Therefore, the encoding genes from both types were sequenced, and a secondary mutation (Cys163Asn-Thr110Ile) was detected in one of the clones. Therefore, this mutant was also included in the following analysis. The recombinant clones were cultivated, the nitrilases induced by the addition of rhamnose, and resting cells analyzed for the conversion of the model substrates 2-phenylpropionitrile (2-PPN) and mandelonitrile (MN) with respect to the enzyme activities and enantioselectivities of the reactions.
Conversion of 2-phenylpropionitrile and mandelonitrile by the mutants.
The Cys163Ser and Cys163Ala variants demonstrated no significant differences in the enzyme activities compared to the wild type (wt), but it was observed that the Cys163Ala enzyme variant showed an extremely low degree of enantioselectivity for the formation of (R)-mandelic acid (Table 1; see also Fig. S1 and S2 in the supplemental material). In contrast, both mutations resulted in significant decreases in the relative amounts of mandeloamide formed from mandelonitrile (Table 1; see also Fig. S2 in the supplemental material). The importance of this amino acid residue for amide formation was confirmed by the analysis of the Cys163Gln, Cys163Asn, and Cys163Asn-Thr110Ile mutants. These enzyme variants formed up to 59% of mandeloamide from the total amount of mandelonitrile converted (Table 1). The increase in the amide-forming ability of this group of mutants became especially evident when the formation of 2-phenylpropionamide (2-PPAA) from 2-PPN was analyzed. Thus, the Cys163Asn-Thr110Ile enzyme variant formed about 100-fold more 2-PPAA (in relation to the total amount of 2-PPN converted) than the wild type, although the relative activity of this mutant for the conversion of 2-PPN to 2-phenylpropionic acid (2-PPA) was only 4% of that observed for the wild type. Furthermore, it was found that the mutants carrying an asparagine or glutamine at the relevant position in the enzyme also demonstrated an increased enantioselectivity for the formation of (R)-mandelic acid.
TABLE 1.
Comparison of different variants of the nitrilase from P. fluorescens EBC191 carrying different amino acid exchanges in different positions
| Characteristice | Value for indicated variant |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wt | C163A | C163S | C163Q | C163N | T110I- C163N | T110I | T110F | L111F | L113F | A116F | A116C | Q115F | I117F | |
| Formation of 2-PPA (%)a | 100 | 106 | 30 | 93 | 35 | 4 | 18 | 9 | 10 | 56 | 3 | 86 | 26 | 26 |
| ee 2-PPA (%)b (preferentially formed enantiomer) | 65 (S) | 66 (S) | 51 (S) | 75 (S) | 63 (S) | 33 (S) | 18 (S) | 13 (S) | 62 (S) | 76 (S) | 66 (S) | 66 (S) | 68 (S) | 66 (S) |
| % 2-PPAAc | 0.2 | 0.2 | <0.1 | 3.6 | 3.6 | 20.3 | 6 | 5.8 | 0.1 | 0.2 | 0.2 | 0.5 | 0 | 0.5 |
| ee 2-PPAA (%)b (preferentially formed enantiomer) | 7 (R) | 88 (R) | 69 (R) | 3 (R) | 38 (R) | 78 (R) | 100 (R) | 82 (R) | 11 (R) | 65 (R) | 41 (R) | 36 (R) | 42 (R) | |
| Relative activity of MN/2-PPN | 7.3 | 4.2 | 11.0 | 1.1 | 4.0 | 5.5 | 12 | 6.8 | 8.3 | 6.7 | 8.3 | 5.8 | 13 | 6.2 |
| Formation of MA (%)a | 100 | 61 | 46 | 14 | 19 | 3 | 6 | 8 | 11 | 51 | 4 | 68 | 9 | 22 |
| ee MA (%)b (preferentially formed enantiomer) | 31 (R) | 11 (R) | 32 (R) | 66 (R) | 59 (R) | 86 (R) | 73 (R) | 70 (R) | 26 (R) | 10 (R) | 28 (R) | 33 (R) | 4 (S) | 33 (R) |
| % MAAd | 16 | 3 | 2 | 51 | 43 | 59 | 45 | 43 | 12 | 9 | 13 | 19 | 3 | 19 |
| ee MAA (%)b (preferentially formed enantiomer) | 84 (S) | 78 (S) | 68 (S) | 76 (S) | 62 (S) | 42 (S) | 48 (S) | 68 (S) | 67 (S) | 92 (S) | 77 (S) | 77 (S) | 15 (S) | 79 (S) |
In comparison to a resting cell suspension of E. coli JM109(pIK9) induced under the same conditions and calculated for cell suspensions with the same optical densities (activity = 100%).
Compared at about 30% substrate conversion.
2-PPAA/(2-PPA + 2-PPAA).
MAA/(MA + MAA).
ee, enantiomeric excess; 2-PPA, 2-phenylpropionic acid; 2-PPAA, 2-phenylpropionamide; 2-PPN, 2-phenylpropionitrile; MN, mandelonitrile; MA, mandelic acid; MAA, mandeloamide.
Analysis of the function of individual amino acid residues within the proposed β-fold S4.
It was shown in the previous paragraph that the additional introduction of the Thr110Ile mutation into the Cys163Asn enzyme variant resulted in an almost 6-fold increase in the relative amount of 2-PPAA formed from 2-PPN. The previously performed sequence alignments and comparisons to related proteins with solved structures suggested that this amino acid is part of (or at least directly adjacent to) a β-fold structure labeled S4 (9). It has been suggested that this region forms in the nitrilase of Rhodococcus rhodochrous J1 a loop structure, which takes part in the formation of the “C-surface,” which is essential for the formation of the spiral quaternary structure of nitrilases (30, 31). It was therefore decided that individual amino acids within the relevant stretch of the amino acid sequence would be mutated and that a phenylalanine residue would be introduced at each available position in order to analyze the influence of a bulky residue on the substrate specificity and enantioselectivity. Thus, the Gly109Phe, Thr110Phe, Leu111Phe, Leu113Phe, Ala114Phe, Gln115Phe, Ala116Phe, and Ile117Phe mutants were generated. Furthermore, the Ala116Cys mutant was also generated, because this substitution was the only significant difference in the relevant sequence space between the sequences of the nitrilase from P. fluorescens and that of the nitrilase from Alcaligenes faecalis [which is highly enantiospecific for the formation of (R)-mandelic acid] (35, 36). Initial experiments with the Gly109Phe and Ala114Phe variants showed that these mutants were inactive, and these mutants were therefore not further analyzed. It was demonstrated by SDS-gel electrophoresis that the Thr110Phe, Leu113Phe, and Ala116Cys nitrilase variants were expressed with intensities similar to those of the wild-type enzyme. In contrast, the Leu111Phe, Ala116Phe, and Ile117Phe enzyme variants were expressed with only about half this intensity.
The conversion of (R,S)-mandelonitrile and (R,S)-2-PPN by these mutant variants clearly demonstrated that only the mutation at position 110 resulted in the formation of increased amounts of the respective amides (Table 1; see also Fig. S3 and S4 in the supplemental material). In addition, the Thr110Phe mutant showed a significantly reduced enantioselectivity for the formation of (S)-PPA and thus confirmed the observations regarding the effects of mutations at position 110 that had been made during the comparison of the Cys163Asn and Cys163Asn-Thr110Ile mutants.
The analysis of the reactions with mandelonitrile confirmed for most mutants the trends observed during the turnover of 2-PPN. The only significant difference was observed for the Leu113Phe mutant. This enzyme demonstrated a significantly decreased enantioselectivity for the formation of (R)-mandelic acid in combination with an increased enantioselectivity for the formation of (S)-mandeloamide.
Influence of a free amide group in direct neighborhood to the catalytic active cysteine residue on amide formation.
The results presented above demonstrated a pronounced increase in amide formation when an asparagine or glutamine residue was introduced at position 163. Similarly, it was previously found that among various mutants (the Ala165Gly, Ala165Phe, Ala165Tyr, Ala165Trp, Ala165His, Ala165Glu, and Ala165Arg mutants) which had been generated at position 165 (thus on the “other side” of the catalytic active cysteine residue), only the Ala165Arg mutant demonstrated an increase in amide formation (10). This suggested that a free amino group in close proximity to the catalytic center (which could somehow “mimic” the amide function of the enzyme-bound reaction intermediate after the addition of the first water molecule) could result in an increased level of amide formation. Therefore, the Cys163Glu and Cys163Asp mutants (as “negative controls”) and the Ala165Asn and Ala165Gln mutants were generated, and it was found that the mutants indeed showed the expected phenotypes. Thus, the Cys163Glu and Cys163Asp variants formed from mandelonitrile (after the complete conversion of the nitrile) only 1 and 9% amide, respectively. In contrast, the Ala165Asn and Ala165Gln mutants formed about twice as much mandeloamide as the wild-type enzyme (Table 2).
TABLE 2.
Comparison of different mutant variants of the nitrilase from P. fluorescens EBC191 with increased amide-forming capacities
| Characteristic | Value for indicated variant |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| wt | C163D | C163E | A165Q | A165N | Nit(DelC-60) (pCK105) | Nit(DelC-60)- C163N | Nit(DelC-60)- C163N- A165R | Nit(DelC-60)- C163N- A165R-T110I | |
| Formation of 2-PPA (%)a | 100 | 44 | 88 | 2 | 62 | 10 | 14 | 6 | 4 |
| ee 2-PPA (%)b (preferentially formed enantiomer) | 63 (S) | 3 (S) | 67 (S) | 13 (S) | 12 (R) | 23 (S) | 34 (S) | 57 (R) | 53 (R) |
| % 2-PPAAc | ≥0.2 | 0 | 0 | 0 | 0 | 8 | 25 | 39 | 43 |
| ee 2-PPAA (%)b (preferentially formed enantiomer) | 7 (R) | 93 (R) | 85 (R) | 96 (R) | 99 (R) | ||||
| Formation of MA (%)a | 100 | 19 | 31 | 18 | 4 | 7 | 3 | 5 | 4 |
| ee MA (%)b (preferentially formed enantiomer) | 24 (R) | 47 (R) | 2 (R) | 70 (R) | 70 (R) | 80 (R) | 79 (R) | 100 (R) | 75 (R) |
| % MAAd | 16 | 1 | 9 | 31 | 38 | 48 | 70 | 70 | 72 |
| ee MAA (%)b (preferentially formed enantiomer) | 63 (S) | 11 (R) | 52 (S) | 1 (S) | 58 (S) | 48 (S) | 34 (S) | 2 (R) | 3 (R) |
In comparison to a resting cell suspension of E. coli JM109(pIK9) induced under the same conditions and calculated for cell suspensions with the same optical densities (activity = 100%).
Compared at about 30% substrate conversion. ee, enantiomeric excess.
2-PPAA/(2-PPA + 2-PPAA).
MAA/(MA + MAA).
Combination of different mutations in order to increase the amide formation capacity.
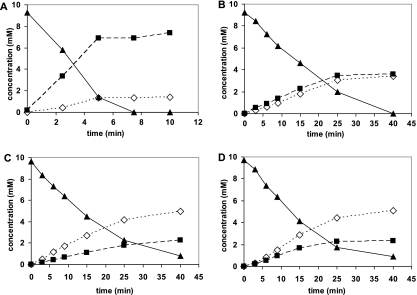
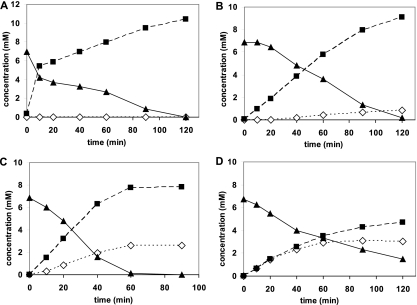
It was previously found that deletions of 47 to 67 aa from the C terminus and also the replacement of the alanine residue at position 165 with an arginine residue resulted in increased amide formation (9, 10). In addition, it was shown in the present study that the replacement of the cysteine residue at position 163 with an asparagine residue caused the same effect. It was therefore decided that the effects of combinations of these mutations would be tested. Therefore, the Nit(DelC-60)-Cys163Asn and Nit(DelC-60)-Cys163Asn-Ala165Arg mutants were constructed and the variants applied for the conversion of (R,S)-mandelonitrile and (R,S)-2-PPN. These experiments clearly demonstrated that the combination of the independently obtained mutations resulted in enzyme variants with further increased amide-forming capacities, but some differences for the conversion of both substrates were also observed (Fig. 1 and 2). Thus, it was found with (R,S)-mandelonitrile as a substrate that the combination of the C-terminal deletion with the secondary mutation Cys163Asn resulted in an approximately 1.5-fold increase in the relative amount of mandeloamide formed (Fig. 1B and C). This mutant formed more than twice as much mandeloamide than mandelic acid from (R,S)-mandelonitrile and thus demonstrated with this substrate the highest level of nitrile hydratase activity of all variants of the nitrilase from P. fluorescens ever observed. With (R,S)-mandelonitrile as a substrate, the introduction of the tertiary mutation Ala165Arg did not further enhance the relative amount of amide formed (Fig. 1D). In contrast, this triple mutant demonstrated a significantly increased formation of amide from 2-PPN compared to the double mutant and converted this substrate to almost equimolar amounts of 2-PPA and 2-PPAA (Fig. 2D). Thus, this mutant proved an almost 200-fold increase in relative amide formation compared to the wild type. Subsequently, a “4-fold” mutant which additionally also contained the amino acid exchange Thr110Ile was constructed, but this mutation resulted in no significant further increase in amide formation (Table 2).
FIG. 1.
Conversion of mandelonitrile by the wild-type nitrilase from P. fluorescens EBC191 or different nitrilase variants expressed in E. coli JM109. The E. coli strains were grown in LB medium with ampicillin and the nitrilase induced by the addition of l-rhamnose (see Materials and Methods). The cells were harvested by centrifugation and resuspended to different optical densities (ODs) in 950 μl Na-citrate buffer (100 mM, pH 5) in order to obtain similar substrate turnover rates. The cell suspensions were incubated for 10 min at 30°C in a thermo shaker. The reactions were started by the addition of 50 μl of a stock solution of mandelonitrile (200 mM in methanol). Aliquots (90 μl each) were taken at different time intervals, and the reactions were terminated by the addition of 10 μl of 1 M HCl. The concentrations of mandelonitrile (▴), mandelic acid (▪), and mandeloamide (⋄) in the supernatants were determined by reversed-phase HPLC. (A) E. coli JM109(pIK9) (wild-type nitrilase [OD600 = 0.7 in the reaction mixture]); (B) E. coli JM109(pCK105) (C-terminally deleted nitrilase variant [OD600 = 1.2]); (C) E. coli JM109(pCK105/Cys163Asn) (OD600 = 1.1); (D) E. coli JM109(pCK105/Cys163Asn-Ala165Arg) (OD600 = 1.3).
FIG. 2.
Conversion of (R,S)-2-phenylpropionitrile by the nitrilase from P. fluorescens EBC191 and different nitrilase variants. Cells were cultivated as described in the legend to Fig. 1. The resting cell experiments were performed with 50 mM Tris-HCl buffer (pH 7.5) as described in the legend to Fig. S1 in the supplemental material. The concentrations of 2-phenylpropionitrile (▴), 2-phenylpropionic acid (▪), and 2-phenylpropionamide (⋄) in the supernatants were determined by reversed-phase HPLC. (A) E. coli JM109(pIK9) (OD600 = 5.5 in the reaction mixture); (B) E. coli JM109(pCK105) (OD600 = 21); (C) E. coli JM109(pCK105/Cys163Asn) (OD600 = 22); (D) E. coli JM109(pCK105/Cys163Asn-Ala165Arg) (OD600 = 22).
The chiral analysis of the produced acids and amides demonstrated that the Nit(DelC-60) mutant and all further developed mutants combined the increased amide formation with an increased tendency to form the (R)-enantiomers of the acids and amides (Table 2).
Application of the nitrilase mutants for the construction of “bienzymatic catalysts” which form either (S)-mandelic acid or (S)-mandeloamide.
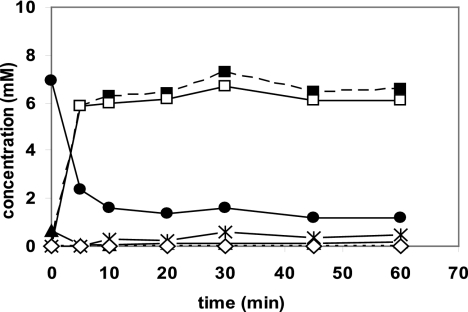
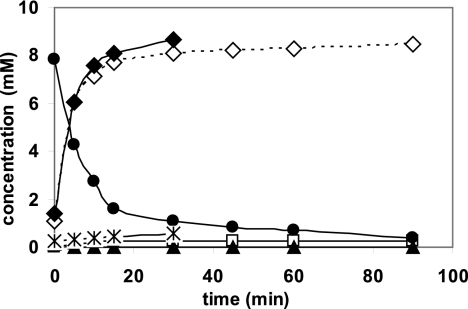
It was previously demonstrated that the simultaneous expression of the nitrilase from P. fluorescens together with the (S)-specific oxynitrilase from cassava (Manihot esculenta) in a single recombinant E. coli strain allowed the construction of efficient “bienzymatic whole-cell catalysts” for the synthesis of (S)-mandelic acid and (S)-mandeloamide from benzaldehyde and cyanide. In these experiments, either the wild-type nitrilase or a C-terminally deleted nitrilase variant have been used. These constructions resulted in biocatalysts that formed (S)-mandelic acid and (S)-mandeloamide at a ratio of either about 8:5 (using the wild-type nitrilase) or 1:9 (using the C-terminally deleted mutant) (28). These experiments demonstrated the principal feasibility of this concept but still resulted in the synthesis of mixtures of (S)-mandelic acid and (S)-mandeloamide. Therefore, the new nitrilase variants were tested in this system, and “double clones” which combined the heterologously expressed (S)-oxynitrilase either with the Cys163Ala nitrilase mutant (encoded by plasmid pCK201), which formed small amounts of amide and showed only a very low level of enantioselectivity during the conversion of mandelonitrile, or with the Nit(DelC-60)-Cys163Asn nitrilase variant (encoded by plasmid pOS105), which produced about 70% of amide from (R,S)-mandelonitrile, were constructed. Subsequently, resting cells of the “double clones” were incubated with benzaldehyde and cyanide, and it was demonstrated that the application of the nitrilase variants which were generated in the course of the present study indeed allowed the construction of “double clones” which converted benzaldehyde and cyanide to almost stoichiometric amounts of (S)-mandelic acid (Fig. 3) or (S)-mandeloamide (Fig. 4).
FIG. 3.
Conversion of benzaldehyde and KCN by resting cells of E. coli JM109(pCK201)(pJOE5361.1). The cells were grown in LB medium supplemented with the relevant antibiotics and induced with rhamnose as described in Materials and Methods. The reaction mixtures contained in a total volume of 2 ml 100 mM Na-citrate buffer (pH 5.0), 10 mM benzaldehyde, 10 mM KCN, and resting cells (OD546 = 20). Aliquots (200 μl each) were taken at the indicated time intervals and the reactions terminated by the addition of 20 μl 1 M HCl. The cells were removed by centrifugation in an Eppendorf centrifuge (5 min at 14,000 × g), and the concentrations of benzaldehyde (•), (R,S)-mandelonitrile (▴), (R,S)-mandeloamide (⋄), and (R,S)-mandelic acid (▪) in the supernatants were analyzed by reversed-phase HPLC. The ratio of (S)-mandelic acid (□) and (R)-mandelic acid ( ) in the individual samples was analyzed after reneutralization by chiral HPLC.
) in the individual samples was analyzed after reneutralization by chiral HPLC.
FIG. 4.
Conversion of benzaldehyde and KCN by resting cells of E. coli JM109(pOS105)(pJOE5361.1). The cells were cultivated as described in the legend to Fig. 1 and resuspended to an optical density (OD600) of 22. The concentrations of benzaldehyde (•), (R,S)-mandelonitrile (▴), (R,S)-mandeloamide (⋄), (R,S)-mandelic acid (□), (S)-mandeloamide (♦), and (R)-mandeloamide ( ) were determined as described in the legend to Fig. 3.
) were determined as described in the legend to Fig. 3.
DISCUSSION
The conversion of nitriles to amides is in nature generally catalyzed by nitrile hydratases, which are noncorrinoid iron- or cobalt-containing lyases which do not share any homology to nitrilases with respect to their reaction mechanism, enzyme structure, and amino acid sequences (13, 14). Nevertheless, there is evidence that for certain amide forming reactions, nature uses enzymes, which are closely related to nitrilases and which belong evolutionarily to the same subgroup within the nitrilase superfamily as true nitrilases. The best-characterized examples for this type of reactions are the cyanide hydratases from fungal species such as Fusarium oxysporum and Gloeoecercospora Y, which convert cyanide to formamide (20). In addition, it has recently been demonstrated that a certain group of plant-derived nitrilases (“NIT4”) preferentially catalyze the transformation of 4-cyanoalanine, which is a typical cyanide detoxification product of plants, to the amide asparagine and not to the expected product aspartate (7, 23). Thus, it is obvious that the enzymes of the nitrilase superfamily in principle possess the catalytic potential to hydrate nitriles to amides. In this context, it is an interesting observation that true nitrile hydratases are found almost exclusively in procaryotes and that only recently has there been a single report which suggested (from a purely genome sequence-based approach) that genes for a nitrile hydratase might also be present in a eukaryote (a protist) (4). In addition, it has been suggested that the nitrilase superfamily has evolved in eukaryotes and that members of this family have only subsequently been transferred by horizontal gene transfer to prokaryotes (21). Thus, it might be speculated that the absence of true nitrile hydratases in eukaryotes has been the driving force for eukaryotes to adapt members of the nitrilase family to the synthesis of amides.
It was demonstrated in this and previous publications that different types of mutations can result in the formation of increased amounts of amides by the nitrilase from P. fluorescens EBC191. It was previously shown that deletion mutants with 47 to 67 amino acids missing at the C terminus of the enzyme and also mutants which carried an exchange of the alanine residue C-terminally adjacent to the catalytic active cysteine residue (Ala165) for an arginine residue each formed at least twice as much mandeloamide from racemic mandelonitrile as the wild type (9, 10). In addition, it was shown in the present study that modifications at position 163 of the nitrilase also had an important influence on the amide-forming process. Thus, it was found that the replacement of the Cys163 residue with a glutamine residue resulted in the transformation of more than half of the converted mandelonitrile to mandeloamide. This is the largest amount of amide formation that has been found in any of the mutants generated from the nitrilase of P. fluorescens EBC191 by a single amino acid exchange yet. The observed increase in amide formation by the introduction of an asparagine or glutamine residue at this position correlates with the results of our previous study which had shown that from different mutations generated at amino acid position 165 (thus “on the other side” of the catalytic active cysteine residue), only the introduction of an arginine residue resulted in significantly increased amide formation rates (10). This might indicate that the presence of free NH2 groups in the amino acid side chains of the protein in close proximity to the catalytic center might favor the release of amides as reaction products. It could be further speculated that in this type of enzyme variants the protein-derived NH2 groups compete with the NH2 group present in the enzyme-bound tetrahedral reaction intermediate derived from the nitrile substrate for protonation, which is essential for the release of ammonia from the reaction intermediate and thus the formation of the acid as a product (3). Nevertheless, it is also evident that the presence of a glutamine or asparagine residue at the relevant position is not indispensable for efficient amide production, because the NIT4 nitrilases from plants (e.g., those from Arabidopsis thaliana or Nicotina tabacum) do not possess these amino acid residues at the homologous positions.
The complexity of the amide formation process is also stressed by the previously demonstrated dependence of amide formation from the chemical character of the substituents which are present on the alpha-carbon atom. Thus, it was demonstrated for a nitrilase (Nit1) from A. thaliana and also for the enzyme from P. fluorescens EBC191 that substituents with a strongly electronegative character (especially halogens) in the alpha position in relationship to the nitrile group result in extraordinarily large amounts of amides formed (2, 3).
The formation of amides is generally regarded as an unwanted side reaction when nitrilases are applied for biotransformation reactions, because this side reaction reduces the yield of the desired carboxylic acids and might also cause additional costs during downstream processing. In contrast, it could be demonstrated in the present work that also the amide-forming capacity of nitrilases can be a useful synthetic characteristic, if it is possible to largely suppress the acid formation at all and thus to use nitrilase variants as (preferentially enantioselective) nitrile hydratases. The progress that was achieved in the present study by demonstrating that the effect of independently generated mutations can be accumulated to a certain extent and thus allow previously unmatched amounts of amides to be formed from different substrates might offer the possibility of finally converting nitrilases into true nitrile hydratases which form stoichiometric amounts of amides from their substrates.
Supplementary Material
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brenner, C. 2002. Catalysis in the nitrilase superfamily. Curr. Opin. Struct. Biol. 12:775-782. [DOI] [PubMed] [Google Scholar]
- 2.Effenberger, F., and S. Osswald. 2001. Enantioselective hydrolysis of (RS)-2-fluoroacetonitriles using nitrilase from Arabidopsis thaliana. Tetrahedron Asymmetry 12:279-285. [Google Scholar]
- 3.Fernandes, B. C. M., C. Mateo, C. Kiziak, J. Wacker, A. Chmura, F. van Rantwijk, A. Stolz, and R. A. Sheldon. 2006. Nitrile hydratase activity of a recombinant nitrilase. Adv. Synth. Catal. 348:2597-2603. [Google Scholar]
- 4.Foerstner, K. U., T. Doerks, J. Muller, J. Raes, and P. Bork. 2008. A nitrile hydratase in the eukaryote Monosiga brevicollis. PLoS One 3:e3976. doi: 10.1371/journal.pone.0003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinemann, U., C. Kiziak, S. Zibek, N. Layh, M. Schmidt, H. Griengl, and A. Stolz. 2003. Conversion of aliphatic 2-acetoxynitriles by nitriles hydrolysing bacteria. Appl. Microbiol. Biotechnol. 63:274-281. [DOI] [PubMed] [Google Scholar]
- 6.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering of hybrid genes without use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 7.Jenrich, R., I. Trompetter, S. Bak, C. E. Olsen, B. Lindberg Møller, and M. Piotrowski. 2007. Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc. Natl. Acad. Sci. U. S. A. 104:18848-18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiziak, C., D. Conradt, A. Stolz, R. Mattes, and J. Klein. 2005. Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639-3648. [DOI] [PubMed] [Google Scholar]
- 9.Kiziak, C., J. Klein, and A. Stolz. 2007. Influence of different carboxy-terminal mutations on the substrate-, reaction-, and enantiospecificity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Protein Eng. Des. Sel. 20:385-396. [DOI] [PubMed] [Google Scholar]
- 10.Kiziak, C., and A. Stolz. 2009. Identification of amino acid residues which are responsible for the enatioselectivity and amide formation capacity of the arylacetonitrilase from Pseudomonas fluorescens EBC 191. Appl. Environ. Microbiol. 75:5592-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi, M., H. Izui, T. Nagasawa, and H. Yamada. 1993. Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc. Natl. Acad. U. S. A. 90:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi, M., H. Komeda, N. Yanaka, T. Nagasawa, and H. Yamada. 1992. Nitrilase from Rhodococcus rhodochrous J1. Sequencing and overexpression of the gene and identification of an essential cysteine residue. J. Biol. Chem. 267:20746-20751. [PubMed] [Google Scholar]
- 13.Kobayashi, M., and S. Shimizu. 1994. Versatile nitrilases: nitrile-hydrolysing enzymes. FEMS Microbiol. Lett. 120:217-224. [Google Scholar]
- 14.Kobayashi, M., and S. Shimizu. 1998. Metalloenzyme nitrile hydratase: structure, regulation, and application to biotechnology. Nat. Biotechnol. 16:733-736. [DOI] [PubMed] [Google Scholar]
- 15.Layh, N., A. Stolz, S. Förster, F. Effenberger, and H.-J. Knackmuss. 1992. Enantioselective hydrolysis of O-acetylmandelonitrile to O-acetylmandelic acid by bacterial nitrilases. Arch. Microbiol. 158:405-411. [Google Scholar]
- 16.Liese, A., K. Seelbach, and C. Wandrey. 2006. Industrial biotransformations, 2nd ed. Wiley-VCH, Weinheim, Germany.
- 17.Martinková, L., and V. Křen. 2002. Nitrile- and amide-converting microbial enzymes: stereo-, regio- and chemoselectivity. Biocatal. Biotrans. 20:79-93. [Google Scholar]
- 18.Martinková, L., and V. Mylerová. 2003. Synthetic applications of nitrile-converting enzymes. Curr. Org. Chem. 7:1279-1295. [Google Scholar]
- 19.Mateo, C., A. Chmura, S. Rustler, F. van Rantwijk, A. Stolz, and R. A. Sheldon. 2006. Synthesis of enantiomerically pure (S)-mandelic acid using an oxynitrilase-nitrilase bienzymatic cascade. A nitrilase surprisingly shows nitrile hydratase activity. Tetrahedron Asymmetry 17:320-323. [Google Scholar]
- 20.O'Reilly, C., and P. D. Turner. 2003. The nitrilase family of CN hydrolysing enzymes—a comparative study. J. Appl. Microbiol. 95:1161-1174. [DOI] [PubMed] [Google Scholar]
- 21.Pace, H. C., and C. Brenner. 2001. The nitrilase superfamily: classification, structure and function. Genome Biol. 2:0001.1-0001.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panova, A., L. J. Mersinger, Q. Liu, T. Foo, D. C. Roe, W. L. Spillan, A. E. Sigmund, A. Ben-Bassat, L. W. Wagner, D. P. O'Keefe, S. Wu, K. L. Petrillo, M. S. Payne, S. T. Breske, F. G. Gallagher, and R. DiCosimo. 2007. Chemoenzymatic synthesis of glycolic acid. Adv. Synth. Catal. 349:1462-1474. [Google Scholar]
- 23.Piotrowski, M., S. Schönfelder, and E. W. Weiler. 2001. The Arabidopsis thaliana isogene NIT4 and its orthologs in tobacco encode beta-cyano-l-alanine hydratase/nitrilase. J. Biol. Chem. 276:2616-2621. [DOI] [PubMed] [Google Scholar]
- 24.Rustler, S., A. Müller, V. Windeisen, A. Chmura, B. C. M. Fernandes, C. Kiziak, and A. Stolz. 2007. Conversion of mandelonitrile and phenylglycinenitrile by recombinant E. coli cells synthesizing a nitrilase from Pseudomonas fluorescens EBC191. Enzyme Microb. Technol. 40:598-606. [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Schulze, B. 2002. Hydrolysis and formation of C-N bonds, p. 699-715. In K. Drauz and H. Waldmann (ed.), Enzyme catalysis in organic synthesis, vol. II. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 27.Singh, R., R. Sharma, N. Tewari, Geetanjali, and D. S. Rawat. 2006. Nitrilase and its application as a green catalyst. Chem. Biodivers. 3:1279-1297. [DOI] [PubMed] [Google Scholar]
- 28.Sosedov, O., K. Matzer, S. Bürger, C. Kiziak, S. Baum, J. Altenbuchner, A. Chmura, F. van Randwijk, and A. Stolz. 2009. Construction of recombinant Escherichia coli catalysts which simultaneously express an (S)-oxynitrilase and different nitrilase variants for the synthesis of (S)-mandelic acid and (S)-mandeloamide from benzaldehyde and cyanide. Adv. Synth. Catal. 351:1531-1538. [Google Scholar]
- 29.Stevenson, D. E., R. Feng, and A. C. Storer. 1990. Detection of covalent enzyme-substrate complexes of nitrilase by ion-spray mass spectroscopy. FEBS Lett. 277:112-114. [DOI] [PubMed] [Google Scholar]
- 30.Thuku, R. N., D. Brady, M. J. Benedik, and B. T. Sewell. 2009. Microbial nitrilases: versatile, spiral forming, industrial enzymes. J. Appl. Microbiol. 106:703-727. [DOI] [PubMed] [Google Scholar]
- 31.Thuku, R. N., B. W. Weber, A. Varsani, and B. T. Sewell. 2007. Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J. 274:2099-2108. [DOI] [PubMed] [Google Scholar]
- 32.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 33.Wu, S., A. J. Fogiel, K. L. Petrillo, E. C. Hann, L. J. Mersinger, R. DiCosimo, D. P. O′Keefe, A. Ben-Bassat, and M. S. Payne. 2007. Protein engineering of Acidovorax facilis 72W nitrilase for bioprocess development. Biotechnol. Bioeng. 97:689-693. [DOI] [PubMed] [Google Scholar]
- 34.Wu, S., A. J. Fogiel, K. L. Petrillo, R. E. Jackson, K. N. Parker, R. DiCosimo, A. Ben-Bassat, D. P. O′Keefe, and M. S. Payne. 2008. Protein engineering of nitrilase for chemoenzymatic production of glycolic acid. Biotechnol. Bioeng. 99:717-720. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto, K., I. Fujimatsu, and K.-I. Komatsu. 1992. Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J. Ferment. Bioeng. 73:425-430. [Google Scholar]
- 36.Yamamoto, K., K. Oishi, I. Fujimatsu, and K.-I. Komatsu. 1991. Production of R-(−)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl. Environ. Microbiol. 57:3028-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.