Abstract
Due to the possibility of a biothreat attack on civilian or military installations, a need exists for technologies that can detect and accurately identify pathogens in a near-real-time approach. One technology potentially capable of meeting these needs is a high-throughput mass spectrometry (MS)-based proteomic approach. This approach utilizes the knowledge of amino acid sequences of peptides derived from the proteolysis of proteins as a basis for reliable bacterial identification. To evaluate this approach, the tryptic digest peptides generated from double-blind biological samples containing either a single bacterium or a mixture of bacteria were analyzed using liquid chromatography-tandem mass spectrometry. Bioinformatic tools that provide bacterial classification were used to evaluate the proteomic approach. Results showed that bacteria in all of the double-blind samples were accurately identified with no false-positive assignment. The MS proteomic approach showed strain-level discrimination for the various bacteria employed. The approach also characterized double-blind bacterial samples to the respective genus, species, and strain levels when the experimental organism was not in the database due to its genome not having been sequenced. One experimental sample did not have its genome sequenced, and the peptide experimental record was added to the virtual bacterial proteome database. A replicate analysis identified the sample to the peptide experimental record stored in the database. The MS proteomic approach proved capable of identifying and classifying organisms within a microbial mixture.
The detection and accurate identification of pathogens of biological origin are of great importance to the armed forces and civilian sectors. Achieving these tasks is vital in the response to manmade or natural biothreat attacks in a proper and efficient manner to minimize the outbreak of epidemic cases. Several approaches reported in the literature have addressed the detection and identification of microorganisms based on the characterization of metabolites (1, 17) and genomic contents of bacterial cells (16). In these studies, the genomic sequence similarities generated from PCR were used to group bacteria at the genus/species level (27). Prior knowledge of the sample, or the targeting of one or a group of biological substances, is required in PCR techniques for proper primer utilization. However, proteins constitute greater than 60% of the dry weight of microorganism cellular components (4, 8, 12, 13, 22) and could provide in-depth information for the bacterial differentiation of species and their strains. Moreover, advancements in mass spectrometry (MS) ionization, detection methods, and data processing make MS a suitable analytical technique for the differentiation of microorganisms (5-7).
Using MS techniques for bacterial differentiation relies on the comparison of the proteomic information generated from the analysis of either intact protein profiles (top down) or the product ion mass spectra of digested peptide sequences (bottom up) (24, 26). For top-down analysis, bacterial differentiation is accomplished through the comparison of the MS data of intact proteins to those of an experimental mass spectral database containing the mass spectral fingerprints of the studied microorganisms (6, 7). Conversely, bacterial differentiation using the product ion mass spectral data of digested peptide sequences is accomplished through the utilization of search engines for publicly available sequence databases to infer identification (25, 29). Several peptide-searching algorithms (i.e., SEQUEST and MASCOT) have been developed to address peptide identification using proteomics databases that were generated from either fully or partially genome-sequenced organisms (6, 11, 19). Thus, our approach is based on a cross-correlation between the generated product ion mass spectra of tryptic peptides and their corresponding bacterial proteins resident in an in-house comprehensive proteome database from online databases of the sequences of microorganism genomes (30).
Recent developments in the microbial differentiation field have focused on improving the selectivity of MS data processing. The product ion mass spectrum-SEQUEST approach was reported for the identification of specific bacteria using a custom-made, limited database of sequences (14, 23). Another approach used open reading frame (ORF) translator programs to predict possible protein sequences from all probable ORFs and correlate them with the genomic sequences to establish an identification of microorganisms (5). This approach did not show advantages over the product ion mass spectrum method with regard to strain level discrimination (28). However, a recent advancement in proteomic approaches to bacterial differentiation reported a hybrid approach combining protein profiling and sequence database searching using accurate mass tags (15, 18). This approach was used to probe defined mixtures of bacteria to evaluate its capabilities.
Alternatively, our approach is based on a cross-correlation between the product ion spectra of the tryptic peptides and their corresponding bacterial proteins derived from an in-house comprehensive proteome database from genome-sequenced microorganisms (9, 10). The exploitation of this proteome database approach allowed for a faster search of the product ion spectra than that using genomic database searching. Also, it eliminates inconsistencies observed in publicly available protein databases due to the utilization of nonstandardized gene-finding programs during the process of constructing the proteome database. The proposed approach uses an ensemble of bioinformatic tools for the classification and potential identification of bacteria based on the peptide sequence information. This information is generated from the liquid chromatography-tandem mass spectrometry (LC-MS-MS) analysis of tryptic digests of bacterial protein extracts and the subsequent profiling of the sequenced peptides to create a matrix of sequence-to-bacterium (STB) assignments. This proteomic approach is an unsupervised approach to reveal the relatedness between the analyzed samples and the database of microorganisms using a binary matrix approach. The binary matrix is analyzed using diverse visualization and multivariate statistical techniques for bacterial classification and identification.
This study investigated the capability of the aforementioned MS-based proteomic approach to identify biological agents using double-blind (hereafter referred to as blind) samples that consisted of various microorganisms of interest to civilian and military installations. The present study included category A biological agents, mixtures of organisms, and negative controls without prior knowledge of the identity of the microorganisms. The in-house database consists of 881 microbial genomes as of 2 May 2009. The identification process for all samples revealed that several samples consisted of a mixture of bacterial species. The results of the blind studies showed a promising outlook for applying this MS-based proteomic approach to the classification of unknown bacterial mixtures at the species and strain level depending on the availability of complete genome sequences.
MATERIALS AND METHODS
Materials and reagents.
Ammonium bicarbonate, dithiothreitol, urea, acetonitrile (ACN; high-performance liquid chromatography [HPLC] grade), and formic acid were purchased from Burdick and Jackson (St. Louis, MO). Sequencing-grade modified trypsin was purchased from Promega (Madison, WI).
Biological sample preparation.
Twenty-one blind biological samples were prepared by streaking cells from cryopreserved stocks onto appropriate agar. Bacillus subtilis, Bacillus thuringiensis, Staphylococcus aureus, Enterococcus faecalis, and Pseudomonas aeruginosa were streaked onto tryptic soy agar (TSA; catalog number CM100; Culture Media and Supplies, Oswego, IL) plus 5% sheep blood. Burkholderia thailandensis and Clostridium phytofermentans ISDg were streaked onto nutrient agar (NA; catalog number CM145; Culture Media and Supplies, Oswego, IL). All plates were incubated for approximately 18 h at 37°C and stored at 4°C for no longer than 10 days. Cells from plate cultures were used to inoculate liquid cultures consisting of 10 ml of tryptic soy broth (TSB; catalog number CM104; Culture Media and Supplies, Oswego, IL) for B. subtilis, B. thuringiensis, S. aureus, E. faecalis, P. aeruginosa, and nutrient broth (NB; catalog number CM146; Culture Media and Supplies, Oswego, IL) for B. thailandensis. All liquid cultures were incubated for approximately 18 h at 37°C with rotary aeration at 180 rpm. After incubation, bacteria from liquid cultures were harvested by centrifugation (2,300 relative centrifugal force [RCF] at 4°C for 10 min), washed, and resuspended in an equal volume of phosphate-buffered saline (PBS). The Bacillus species were observed under a microscope to consist predominately of spores. Samples provided for analysis consisted of either a single bacterium or multiple bacteria mixed together. For mixed samples, all bacteria were added in a ratio of 1:1 by volume. All bacteria were present at a concentration between 10E7 to 10E9 CFU/ml as determined by serial dilution and plating onto appropriate agar. All samples were produced at the microbiology laboratory at the U.S. Army Edgewood Chemical Biological Center in a blind format and were assigned number codes for processing and analysis. The identities of all blind samples were revealed upon the completion of all analyses. A negative control sample also was included that consisted of PBS only (no bacteria).
Processing of blind biological samples.
The lysis of all blind samples was performed using a modified sonication method (2, 20, 21). All blind samples, including any sporulated bacteria, were lysed by microprobe ultrasonication (Branson 450 digital sonifier; Branson, Danbury, CT). The blind samples were placed on ice and lysed with a 20-s pulse on and 5-s pulse off (cooling time) and 25% amplitude for a 5-min duration. To verify that the cells were disrupted, a small portion of the lysate was examined under confocal microscopy, and another portion was reserved for one-dimensional gel analysis.
The lysate was centrifuged at 14,100 × g for 30 min to remove all cellular debris. The supernatant then was added to a Microcon YM-3 filter unit (catalog number 42404; Millipore) and centrifuged at 14,100 × g for 30 min. The effluent was discarded. The filter membrane was washed with 100 mM ammonium bicarbonate (ABC) and centrifuged for 15 to 20 min at 14,100 × g. Cellular proteins were denatured by adding 8 M urea and 3 μg/μl dithiothreitol (DTT) to the filter and incubating it overnight at 37°C on an orbital shaker set to 60 rpm. Twenty microliters of 100% acetonitrile was added to the tubes and allowed to incubate at room temperature for 5 min. The tubes then were centrifuged at 14,100 × g for 30 to 40 min and washed three times using 150 μl of 100 mM ABC solution. On the last wash, the ABC solution was shaken for 20 min, followed by centrifugation at 14,100 × g for 30 to 40 min. The filter unit then was transferred to a new receptor tube, and proteins were digested with 5 μl of trypsin in 240 μl of ABC solution plus 5 μl ACN. Protein digestion occurred overnight at 37°C on an orbital shaker set to 55 rpm. Sixty microliters of 5% ACN-0.5% formic acid (FA) was added to each filter to quench the trypsin digestion, followed by 2 min of vortexing for sample mixing. The tubes were centrifuged for 20 to 30 min at 14,100 × g. An additional 60 μl 5% ACN-0.5% FA mixture was added to the filter and centrifuged. Alternative protocols were used in which the denaturation step was eliminated, and the digestion time was reduced using various amounts of trypsin and different digestion temperatures. The effluent then was analyzed using LC-MS-MS.
LC-MS-MS analysis of peptides.
The tryptic peptides were separated using a capillary Hypersil C18 column (300 Å; 5 μm; 0.1 mm [inner diameter] by 100 mm) by using the Surveyor LC from ThermoFisher (San Jose, CA). The elution was performed using a linear gradient from 98% A (0.1% FA in water) and 2% B (0.1% FA in ACN) to 60% B for 60 min at a flow rate of 200 μl/min, followed by 20 min of isocratic elution. The resolved peptides were electrosprayed into a linear ion trap mass spectrometer (LTQ; Thermo Scientific, San Jose, CA) at a flow rate of 0.8 μl/min. Product ion mass spectra were obtained in the data-dependent acquisition mode that consisted of a survey scan across the m/z range of 400 to 2,000, followed by seven scans on the most intense precursor ions activated for 30 ms by an excitation energy level of 35%. A dynamic exclusion was activated for 3 min after the first MS-MS spectrum acquisition for a given ion. Uninterpreted product ion mass spectra were searched against a microbial database with TurboSEQUEST (Bioworks 3.1; Thermo Scientific, San Jose, CA) followed by the application of an in-house proteomic algorithm for bacterial identification.
Protein database and database search engine.
A protein database was constructed in a FASTA format using the annotated bacterial proteome sequences derived from the sequenced chromosomes of 881 bacteria, including their sequenced plasmids (as of May 2009). A PERL program (ActiveState) was written to automatically download these sequences from the National Institutes of Health National Center for Biotechnology (NCBI) site (http://www.ncbi.nlm.nih.gov). Each database protein sequence was supplemented with information about the source organism and the genomic position of the respective ORF embedded into a header line. The database of bacterial proteomes was constructed by translating putative protein-coding genes and consists of tens of millions of amino acid sequences of potential tryptic peptides obtained by the in silico digestion of all proteins (assuming up to two missed cleavages).
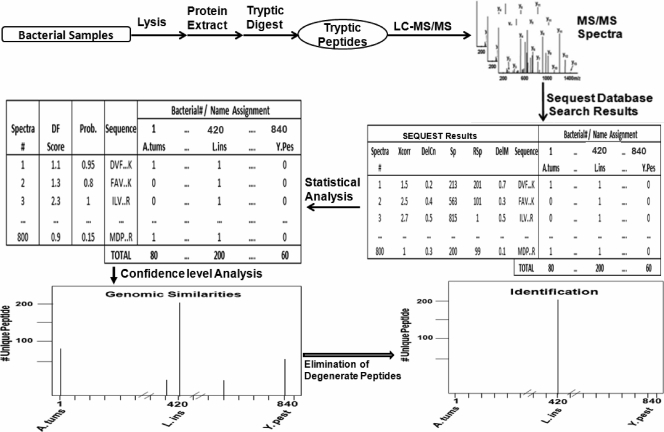
The experimental product ion mass spectral data of bacterial peptides were searched using the SEQUEST (11) algorithm against a constructed proteome database of microorganisms. The SEQUEST thresholds for searching the product ion mass spectra of peptides were Xcorr, deltaCn (DelCn), Sp, RSp, and deltaMpep (DelM). These parameters provided a uniform matching score for all candidate peptides. The generated outfiles of these candidate peptides then were validated using the PeptideProphet algorithm (14). Peptide sequences with a probability score of 95% and higher were retained in the data set and used to generate a binary matrix of STB assignments. The binary matrix assignment was populated by matching the peptides with corresponding proteins in the database and assigning them a score of one. A score of zero was assigned for a nonmatch. The column in the binary matrix represents the proteome of a given bacterium, and each row represents a tryptic peptide sequence from the LC-MS-MS analysis. Microorganisms in a blind sample were matched with the bacterium/bacteria based on the number of unique peptides that remained after the filtering of degenerate peptides from the binary matrix. The verification of the classification and identification of candidate microorganisms was performed through hierarchical clustering analysis and taxonomic classification (Fig. 1).
FIG. 1.
Schematic of the LC-MS-MS analysis and data-processing steps for the identification of microorganisms using the proteomic approach. A. tum, Agrobacterium tumefaciens; CID, collision-induced dissociation; df, discriminant factor; L. inn., Listeria innocua; Y. pest, Yersinia pestis.
Data analysis and algorithms.
The SEQUEST-processed product ion mass spectra of the peptide ions were compared to an NCBI protein database using the in-house-developed software (BACid). BACid (10) provided a taxonomically meaningful and easy-to-interpret output. BACid calculates the probabilities that a peptide sequence assignment to a product ion mass spectrum is correct and uses accepted spectrum-to-sequence matches to generate an STB binary matrix of assignments. Validated peptide sequences, either present or absent in various strains (STB matrices), were visualized as assignment bitmaps and analyzed by a BACid module that used phylogenetic relationships among bacterial species as part of a decision tree process. The bacterial classification and identification algorithm used assignments of organisms to taxonomic groups (phylogenetic classification) based on an organized scheme that begins at the phylum level and follows through the class, order, family, genus, and strain levels. BACid was developed in-house using PERL, MATLAB, and Microsoft Visual Basic.
RESULTS AND DISCUSSION
The capabilities, and possible limitations, of the proteomic approach with regard to the identification of biological agents were evaluated using blind biological samples. Twenty-one blind microbial samples were provided and analyzed by the LC-MS-MS proteomic approach. The composition of the blind samples varied, with some samples having only one bacterium and others having as many as five different bacterial species or strains.
Bacillus subtilis sample.
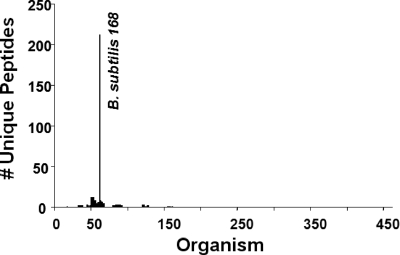
An example of the resultant data from the BACid program for one blind sample is shown in Fig. 2. Blind sample 20 was identified as B. subtilis 168 using the BACid data-processing algorithm. This identification algorithm eliminated all of the unwanted and degenerate peptides and retained only the unique peptides that represent a 99% probability for correct identification. In this case, 212 unique peptides were identified and associated with proteins from the B. subtilis 168 strain. The 212 B. subtilis 168 unique peptides represented 89% of the total number of unique peptides in the blind sample. Table 1 shows a select set of unique peptides and their corresponding proteins that are associated with B. subtilis 168. These bacterial proteins have different cellular functions, such as transcription, translation, and cellular signaling. They represent a set of unique biomarkers that could be utilized to establish strain-level discrimination between B. subtilis 168 and other members of the Bacillus genus.
FIG. 2.
Histogram of the BACid output for the processing of the LC-MS-MS analysis data set for blind bacterial sample 20. The ordinate provides the actual number of SEQUEST-generated and -filtered unique peptides. The abscissa represents the bacteria found at least once in the 21 experimental samples.
TABLE 1.
Partial list of peptides in the double-blind proteomic processing of sample 20, B. subtilis 168
| Peptide no. | Peptide sequence | Protein | Accession no. |
|---|---|---|---|
| 30 | VLDVNENEER | 30S ribosomal protein S1 | NP_390169.1 |
| 39 | AYDVSEAVALVK | 50S ribosomal protein L1 | NP_387984.1 |
| 38 | NVAVTSTMGPGVK | 50S ribosomal protein L1 | NP_387984.1 |
| 31 | GLNVSEVTELR | 50S ribosomal protein L10 | NP_387985.1 |
| 53 | GLNVSEVTELRK | 50S ribosomal protein L10 | NP_387985.1 |
| 41 | TTPMANASTIER | 50S ribosomal protein L13 | NP_388030.1 |
| 85 | GVEMDAYEVGQEVK | 50S ribosomal protein L3 | NP_387997.1 |
| 51 | VESPDQLADVLR | Alpha-acetolactate synthase | NP_391482.1 |
| 71 | EMADFFEETVQK | Aspartyl/glutamyl-tRNA amidotransferase | NP_388551.2 |
| 37 | EAQQLIEEQR | ATP synthase (subunit b) | NP_391566.1 |
| 70 | ENTTIVEGAGETDK | Chaperonin GroEL | NP_388484.1 |
| 74 | AILVMPDTMSMER | Cysteine synthetase A | NP_387954.1 |
| 50 | LADENSADVYLK | Cysteine synthetase A | NP_387954.1 |
| 49 | ALSLNETDGFMK | Dihydrolipoamide dehydrogenase | NP_389344.1 |
| 64 | IGADFLYSVGTLR | Elongation factor EF-2 | NP_387993.1 |
| 72 | SEHGLLFGMPIGVK | Glutamyl-tRNA amidotransferase subunit A | NP_388550.1 |
| 100 | GILGYSEEPLVSGDYNGNK | Glyceraldehyde-3-phosphate dehydrogenase | NP_391274.1 |
| 99 | NSSTIDALSTMVMEGSMVK | Glyceraldehyde-3-phosphate dehydrogenase | NP_391274.1 |
| 47 | TIEVSAERDPAK | Glyceraldehyde-3-phosphate dehydrogenase | NP_391274.1 |
| 92 | VISWYDNESGYSNR | Glyceraldehyde-3-phosphate dehydrogenase | NP_391274.1 |
| 55 | TNPDYLFVIDR | Hypothetical protein BSU03830 | NP_388265.1 |
| 91 | AAGATDIYAVELSPER | Hypothetical protein BSU06240 | NP_388505.1 |
| 35 | DIFPAVLSLMK | Hypothetical protein BSU06240 | NP_388505.1 |
| 44 | GAEIHPNDIVIK | Hypothetical protein BSU06240 | NP_388505.1 |
| 75 | IEHIEEPKTEPGK | Hypothetical protein BSU06240 | NP_388505.1 |
| 89 | EMGHTELPFYQQR | Hypothetical protein BSU12410 | NP_389123.1 |
| 73 | QEETETDLNVLAK | Hypothetical protein BSU12410 | NP_389123.1 |
| 54 | GELEGINFGESAK | Hypothetical protein BSU14180 | NP_389301.1 |
| 66 | SIGVSNFSLEQLK | Inositol utilization protein S | NP_391857.1 |
| 79 | LISFLQNELNVNK | Isocitrate dehydrogenase | NP_390791.1 |
| 5 | AVAEALAEAK | Phosphoglycerate kinase | NP_391273.1 |
| 94 | AVSNPDRPFTAIIGGAK | Phosphoglycerate kinase | NP_391273.1 |
| 36 | AIQISNTFTNK | Phosphoglyceromutase | NP_391271.1 |
| 26 | NETVGNAVALAK | Phosphoglyceromutase | NP_391271.1 |
| 48 | TASVINPAIAFGR | Phosphotransferase system (PTS) fructose-specific enzyme | NP_389323.1 |
| 87 | IANFETAEPLYYR | Putative manganese-dependent inorganic pyrophosphatase | NP_391935.1 |
| 60 | LFANLLETAGATR | Ribose-phosphate pyrophosphokinase | NP_387932.1 |
| 29 | TYAQNVISNAK | Serine hydroxymethyltransferase | NP_391571.1 |
| 81 | FWLSQDKEELLK | S-Ribosylhomocysteinase | NP_390945.1 |
| 24 | GGPVTLVGQEVK | Thiol peroxidase | NP_390827.1 |
| 63 | TLGEAVSFVEEVK | Triosephosphate isomerase | NP_391272.1 |
| 11 | TNDLVADQVK | Triosephosphate isomerase | NP_391272.1 |
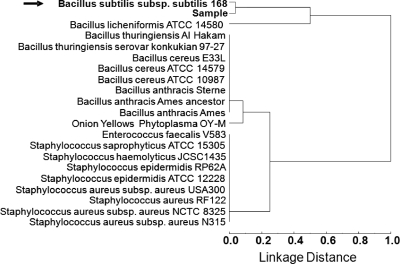
To ensure confidence in the assignment of the candidate bacterium, a similarity analysis was performed on the nearest-neighbor species and strains. In this similarity analysis, all sequenced strains of B. subtilis and Bacillus species that are genetically related to the candidate bacterium were included in the Euclidean distance dendrogram. Figure 3 shows a dendrogram of the similarity analysis of the blind sample identified as B. subtilis 168. In Fig. 3, the sample was identified as being most similar to B. subtilis 168 using the unique peptides that were associated with this bacterial candidate. The next closest bacterium to the candidate was determined to be Bacillus licheniformis ATCC 14580. According to these similarities, a comparison of B. licheniformis and B. subtilis 168 analyses showed a difference of almost 50% in the unique proteins identified by the BACid algorithm. Based on these significant differences and a lower degree of confidence assigned, B. licheniformis was not included as a candidate bacterium. Therefore, the identity of sample 20 was assigned to B. subtilis 168 using the BACid algorithm. This assignment was correct as later revealed at the completion of the tests.
FIG. 3.
Dendrogram of the multivariate cluster analysis using Euclidean distances of the sequence-to-bacterium assignment matrices for blind sample 20. The dendrogram is the result of the complete linkage construction in multivariate dataspace of the furthest neighbor with the 221 unique peptide sequences shown in Fig. 2.
Blind mixture analysis.
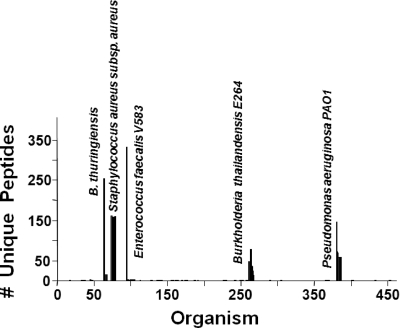
The BACid analysis of sample 18 is shown in Fig. 4. BACid eliminated all of the unwanted and degenerate peptides, and only the unique peptides that represented a 99% confidence level and above were retained for each organism. In this case, the number of unique peptides varied for the different bacterial candidates. E. faecalis had the highest number of unique peptides followed by B. thuringiensis, and B. thailandensis had the least number of unique peptides. Interestingly, it was revealed that after the tests the blind samples had approximately equivalent bacterial concentrations for each organism, yet the number of unique peptides differed. This variation in the number of unique peptides in the output of the BACid could be due to the dynamic nature of the bacterial species during sample processing. Some bacteria could have a larger number of lysed proteins that were suspended in the extraction buffer than that of other species in the sample. This difference in bacterial protein concentrations is shown in the histogram in Fig. 4 generated from the BACid output, where the relative number of peptides for each species is compared to that of the other species. This feature in the BACid algorithm could be used as a pseudoquantitative technique in the determination of lysed bacterial proteins in a biological sample and thus aid in evaluating sample-processing modules. Also shown in Fig. 4 are six bacterial candidates near the cutoff threshold within the Staphylococcus genus. This pattern is due to the fact that the Staphylococcus aureus ATCC 3359 strain present in the blind sample has not been sequenced or reported in the public domains, and thus it was not part of the constructed proteome database. However, the BACid was capable of providing a nearest-neighbor match to the species level (S. aureus) and thus identified the bacterium correctly as S. aureus subsp. aureus. It is noteworthy that this bacterial strain, which is not genomically sequenced, could be identified only to the species level. The rapid increase in the number of sequenced bacteria will benefit this proteomic approach and enhance its robustness in the identification process of biological samples. However, a significant advantage of the approach is that if a particular strain has not been sequenced but the species is represented in the database, it is highly likely that the unsequenced sample strain will be identified to the species level. The appearance of the histogram from a BACid analysis indicates the degree of the accuracy of the identification process. Strain-level experimental identification is indicated by a single line (Fig. 4) in the histogram (Enterococcus faecalis V538) or by a grouping of lines, where one line clearly dominates (e.g., Burkholderia thailandensis E264 and Pseudomonas aeruginosa PAO1) with respect to the number of unique peptides. B. thuringiensis has two strains resident in the database, and both provide a similar set of peptides. This occurs because the two strains do not display peptides that clearly distinguish themselves. The fifth bacterium in the sample 18 mixture was S. aureus strain ATCC 3359, and this organism's genome has not been sequenced. However, the species-level identification (S. aureus) of this strain is indicated by a grouping of lines (Fig. 4) that does not display a significant difference in the number of unique peptides. This blind sample was correctly identified as a mixture of five bacteria: B. thuringiensis, S. aureus subsp. aureus, E. faecalis V583, B. thailandensis E264, and P. aeruginosa PA01, where S. aureus and B. thuringiensis were identified to the species level, and the other three were identified to the strain level.
FIG. 4.
Histogram of the BACid output for the processing of the LC-MS-MS data set for the biological mixture in sample 18. Refer to the legend of Fig. 2 for details.
Suite of bacterial samples.
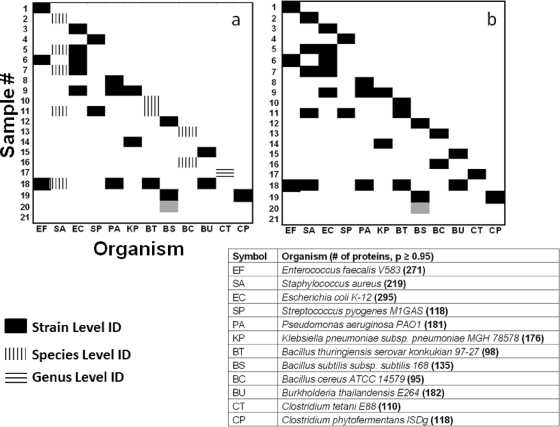
The in-house database originated from 881 genomically sequenced bacterial strains. The blind sample suspensions consisted of bacteria in single and mixture forms, and their genomes were sequenced or not sequenced. The bacterial strains found in experimental samples that do not have a sequenced genome, therefore, cannot be found in available public databases and the in-house database. Figure 5a shows the classification map of the 21 experimentally processed blind samples, and Fig. 5b shows that of the bacterial strain sample identities (sample key). In Fig. 5a, the bacteria on the abscissa reflect every bacterium found at least once in the 21 experimentally determined samples. The bacteria listed in Fig. 5a were not disclosed in advance; rather, all 21 experiments produced the bacterial identities from the BACid algorithm (10). Figure 5b represents the sample key or actual bacterial species and strains in the blind samples. This information was not released to the authors until the Fig. 5a results were turned in for experimental performance verification. A comparison between Fig. 5a and b shows that bacterial discrimination was achieved by relying on the unique peptides corresponding to the bacteria in the blind samples. An identification was based on the matching probability of the unique peptides from a blind sample with a bacterial entry in the bacterial proteome database at more than a P = 0.95 confidence level. The strain-level identification, indicated by the filled red boxes in Fig. 5a, was assigned due to a close match with the analyzed microorganisms' unique peptides and their nearest-neighbor strains.
FIG. 5.
(a) Classification map of the experimentally processed samples. The bacteria on the abscissa indicate that they were found at least once in the 21 samples. (b) Actual or sample key of bacteria present in all 21 samples. The dark gray coloring for sample 20 represents Bacillus atrophaeus, which was identified as B. subtilis in panel a. Sample 21 was a blank. In the table, the numbers in parentheses for each organism signify the number of proteins identified with a P ≥ 0.95 probability match. Solid box, strain-level identification; vertically hatched box, species-level identification; horizontally hatched box, genus-level identification.
Figure 4 shows the analysis of sample 18 and provides an example of identification to the strain level as well as classification to the species level (as described above) for Staphylococcus aureus strain ATCC 3359, which is not currently sequenced. A correct species level of identification was experienced with all bacteria in the blind samples that are unsequenced and are indicated by a vertical hashed box in Fig. 5a. Thus, the classification probability was statistically high enough based on a comparison of the virtual proteome of a database strain and the experimental unique proteins of the unsequenced-genome bacterial sample. Therefore, identification was reported at the species level. Blind sample 20 (Fig. 2) was identified as B. subtilis; however, the sample key reported it as B. atrophaeus. This difference is due to the lack of a proteome for B. atrophaeus, which taxonomically is considered B. subtilis. Our data support the proposition that B. atrophaeus should be reclassified as a strain of B. subtilis (3) (the gray square for sample 20 in Fig. 5a and b).
Genus-level identification.
Blind sample 17 was investigated for BACid characterization. The experimental set of peptides could provide results only to the Clostridium genus level, because all nine clostridial bacteria (species and strains) resident in the database produced a histogram (data not shown) similar to that of Staphylococcus aureus, which is shown in Fig. 4. The experimental peptides matched that portion of the virtual proteome common to all Clostridia. Therefore, the complete experimentally derived, tryptic peptide information record was stored as a separate bacterial line item as Clostridium species 1 in the database of 881 bacteria. Another aliquot of the blind sample was processed with data reduction and searching in the new hybrid database. The highest match was with the Clostridium species 1 entry. After the results were submitted, the identity of sample 17 was revealed to be Clostridium phytofermentans ISDg. This strain does not have its genome sequenced, yet BACid was able to match the virtual proteins that are similar to those of the Clostridium genus to the experimentally observed peptides. Thus, BACid was able to characterize sample 17 as Clostridium without choosing one of the nine clostridia strains resident in the database or other bacterial genera. BACid instead matched Clostridium species 1 to the experimental peptides, which indicated that there is sufficient information in the experimental peptides to differentiate Clostridium phytofermentans ISDg from the nine database clostridia strains. It is tempting to consider that this approach, when combined with the accurate mass tag approach of Lipton et al. (15), has the potential to diminish the impact of genome-sequencing deficiencies for some bacterial strains. The rapid advancement in genome-sequencing projects will enhance the robustness of this approach through the expansion of the proteome database. This expansion in the proteome database is anticipated to include the cellular proteins that can be utilized for strain-level differentiation.
The results showed that the method was effective in identifying bacteria whether the sample was composed of one organism or a mixture, or even if the sample is not resident in the database. No false positives were observed for any of the blind samples that were analyzed, including the blank sample. The proteomic MS approach reported herein is not meant as a replacement for DNA-based identification methods. We envision this approach as a second, confirmatory approach to pathogen identification. Additionally, there are some major advantages to the proteomic method over other molecular biology methods such as the DNA-based methods, in that (i) no prior information about the sample is required for analysis; (ii) no specific reagents are needed in the analysis process; (iii) proteomic MS is capable of identifying an organism when a primer/probe set is not available; (iv) proteomic MS requires less rigorous sample preparation than PCR; and (v) proteomic MS can provide a presumptive identification of a true unknown organism by mapping its phylogenetic relationship with other, known pathogens. The proteomic method also could be applied to identify viruses and toxins, because viruses and toxins are included in the proteome database.
Naturally occurring environmental samples usually contain a great deal of organisms at very low concentrations in addition to the target species. The total amount of background organisms may consist of greater numbers than that of the target organism. Therefore, this is a topic that would challenge the method reported herein. This is being addressed by spiking a target organism in several environmental matrices at different applied amounts.
Improvement in sample preparation and mass spectrometry technologies will enhance and increase the number of peptides identified compared to those of the current methods. This can allow for MS proteomics being a valuable tool in conjunction with genomic approaches to address the issue of the identification and classification of microorganisms. Overall, these studies showed that the proposed MS-based proteomic approach is a useful method that may be applied to diverse biothreat scenarios and has the potential for bacterial differentiation and identification at species and strain levels of individual bacteria or their mixtures.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Akoto, L., R. Pel, H. Irth, U. A. T. Brinkman, and R. J. J. Vreuls. 2005. Automated GC-MS analysis of raw biological samples-application to fatty acid profiling of aquatic micro-organisms. J. Anal. Appl. Pyrolysis 73:69-75. [Google Scholar]
- 2.Brown, S. D., M. R. Thompson, N. C. VerBerkmoes, K. Chourey, M. Shah, J. Zhou, R. L. Hettich, and D. K. Thompson. 2006. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol. Cell Proteomics 5:1054-1071. [DOI] [PubMed] [Google Scholar]
- 3.Burke, S. A., J. D. Wright, M. K. Robinson, B. V. Bronk, and R. L. Warren. 2004. Detection of molecular diversity in Bacillus atrophaeus by amplified fragment length polymorphism analysis. 2004. Appl. Environ. Microbiol. 70:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cain, T. C., D. M. Lubman, and W. J. Weber. 1994. Differentiation of bacteria using protein profiles from MALDI-TOF/MS. Rapid Commun. Mass Spectrom. 8:1026-1030. [Google Scholar]
- 5.Chen, W., K. E. Laidig, Y. Park, K. Park, J. R. Yates III, R. J. Lamont, and M. Hackett. 2001. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst 126:52-57. [DOI] [PubMed] [Google Scholar]
- 6.Craig, R., and R. Beavis. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466-1467. [DOI] [PubMed] [Google Scholar]
- 7.Demirev, P., J. Ramirez, and C. Fenselau. 2001. Tandem mass spectrometry of intact proteins for characterization of biomarkers from Bacillus cereus T spores. Anal. Chem. 73:5725-5731. [DOI] [PubMed] [Google Scholar]
- 8.Duché, O., F. Trémoulet, A. Namane, and J. C. Labadie. 2002. A proteomic analysis of the salt stress response of Listeria monocytogenes. FEMS Microbiol. Lett. 215:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Dworzanski, J. P., A. P. Snyder, R. Chen, H. Zhang, D. Wishart, and L. Li. 2004. Identification of bacteria using tandem mass spectrometry combined with a proteome database and statistical scoring. Anal. Chem. 76:2355-2366. [DOI] [PubMed] [Google Scholar]
- 10.Dworzanski, J. P., S. V. Deshpande, R. Chen, R. E. Jabbour, A. P. Snyder, C. H. Wick, and L. Li. 2006. Mass spectrometry-based proteomics combined with bioinformatics tools for bacterial classification. J. Proteome Res. 5:76-87. [DOI] [PubMed] [Google Scholar]
- 11.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass-spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 12.Hesketh, A. R., G. Chandra, A. D. Shaw, J. J. Rowland, D. B. Kell, M. J. Bibb, and K. F. Chater. 2002. Primary and secondary metabolism, and post-translational protein modifications, as portrayed by proteomic analysis of Streptomyces coelicolor. Mol. Microbiol. 46:917-932. [DOI] [PubMed] [Google Scholar]
- 13.Holland, R. D., J. G. Wilkes, F. Rafii, J. B. Sutherland, C. C. Persons, K. J. Voorhees, and J. O. Lay, Jr. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227-1232. [DOI] [PubMed] [Google Scholar]
- 14.Keller, A., A. I. Nesvizhskii, I. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS-MS and database search. Anal. Chem. 74:5383-5392. [DOI] [PubMed] [Google Scholar]
- 15.Lipton, M. S., L. Pasa-Tolic, G. A. Anderson, D. J. Anderson, D. L. Auberry, J. R. Battista, M. J. Daly, J. Fredrickson, K. K. Hixson, H. Ksotandarithes, C. Masselon, L. M. Markillie, R. J. Moore, M. F. Romine, Y. Shen, E. Stritmatter, N. Tolic, H. R. Udseth, A. Venkaeswaran, K. K. Wong, R. Zhao, and R. D. Smith. 2002. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. U. S. A. 99:11049-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr, B., U. Kobold, M. Moczko, A. Nyeki, T. Koch, and C. Huber. 2005. Identification of bacteria by polymerase chain reaction followed by liquid chromatography-mass spectrometry. Anal. Chem. 77:4563-4570. [DOI] [PubMed] [Google Scholar]
- 17.Moe, M. K., T. Anderssen, M. B. Strom, and E. Jensen. 2005. Total structure characterization of unsaturated acidic phospholipids provided by vicinal di-hydroxylation of fatty acid double bonds and negative electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 16:46-59. [DOI] [PubMed] [Google Scholar]
- 18.Norbeck, A. D., S. J. Callister, M. E. Monroe, N. Jaitly, D. A. Elias, M. S. Lipton, and R. D. Smith. 2006. Proteomic approaches to bacterial differentiation. J. Microbiol. Methods 67:473-486. [DOI] [PubMed] [Google Scholar]
- 19.Perkins, D., D. Pappin, D. Creasy, and J. Cottrell. 1999. Probability-based protein identification by searching sequence database using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 20.Ryzhov, V., Y. Hathout, and C. Fenselau. 2000. Rapid characterization of spores of Bacillus cereus group bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Appl. Environ. Microbiol. 66:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saikaly, P. E., M. A. Barlaz, and F. L. de los Reyes III. 2007. Development of quantitative real-time PCR assays for detection and quantification of surrogate biological warfare agents in building debris and leachate. Appl. Environ. Microbiol. 73:6557-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan, Y. P., Q. Lin, X. H. Wang, S. Joshi, C. L. Hew, and K. Y. Leung. 2002. Comparative proteomic analysis of extracellular proteins of Edwardsiella tarda. FEMS Infect. Immun. 215:6475-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VerBerkmoes, N. C., W. J. Hervey, M. Shah, M. Land, L. Hauser, F. W. Larimer, G. J. van Berkel, and D. E. Goeringer. 2005. Evaluation of “shotgun” proteomics for identification of biological threat agents in complex environmental matrices: experimental simulations. Anal. Chem. 77:923-932. [DOI] [PubMed] [Google Scholar]
- 24.Warscheid, B., and C. Fenselau. 2003. Characterization of Bacillus spore species and their mixtures using post source decay with a curved-field reflectron. Anal. Chem. 75:5618-5627. [DOI] [PubMed] [Google Scholar]
- 25.Warscheid, B., and C. Fenselau. 2004. A targeted proteomics approach to the rapid identification of bacterial cell mixtures by matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 4:2877-2892. [DOI] [PubMed] [Google Scholar]
- 26.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 27.Woese, C. R., E. Stackebrandt, T. J. Macke, and G. E. Fox. 1985. A phylogenetic definition of the major eubacterial taxa. Syst. Appl. Microbiol. 61:143-151. [DOI] [PubMed] [Google Scholar]
- 28.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683-5690. [DOI] [PubMed] [Google Scholar]
- 29.Xiang, F., G. Anderson, T. Veenstra, M. Lipton, and R. Smith. 2000. Characterization of microorganisms and biomarker development from global ESI-MS-MS analyses of cell lysates. Anal. Chem. 72:2475-2481. [DOI] [PubMed] [Google Scholar]
- 30.Yates, J. R., III, and J. K. Eng. January 2000. Identification of nucleotides, amino acids, or carbohydrates by mass spectrometry. U.S. patent 6,017,693.