Abstract
Recently, Salmonella enterica subsp. enterica serovar Saintpaul has increasingly been observed in several countries, including Germany. However, the pathogenic potential and epidemiology of this serovar are not very well known. This study describes biological attributes of S. Saintpaul isolates obtained from turkeys in Germany based on characterization of their pheno- and genotypic properties. Fifty-five S. Saintpaul isolates from German turkeys and turkey-derived food products isolated from 2000 to 2007 were analyzed by using antimicrobial agent, organic solvent, and disinfectant susceptibility tests, isoelectric focusing, detection of resistance determinants, plasmid profiling, pulsed-field gel electrophoresis (PFGE), and hybridization experiments. These isolates were compared to an outgroup consisting of 24 S. Saintpaul isolates obtained from humans and chickens in Germany and from poultry and poultry products (including turkeys) in Netherlands. A common core resistance pattern was detected for 27 German turkey and turkey product isolates. This pattern included resistance (full or intermediate) to ampicillin, amoxicillin-clavulanic acid, gentamicin, kanamycin, nalidixic acid, streptomycin, spectinomycin, and sulfamethoxazole and intermediate resistance or decreased susceptibility to ciprofloxacin (MIC, 2 or 1 μg/ml, respectively) and several third-generation cephalosporins (including ceftiofur and cefoxitin [MIC, 4 to 2 and 16 to 2 μg/ml, respectively]). These isolates had the same core resistance genotype, with blaTEM-1, aadB, aadA2, sul1, a Ser83→Glu83 mutation in the gyrA gene, and a chromosomal class 1 integron carrying the aadB-aadA2 gene cassette. Their XbaI, BlnI, and combined XbaI-BlnI PFGE patterns revealed levels of genetic similarity of 93, 75, and 90%, respectively. This study revealed that a multiresistant S. Saintpaul clonal line is widespread in turkeys and turkey products in Germany and was also detected among German human fecal and Dutch poultry isolates.
Over the last few decades, the emergence and spread of antimicrobial agent-resistant zoonotic bacteria has become a serious public health concern (2, 23). The widespread use of antimicrobial agents for disease control, including at the farm level, has increased selection of antimicrobial agent-resistant Salmonella isolates (1, 23, 44). Food animals are considered an important reservoir for resistant bacteria. These animals and food products derived from them are traded worldwide, which contributes to the global spread of zoonotic agents and antimicrobial resistance. In the last few years, several monitoring activities were initiated in order to generate baseline data on antimicrobial resistance in bacteria isolated from livestock and food derived from animals that could be used in future assessments of the risk of antimicrobial resistance (10).
According to European Union (EU) Zoonoses Regulation (EC) no. 2160/2003 on the control of Salmonella and other specified food-borne zoonotic agents, a European Community target for reducing the prevalence of Salmonella in turkey flocks had to be established. Consequently, EU Commission decision 2006/662/EC was released, and a baseline survey of the prevalence of Salmonella in turkey flocks was carried out in all European countries, including Germany, over a 1-year period starting on 1 October 2006 (http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178706574172.htm). The main objective of this study was to estimate the prevalence of Salmonella in commercial flocks of turkeys. The data showed that at the EU level Salmonella enterica serovar Bredeney was the serovar reported most frequently for fattening turkey flocks and occurred in 17.2% of the samples from Salmonella-positive flocks (1,084 of 3,702 flocks were positive), followed by S. enterica serovar Hadar, S. enterica serovar Derby, and then S. enterica serovar Saintpaul (14.0%, 11.3%, and 10.4% of the samples from positive flocks, respectively). In this study, S. Saintpaul was detected in fattening turkeys in 12 countries, reflecting the wide spread of this serovar. Recently, S. Saintpaul has been increasingly observed in several countries, including Germany. According to Enter-Net reports (data on Salmonella human isolates identified by European national reference centers), for the last quarter of the year 2006 S. Saintpaul was the fourth most common serovar (1.6%) and, in contrast to the data for previous years, was one of the most frequent causes of human salmonellosis in Europe. After this, its prevalence was 1.2% and 0.6% in the first quarters of 2007 and 2008, respectively, among the Salmonella serotypes implicated in human disease (http://ecdc.europa.eu/en/publications/Pages/Surveillance_Reports.aspx). During the period from 2001 to 2009 in Germany, 463 cases of human salmonellosis related to S. Saintpaul (0.09% of all cases; the maximum prevalence was 0.15% in 2008, the prevalence was 0.1% in 2002, 2005, 2006, and 2009 and 0.06% in 2004, and the minimum prevalence was 0.05% in 2007) were reported to the Robert Koch Institute (Berlin, Germany) (www3.rki.de/SurvStat). In Germany, S. Saintpaul attracted public attention particularly in 1993, when it caused a nationwide food-borne outbreak (27). This serotype has often been related to outbreaks in other countries, and in 2008 it was implicated in a large multistate human outbreak associated with various vegetables in the United States (4).
Previous studies showed that isolates of S. Saintpaul are often multidrug resistant (33, 35), but little is known about the mechanisms underlying antimicrobial resistance or about the pathogenic potential and epidemiology of isolates belonging to this serotype. The goals of this study were to obtain information about the resistance characteristics of isolates collected between 2000 and 2007 in Germany and to assess possible clonal relationships. The isolates used originated from turkey feces collected during the German Salmonella baseline study (in 2006 and 2007) or from diagnostic samples, including samples of turkey feces and turkey-related food products. These isolates were compared with German strains isolated from humans and chickens and with poultry strains isolated in Netherlands.
MATERIALS AND METHODS
Bacterial isolates and selection of isolates.
Fifty-five S. Saintpaul isolates collected in Germany from 2000 to 2007 were analyzed. All of the isolates originated from turkey (33 feces isolates) or turkey meat (22 food isolates). They were selected from isolates submitted to the Salmonella Reference Laboratory (NRL-Salm of the Federal Institute for Risk Assessment) for typing. Duplicate isolates that were putative siblings (isolates obtained from the same place on the same date and exhibiting the same resistance profile) were not included in the study. All isolates included had routinely been tested previously for susceptibility to 17 antimicrobial agents by the broth microdilution method by following the Clinical and Laboratory Standards Institute (CLSI) guidelines (13) as described by Schroeter et al. (37). The antimicrobials tested and MIC breakpoints are shown in Table 1.
TABLE 1.
Sources and characteristics of German S. Saintpaul isolates (n = 55)
| Group | Origin | No. of strains | Resistance phenotypea | Resistance genotype | Class 1 integronb | Origin (no. of isolates)c | PFGE profile |
Plasmid sizes (no. of isolates)d | ||
|---|---|---|---|---|---|---|---|---|---|---|
| XbaI | BlnI | No. | ||||||||
| 1 | Turkey baseline study feces samples (2006 and 2007) | 16 | [R]e | [R′]f | [I]g | NI (13) | X1 | B1 | 4 | 7 kb, two ≤6 kb (2); 7 kb, ≤6 kb; NP |
| X1 | B2 | 8 | NP (7); 41 kb, 16 kb | |||||||
| X1 | B1b | 1 | 28 kb | |||||||
| [R]-TET | [R′]-tet(A) | [I] | MV | X1 | B1 | 1 | ≤6 kb | |||
| NI | X5 | B2 | 1 | NP | ||||||
| [R]-TET | [R′]-tet(A) | [I] + 1,000 bp/aadA1 | NI | X1 | B2a | 1 | 100 kb, 16 kb | |||
| 2 | Turkey diagnostic feces samples (2002 and 2003, 2005 and 2006) | 17 | [R] | [R′] | [I] | BW | X1 | B1 | 1 | 16 kb |
| BW, ST (3) | X1 | B2 | 4 | NP (3); 54 kb | ||||||
| Unknown | X1a | B2 | 1 | ≤6 kb | ||||||
| B | X14 | B3 | 1 | NP | ||||||
| HE | X3 | B10 | 1 | ≤6 kb | ||||||
| B | X4 | B4 | 1 | 16 kb | ||||||
| [R] | [R′] | [I] + 1,000 bp/aadA1 | B | X4 | B4 | 1 | 100 kb, 49 kb | |||
| [R]-TET | [R′]-tet(B) | [I] | NRW | X1 | B2 | 1 | 47 kb, ≤6 kb | |||
| BW, NI | X4 | B4 | 2 | 49 kb, ≤6 kb; 62 kb, ≤6 kb | ||||||
| [R]-CHL | [R′]-sul3-cmlA | [I] | SN | X1 | B2 | 1 | 69 kb, 16 kb | |||
| [R]-CHL-TMP/SXT | [R′]-blaPSE-1-sul3-cmlA-(unknown)h | [I] | B | X2 | B2c | 1 | 100 kb, 16 kb | |||
| Susceptible | BW (2) | X13a | B5 | 2 | 12 kb, three ≤6 kb (2) | |||||
| 3 | Turkey-derived food diagnostic samples (2000 to 2007) | 22 | [R] | [R′] | [I] | NI | X1 | B1 | 1 | NP |
| NRW | X1 | B1a | 1 | NP | ||||||
| NI, NRW | X1 | B2 | 2 | NP (2) | ||||||
| [R]i | [R′] | 1,600 bp/dfrA1-aadA1 | ST | X4 | B4 | 1 | ≤6 kb | |||
| NI | X4 | B14 | 1 | ≤6 kb | ||||||
| [R] | [R′] | [I] + 1,000 bp/aadA1 | B | X4 | B4 | 1 | 100 kb, 16 kb | |||
| [R]-TET | [R′]-aphA1-strA-(unknown)j | [I] | B | X11 | B11 | 1 | 138 kb, 70 kb, 9 kb, 7 kb, three ≤6 kb | |||
| [R]-TET-TMP/SXT | [R′]-aphA1-sul2-tet(A)-dfrA14 | [I] | B | X11 | B11 | 1 | 174 kb, 77 kb, 37 kb | |||
| [R]-CHL-TET-TMP/SXT | [R′]-catA1-tet(B)-dfrA1-like | [I] | BW, RP | X8 | B2b | 2 | 28 kb, ≤6 kb; ≤6 kb | |||
| AMP-TET-STR/SPE-SUL-TMP/SXT | blaTEM-1-tet(A)/tet(B)-aadA1-like-sul1-dfrA1-like | 1,950 bp/estX-aadA1 + 700 bp/estX | B | X6 | B6 | 1 | 41 kb, three ≤6 kb | |||
| TET-STR/SPE-SUL | tet(B)-aadA1-like-sul1 | 1,950 bp/estX-aadA1 + 700 bp/estX | NRW | X6 | B6 | 1 | Three ≤6 kb | |||
| TET-STR/SPE-SUL | tet(B)-aadA1-like-sul1 | 1,950 bp/estX-aadA1 | B (2) | X6 | B6 | 2 | Three ≤6 kb; 70 kb, three ≤6 kb | |||
| AMP-CHL-TET-STR-SUL-TMP/SXT | blaTEM-1- catA1-tet(A)-strA/aadA1-like-sul1/sul2-dfrA1-like | 1,600 bp/dfrA1-aadA1 | SH | X10 | B8 | 1 | NP | |||
| AMP/AMC(i/r)-NAL-TET-STR-SUL | blaTEM-1-gyrASer83→Tyr83-tet(A)-strA-sul2 | 200 bp/without gene cassettes | NRW | X7 | B7 | 1 | 100 kb, two ≤6 kb | |||
| NAL | gyrASer83→Tyr83 | BW | X7a | B7 | 1 | 16 kb, two ≤6 kb | ||||
| GEN-NAL-TET-STR/SPE-SUL | gyrASer83→Glu83-tet(B)-aadA1-like-sul1 | 1,000 bp/aadA1 | TH | X9 | B9 | 1 | 138 kb, 70 kb | |||
| Susceptible | RP | X13a | B5 | 1 | 12 kb, three ≤6 kb | |||||
| NRW | X12 | B12 | 1 | 74 kb | ||||||
| SN | X13 | B13 | 1 | Two ≤6 kb | ||||||
Resistance phenotypes were assessed by the broth microdilution method as described by Schroeter et al. (37) by following the guidelines of the CLSI (13). The breakpoints used for ampicillin (AMP), amoxicillin-clavulanic acid (AMC), ceftiofur (XNL), chloramphenicol (CHL), ciprofloxacin (CIP), gentamicin (GEN), kanamycin (KAN), sulfonamides (SUL), and tetracycline (TET) were the CLSI breakpoints (14). For ciprofloxacin, the MIC breakpoints were as follows: resistant, ≥4 μg/ml; intermediate, 2 μg/ml; and susceptible, ≤1 μg/ml (decreased susceptibility, 1 to 0.12 μg/ml; fully susceptible, ≤0.06 μg/ml). The breakpoints used for colistin (COL) (resistant, ≥16 μg/ml; susceptible, ≤8 μg/ml), florfenicol (FLO) (resistant, ≥32 μg/ml; susceptible, ≤8 μg/ml), nalidixic acid (NAL) (resistant, ≥32 μg/ml; susceptible, ≤16 μg/ml), spectinomycin (SPE) (resistant, ≥128 μg/ml; susceptible, ≤64 μg/ml), and trimethoprim (TMP) (resistant, ≥16 μg/ml; susceptible, ≤8 μg/ml) were the DANMAP breakpoints (17). And the breakpoints used for neomycin (NEO) (resistant, ≥16 μg/ml; susceptible, ≤4 μg/ml), streptomycin (STR) (resistant, ≥32 μg/ml; susceptible, ≤8 μg/ml), and trimethoprim-sulfamethoxazole (SXT) (resistant, ≥4 and ≥76 μg/ml respectively; susceptible, ≤2 and ≤38 μg/ml, respectively) were the breakpoints of Schroeter et al. (37).
Variable regions of class 1 integrons; indicated are their lengths (determined using the 5′CS and 3′CS primers [28]) and the inserted gene cassettes.
B, Berlin; BW, Baden-Württemberg; HE, Hesse; MV, Mecklenburg Western Pomerania; NI, Lower Saxony; NRW, North Rhine-Westphalia; RP, Rhineland-Palatinate; SH, Schleswig-Holstein; SN, Saxony; ST, Saxony-Anhalt; TH, Thuringia. Unless indicated otherwise, the number of isolates was one.
NP, no plasmid detected. Unless indicated otherwise, the number of isolates was one.
[R], core resistance phenotype [AMP/AMC(i/r)-GEN(i/r)-KAN(i/r)-NAL-CIP(i/ds)-STR/SPE-SUL], where (i/r) indicates intermediate or full resistance and (i/ds) indicates intermediate or decreased susceptibility.
[R′], core resistance genotype [blaTEM-1-aadB-gyrASer83→Glu83-aadA1-like/aadA2-sul1].
[I], class 1 integron with the 1,700 bp/aadB-aadA2 variable region.
None of the genes encoding trimethoprim resistance tested were detected in the strain.
The strain exhibited the core resistance pattern, but the class 1 integron with the 1,700 bp/aadB-aadA2 variable region was not detected by PCR.
None of the genes encoding tetracycline resistance tested were detected in the strain.
The strains included in this study were randomly selected and represented the different resistance phenotypes found in each of the three defined sampling groups (Tables 1 and 2). Group 1 consisted of 16 isolates obtained from fecal samples from the 2006-2007 EU turkey baseline study (11 S. Saintpaul isolates and 5 rough isolates confirmed to be S. Saintpaul isolates by molecular methods, which represented [without siblings] the 19 S. Saintpaul isolates and 12 S. rough isolates collected in the baseline study). Group 2 comprised 17 turkey feces isolates randomly selected from routine submissions (11 of 167 S. Saintpaul isolates from turkey feces submitted in 2002 and 2003 and 6 of 46 isolates from turkey feces submitted in 2005 and 2006). Group 3 included 22 turkey-derived food isolates randomly selected from routine submissions of 147 S. Saintpaul isolates from turkey food products from 2000 to 2007.
TABLE 2.
Resistance to individual antibiotics and prevalence of selected resistance genes in German S. Saintpaul isolates (n = 55)a
| Antimicrobialb | No. (%) of resistant isolates | Resistance gene(s) | Isolates containing resistance gene(s) |
|
|---|---|---|---|---|
| No. (%) | Origin (no. of isolates)c | |||
| β-Lactams | ||||
| Ampicillin | 45 (82) | blaPSE-1 | 1 (2) | G2 (1) |
| blaOXA-1-like | 0 | |||
| blaTEM-1-like | 45 (100) | G1 (16), G2 (15), G3 (14) | ||
| blaCTX-M | 0 | |||
| blaSHV | 0 | |||
| Aminoglycosides | ||||
| Streptomycin | 49 (89) | strA | 3 (6) | G3 (3) |
| aadA1-like | 48 (98) | G1 (16), G2 (15), G3 (17) | ||
| aadA2 | 42 (86) | G1 (16), G2 (15), G3 (11) | ||
| Gentamicin | 43 (78)d | aacC2 | 0 | |
| aacC4 | 0 | |||
| aadBe | 42 (98) | G1 (16), G2 (15), G3 (11) | ||
| armA | 0 | |||
| Othersf | 1 (2) | G3 (1) | ||
| Kanamycin | 42 (76)d | aphA1 | 2 (5) | G3 (2) |
| aphA2 | 0 | |||
| aac(6)-1b | 0 | |||
| Phenicols | ||||
| Chloramphenicol | 5 (9) | catA1 | 3 (3/5)g | G3 (3) |
| cmlA1 | 2 (2/5)g | G2 (2) | ||
| floR | 0 | |||
| Folate pathway inhibitors | ||||
| Sulfamethoxazole | 49 (89) | sul1 | 48 (98) | G1 (16), G2 (15), G3 (17) |
| sul2 | 3 (6) | G3 (3) | ||
| sul3 | 2 (4) | G2 (2) | ||
| Trimethoprim | 6 (11) | dfrA1-like | 4 (4/6)g | G3 (4) |
| dfrA7 | 0 | |||
| dfrA12 | 0 | |||
| dfrA14 | 1 (1/6)g | G3 (1) | ||
| dfrA17 | 0 | |||
| Othersf | 1 (1/6)g | G2 (1) | ||
| Tetracyclines | ||||
| Tetracycline | 17 (31) | tet(A) | 5 (29) | G1 (1), G3 (4) |
| tet(B) | 10 (59) | G2 (3), G3 (7) | ||
| tet(G) | 0 | |||
| Othersf | 3 (18) | G1 (2), G3 (1) | ||
| Quinolones and fluoroquinolones | ||||
| Nalidixic acid and ciprofloxacin | 45 (82), 16 (29)h | qnrA | 0 | |
| qnrB | 0 | |||
| qnrC | 0 | |||
| qnrD | 0 | |||
| qnrS | 0 | |||
Five of the 55 isolates selected were susceptible to all of the antimicrobials tested.
The breakpoints used are shown in Table 1. Seven other antimicrobials were also tested: no resistance to florfenicol, ceftiofur, or colistin was detected; 46 (84%) of the isolates were resistant to spectinomycin; 6 (11%) of the isolates were resistant to trimethoprim-sulfamethoxazole; 13 (24%) of the isolates were resistant to amoxicillin-clavulanic acid; and 2 (4%) of the isolates were resistant to neomycin.
G1, turkey baseline study (feces) performed in 2006 and 2007; G2, turkey diagnostic isolates (feces) collected in 2002 and 2003 and in 2005 and 2006; G3, turkey-derived food diagnostic isolates collected in 2000 to 2007.
Intermediate or full resistance.
The aadB gene also confers a low resistance to kanamycin according to Zhao et al. (45).
The resistance mechanisms were not identified in this study.
Because of the low number of resistant isolates (<10), only the absolute number is indicated.
For ciprofloxacin, only isolates with intermediate resistance (MIC, 2 μg/ml) are included. An additional 29 isolates (53%) showed reduced susceptibility (MIC, 1 to 0.25 μg/ml).
Additionally, eight S. Saintpaul isolates from human stool samples (isolated in 2006 and 2007 and provided by the Robert Koch Institute, Wernigerode, Germany) and two S. Saintpaul isolates from chickens (2006), all isolated in Germany, as well as 14 Dutch S. Saintpaul isolates (provided by the Central Veterinary Institute of Wageningen, Lelystad, Netherlands) that originated from poultry and poultry products (in 2006 to 2008), were added as outgroup strains for comparison.
Antimicrobial susceptibility testing.
All isolates were tested by the disk diffusion method (15) to determine their susceptibilities to a panel of 14 β-lactams (Oxoid Ltd., London, England). The antimicrobial agents, concentrations, and breakpoints used (16) are shown in Table 3. A phenotypic test to confirm the presence of extended-spectrum beta-lactamases (ESBLs) was performed using the double-disk technique by placing amoxicillin-clavulanic acid disks (30 μg) 30 mm from ceftazidime (30 μg) and cefotaxime (30 μg) disks. An increase in the size of the cephalosporin zone close to the disk containing the β-lactamase inhibitor (synergy) indicates the presence of ESBLs (www.medvetnet.org/pdf/Reports/Appendix_2_Workpackage_9.doc).
TABLE 3.
β-Lactam susceptibility tests with S. Saintpaul isolates (n = 55)
| Susceptibility testa | Antimicrobial agents |
Concn tested | No. (%) of isolates |
|||||
|---|---|---|---|---|---|---|---|---|
| Subclass | Antibiotic | Resistant | Intermediate | Susceptibleb | Decreased susceptibility | Fully susceptible | ||
| Agar dilution (MIC) | First-generation cephalosporins | Cephalothin | 1-32 μg/ml | 41 (74.5) | 4 (7.3) | 10 (18.2) | ||
| Second-generation cephalosporins | Cefuroxime | 1-32 μg/ml | 35 (63.6) | 10 (18.2) | 10 (18.2) | |||
| Third-generation cephalosporins | Cefotaxime | 1-64 μg/ml | 55 (100) | |||||
| Ceftazidime | 1-32 μg/ml | 5 (9.1) | 50 (90.1) | |||||
| Cefoxitin | 1-32 μg/ml | 8 (14.5) | 40 (72.7) | 7 (12.7) | ||||
| Ceftiofur | 0.5-8 μg/ml | 19 (34.5) | 31 (56.4) | 5 (9.1) | ||||
| Disk diffusion (zone diam) | Penicillins | Ampicillin | 10 μg | 45 (81.8) | 10 (18.2) | |||
| Carboxypenicillins | Ticarcillin | 75 μg | 45 (81.8) | 10 (18.2) | ||||
| Acylaminopenicillins | Piperacillin | 100 μg | 45 (81.8) | 10 (18.2) | ||||
| β-Lactamase inhibitors | Amoxicillin-clavulanic acid | 30 μg | 13 (23.6) | 27 (49.1) | 15 (27.3) | |||
| Third-generation cephalosporins | Cefpodoxime | 10 μg | 6 (10.9) | 49 (89.1) | ||||
| Ceftriaxone | 30 μg | 55 (100) | ||||||
| Fourth-generation cephalosporins | Cefepime | 30 μg | 55 (100) | |||||
| Carbapenems | Imipenem | 10 μg | 55 (100) | |||||
| Monobactams | Aztreonam | 30 μg | 55 (100) | |||||
The MIC breakpoints and zone diameter standards were defined as described by the CLSI (16). The MIC breakpoints for cephalothin, cefuroxime, ceftazidime, and cefoxitin were as follows: resistant, ≥32 μg/ml; intermediate, 16 μg/ml; and susceptible, ≤8 μg/ml (decreased susceptibility, 8 to 2 μg/ml; fully susceptible, <2 μg/ml). The MIC breakpoints for cefotaxime were as follows: resistant, ≥64 μg/ml; intermediate, 32 to 16 μg/ml; and susceptible, ≤8 μg/ml (decreased susceptibility, 8 to 2 μg/ml; fully susceptible, <2 μg/ml). The MIC breakpoints for ceftiofur were as follows: resistant, ≥8 μg/ml; intermediate, 4 μg/ml; and susceptible, <4 μg/ml (decreased susceptibility, 2 to 1 μg/ml; fully susceptible, <1 μg/ml). The zone diameter standards for cefpodoxime and piperacillin were as follows: resistant, ≤17 mm; intermediate, 18 to 20 mm; and susceptible, ≥21 mm. The zone diameter standards for ampicillin were as follows: resistant, ≤13 mm; intermediate, 14 to 16 mm; and susceptible, ≥17 mm. The zone diameter standards for ceftriaxone were as follows: resistant, ≤13 mm; intermediate, 14 to 20 mm; and susceptible, ≥21 mm. The zone diameter standards for amoxicillin-clavulanic acid were as follows: resistant, ≤13 mm; intermediate, 14 to 17 mm; and susceptible, ≥18 mm. The zone diameter standards for cefepime were as follows: resistant, ≤14 mm; intermediate, 15 to 17 mm; and susceptible, ≥18 mm. The zone diameter standards for ticarcillin were as follows: resistant, ≤14 mm; intermediate, 15 to 19 mm; and susceptible, ≥20 mm. The zone diameter standards for imipenem were as follows: resistant, ≤13 mm; intermediate, 14 to 15 mm; and susceptible, ≥16 mm. And the zone diameter standards for aztreonam were as follows: resistant, ≤15 mm; intermediate, 16 to 21 mm; and susceptible, ≥22 mm.
Includes isolates with decreased and full susceptibility.
Further characterization of the MIC values of six selected β-lactams (Sigma-Aldrich, Taufkirchen, Germany) was performed only for the 55 S. Saintpaul isolates from German turkey or turkey food products by using the agar dilution method (13). The antimicrobials tested (Table 3) included first-generation (cephalothin), second-generation (cefuroxime), and third-generation (cefoxitin, cefotaxime, ceftiofur, and ceftazidime) cephalosporins. The antimicrobial concentrations and MIC breakpoints used (16) are shown in Table 3. Escherichia coli ATCC 25922 was used as the control strain.
Susceptibility to organic solvents and triclosan.
All S. Saintpaul isolates were tested to determine their susceptibilities to organic solvents and the disinfectant triclosan. The susceptibility to n-hexane and cyclohexane was analyzed as previously described (8, 29). E. coli AG100 (wild-type E. coli K-12) and AG102 (marR1 mutant of AG100) were used as controls (42). Triclosan (Irgasan; Sigma-Aldrich, Taufkirchen, Germany) susceptibility tests were carried out by using the CLSI agar dilution method (13). The concentrations tested in Mueller-Hinton agar ranged from 0.015 to 64 μg/ml. Isolates were considered to have a high level of resistance (MIC, >32 μg/ml), a medium level of resistance (MIC, 16 to 32 μg/ml), or a low level of resistance (MIC, 4 to 8 μg/ml) or to be susceptible (MIC, <4 μg/ml) as described by Webber et al. (41). Isolates with MIC values between 0.125 and 2 μg/ml were considered to have decreased susceptibility. Three S. enterica serovar Typhimurium strains, L700, L701, and L702, were used as controls.
Detection of resistance determinants and isoelectric focusing.
The isolates were analyzed to determine the presence of the following resistance genes related to their resistance phenotypes: blaPSE-1, blaOXA-1-like, blaTEM-1-like, blaCTX-M, and blaSHV (encoding β-lactam resistance); aadA1-like, aadA2, strA, aacC2, aacC4, aadB, armA, aphA1, aphA2, and aac(6)-1b (aminoglycoside resistance); catA1, cmlA1, and floR (chloramphenicol resistance); sul1, sul2, and sul3 (sulfamethoxazole resistance); tet(A), tet(B), and tet(G) (tetracycline resistance); dfrA1-like, dfrA7-17, dfrA12, dfrA5-14, and dfrA17 (trimethoprim resistance); and qnrA, qnrB, qnrS, qnrC, and qnrD (decreased susceptibility to fluoroquinolone) (12, 21, 36). Mutations in the gyrA gene in the nalidixic acid-resistant isolates was determined by PCR sequencing, as previously described (20, 31). All isolates were screened for the presence of class 1 and 2 integrons by PCR sequencing using primer pairs 5′CS/3′CS (28) and Hep74/Hep51 (43), respectively. Sequencing was performed by AGOWA GmbH (Berlin, Germany), and the sequences obtained were compared with sequences in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html).
For 19 ampicillin-resistant isolates (German turkey, turkey meat, and chicken isolates) the β-lactamase isoelectric point (pI) was determined by isoelectric focusing using a Phast System (GE Healthcare, Freiburg, Germany) as previously described (36).
Plasmid profile typing, macrorestriction analysis, and hybridization experiments.
Bacterial plasmid DNA was obtained from the 55 German S. Saintpaul isolates (from turkey and turkey food products) by using the procedure of Kado and Liu (25) and was separated on 0.8% vertical agarose gels. E. coli reference plasmids R27 (169 kb), R1 (94 kb), RP4 (55 kb), and ColE1 (6 kb) were used as size standards.
Macrorestriction analysis of genomic DNA was carried out for all isolates using restriction endonucleases XbaI and BlnI (Roche Diagnostics, Mannheim, Germany). The fragments obtained were separated by pulsed-field gel electrophoresis (PFGE) using a CHEF-DRIII SYS220/240 system (Bio-Rad). Agarose gel plugs were prepared and PFGE was performed using the PulseNet standardized protocol (www.pulsenet-europe.org). The resulting profiles were analyzed by recording the presence or absence of fragments larger than 33 kb. Profiles in which there were differences in two or more bands were designated using numbers (e.g., X1, X2, etc., or B1, B2, etc.). Similar patterns in which only one band was different were designated using letters (e.g., X1, X1a, and X1b). The genetic similarity between profiles (XbaI, BlnI, or combined XbaI-BlnI PFGE profiles) was determined with the unweighted-pair method with arithmetic averages and Jaccard's coefficient, using the MVSP software (version 3.1; Multivariate Statistics Package for PCs; RockWare Inc., United States).
DNA from XbaI and BlnI PFGE and plasmid gels were transferred by Southern blotting onto membranes and hybridized with a probe for the aadB gene, using methods described previously (21). Plasmid gels were also hybridized with a blaTEM-1 probe.
Statistical methods.
To assess the discriminatory power of PFGE with XbaI and BlnI and a combination of the two methods, discrimination indices (DI) and their 95% confidence intervals (CI) were calculated using the Simpson's index of diversity as previously described (19, 24). This index of diversity determined the probability that two unrelated strains from the test population would be placed in different typing groups. The level of acceptance of discrimination of a typing method depends on several factors, and a DI greater than 0.9 is desirable.
RESULTS
Phenotypic antimicrobial resistance.
The 55 S. Saintpaul isolates were tested to determine their resistance to several β-lactams by using agar diffusion and/or disk diffusion. The results and corresponding breakpoints are shown in Table 3. Eighty-two percent of the isolates showed resistance to penicillins, 75% of the isolates showed resistance to cephalothin, and 64% of the isolates showed resistance to cefuroxime (an additional 36% of the isolates showed intermediate or decreased susceptibility to this antimicrobial agent). No isolate showed full resistance to the different third-generation cephalosporins, but intermediate resistance was detected (14.5% of the isolates showed intermediate resistance to cefoxitin, 34.5% of the isolates showed intermediate resistance to ceftiofur, and 11% of the isolates showed intermediate resistance to cefpodoxime), as was decreased susceptibility (73% of the isolates showed decreased susceptibility to cefoxitin, 56% of the isolates showed decreased susceptibility to ceftiofur, and 9% of the isolates showed decreased susceptibility to ceftazidime). The double-disk synergy test using different third-generation cephalosporins and clavulanic acid was negative for all isolates.
The 16 isolates in group 1 (from the Salmonella baseline study of turkeys) showed two different antimicrobial resistance profiles with the core resistance pattern, AMP/AMC(i/r)-GEN(i/r)-KAN(i/r)-NAL-CIP(i/ds)-STR/SPE-SUL (Table 1), which includes full resistance to ampicillin, streptomycin/spectinomycin, sulfamethoxazole, and nalidixic acid, full (r) or intermediate (i) resistance to amoxicillin-clavulanic acid (MIC, 16 to 32 μg/ml), gentamicin (MIC, 8 to 32 μg/ml), and kanamycin (MIC, 32 to 64 μg/ml), intermediate or decreased susceptibility (ds) to ciprofloxacin (MIC, 2 or 1 μg/ml, respectively), and intermediate or decreased susceptibility to ceftiofur (MIC, 4 to 2 μg/ml). This pattern was also the pattern most frequently observed for groups 2 (diagnostic feces isolates) and 3 (diagnostic food isolates). In addition, this core resistance pattern was also connected with other resistance patterns. In group 3 five other resistance phenotypes were also detected (Table 1). This core resistance pattern was also found in 15 of the 24 outgroup isolates (German chicken and human isolates and Dutch poultry and poultry food isolates).
Determination of resistance to organic solvents and disinfectants.
E. coli K-12 wild-type control strain AG100 produced confluent growth with n-hexane and did not grow in the presence of cyclohexane, whereas E. coli AG102 (resistant control) grew in the presence of both n-hexane and cyclohexane (42). All 55 S. Saintpaul isolates were tolerant to n-hexane, whereas 43 of them (78%) exhibited tolerance to cyclohexane. Furthermore, all isolates (including the outgroup isolates) were susceptible to triclosan, determined as described by Webber et al. (41), and the MIC values were 0.06 μg/ml (13 isolates), 0.125 μg/ml (8 isolates), 0.25 μg/ml (50 isolates), 0.5 μg/ml (7 isolates), and 1 μg/ml (1 isolate).
Antimicrobial resistance determinants.
The prevalence of resistance genes and their distribution in the different groups are shown in Table 2. The blaTEM-1-like, aadB, aadA1-like, aadA2, and sul1 genes conferring resistance to ampicillin, gentamicin and kanamycin, streptomycin and spectinomycin, and sulfamethoxazole, respectively, were the genes that were found most frequently. The aadB gene was detected in 42 of 43 isolates with intermediate or full resistance to gentamicin. Forty of these 42 isolates exhibited intermediate or low resistance to kanamycin (MIC for 36 isolates, 32 μg/ml; MIC for 4 isolates, 64 μg/ml), and 2 isolates, which also contained the aphA1 gene, exhibited full resistance to kanamycin (MIC, >64 μg/ml).
All 45 ampicillin-resistant isolates (82% of the study group) (Tables 2 and 3) contained blaTEM-1-like genes (these genes were not present in the ampicillin-susceptible isolates). The isoelectric focusing method revealed the presence of a β-lactamase with an isoelectric point (pI) of 5.4 (characteristic of TEM-1) in the 19 ampicillin-resistant S. Saintpaul isolates tested. Only one of these isolates also produced a band at approximately pI 5.7, which is characteristic of PSE-1. Sequencing of the blaTEM-1-like PCR products confirmed the presence of the blaTEM-1 gene.
As shown in Table 2, 45 of 55 (82%) isolates were resistant to nalidixic acid (MIC, >128 μg/ml) and showed intermediate or decreased susceptibility to ciprofloxacin (MIC, 2 to 0.25 μg/ml). The other 10 isolates were susceptible to nalidixic acid (MIC, ≤8 μg/ml), and the MIC of ciprofloxacin were ≤0.03 μg/ml. Sequence analysis revealed that in all 45 nalidixic acid-resistant isolates there was a point mutation in the gyrA gene (resulting in a change from Ser83 to Glu83 in 43 isolates [96%] and in a change from Ser83 to Thr83 in 2 food isolates [4%]). All 55 isolates were negative for the plasmid quinolone resistance genes qnrA, qnrB, qnrC, qnrD, and qnrS.
Forty-nine isolates contained class 1 integrons. Six types of class 1 integrons were identified based on the amplicon size and the gene cassette content in the variable region (Table 1). In 37 isolates (64% of the series) class 1 integrons with a 1,700-kb variable region contained aadB-aadA2 gene cassettes. All isolates containing class 1 integrons were also positive for qacEΔ1 (conferring resistance to quaternary ammonium compounds) and, with the exception of one isolate (with a variable-region amplicon consisting of only 200 bp), for sul1. Class 2 integrons were not detected.
To locate the 1,700-bp class 1 integron, PFGE and plasmid DNA Southern blot hybridization using an aadB probe were performed. The aadB-containing integron mapped on chromosomal XbaI fragments ranging from 150 to 245 kb long. In all isolates that produced PFGE patterns similar to the X1 pattern described below (Fig. 1), the integron was always inserted into the smaller, ∼150-kb fragment. The same hybridization experiment was performed with BlnI-digested chromosomal DNA, and it showed that aadB was in fragments ranging from 470 to 600 kb long. Plasmid DNA hybridization with aadB or blaTEM-1 was negative.
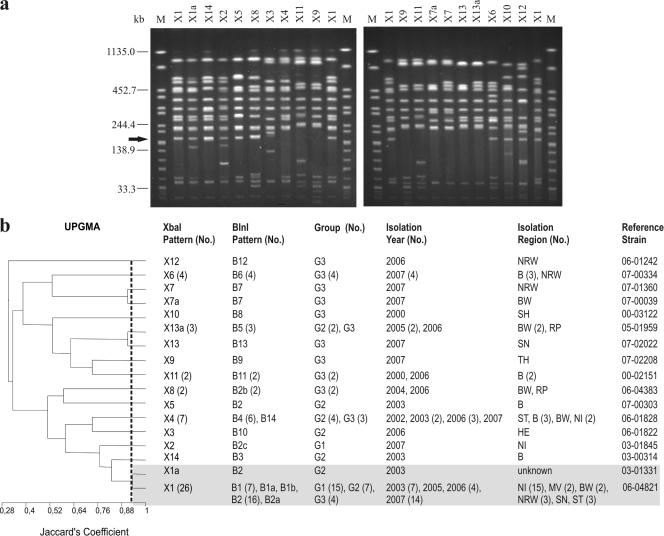
FIG. 1.
(a) XbaI PFGE profiles of representative S. Saintpaul isolates. To define the profiles, only bands at >33 kb were considered. Similar profiles with only a one-band difference were designated by letters. Profiles with differences in two or more bands were designated by numbers. Profiles X1 and X1a are the profiles of the clonal line. The arrow indicates the position of the aadB gene. Lanes M contained XbaI-digested DNA of S. enterica serovar Braenderup H9812, which was used as a size standard. (b) Dendrogram showing the genetic similarity between XbaI profiles determined by using the unweighted-pair group method with arithmetic averages (UPGMA) and Jaccard's coefficient (J). The numbers in parentheses are the numbers of isolates when there was more than one isolate. G1, group 1 (baseline study); G2, group 2 (diagnostic feces study); G3, group 3 (diagnostic food study). Abbreviations for isolation regions in Germany are explained in Table 1. The shading indicates related clonal patterns at a Jaccard's coefficient of 0.93.
Molecular typing.
Thirty-seven (67%) of the isolates harbored plasmids (26 different patterns, with one to seven plasmids ranging from 6 to 174 kb) (Table 1). Only 13 isolates (seven of them from food) harbored plasmids larger than 50 kb. The other 18 isolates (33%), one-half of them (9 isolates) originating from the turkey baseline study (group 1), did not harbor any plasmid.
As determined by PFGE typing, the 55 isolates exhibited 17 XbaI patterns (DI, 0.76 [CI, 0.64 to 0.87]), 19 BlnI patterns (DI, 0.86 [CI, 0.80 to 0.93]), and 22 combined XbaI-BlnI patterns (DI, 0.89 [CI, 0.83 to 0.95]) (Fig. 1 to 3). Eleven XbaI profiles and 12 BlnI profiles were represented by only one isolate, whereas all other profiles were represented by at least two isolates. For the baseline study isolates (group 1) two XbaI patterns and four BlnI patterns were found. For the diagnostic fecal isolates (group 2) the two enzymes each produced seven patterns. The greatest heterogeneity was found for the food isolates (group 3), which exhibited 12 XbaI patterns and 14 BlnI patterns. Information about PFGE patterns, groups, and isolation regions and years is shown in Fig. 1 to 3 and Table 1. The XbaI profile detected most frequently was profile X1 (26 isolates [47%]), which was produced by 15, 7, and 4 isolates belonging to groups 1, 2, and 3, respectively. The most frequent profiles obtained by BlnI digestion were profiles B2 (18 isolates) and B1 (8 isolates). Profile B2 was detected for 9, 7, and 2 isolates belonging to groups 1, 2, and 3, respectively.
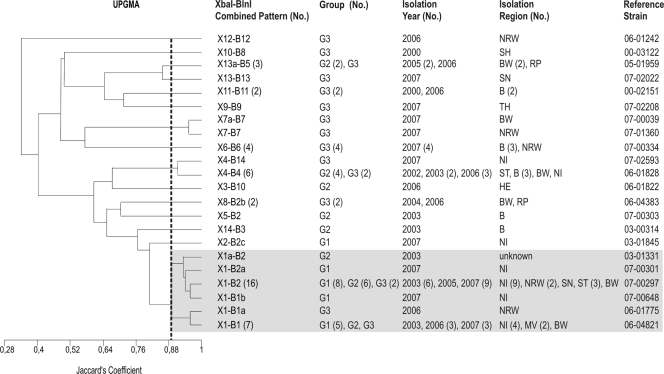
FIG. 3.
Dendrogram showing the genetic similarity between combined XbaI-BlnI profiles determined by using the unweighted-pair group method with arithmetic averages (UPGMA) and Jaccard's coefficient (J). The numbers in parentheses are the numbers of isolates when there was more than one isolate. G1, group 1 (baseline study); G2, group 2 (diagnostic feces study); G3, group 3 (diagnostic food study). Abbreviations for isolation regions in Germany are explained in Table 1. The shading indicates related clonal patterns at a Jaccard's coefficient of 0.90.
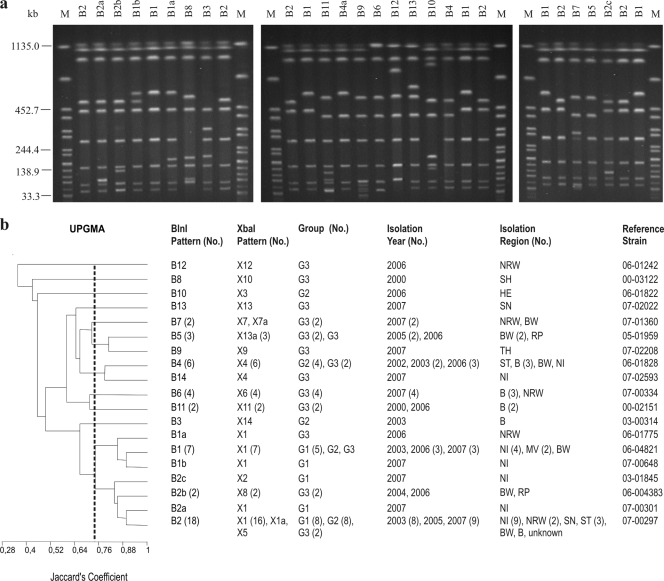
Based on the XbaI analysis, a cluster that included the X1 and X1a profiles (93% similarity; 27 isolates, including 15 isolates belonging to group 1, 8 isolates belonging to group 2, and 4 isolates belonging to group 3) was recognized (Fig. 1). As determined by BlnI analysis, 7 BlnI profiles (B1, B1a, B1b, B2, B2a, B2b, and B2c) clustered together (75% similarity; 31 isolates, including 16 isolates belonging to group 1, 10 isolates belonging to group 2, and 5 isolates belonging to group 3) (Fig. 2). There was a clonal relationship among the 27 isolates that clustered together at 90% similarity using the combinations of profiles (profiles X1 and X1a combined with profiles B1, B1a, B1b, B2, and B2a; 15 isolates belonging to group 1, 8 isolates belonging to group 2, and 4 isolates belonging to group 3) or, as shown above, at 93% similarity using the XbaI profiles (Fig. 1).
FIG. 2.
(a) BlnI PFGE profiles of representative S. Saintpaul isolates. To define profiles, only bands at >33 kb were considered. Similar profiles with only a one-band difference were designated by letters. Profiles with differences in two or more bands were designated by numbers. Profiles B1, B1a, B1b, B2, and B2a are the profiles of the clonal line. Lane M contained XbaI-digested DNA of S. enterica serovar Braenderup H9812, which was used as a size standard. (b) Dendrogram showing the genetic similarity between BlnI profiles determined by using the unweighted-pair group method with arithmetic averages (UPGMA) and Jaccard's coefficient (J). The numbers in parentheses are the numbers of isolates when there was more than one isolate. G1, group 1 (baseline study); G2, group 2 (diagnostic feces study); G3, group 3 (diagnostic food study). Abbreviations for isolation regions in Germany are explained in Table 1. The B1, B1a, B1b, B2, B2a B2b, and B2c profiles clustered together at a Jaccard's coefficient of 0.75.
The outgroup isolates were also analyzed by PFGE with XbaI. Profile X1 was detected for one Dutch food isolate and two German human isolates. Several of the other profiles found previously were also detected for these isolates, including profile X8 for both German chicken isolates and 10 Dutch poultry isolates, profile X4 for one Dutch poultry isolate, profile X13a for three German human isolates, and profile X6 for one German human isolate.
DISCUSSION
Recently, S. Saintpaul has become more important, and in 2006, it ranked among the top five causes of human salmonellosis in Europe (http://ecdc.europa.eu/en/publications/Pages/Surveillance_Reports.aspx). This development has been noticed not only in European countries but also worldwide. Exceptionally eye-catching were two multistate S. Saintpaul outbreaks, one in 2006 in Australia that was associated with cantaloupe consumption (34) and the other in the United States that affected around 1,500 people in the summer of 2008 and was due to vegetables such as jalapeño peppers and tomatoes (4). The latest outbreak, in February 2009 in the United States, was associated with alfalfa sprouts (6). In Germany an important nationwide outbreak in 1993 was linked to contaminated paprika (27), and the isolates recovered showed a high degree of similarity to strains isolated from turkeys, which were suspected to have caused sporadic human infections (9). Most of the outbreaks reported to date in which S. Saintpaul isolates were implicated were related to the consumption of vegetables (paprika, tomatoes, alfalfa) and to the consumption of turkey products (i.e., ready-to-eat meat products) (22, 26), indicating that both kinds of foods have important roles as vehicles of infection. Whereas antimicrobial agent-susceptible strains were involved in most of the vegetable-related outbreaks, the isolates obtained from turkey-related outbreaks were multidrug resistant.
The European Food Safety Authority report on the baseline survey in 2006 and 2007 on the prevalence of Salmonella in turkey flocks indicated that S. Saintpaul was one of the most frequent serotypes (10.4% of isolates) in positive flocks of fattening turkeys in Europe and the leading serotype in Poland, Slovakia, and Netherlands (http://www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178706574172.htm). In Germany 98 flocks of breeding turkeys and 295 flocks of fattening turkey were sampled, and 111 Salmonella isolates were obtained. Nineteen (17%) of these isolates were S. Saintpaul strains, and this serovar was the third most common serovar (5). All S. Saintpaul isolates collected in the study showed resistance to at least five antimicrobials and were included (except for siblings) in the present study.
Of the 52,318 Salmonella isolates typed at NRL-Salm from 1998 to September 2009, 799 were S. Saintpaul isolates, and 533 (67%) of these S. Saintpaul isolates originated from turkeys or turkey products. More than 90% of these isolates were resistant to 2 to 12 antimicrobials. Similar results were described by other workers (33, 35). The most common types of resistance found for these S. Saintpaul isolates were resistance to ampicillin (85% of the isolates), resistance to sulfamethoxazole (82%), resistance to nalidixic acid (79%), and resistance to streptomycin (75%); 34% and 25% of the isolates were resistant to gentamicin and kanamycin, whereas 44% and 49% of the isolates exhibited intermediate resistance to these antimicrobials. When the strains used in the study were selected, the intention was to use representative isolates obtained from active and passive monitoring over a defined period of time and to compare them to human and other European isolates. As demonstrated in the present study and in previous studies (32), some of the types of resistance observed for S. Saintpaul isolates are related to the presence of class 1 integrons (Table 1), which are genetic structures that confer multidrug resistance. The presence of integrons enhances the coselection of different types of resistance.
For the organisms analyzed in this study, the percentage of isolates resistant to nalidixic acid in both feces and food samples was also high (82%). The percentage of nalidixic acid-resistant isolates was much higher than the percentages found in other poultry studies (7, 11, 30, 31). These isolates displayed intermediate resistance to ciprofloxacin (16 isolates [29%] with an MIC of 2 μg/ml) or decreased susceptibility to this agent (25 isolates [46%] with an MIC of 1 μg/ml and 4 isolates [7%] with MIC of 0.5 to 0.25 μg/ml). Using the harmonized cutoff values defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (www.eucast.org), as required in the EU guidelines for monitoring antimicrobial resistance (10), all these isolates are now considered resistant (cutoff value, >0.06 μg/ml). Because of this, the results are worrying since fluoroquinolones are antimicrobial agents that are critically important in human and veterinary medicine (3). As described by Malorny et al. (31), nalidixic acid-resistant isolates were isolated more frequently from turkeys than from other poultry. Although qnr genes were found in S. Saintpaul isolates analyzed in other studies (12, 18; B. Guerra, unpublished data), no qnrA, qnrB, qnrS, qnrC, or qnrD genes were detected in the 55 isolates tested. However, these isolates had mutations in the quinolone resistance-determining region of the gyrA gene and probably had enhanced active efflux mechanisms that could be implicated in the resistance to cyclohexane and the decreased susceptibility to triclosan and fluoroquinolones (29, 41, 42).
A common core resistance pattern, AMP/AMC(i/r)-GEN(i/r)-KAN(i/r)-NAL-CIP(i/ds)-STR/SPE-SUL, was found preferentially in the isolates from feces samples. This pattern was associated with intermediate resistance or decreased susceptibility to ciprofloxacin, resistance to first-generation cephalosporins, resistance or intermediate resistance to second-generation cephalosporins, and intermediate or decreased susceptibility to some third-generation cephalosporins. From a public health perspective, this finding is also worrisome, because these antimicrobials, together with the fluoroquinolones, are drugs of choice for clinical treatment of systemic Salmonella infections (3, 23). However, the results of screening for resistance-conferring β-lactam genes, such as blaCTX-M, blaSHV, and blaOXA-1-like, were negative, and only the blaTEM-1 gene, located in the chromosome, was detected. Therefore, we speculate that the negative isolates possess other molecular mechanisms (e.g., loss of porins, genes that were not detected, or multidrug resistance pumps) (38) that are responsible for resistance to β-lactams, which could not be determined in the present work. In order to examine this possibility, these isolates are being investigated further.
PFGE typing proved that 15 of 16 isolates used in the baseline survey (isolated from five farms in two federal regions) exhibiting the core resistance pattern described above belonged to the same clonal line (XbaI PFGE patterns with 93% genetic similarity). All of the isolates belonging to this clonal line had the same core resistance genotype (blaTEM-1, aadB, aadA2, sul1, and the Ser83→Glu83 mutation in the gyrA gene), including a class 1 integron conferring resistance to gentamicin, kanamycin, streptomycin/spectinomycin, and sulfonamides. Overall, the distribution of plasmids in S. Saintpaul was highly variable, although the majority of isolates that belonged to the clonal line contained only small plasmids or no plasmids. In fact, hybridization experiments showed that the class 1 integron carrying the aadB gene was located on the chromosome. This integron, which was also present in 9 other isolates included in our work that do not belong to the same clonal line, was described in previous studies (32, 45), but to our knowledge, so far the aadB gene has been detected only on plasmids in Salmonella (39) and a chromosomal location has not been described previously. In some of the poultry- and turkey-rearing facilities where the S. Saintpaul strains were isolated, antimicrobials like penicillin and enrofloxacin were used (unpublished data). Consequently, the presence of chromosomally located resistance determinants (i.e., genes and mutations) in the S. Saintpaul isolates conferred a selective advantage in the presence of the antimicrobial selection pressure on these farms.
Derivatives of the multiresistant S. Sainpaul strains described above were found throughout Germany not only among isolates examined in the baseline study performed in 2006 and 2007 but also in diagnostic feces samples obtained previously in 2003 to 2005. Isolates belonging to this clonal line were also found among S. Saintpaul isolates that originated from poultry in neighboring countries, like Netherlands, which suggests that this line might be widespread. This clonal line was also represented by German food isolates, making transmission to humans likely. The analysis of human isolates provided by the Robert Koch Institute confirmed this hypothesis. The results presented here describe the characteristics of multiresistant S. Saintpaul isolates originating from turkeys whose spread and molecular development should be examined further for protection of public health.
Acknowledgments
We thank W. Barownick, G. Berendonk, M. Jaber, K. Thomas, P. Bahn, and R. Dieckmann for their invaluable help and advice. We also gratefully acknowledge D. Mevius and K. Veldman (Central Veterinary Institute of Wageningen, Lelystad, Netherlands) for providing the Dutch S. Saintpaul isolates, W. Rabsch (Robert Koch Institute, Wernigerode, Germany) for providing the German S. Saintpaul isolates from humans, E. Liebana (European Food Safety Authority, Parma, Italy) and M. Webber (University of Birmingham, Birmingham, United Kingdom) for providing the control strains used for organic solvent and disinfectant susceptibility analysis, and M. A. Argudín (University of Oviedo, Spain) for providing advice.
This work was supported by the Federal Institute for Risk Assessment (grants BfR-45-004, BfR-45-005, and BfR-46-001). I. Rodríguez was a recipient of a postdoctoral grant from the “Fundación Ramón Areces,” Spain.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Acar, J. F., and G. Moulin. 2006. Antimicrobial resistance at farm level. Rev. Sci. Tech. 25:775-792. [PubMed] [Google Scholar]
- 2.Anonymous. 2003. Joint FAO/OIE/WHO Expert Workshop on Non-Human Antimicrobial Usage and Antimicrobial Resistance: Scientific Assessment. Report of a meeting held in Geneva, Switzerland, 1 to 5 December 2003. World Health Organization, Geneva, Switzerland.
- 3.Anonymous. 2007. Joint FAO/WHO/OIE Expert Meeting on Critically Important Antimicrobials. Report of a meeting held in Rome, Italy, 26 to 30 November 2007. Food and Agriculture Organization of the United Nations Headquarters, Rome, Italy.
- 4.Anonymous. 2008. Outbreak of Salmonella serotype Saintpaul infections associated with multiple raw produce items—United States, 2008. Centers for Diseases Control (CDC). MMWR Morb. Mortal. Wkly. Rep. 57:929-934. [PubMed] [Google Scholar]
- 5.Anonymous. 2008. Grundlagenstudie zur Erhebung der Prävalenz von Salmonellen in Truthühnerbeständen. Bericht des BfR vom 04. März 2008. Bundesinstitut für Risikobewertung, Berlin, Germany.
- 6.Anonymous. 2009. Outbreak of Salmonella serotype Saintpaul infections associated with eating alfalfa sprouts—United States, 2009. Centers for Diseases Control (CDC). MMWR Morb. Mortal. Wkly. Rep. 58:500-503. [PubMed] [Google Scholar]
- 7.Antunes, P., C. Reu, J. C. Sousa, L. Peixe, and N. Pestana. 2003. Incidence of Salmonella from poultry products and their susceptibility to antimicrobial agents. Int. J. Food Microbiol. 82:97-103. [DOI] [PubMed] [Google Scholar]
- 8.Asako, H., H. Nakajima, K. Kobayashi, M. Kobayashi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 63:1428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beyer, W., F. M. Mukendi, P. Kimmig, and R. Bohm. 1998. Suitability of repetitive-DNA-sequence-based PCR fingerprinting for characterizing epidemic isolates of Salmonella enterica serovar Saintpaul. J. Clin. Microbiol. 36:1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bronzwaer, S., F. Aarestrup, A. Battisti, B. Bengtsson, S. Piriz Duran, H. D. Emborg, G. Kahlmeter, D. Mevius, G. Regula, P. Sanders, C. Teale, D. Wasyl, K. De Smet, J. Torren Edo, P. Tüll, H. Deluyker, and P. Mäkelä. 2008. Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin. Microbiol. Infect. 14:522-533. [DOI] [PubMed] [Google Scholar]
- 11.Carramiñana, J. J., C. Rota, I. Agustín, and A. Herrera. 2004. High prevalence of multiple resistance to antibiotics in Salmonella serovars isolated from a poultry slaughterhouse in Spain. Vet. Microbiol. 104:133-139. [DOI] [PubMed] [Google Scholar]
- 12.Cavaco, L. M., H. Hasman, S. Xia, and F. M. Aarestrup. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 7th ed. (M7-A7), vol. 26, no. 2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 14.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement (M100-S16), vol. 26, no. 3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 15.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 9th ed. (M2-A9), vol. 26, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 16.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing, 18th informational supplement (M100-S18), vol. 28, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 17.Emborg, H.-D., and A. M. Hammerum (ed.). 2007. DANMAP 2006. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. National Food Institute, Technical University of Denmark, Søborg, Denmark. http://www.danmap.org/pdfFiles/Danmap_2006.pdf.
- 18.García-Fernández, A., D. Fortini, K. Veldman, D. Mevius, and A. Carattoli. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63:274-281. [DOI] [PubMed] [Google Scholar]
- 19.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerra, B., B. Malorny, A. Schroeter, and R. Helmuth. 2003. Multiple resistance mechanisms in fluoroquinolone-resistant Salmonella isolates from Germany. Antimicrob. Agents Chemother. 47:2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerra, B., E. Junker, A. Miko, R. Helmuth, and M. C. Mendoza. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83-91. [DOI] [PubMed] [Google Scholar]
- 22.Helmuth, R., O. Pietzsch, R. Stephan, T. Chakraborty, and E. Bulling. 1984. Gentamicin-resistant salmonellae in turkey rearing, p. 237-242. In M. Woodbine (ed.), Antimicrobials and agriculture. Proceedings of the 4th International Symposium. Butterworths, London, United Kingdom.
- 23.Helmuth, R., and A. Hensel. 2004. Towards the rational use of antibiotics: results of the first International Symposium on the Risk Analysis of Antibiotic Resistance. J. Vet. Med. Ser. B 51:357-360. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson′s index of diversity. J. Clin. Microbiol. 26:2464-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaitsa, M. L., R. B. Kegode, and D. K. Doetkott. 2007. Occurrence of antimicrobial-resistant Salmonella species in raw and ready to eat turkey meat products from retail outlets in the midwestern United States. Foodborne Pathog. Dis. 4:517-525. [DOI] [PubMed] [Google Scholar]
- 27.Lehmacher, A., J. Bockemühl, and S. Aleksic. 1995. Nationwide outbreak of human salmonellosis in Germany due to contaminated paprika and paprika-powdered potato chips. Epidemiol. Infect. 115:501-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebana, E., C. Clouting, C. A. Cassar, L. P. Randall, R. A. Walker, E. J. Threlfall, F. A. Clifton-Hadley, A. M. Ridley, and R. H. Davies. 2002. Comparison of gyrA mutations, cyclohexane resistance, and the presence of class I integrons in Salmonella enterica from farm animals in England and Wales. J. Clin. Microbiol. 40:1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logue, C. M., J. S. Sherwood, P. A. Olah, L. M. Elijah, and M. R. Dockter. 2003. The incidence of antimicrobial-resistant Salmonella spp. on freshly processed poultry from US midwestern processing plants. J. Appl. Microbiol. 94:16-24. [DOI] [PubMed] [Google Scholar]
- 31.Malorny, B., A. Schroeter, B. Guerra, and R. Helmuth. 2003. Incidence of quinolone resistance in strains of Salmonella isolated from poultry, cattle and pigs in Germany between 1998 and 2001. Vet. Rec. 153:643-648. [DOI] [PubMed] [Google Scholar]
- 32.Miko, A., K. Pries, A. Schroeter, and R. Helmuth. 2005. Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany. J. Antimicrob. Chemother. 56:1025-1033. [DOI] [PubMed] [Google Scholar]
- 33.Molla, B., A. Miko, K. Pries, G. Hildebrandt, J. Kleer, A. Schroeter, and R. Helmuth. 2007. Class 1 integrons and resistance gene cassettes among multidrug resistant Salmonella serovars isolated from slaughter animals and foods of animal origin in Ethiopia. Acta Trop. 103:142-149. [DOI] [PubMed] [Google Scholar]
- 34.Munnoch, S. A., K. Ward, S. Sheridan, G. J. Fitzsimmons, C. T. Shadbolt, J. P. Piispanen, Q. Wang, T. J. Ward, T. L. Worgan, C. Oxenford, J. A. Musto, J. McAnulty, and D. N. Durrheim. 2009. A multi-state outbreak of Salmonella Saintpaul in Australia associated with cantaloupe consumption. Epidemiol. Infect. 137:367-374. [DOI] [PubMed] [Google Scholar]
- 35.Nde, C. W., and C. M. Logue. 2008. Characterization of antimicrobial susceptibility and virulence genes of Salmonella serovars collected at a commercial turkey processing plant. J. Appl. Microbiol. 104:215-223. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez, I., W. Barownick, R. Helmuth, M. C. Mendoza, M. R. Rodicio, A. Schroeter, and B. Guerra. 2009. Extended-spectrum β-lactamases and AmpC β-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003-07. J. Antimicrob. Chemother. 64:301-309. [DOI] [PubMed] [Google Scholar]
- 37.Schroeter, A., B. Hoog, and R. Helmuth. 2004. Resistance of Salmonella isolates in Germany. J. Vet. Med. Ser. B 51:389-392. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz, S., and E. Chaslus-Dancla. 2001. Use of antimicrobials in veterinary medicine and mechanisms of resistance. Vet. Res. 32:201-225. [DOI] [PubMed] [Google Scholar]
- 39.Tosini, F., P. Visca, I. Luzzi, A. M. Dionisi, C. Pezzella, A. Petrucca, and A. Carattoli. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, M., Q. Guo, X. Xu, X. Wang, X. Ye, S. Wu, D. C. Hooper, and M. Wang. 2009. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53:1892-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webber, M. A., L. P. Randall, S. Cooles, M. J. Woodward, and L. J. Piddock. 2008. Triclosan resistance in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 62:83-91. [DOI] [PubMed] [Google Scholar]
- 42.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, D. G., S. Zhao, S. Simjee, D. D. Wagner, and P. F. McDermott. 2002. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 4:405-412. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, S., P. J. Fedorka-Cray, S. Friedman, P. F. McDermott, R. D. Walker, S. Qaiyumi, S. L. Foley, S. K. Hubert, S. Ayers, L. English, D. A. Dargatz, B. Salamone, and D. G. White. 2005. Characterization of Salmonella Typhimurium of animal origin obtained from the National Antimicrobial Resistance Monitoring System. Foodborne Pathog. Dis. 2:169-181. [DOI] [PubMed] [Google Scholar]