Abstract
To elucidate the function of an unknown regulator in Streptomyces, differences in phenotype and antibiotic production between a deletion mutant and a wild-type strain (WT) were compared. These differences are easily hidden by complex media. To determine the specific nutrient conditions that reveal such differences, we used a multiwell method containing different nutrients along with bromothymol blue. We found several nutrients that provide key information on characterization conditions. By comparing the growth of wild-type and mutant strains on screened nutrients, we were able to measure growth, organic acid production, and antibiotic production for the elucidation of regulator function. As a result of this method, a member of the MarR-like regulator family, SCO5405 (AbsC), was newly characterized to control pyruvate dehydrogenase in Streptomyces coelicolor. Deletion of SCO5405 increased the pH of the culture broth due to decreased production of organic acids such as pyruvate and α-ketoglutarate and increased extracellular actinorhodin (ACT) production in minimal medium containing glucose and alanine (MMGA). This method could therefore be a high-throughput method for the characterization of unknown regulators.
The rapid increase in genome sequencing data, combined with proteomics techniques and a PCR targeting method using λ undecylprodigiosin (λ RED)-mediated recombination (12), has made available abundant physiological and genetic information on Streptomyces species, the major producers of antibiotics (2, 16). These technical advances are time-saving with the help of bioinformatics and therefore can contribute to elucidating the individual and exact functions of genes (8). Despite these developments, the rapid functional identification of any unknown gene is still difficult and time-consuming when the gene is located between uncharacterized, neighboring genes or when it does not show any homology to other species. Especially for Streptomyces, which has many more regulators than any other bacterium (42), the existence of unknown or putative regulators prevents the full understanding of antibiotic production and regulator interaction under various conditions. Even worse, detection of differences in gene expression levels is difficult, since Streptomyces bacteria express biosynthetic and catabolic enzymes at relatively low constitutive levels (15). Thus, elucidating the relationship between regulatory function and nutrient composition takes more time than finding novel regulators using various approaches.
One of the easiest ways to characterize the function of a regulator is to delete the gene and monitor differences that occur in phenotype or antibiotic production compared to those of the wild type (WT) under various nutrient conditions. This is effective because the primary and secondary metabolisms of Streptomyces are tremendously affected by nutrients, as are various other cellular events (27, 28) which involve many regulators either directly and/or indirectly (Fig. 1) (42). However, there are also many regulators that affect primary metabolism only under a specific nutrient state without dramatically altering antibiotic production or having only a small effect. Even more, their functions were easily masked by the numerous nutrients in complex media, which are the conventional choice for growing Streptomyces cells.
FIG. 1.
Schematic of transcriptional regulator characterization. Application of pH indicator can complement the finding of nutrient information.
To overcome these obstacles, we developed an improved method to sensitively monitor pH differences caused by varying the production or consumption of organic acids between the WT and the deletion mutant. This screening method clearly complements general nutritional screening and shows a color change depending on the pH of the medium. Such a change in color cannot be easily observed in normal growth media in a 96-well plate without bromothymol blue. A different color suggests that deletion of a regulator caused an abnormal pH change compared to the pH of the WT when under specific nutrient conditions. The regulator is expected to be involved in utilizing these nitrogen or carbon sources for the production of organic acids such as pyruvate, acetate, and α-ketoglutarate. As a result, growth of both the WT and the deletion mutant with screened nutrients amplifies the small differences between them that were previously hidden by complex media. This allows the easy identification of phenotypic and antibiotic differences, which in turn provide critical information for the characterization of a regulator of unknown function and for the complementation of a deleted gene. Overall, these methods can accelerate the characterization of unknown regulators (Fig. 1).
Previous efforts to find new regulators affecting antibiotic production have utilized DNA affinity capture assay (DACA) (25, 26, 41), which found a new MarR-like regulator (SCO5405) that is involved in both actinorhodin (ACT) and undecylprodigiosin (RED). This regulator has been separately named AbsC by another group (34), but one of its functions had just been reported when we revised our manuscript (13). In general, the MarR family transcriptional regulatory system (31) is known to provide resistance to multiple antibiotics, it detects environmental chemicals such as organic solvents and aromatic compounds as well as oxidative stress agents like organic hydroperoxides (4, 5), and it regulates the expression of virulence factors and adaptation to environmental changes. These MarR regulators widely exist in various bacteria (11), such as Streptomyces coelicolor, which alone contains 42 putative MarR-like regulators (http://strepdb.streptomyces.org.uk/) (6); however, most of their specific gene functions are still unknown in Streptomyces. Therefore, a homology search with the MarR family did not give any further clues for the identification of SCO5405 function.
In this report, the function of the MarR regulator, SCO5405, in S. coelicolor was elucidated. It was shown that SCO5405 controls the production of organic acids such as pyruvate and α-ketoglutarate, resulting in a pH increase in the liquid culture media. It also binds to the upstream promoter region of genes encoding pyruvate dehydrogenase units (aceE1, aceE2). SCO5405 is named AbsC, antibiotic synthesis-deficient regulator in S. coelicolor.
MATERIALS AND METHODS
Bacterial strains.
All Escherichia coli and Streptomyces strains used in this work are listed in Table 1. The culture method for strain M145 followed standard procedures (18). Briefly, fresh spores of the M145 strain (7.92 × 105 colonies/μl) and the SCO5405 deletion mutant (BG711) (5.32 × 105 colonies/μl) were collected on R5− medium, composed of 103 g sucrose, 0.25 g K2SO4, 10.12 g MgCl2·6H2O, 10 g glucose, 0.1 g Difco Casamino Acids, 2 ml of a trace element solution composed of 40 mg ZnCl2, 200 mg FeCl3·6H2O, 10 mg CuCl2·2H2O, 10 mg MnCl2·4H2O, 10 mg Na2B4O7·10H2O, 10 mg (NH4)6Mo7O24·4H2O in 1 liter of distilled water, 5 g yeast extract, 5.73 g TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer, and 7 ml of 1 N NaOH in 1 liter of distilled water. Minimal medium with glucose and alanine (MMGA) contained 0.5 g l-alanine as a nitrogen source, 0.5 g K2HPO4, 0.2 g MgSO4·7H2O, 0.01 g FeSO4·7H2O, 22 g agar, and 10 g glucose as a carbon source in 1 liter distilled water. To monitor the growth curve, 1 ml of each sample was collected at specified times and the optical density (OD) was measured at 450 nm. The culture supernatant OD was then subtracted from the cell OD.
TABLE 1.
Strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant information | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli strains | ||
| DH5α | F− φ80lacZ M15 endA recA hsdR(rk− mk−) supE thi gyrA relA Δ(lacZYA-argF)U169 | Laboratory stock |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm | Novagen |
| JM110 | dam dcm mutant | Laboratory stock |
| BW25113 | K-12 derivative; ΔaraBAD ΔrhaBAD | 12 |
| S. coelicolor | ||
| A3(2)M145 | SCP1− SCB2− Pgl+ | KCTCa |
| BG711 | absC deletion mutant | This paper |
| BG712 | A3(2) M145 carrying pYH713 | This paper |
| BG713 | absC deletion mutant carrying pYH713 | |
| BG714 | absC deletion mutant carrying pYH714 | |
| Plasmids | ||
| pET24ma | p15A replication origin, T7 lac promoter, C-terminal His-tag coding, Kanr | 40 |
| pIBR25 | pWHM3 carrying ermE* promoter (EcoRI/KpnI) from Saccharopolyspora erythraea | 37 |
| pYH712 | pET24ma carrying PCR product of absC from S. coelicolor | |
| pYH713 | pIBR25 carrying PCR product of absC (SCO5405) from S. coelicolor | |
| pYH714 | pWHM3 carrying PCR product of SCO5405 and SCO5406 together with 200 bp upstream of SCO5406 as the promoter | |
| pIJ773 | aac(3)IV(Aprar) + oriT | 12 |
| Primers | ||
| 5405SCf | CGTGGATCCATGGAGACCGAGACGGCCAC | |
| 5405SCb | ACGAAGCTTTCAACCGTTGCGGCGCAGCG | |
| 5406_200bp | ATATGGATCCGTCTTCGAGGTGCCCATCACC/ATATAAGCTTGATCAGGGACTCAGGGTCGTCb | |
| 5405Ecolif | CGTGGATCCATGGAGACCGAGACGGCCAC | |
| 5405Ecolib | ACGAAGCTTTCAGTGGTGGTGGTGGTGGTGACCGTTGCGGCGCAGCG | |
| Del5405f | TAGTTCGCCTTTCAACCTTTTAGGTAGACTTCATTCATGATTCCGGGGATCCGTCGACC | |
| Del5405b | CCGGGCCGGAACGGCCCGGGACATGTGATCAGGGACTCATGTAGGCTGGAGCTGCTTC | |
| 5405confirm | CGGTAATCTGCGGAGTGTGAC/GCACGCTCACGGTGCACGACGb | |
| Del-scbRf | ATCCTTCCCGGGGGAGACATGAACAAGGAGGCAGGCATGATTCCGGGGATCCGTCGACC | |
| Del-scbRb | ACGGCGGGTCGGTATCCGGTGCGGGCGCTTCGGCGGTCATGTAGGCTGGAGCTGCTTC | |
| EMSA-citA | ACGGTGTCGGCCGGGAGGCGG/GTCTTCCCTCACCGACGTAGTb | 300 bp (−300∼−1) |
| EMSA-sacA | CCCCTTCCGTGTCGCGCCGGG/GACAGTCTCCTTCATTTATGTb | 300 bp (−300∼−1) |
| EMSA-aceE1 | CCCTGGAGAAGGGCTGCGGAC/GTCGCCGCCTTCCTCAGTCGAb | 300 bp (−300∼−1) |
| EMSA-aceE2 | GGTGACGATCCCGGCTGTGTC/GCTGTTCCTTCGCTGTCAGAGb | 300 bp (−300∼−1) |
KCTC, Korean Collection for Type Culture.
Forward/reverse.
To construct pYH712 and pYH713, the coding region of absC was amplified by PCR using primers (Table 1) from the genomic DNA of S. coelicolor A3 (2) M145. The PCR amplifications were carried out for 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, with a final extension stage at 60°C for 5 min using PFU polymerase (Genenmed, South Korea). The absC fragments were digested with BamHI/HindIII (Roche, Germany) and inserted into the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression vector pET24ma, which was kindly donated by Hiroshi Sakamoto (Pasteur Institute, France), and pIBR25 (Table 1). Other DNA manipulations, including preparation of plasmids, restriction enzyme digestion, ligation, and transformation of Escherichia coli, were followed as previously described (29).
Preparation of 96-well experiments.
Fresh spores of the wild-type strain (M145), scbR deletion mutant (ΔscbR), and BG711 were collected on R5− medium (18). To prepare 96-well plates, eight carbon sources (glucose, mannose, N-acetylglucosamine, glucosamine, xylose, fructose, maltose, and glycerol) were prepared at vertical-axis wells and 12 amino acids as nitrogen sources (glutamate, aspartate, serine, threonine, phenylalanine, alanine, valine, methionine, asparagine, glutamine, leucine, and glycine) were prepared at horizontal-axis wells in a 96-well plate (Fig. 2). Bromothymol blue (0.2 g/liter MgSO4·7H2O and 0.01 g/liter FeSO4·7H2O, 0.01% [wt/vol]) was added to 180 μl of autoclaved minimal medium containing 0.5 g/liter K2HPO4 (18), which was aliquoted on the 96-well plate using a multipipette. Ten μl of 100 mM carbon source and nitrogen source was added to each well. To characterize the use of nutrients, M145 spores and deletion mutants were inoculated and grown in two 100-ml flasks containing 15 ml R5− medium for 20 h. Cells were then harvested by centrifugation at 13,000 × g and sequentially washed with sterilized water two times before inoculation into minimal medium in the 96-well plate. Organic acid was measured with high-performance liquid chromatography (HPLC) at 210 nm with a CarboPaK PA-1 column (Dionex) (0.7 ml/min, 0.1% H2SO4 in water) isocratically.
FIG. 2.
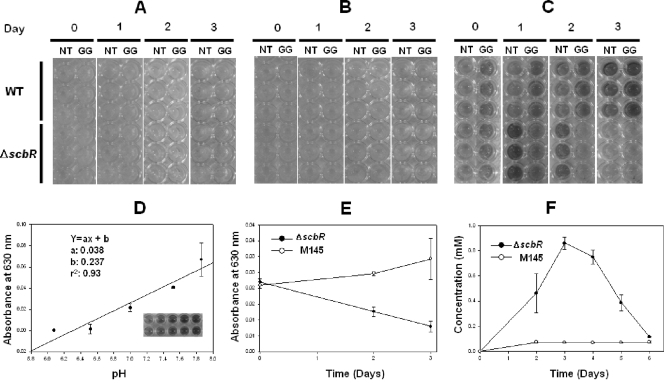
Application of pH indicator for monitoring of sensitive pH differences. Wild-type and ΔscbR cells were grown in media containing N-acetylglucosamine/threonine (NT) and glucose/glycine (GG) without pH indicator (A), with 0.01% (wt/vol) of 4-nitrophenol (B), and with bromothymol blue (C). (D) A linearity curve of bromothymol blue is shown. (E) A630 was measured in glucose/glycine medium supplemented with bromothymol blue from the result shown in panel C. (F) Quantification of pyruvate in glucose/glycine medium using HPLC.
Electrophoretic mobility shift assay (EMSA).
The double-stranded oligonucleotides of the promoter region were radiolabeled with T4 polynucleotide kinase in the presence of [γ-32P]dATP, followed by removal of unreacted [γ-32P]dATP using ProbeQuant G-50 microcolumns (GE Healthcare). The labeled probes were incubated with purified His-tagged proteins for 30 min at room temperature in 20 μl of 20 mM HEPES buffer (pH 7.8) containing 10% (wt/vol) glycerol, 100 mM KCl, 0.05 mM EDTA, 5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.01% Nonidet P-40, and 2 μg sheared salmon sperm DNA (sssDNA). Protein-bound and free DNAs were resolved on 5% acrylamide gels in 0.5× Tris-borate-EDTA (TBE) buffer at room temperature. The gels were dried and analyzed with Typhoon 8600 (GE Healthcare).
Construction of deletion mutant.
The complete coding region of SCO5405 was deleted from the corresponding cosmid (ST8F4) by PCR targeting using oligonucleotide primers (Del5405 in Table 1) with 5′ ends overlapping the upstream (36 bp) and downstream (36 bp) ends of the SCO5405 coding sequence and 3′ (priming) ends containing start and stop codons. These primers were designed to amplify the pIJ773 cassette containing the apramycin resistance gene (Table 1). The mutant was constructed as described elsewhere (12). The deletion of SCO5405 was confirmed by sequencing of corresponding PCR products. In this study, BG713 and BG714 were constructed to check if the SCO5405 is complemented (Table 1). BG713 is cloned in pIBR25, which contains the ermE* promoter, whereas BG714 is cloned in the promoterless pWHM3. For the promoter of BG714, 200 bp upstream of SCO5406 is cloned together with SCO5405 and SCO5406. It is also shown that SCO5405 and SCO5406 can complement the SCO5405 point-mutated strain identified by another research group (13).
Measurement of RED and ACT production.
To determine the amount of extracellular ACT, 1 ml of each culture supernatant was treated with KOH (1 N final concentration) and the A630 was measured for each one. The remaining cells were washed twice with double-distilled water (DDW), and RED was measured at 530 nm after extraction with HCl-acidified methanol (pH 2). Absorbance was measured using a 96-well plate in a multiscanner (Thermo Scientific) (10, 18).
Assay of enzyme activities and organic acids.
WT and BG711 were grown for 20 h in R5− medium by inoculating the spores prepared in R5− medium. The cells were then harvested by centrifugation at 13,000 × g and sequentially washed with sterilized water two times before duplicate inoculation into 20 ml of MMGA. After 2 days and 4 days of growth in MMGA, 1 ml of each sample was taken and immediately treated with RNAlater (Sigma) and preserved in 4°C for later use in reverse transcription-PCR (RT-PCR) analysis. The remaining cells were harvested and washed twice with phosphate-buffered saline (PBS), subsequently resuspended in 4 ml of 50 mM Tri-HCl buffer (pH 7.8) containing 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.01% (vol/vol) 2-mercaptoethanol, and 1 mM dithiothreitol, and then subjected to ultrasonic disruption for 5 min (3 s on and 8 s off). The supernatant was obtained immediately after centrifugation. The amount of cytosolic total protein was measured by Bradford assay. Pyruvate dehydrogenase activity was measured at 340 nm using 50 mM Tris, pH 7.8, 1 mM MgCl2, 0.2 mM thiamine pyrophosphate, 2 mM sodium pyruvate, 1 mM cysteine, 0.5 mM NAD+, 0.1 mM coenzyme A, and 10 μg of cell extracts (43). Citrate synthase activity was measured at 412 nm with 50 mM Tris-HCl buffer (pH 7.8), 0.1 mM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), 5 mM acetyl coenzyme A (acetyl-CoA), 0.5 mM oxaloacetate, and 10 μg of cell extracts (43). Aconitase activity was measured at 340 nm with 50 mM Tris-HCl buffer (pH 7.8), 5 mM citrate, 1 mM MgCl2, 0.2 mM NADP+, 1 unit of isocitrate dehydrogenase, and 10 μg of cell extracts (43). All activities were measured in a 96-well plate using a multiscanner (Thermo Scientific). The concentration of ammonium ion was measured using an ammonium kit (Merck, Germany), and the organic acids were measured by HPLC with a CarboPaK PA-1 column (Dionex) using 0.1% H2SO4 in water as the isocratic mobile phase.
RT-PCR.
RNA extraction from RNAlater-treated cells was carried out using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The concentration of RNA was determined by measuring the A260 using a NanoDrop ND-1000 spectrophotometer. The purity of RNA was estimated from the absorbance ratio at 260 nm and 280 nm (A260/A280). RNA was reverse transcribed into first-strand cDNA in the presence of 1 μg total RNA, 5× first-strand buffer, 0.5 mM deoxynucleoside triphosphate (dNTP) mix, 200 units SuperScript III reverse transcriptase (Invitrogen), 40 units RNaseOUT recombinant RNase inhibitor, 3 μg of random hexamer, and 5 mM DTT in a final reaction volume of 20 μl. Reactions were carried out in a water bath at 25°C for 5 min and 50°C for 30 min, followed by heating to 70°C for 15 min and then cooling to 4°C. The synthesized cDNA was amplified by PCR in a total volume of 50 μl containing 10 μM each primer, 2 μl cDNA, 4 units LA Taq DNA polymerase (TAKARA, Japan), 2× GC II buffer, and 200 μM dNTP mix. 16S rRNA genes were used as a control to normalize the expression level of each mRNA as well as sample-to-sample variations. Gel band intensities of absC, aceE1, aceE2, actII-ORF4, and redD were compared after 25 cycles of PCR to ensure that DNA amplification was in the linear range for each template on kinetic analysis. The PCR products were visualized by 2% (wt/vol) agarose gel electrophoresis with ethidium bromide staining.
Two-dimensional gel electrophoresis.
WT and BG711 were grown for 20 h in R5− medium by spore inoculation, and cells were harvested by centrifugation at 13,000 × g and sequentially washed with sterilized water two times before duplicate inoculation into 20 ml of MMGA. The mycelial broths cultured in MMGA were harvested at 96 h, and cell pellets were disrupted by sonication. The concentration of cytosolic proteins was determined by Bradford assay (Bio-Rad). Protein extracts (300 μg) were mixed with 350 μl of rehydration solution containing 8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM DTT, and 0.5% Ampholine (pH 4 to 7), followed by isoelectric focusing as the first dimensional separation using 18-cm immobilized pH gradient (IPG) strips with a linear pH gradient from 4 to 7 (GE Healthcare). Isoelectric focusing was carried out to a total of 87,000 Vh with a maximum voltage of 5,000 V followed by equilibration. The first equilibration was carried out in an equilibration solution containing 6 M urea, 2% SDS, 50 mM Tris-HCl, 30% glycerol, 0.002% bromophenol blue, and 65 mM DTT (pH 8.8), and the second equilibration was carried out in the same equilibration solution with 135 mM iodoacetamide in place of 65 mM DTT for 15 min.
Second-dimensional separation was performed using 12.5% polyacrylamide gels. In order to minimize experimental variation, three gels were run for each triplicate culture. The gels were stained with colloidal Coomassie brilliant blue (23), scanned with a POWERLook 1100 densitometer (UMAX), and analyzed by ImageMaster 2D Platinum software (GE Healthcare). Normalized percentage integrated optical density (percent volume) was calculated for each protein spot detected. Student's t test analysis (P < 0.05) was carried out to determine statistically significant protein fold changes in these two strains.
Protein identification by nLC-MS/MS.
Spots upregulated by more than 2-fold were excised from gels treated with M145 and ΔabsC mutant (BG711) using the tip of a clean polypropylene micropipette. Each gel piece was then trypsin digested in-gel for analysis by nano liquid chromatography-tandem mass spectrometry (nLC-MS/MS) (Ultimate 3000; Dionex; and LTQ-Orbitrap ESI mass spectrometer; Thermo Scientific). A fused silica capillary column (360-μm outer dimension, 100-μm inner dimension) was pulled with a P-2000 laser puller (Sutter Instrument) to make nanospray tips. Most proteins searched for in the S. coelicolor database using SEQUEST Sorcerer (Thermo Scientific) were specifically identified, as two or more peptides had cross-correlation scores (Xcorr) equal or greater than 1.7 for single-charged, 2.5 for double-charged, and 3.0 for triple-charged peptides.
Many proteins were identified as multiple spots at different isoelectric points and/or molecular weights, with most being isoforms of metabolic enzymes (14). Some proteins showed differences between experimental and theoretical molecular weights due to truncation of the protein by proteolytic processing. In vivo fragmentation of expressed proteins was already reported (14). The most prominent examples in this report are glyceraldehyde-3-phosphate dehydrogenase, dihydrolipoamide dehydrogenase, and glutamine synthetase. This explains why there is more than one spot ID in Table 2.
TABLE 2.
Differentially expressed spots of absC mutant (BG711) in two-dimensional gela
| Protein function | Regulator | Characteristic | Spot ID no. | Fold changeb | No. of matched peptides |
|---|---|---|---|---|---|
| Downregulated | |||||
| Purine metabolism | SCO4928 | Adenylate cyclase, CyaA | 474 | O | 2 |
| SCO4068 | Phosphoribosylamine-glycine ligase, PurD | 407 | O | 10 | |
| SCO4087 | Phosphoribosylaminoimidazole synthetase, PurM | 529 | O | 16 | |
| Pyrimidine metabolism | SCO4041 | Uracil phosphoribosyltransferase, UraP | 675 | O | 2 |
| Pentose phosphate pathway | SCO2627 | Ribose-5-phosphate isomerase B | 822 | O | 10 |
| Fatty acid metabolism/biosynthesis | SCO2387 | ACP S-malonyltransferase, FabD | 594 | 5.5 | 11 |
| SCO3079 | Putative thiolase | 442 | O | 9 | |
| SCO5144 | Acyl-CoA isomerase | 654 | 7.7 | 2 | |
| Vitamin B6 metabolism | SCO1523 | Pyridoxine biosynthesis protein, YaaD | 586 | O | 2 |
| SCO4293 | Threonine synthase | 413 | O | 17 | |
| SCO4366 | Phosphoserine aminotransferase | 453 | O | 8 | |
| Folate metabolism | SCO3401 | Hydroxymethyldihydropteridine pyrophosphokinase, FolK | 474 | O | 11 |
| SCO4824 | Methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase, FolD | 570 | O | 2 | |
| Glycolysis and TCA | SCO1945 | Triosephosphate isomerase, TpiA | 632 | 2.8 | 19 |
| SCO1947 | Glyceraldehyde-3-phosphate dehydrogenase, Gap1 | 453, 474, 483 | O | 8, 5, 4 | |
| SCO2180 | Dihydrolipoamide dehydrogenase, PdhL | 389 | O | 38 | |
| SCO2181 | Dihydrolipoamide succinyltransferase- SucB | 663 | 11 | 9 | |
| SCO4808 | Succinyl-CoA synthetase subunit beta, SucC | 492 | 4.9 | 2 | |
| SCO4827 | Malate dehydrogenase, Mdh | 545 | O | 20 | |
| SCO7511 | Glyceraldehyde 3-phosphate dehydrogenase, Gap2 | 511 | O | 6 | |
| Antioxidant mechanism | SCO2633 | Superoxide dismutase [Fe-Zn], SodF | 689 | 2.4 | 6 |
| SCO4164 | Thiosulfate sulfurtransferase, CysA | 578 | 7.5 | 13 | |
| SCO4394 | Iron repressor, DesR | 633 | O | 2 | |
| SCO5419 | Thioredoxin, TrxA4 | 511 | O | 12 | |
| Type I polyketide biosynthesis | SCO6282 | 3-Oxoacyl-[acyl-carrier protein] reductase | 653 | 2.8 | 11 |
| CDA synthesis | SCO3231 | CDA peptide synthetase II, CdaPS2 | 569 | O | 2 |
| SCO3247 | Acyl-CoA oxidase | 693 | 3 | 2 | |
| Upregulated | |||||
| Glycolysis and TCA | SCO1946 | Phosphoglycerate kinase, Pgk | 317 | 5.3 | 5 |
| SCO1947 | Glyceraldehyde-3-phosphate dehydrogenase, Gap1 | 360, 381 | O | 4, 4 | |
| SCO2180 | Dihydrolipoamide dehydrogenase, PdhL | 282 | O | 19 | |
| SCO4209 | Phosphoglyceromutase, Pgm | 573 | O | 11 | |
| SCO5107 | Succinate dehydrogenase, SdhA | 191 | 25.1 | 2 | |
| SCO5424 | Acetate kinase, AckA | 314 | O | 1 | |
| SCO5425 | Phosphate acetyltransferase, Pta | 228 | 3.3 | 3 | |
| SCO7000 | Isocitrate dehydrogenase, Idh | 177 | O | 27 | |
| SCO7412 | Pyruvate decarboxylase | 556 | 4.4 | 1 | |
| Secondary metabolite signaling | SCO5113 | ABC transport system lipoprotein, BldKB | 189 | 6.2 | 19 |
| SCO5737 | Guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase, GpsI | 173 | O | 11 | |
| Type I polyketide biosynthesis | SCO6275 | Type I polyketide synthase | 320 | O | 2 |
| Amino acid metabolism | SCO1501 | Alanine tRNA synthetase, AlaS | 556 | 4.4 | 1 |
| SCO1595 | Phenylalanyl-tRNA synthetase alpha subunit, PheS | 317 | 5.3 | 2 | |
| SCO2198 | Glutamine synthetase I, GlnA | 253 | 2.7 | 4 | |
| SCO2210 | Glutamine synthetase, GlnII | 556 | 4.4 | 1 | |
| SCO2241 | Probable glutamine synthetase | 253 | 2.7 | 7 | |
| SCO5699 | Prolyl tRNA synthetase, ProS | 189 | 6.2 | 2 | |
| Pentose phosphate pathway | SCO1935 | Transketolase A, TktA1 | 181 | 4 | 5 |
| Antioxidant mechanism | SCO0379 | Catalase, KatA | 257 | O | 14 |
| Other | SCO3820 | Serine/threonine protein kinase | 224 | O | 3 |
Spot ID, fold change, and number of matched peptides determined by mass spectrometric analysis are indicated for each classified protein.
Some protein spots appeared only in M145 or BG711 because they are under the detection limit of image analysis spot recognition criteria. These are indicated as “O” in fold change column.
RESULTS
Selection of indicator and sensitive detection of pH differences in 96-well.
Although 96-well plate-based experiments with Streptomyces allowed examination and monitoring of different nutrient conditions at different time points, the detection was not sensitive enough to confirm differences between all mutants (33). Moreover, there also were some problems such as lack of sufficient amount of cells, evaporation of moisture in the media, and contamination due to the growth period (5 to 7 days) in liquid minimal medium. Therefore, we established a new system using a pH indicator to identify changes in pH caused by altered production of organic acids under specific nutrient conditions and a short time period (3 days), which is too short for antibiotic production when cultured in 96-well plates.
Among various indicators covering pH 6 to 8, bromothymol blue was selected due to its wide use as a conventional organic indicator. 4-Nitrophenol has no color at pH 5.4, although it can show different colors from 6 to 8 (Fig. 2A to C). In addition, a previous study showed that bromothymol blue did not affect cell growth (21) and had good optical linearity from pH 6 to 7.8 (Fig. 2D). Thus, we can easily analyze the color changes in each well of a 96-well plate using bare eyes or a multiscanner (Thermo Electron Corp., Finland) at 630 nm (Fig. 2E).
Initially, the color of minimal medium containing bromothymol blue indicator (pH 7.4) is sky blue, but it changes to a yellow color when glucosamine or aspartate is used as a C or N source. Along with cell growth, a pH change that depends on the amount and type of organic acid produced by Streptomyces is clearly indicated by a yellow (below pH 6.0), green (pH 6.0 to 7.6), or blue (above pH 7.6) color. Differences in color change between the WT and deletion mutant can be compared during the cultivation time when antibiotics are not normally produced.
To demonstrate the feasibility of our proposed method, the WT strain and a known scbR deletion mutant were investigated in media containing various combinations of carbon and nitrogen sources. Since ScbR is a well-known receptor of gamma-butyrolactones that triggers secondary metabolism in Streptomyces, the scbR deletion mutant would result in delay of undecylprodigiosin and actinorhodin production (36). The scbR deletion mutant clearly showed differences in color when cultured in minimal medium containing glucose and glycine (Fig. 2C). However, M145 and the scbR deletion mutant did not produce any antibiotics over three days (Fig. 2A) and did not show large differences in antibiotic production over six days (data not shown). However, the pH of the culture broth of scbR deletion mutant decreased remarkably when cultured in glucose/glycine minimal medium (Fig. 2E). This was mainly caused by the different amount of pyruvate produced, since almost the same amount of acetate production was observed in the culture media through HPLC analysis (Fig. 2F). Since glycine is an important precursor for proline synthesis and hence undecylprodigiosin production, the low pH of the scbR deletion mutant in glycine-containing minimal medium can be correlated with the delayed production of undecylprodigiosin. This shows that the application of a pH indicator can provide helpful information for elucidating the function of a regulator.
Nutrient information for SCO5405.
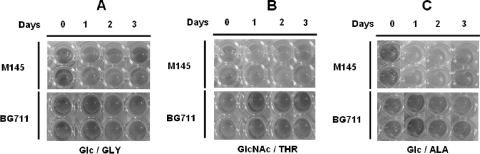
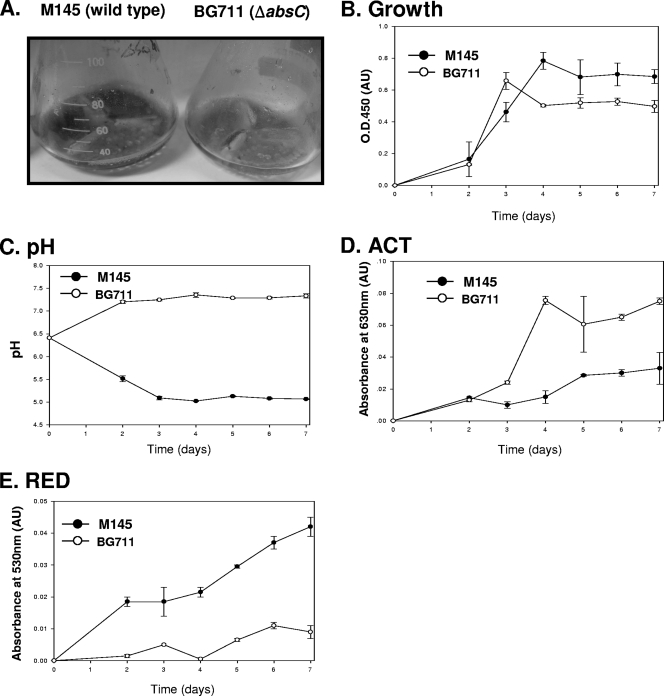
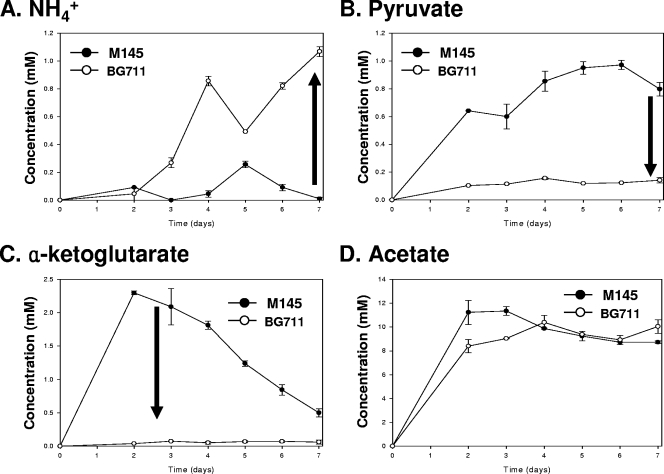
The SCO5405 deletion mutant (BG711) produces little ACT in SMM medium and large amounts in R5− medium. Although it is interesting that BG711 produces ACT in R5− medium, the amount produced is so similar to that of M145, as indicated by a purple color, that no additional information is gained besides the existence of antibiotic production (data not shown). However, by using a multiwell method with bromothymol blue, M145 and BG711 were found to produce different colors in various minimal media containing glucose/glycine (Fig. 3A), N-acetylglucosamine/thereonine (Fig. 3B), or glucose/alanine (Fig. 3C), thus indicating a pH difference between the WT and BG711. In these three types of media, no color change was observed for BG711. However, the color changed to yellow for M145, indicating acidification of the medium. To monitor M145 and BG711 in detail, both were grown in 50 ml of minimal medium in a 250-ml flask, and changes in pH and antibiotic production in minimal medium containing glucose and alanine as C and N sources, respectively, were observed (Fig. 4A). In liquid MMGA medium, M145 was found to grow better than BG711 (Fig. 4B). In addition, the M145 strain could produce RED and ACT in 5 days, as indicated by the purple color in the broth (Fig. 4A). For BG711, the color of the broth was sky blue with no red color. These results are consistent and were confirmed in repeated cultures. The differences in color were mainly due to changes in the pH of culture broth as well as varying levels of antibiotic production. The pH of M145 culture broth was decreased from pH 6.5 to 5 and thereafter remained almost constant. However, the pH of BG711 culture broth was gradually increased up to pH 7.2 (Fig. 4C). The production of ACT from BG711 was slightly higher than that of M145 and RED (Fig. 4D and E). There could be several factors causing the pH increase in BG711, but previous reports on pH changes caused by the deletion of metabolic enzymes provide some clues to the possible role of AbsC (35, 38), leading us to examine the effect of AbsC on metabolic pathways and organic acid production.
FIG. 3.
Comparison of pH changes in M145 and BG711(ΔSCO5405) grown in glucose/glycine (Glc/GLY) (A), N-acetylglucosamine/threonine (GlcNAc/THR) (B), and glucose/alanine minimal medium (Glc/ALA) (C), as monitored by bromothymol blue at 0, 1, 2, and 3 days as indicated.
FIG. 4.
Growth and antibiotic production of M145 and BG711 in minimal medium containing glucose and alanine. (A) Media after four days of culture. The growth curve (B), pH changes (C), and ACT (D) and RED production (E) in M145 (•) and BG711 (○) are shown.
Main causes of the pH increase in BG711.
To determine the causes of the pH changes, the amounts of ammonium ions and organic acids such as acetate, pyruvate, and α-ketoglutarate in the culture broth were measured and compared using the methods described in Materials and Methods. The increased amount of ammonium ions (Fig. 5A) might explain the pH increase in BG711 culture broth, but only partially because the difference in ammonium ion concentration was rather small. Streptomyces species are known to produce pyruvate and α-ketoglutarate as their major organic acids (3, 20), so it was not surprising that M145 mainly produced pyruvate and α-ketoglutarate. Interestingly, only small amounts of pyruvate and α-ketoglutarate were detected in the BG711 culture medium (Fig. 5B and C), suggesting that BG711 either consumes more or produces less pyruvate and α-ketoglutarate. The difference in the amounts of acetate in the media of the two strains was insignificant (Fig. 5D), indicating that the dramatic pH difference between M145 and BG711 was caused mainly by different amounts of organic acids secreted in the broths. Based on the low accumulation of organic acids and varying antibiotic production, a possible role for AbsC in MMGA liquid medium can thus be expected.
FIG. 5.
Concentrations of NH4+ (A), pyruvate (B), α-ketoglutarate (C), and acetate (D) in the media in which M145 (•) and BG711 (○) were grown.
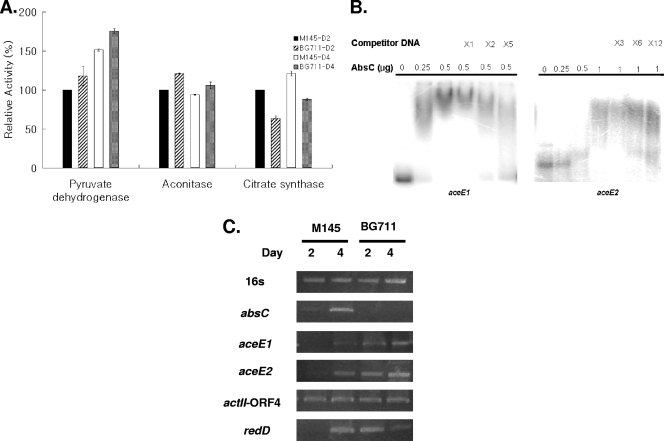
Both pyruvate and α-ketoglutarate are main nodes of various metabolic pathways (28), though the former has been determined to be more directly involved in antibiotic production in S. coelicolor than the latter (9). To examine the effect of absC deletion on the enzyme activities of pyruvate and acetyl-CoA in M145, the activities of pyruvate dehydrogenase, citrate synthase, and aconitase, which are mainly involved in pyruvate synthesis, were measured and compared (Fig. 6A). Pyruvate dehydrogenase and aconitase activities were increased, but citrate synthase activity was decreased in BG711, suggesting that these changes in enzyme activities resulted in more acetyl-CoA in BG711 and also led to more ACT production eventually.
FIG. 6.
(A) Enzymatic assay with cell extracts from M145 and BG711 grown for 2 and 4 days. (B) Gel retardation assay of AbsC with upstream regions of aceE1 and aceE2. Different amounts of proteins as indicated at the top of the figure were used, and 30 ng of each DNA probe was prepared as explained in Materials and Methods. (C) RT-PCR results of M145 and BG711 sampled at 2 and 4 days of culture. Gel band intensities of 16S rRNA genes were set as the control, and the expression levels of absC, aceE1, aceE2, actII-ORF4, and redD were compared.
Since BG711 showed changes in several enzyme activities involved in glycolysis and the tricarboxylic acid (TCA) cycle, it could be speculated that AbsC may directly bind to the promoter regions of those enzymes. To examine whether enzymatic activity directly regulates AbsC by binding to the promoter regions of genes encoding aconitase (SacA, SCO5999), citrate synthase (CitA, SCO2736) and the E1 components of pyruvate dehydrogenase (AceE1, SCO2183, and AceE2, SCO2371), gel retardation experiments were performed with the same His-tag-purified AbsC in E. coli. It was shown that AbsC binds to the upstream regions of aceE1 and aceE2 (Fig. 6B), but not with the same amount of AbsC to the upstream regions of citA and sacA (data not shown). These data indicate that there was direct control of AbsC on pyruvate dehydrogenase units, and this control of AbsC negatively affects the enzyme activity of pyruvate dehydrogenase. Using reverse transcription-PCR (RT-PCR), the function of AbsC was shown as a repressor on aceE1 and aceE2 based on the observation that its deletion increased the amount of mRNAs of both genes (Fig. 6C).
Two-dimensional gel analysis of the effects of the absC deletion.
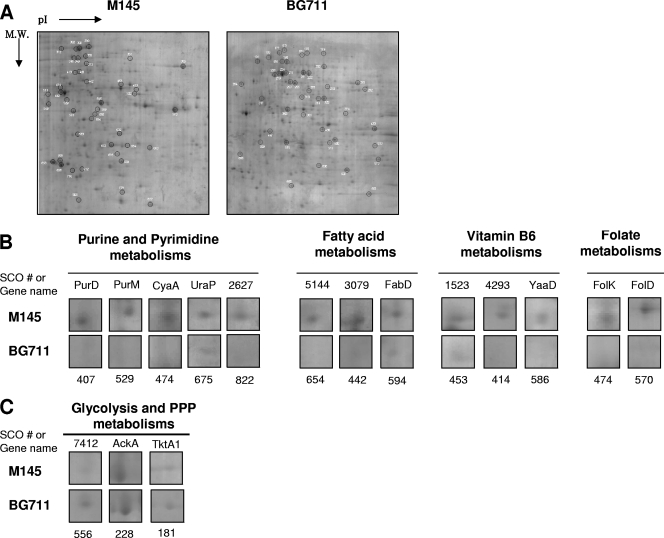
To determine the function of AbsC, the proteomes of 4-day-old cultures of M145 and BG711 in MMGA medium were compared (Fig. 7A). The list of gel spots that were decreased by at least 2-fold in BG711 are shown in Fig. 7B and Table 2, and those increased by more than 2-fold are shown in Fig. 7C and Table 2. Several proteins involved in purine and pyrimidine metabolism (SCO4068 [spot no. 407], SCO4087 [no. 529], SCO4928 [no. 474], and SCO4041 [no. 675]) were significantly downregulated in BG711 (Fig. 7B). In addition, the expressions of several proteins involved in fatty acid metabolism and synthesis (SCO2387 [spot no. 594], SCO3079 [no. 442], and SCO5144 [no. 654]), vitamin B6 metabolism (SCO1523 [no. 453], SCO4293 [no. 414], and SCO4366 [no. 586]), and folate metabolism (SCO3401 [no. 474] and SCO4824 [no. 570]) were decreased in BG711 (Fig. 7B). Considering that nucleotide metabolism, fatty acid metabolism, and vitamin metabolism are the representative indicators of active cell growth, BG711 appeared to display stationary phase cell physiology even in its earlier stages of cell growth. This was also supported by the upregulation of transketolase A (SCO1935 [spot no. 181]) and the downregulation of ribose-5-phosphate isomerase (SCO2627 [no. 822]), both of which are related to a precursor of purine nucleotide sugars, 5-phospho-α-d-ribosyl-1-pyrophosphate (PRPP), in the pentose phosphate pathway (Fig. 7C).
FIG. 7.
(A) Comparative proteomic analysis between M145 and BG711. (B and C) Downregulated (B) and upregulated (C) proteins in BG711.
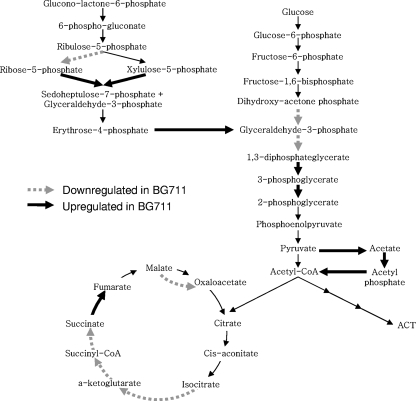
Interestingly, the high level of transketolase (SCO1935) in BG711 indicated that carbon flux toward the pentose phosphate pathway (PPP) was channeled into glycolysis. The high levels of pyruvate decarboxylase (SCO7412 [spot no. 556]), acetate kinase (SCO5424 [no. 314]), and phosphate acetyltransferase (SCO5425 [no. 556]) suggest high carbon flux into the glycolysis pathway, leading to acetyl-CoA (Fig. 8). The putative pyruvate decarboxylase (SCO7412) corresponds to the same EC number (EC 1.2.2.2) as E. coli pyruvate oxidase PoxB, which has been preferentially used at low growth rates over the pyruvate dehydrogenase complex and is involved in the conversion of pyruvate to acetyl-CoA via acetate (1). Finally, compared to M145, many enzymes in the TCA cycle were downregulated in BG711, which explains the smaller amount of α-ketoglutarate in BG711 (Fig. 5C). These results explain the lower levels of pyruvate and α-ketoglutarate in BG711, but it is still unclear whether these proteomes resulted from a pH change or caused the pH change of BG711.
FIG. 8.
Up- and downregulated proteins indicated in glycolysis, TCA cycle, and the pentose phosphate pathway. Broken arrows indicate enzymes that are downregulated in BG711, and thick solid arrows indicate enzymes upregulated in BG711.
DISCUSSION
By culturing Streptomyces in a 96-well plate, we tried to develop a sensitive and fast method to identify the functions of previously unknown genes and save time in finding the conditions that allow characterization. We introduced bromothymol blue to a 96-well system and showed that this was helpful for characterizing an unknown regulator. The advantage of this method is that we can detect differences with the naked eye without any expensive equipment and can compare pH differences between WT and several deletion mutants even when dealing with several hundreds of samples from various combinations of nutrients at different time points. In addition to functional identification in liquid media, this method could be used to confirm the function of a gene by complementation.
Using bromothymol blue, we easily found information for the characterization of a new MarR family regulator in S. coelicolor. This putative regulator was shown to control pyruvate directly by binding to the upstream regions of pyruvate dehydrogenase E1 components; α-ketoglutarate synthesis was regulated indirectly. The role of SCO5405 appears to be as an on-off switch for carbon flux from the TCA cycle to antibiotic production depending on cell growth and nutrient states. This is therefore an interesting example of regulators in Streptomyces being involved in the dual control of both carbon flux from glycolysis to the TCA cycle and secondary metabolism. This shows that secondary metabolism was tightly linked to its primary metabolism.
One possible scenario that explains the effect of SCO5405 in MMGA medium on organic production and antibiotic production is as follows. Considering the lower concentration of pyruvate in BG711, BG711 either consumed pyruvate faster than M145 or simply produced less pyruvate. Normally, pyruvate can be channeled through other pathways such as phosphoenolpyruvate (PEP), RED, and acetyl-CoA production in Streptomyces (9). Since RED synthesis was not increased dramatically in BG711, most pyruvate was probably converted into acetyl-CoA by pyruvate dehydrogenase or other pathways. If so, more acetyl-CoA would be channeled into ACT production than in the wild-type M145 strain, accounting for a similar amount of acetate production (Fig. 5D). In BG711, the pathway from acetyl-CoA to α-ketoglutarate via citrate appeared to be repressed, producing a much smaller amount of α-ketoglutarate. This result is somewhat similar to that observed between M145 and BG711 and is supported by previous reports on the negative correlation between organic acids and production of antibiotics such as anthracycline and macrolide in acidogenic Streptomyces (13, 40). However, a detailed explanation as to why more α-ketoglutarate was present in M145 than in BG711 seems more complex and difficult than for pyruvate.
Although it is very important to understand how gene regulators control primary and secondary metabolism, there have been only a few studies on regulators directly linking primary and secondary metabolism in S. coelicolor (30). In addition, there are not many reports on regulators that simultaneously control primary metabolism as well as antibiotic production by directly binding to corresponding promoter regions such as actII-ORF4, cdaR, and redZ (22). In this regard, finding AbsC appears to be a very worthy endeavor. As of now, there have been several reports of regulators involved only in glycolysis and the TCA cycle by directly binding upstream of pyruvate dehydrogenase, citrate synthase, or aconitase in other bacterial species. A GntR family regulator, i.e., pyruvate dehydrogenase complex regulator (PdhR) from Escherichia coli, binding to the pdh operon (24), and a TetR-type repressor, i.e., AcnR from Corynebacterium glutamicum, binding to the aconitase promoter region (19), have been reported. In addition, a LysR-type transcriptional regulator, CcpC from Bacillus subtilis, and a AraC-type regulator, RipA from Corynebacterium glutamicum (17), are known to regulate aconitase by binding to upstream regions (39), and RamB from Corynebacterium glutamicum is known to activate the aceE gene, i.e., E1p subunit of the pyruvate dehydrogenase complex (7). Our results for AbsC demonstrate a novel type of regulator controlling primary metabolism as well as secondary metabolism in Streptomyces.
Considering that slow-growing bacteria spend most of their time at the transition point between balanced growth and the stationary phase (32), the function of a regulator would be maintaining the transition period by controlling intermediates until certain signals or nutrient states are produced. AbsC appears to be one regulator involved in maintaining the transition point, working as a switch-triggering secondary metabolism.
Acknowledgments
We thank the John Innes Center for providing the cosmid library.
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (no. 20090083035) and a grant funded by the Korea Research Foundation (KRF-2005-005-20090094021), Republic of Korea.
Footnotes
Published ahead of print on 9 April 2010.
REFERENCES
- 1.Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold, R., and M. Mann. 2003. Mass spectrometry-based proteomics. Nature 422:198-207. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, Z. U., S. Shapiro, and L. C. Vining. 1984. Excretion of alpha-keto acids by strains of Streptomyces venezuelae. Can. J. Microbiol. 30:1014-1021. [DOI] [PubMed] [Google Scholar]
- 4.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 5.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat. Struct. Biol. 8:710-714. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.Blombach, B., A. Cramer, B. J. Eikmanns, and M. Schreiner. 2009. RamB is an activator of the pyruvate dehydrogenase complex subunit E1p gene in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 16:236-239. [DOI] [PubMed] [Google Scholar]
- 8.Bochner, B. R. 2003. New technologies to assess genotype-phenotype relationships. Nat. Rev. Genet. 4:309-314. [DOI] [PubMed] [Google Scholar]
- 9.Bruheim, P., M. Butler, and T. E. Ellingsen. 2002. A theoretical analysis of the biosynthesis of actinorhodin in a hyper-producing Streptomyces lividans strain cultivated on various carbon sources. Appl. Microbiol. Biotechnol. 58:735-742. [DOI] [PubMed] [Google Scholar]
- 10.Bystrykh, L. V., M. A. Fernandez-Moreno, J. K. Herrema, F. Malpartida, D. A. Hopwood, and L. Dijkhuizen. 1996. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J. Bacteriol. 178:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison, D. W., and V. L. Miller. 2006. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9:153-159. [DOI] [PubMed] [Google Scholar]
- 12.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesketh, A., H. Kock, S. Mootien, and M. Bibb. 2009. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 74:1427-1444. [DOI] [PubMed] [Google Scholar]
- 14.Hesketh, A. R., G. Chandra, A. D. Shaw, J. J. Rowland, D. B. Kell, M. J. Bibb, and K. F. Chater. 2002. Primary and secondary metabolism, and post-translational protein modifications, as portrayed by proteomic analysis of Streptomyces coelicolor. Mol. Microbiol. 46:917-932. [DOI] [PubMed] [Google Scholar]
- 15.Hood, D. W., R. Heidstra, U. K. Swoboda, and D. A. Hodgson. 1992. Molecular genetic analysis of proline and tryptophan biosynthesis in Streptomyces coelicolor A3(2): interaction between primary and secondary metabolism—a review. Gene 115:5-12. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood, D. A. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40:1-23. [DOI] [PubMed] [Google Scholar]
- 17.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., M. J. Bibb, M. J. Buttner, K. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom.
- 19.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280:585-595. [DOI] [PubMed] [Google Scholar]
- 20.Madden, T., J. M. Ward, and A. P. Ison. 1996. Organic acid excretion by Streptomyces lividans TK24 during growth on defined carbon and nitrogen sources. Microbiology 142(part 11):3181-3185. [DOI] [PubMed] [Google Scholar]
- 21.Magae, Y., K. Akahane, K. Nakamura, and S. Tsunoda. 2005. Simple colorimetric method for detecting degenerate strains of the cultivated basidiomycete Flammulina velutipes (Enokitake). Appl. Environ. Microbiol. 71:6388-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenzie, N. L., and J. R. Nodwell. 2007. Phosphorylated AbsA2 negatively regulates antibiotic production in Streptomyces coelicolor through interactions with pathway-specific regulatory gene promoters. J. Bacteriol. 189:5284-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 24.Ogasawara, H., Y. Ishida, K. Yamada, K. Yamamoto, and A. Ishihama. 2007. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189:5534-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, S. S., B. J. Ko, and B. G. Kim. 2005. Mass spectrometric screening of transcriptional regulators using DNA affinity capture assay. Anal. Biochem. 344:152-154. [DOI] [PubMed] [Google Scholar]
- 26.Park, S. S., Y. H. Yang, E. Song, E. J. Kim, W. S. Kim, J. K. Sohng, H. C. Lee, K. K. Liou, and B. G. Kim. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 36:1073-1083. [DOI] [PubMed] [Google Scholar]
- 27.Rigali, S., H. Nothaft, E. E. Noens, M. Schlicht, S. Colson, M. Muller, B. Joris, H. K. Koerten, D. A. Hopwood, F. Titgemeyer, and G. P. van Wezel. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237-1251. [DOI] [PubMed] [Google Scholar]
- 28.Rokem, J. S., A. E. Lantz, and J. Nielsen. 2007. Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep. 24:1262-1287. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. Fritsch, and T. Maniatis. 1998. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 30.Santamarta, I., M. T. Lopez-Garcia, R. Perez-Redondo, B. Koekman, J. F. Martin, and P. Liras. 2007. Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the ARE(ccaR) sequence of S. clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Mol. Microbiol. 66:511-524. [DOI] [PubMed] [Google Scholar]
- 31.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, I., R. A. Slepecky, and P. Setlow. 1989. Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. American Society for Microbiology, Washington, DC.
- 33.Song, E., Y. H. Yang, B. R. Lee, J. N. Kim, E. J. Kim, S. S. Park, K. Lee, W. S. Kim, S. You, D. Hwang, and B. G. Kim. 2010. An integrative approach for high-throughput screening and characterization of transcriptional regulators in Streptomyces coelicolor. Pure Appl. Chem. 82:57-67. [Google Scholar]
- 34.Stevenson, C. E., H. Kock, S. Mootien, S. C. Davies, M. J. Bibb, and D. M. Lawson. 2007. Crystallization and preliminary X-ray analysis of AbsC, a novel regulator of antibiotic production in Streptomyces coelicolor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Susstrunk, U., J. Pidoux, S. Taubert, A. Ullmann, and C. J. Thompson. 1998. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30:33-46. [DOI] [PubMed] [Google Scholar]
- 36.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 37.Thuy, M. L., M. K. Kharel, R. Lamichhane, H. C. Lee, J. W. Suh, K. Liou, and J. K. Sohng. 2005. Expression of 2-deoxy-scyllo-inosose synthase (kanA) from kanamycin gene cluster in Streptomyces lividans. Biotechnol. Lett. 27:465-470. [DOI] [PubMed] [Google Scholar]
- 38.Viollier, P. H., W. Minas, G. E. Dale, M. Folcher, and C. J. Thompson. 2001. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. J. Bacteriol. 183:3184-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 280:40500-40508. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y. H., H. S. Joo, K. Lee, K. K. Liou, H. C. Lee, J. K. Sohng, and B. G. Kim. 2005. Novel method for detection of butanolides in Streptomyces coelicolor culture broth, using a His-tagged receptor (ScbR) and mass spectrometry. Appl. Environ. Microbiol. 71:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, Y. H., J. N. Kim, E. Song, E. Kim, M. K. Oh, and B. G. Kim. 2008. Finding new pathway-specific regulators by clustering method using threshold standard deviation based on DNA chip data of Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 80:709-717. [DOI] [PubMed] [Google Scholar]
- 42.Yang, Y. H., E. Song, E. J. Kim, K. Lee, W. S. Kim, S. S. Park, J. S. Hahn, and B. G. Kim. 2009. NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 82:501-511. [DOI] [PubMed] [Google Scholar]
- 43.Yarian, C. S., and R. S. Sohal. 2005. In the aging housefly aconitase is the only citric acid cycle enzyme to decline significantly. J. Bioenerg. Biomembr. 37:91-96. [DOI] [PubMed] [Google Scholar]