Abstract
GlnD is a bifunctional uridylyltransferase/uridylyl-removing enzyme (UTase/UR) and is believed to be the primary sensor of nitrogen status in the cell by sensing the level of glutamine in enteric bacteria. It plays an important role in nitrogen assimilation and metabolism by reversibly regulating the modification of PII protein; PII in turn regulates a variety of other proteins. GlnD appears to have four distinct domains: an N-terminal nucleotidyltransferase (NT) domain; a central HD domain, named after conserved histidine and aspartate residues; and two C-terminal ACT domains, named after three of the allosterically regulated enzymes in which this domain is found. Here we report the functional analysis of these domains of GlnD from Escherichia coli and Rhodospirillum rubrum. We confirm the assignment of UTase activity to the NT domain and show that the UR activity is a property specifically of the HD domain: substitutions in this domain eliminated UR activity, and a truncated protein lacking the NT domain displayed UR activity. The deletion of C-terminal ACT domains had little effect on UR activity itself but eliminated the ability of glutamine to stimulate that activity, suggesting a role for glutamine sensing by these domains. The deletion of C-terminal ACT domains also dramatically decreased UTase activity under all conditions tested, but some of these effects are due to the competition of UTase activity with unregulated UR activity in these variants.
GlnD is a bifunctional uridylyltransferase/uridylyl-removing enzyme (UTase/UR; gene product of glnD) that regulates PII proteins by uridylylation or deuridylylation. In enteric bacteria, GlnD is believed to be a primary sensor of intracellular nitrogen status, determined by the level of glutamine in the cell (29, 34-36). PII is one of the most broadly distributed regulatory proteins in nature and directly or indirectly senses nitrogen and carbon signals in the cell. Multiple PII homologs, mainly termed GlnB and GlnK, have been found in many bacteria, and they play overlapping but distinct roles in the cell (7, 46, 54, 79).
In Escherichia coli, PII proteins can interact with a variety of receptor proteins, including NtrB, a sensor protein of the two-component NtrB/NtrC regulatory system. NtrB acts as a histidine kinase that phosphorylates NtrC under nitrogen-limiting conditions and can also act as a phosphatase to dephosphorylate NtrC under conditions of nitrogen excess (55). Under nitrogen excess conditions, PII proteins are deuridylylated by GlnD so that they can interact with NtrB to stimulate its phosphatase activity, resulting in the dephosphorylation of NtrC. However, under nitrogen-limiting conditions, PII proteins become uridylylated, and this uridylylation prevents their interaction with NtrB, so that NtrB is dominated by its kinase activity to phosphorylate NtrC (33). The phosphorylated form of NtrC acts as a transcriptional activator of glnK amtB, glnA, and other operons involved in nitrogen assimilation. PII, together with adenylyltransferase (ATase, encoded by glnE), also controls glutamine synthetase activity by reversible adenylylation (1, 32). AmtB, a gas channel for NH3, is another receptor for PII (11, 16, 31).
In the photosynthetic bacterium Rhodospirillum rubrum, three PII homologs, named GlnB, GlnK, and GlnJ, have been identified (37, 77). Although the amino acid sequences of these three homologs are very similar, they show both distinct and overlapping functions in the cell. They are involved in the regulation of NifA and NtrB activities, the covalent modification of glutamine synthetase, and modulation of the posttranslational regulation of nitrogenase (77). In R. rubrum, NifA activity is tightly controlled through the direct interaction between NifA and the uridylylated form of GlnB in response to NH4+ (78).
Although GlnD is central to the global nitrogen regulatory (Ntr) system, it has not been as well studied as NtrB/NtrC and PII. In 1971, Brown et al. first reported the UTase activity of partially purified GlnD by showing the conversion of PII to PII-UMP in the presence of ATP, 2-oxoglutarate, or α-ketoglutarate (α-KG) and UTP, and this activity was inhibited by glutamine (14). UR activity was reported by Mangum et al. in 1973 (49), and this was followed by several other studies (1, 20, 22, 24). It was found that only Mn2+ supports UR activity, while both Mg2+ and Mn2+ support UTase activity (1, 49). ATP and α-KG are necessary for UTase activity but not for UR activity (14, 20, 49). Glutamine stimulates UR activity while inhibiting UTase activity (20). The most extensive enzymological characterization of E. coli GlnD was done by Jiang et al. in 1998 and the detailed regulation of both UTase and UR activities by small effectors, such as ATP, α-KG, and glutamine, as well as metal ions, was investigated (34). It appears that the uridylylation state of PII is largely dependent on the level of glutamine in the cell (34). Recently, in vitro studies of GlnD activities in other bacteria, such as Azospirillum brasilense, Herbaspirillum seropedicae, and R. rubrum, were also reported (2, 3, 10, 13, 38).
GlnD has at least 4 domains, based on a Pfam search (http://pfam.sanger.ac.uk) (9), as well as on previous domain analyses (4, 5, 15, 27). There is an N-terminal nucleotidyltransferase (NT) domain, a central HD domain, named after the conserved histidine and aspartate residues (6), and two C-terminal ACT domains, named after three of the allosterically regulated enzymes in which this domain is found: aspartokinase, chorismate mutase, and TyrA (prephenate dehydrogenase) (5).
It is clear that UTase activity is localized to the N-terminal NT domain. This domain has a distinct amino acid residue pattern with conserved glycine (G) and aspartate (D) residues (Fig. 1) (4, 27). Enzymes with this conserved domain belong to the eukaryotic DNA polymerase β nucleotidyltransferase superfamily (Polβ superfamily), and they display nucleotidyltransferase activity (4). The structures of this domain have been solved for several family members and are very similar to each other (17, 18, 27, 50, 58, 59, 65, 73). These structures also show that the two conserved aspartate residues (aspartate and glutamate residues in some members) are involved in the direct or indirect binding of metal ions, which are essential for substrate catalysis (18, 62, 65). In kanamycin nucleotidyltransferase (Kan-NT) and DNA polymerase β, conserved glycine and serine residues are located in the helical-turn motif, connecting the first β-sheet and a short helix. The serine residues in some family members contact the phosphate of nucleotides (50, 59, 65).
FIG. 1.
Alignment of nucleotidyltransferase (NT) domains of kanamycin nucleotidyltransferase (Kan-NT), rat DNA polymerase β, RNA-editing terminal uridylyl transferase 2 (TUTase 2) from Trypanosoma brucei, bovine poly(A) polymerase, and GlnD and GlnE from R. rubrum (Rr) and E. coli (Ec). Conserved glycine (serine or lysine in some members) and aspartate (glutamate in Kan-NT) residues are in bold. A gap in the DNA polymerase β sequence is indicated by an asterisk.
Another member of this family is GlnE, which is also a bifunctional enzyme and controls glutamine synthetase activity by adenylylation and deadenylylation. It is interesting that GlnE has two NT domains (Fig. 1), one at the C terminus that is responsible for an adenylyltransferase activity, and another at the N terminus that represents an adenylyl-removing (AR) activity (30). In contrast, GlnD has a single NT domain at the N terminus (Fig. 1). This raised a question about the location of the UR active site in GlnD: does it share the active site with UTase, or is it localized in a different portion of the protein? Based on kinetic analysis and mutational studies, Jiang et al. proposed that UTase and UR reactions likely occur at a single active center on GlnD (34, 56).
In contrast to the wealth of information about NT domains, the roles of the HD domain in the center of GlnD and of the two ACT domains at the C terminus are poorly understood. HD domains have been found in a superfamily of metal-dependent phosphohydrolases (6). A number of HD domain proteins have been characterized biochemically (12, 25, 26, 43, 52, 53, 60, 61, 63, 69, 74). The structures of this domain in several family members have been solved and show that the conserved HD residues chelate metal ions and constitute the catalytic center (26, 28, 44, 82). Based on sequence comparisons, it had been suggested that the HD domain in GlnD might represent the UR activity (6).
The ACT domain consensus sequence has been identified in the sequences of very diverse proteins, and some are involved in amino acid and purine synthesis (5). It has been proposed that the ACT is a ligand-binding domain (5, 15). Though only a few proteins containing this domain have been crystallized (19, 23, 41, 42, 51, 68, 71), their domain structures show a similar fold and a direct involvement in the binding of amino acids and other effectors (19, 41, 51, 71). Given the role of GlnD in sensing glutamine, one supposes that these ACT domains might be relevant to that sensing, though this has not been experimentally tested.
To better understand the role of the uridylylation of the PII proteins in nitrogen fixation in R. rubrum, we previously constructed insertion and deletion mutations in glnD and characterized the roles of GlnD in the regulation of NifA and NtrC activities and the posttranslational regulation of nitrogenase activity (80). Here, we report further functional analysis of the domains in GlnD from both R. rubrum and E. coli.
MATERIALS AND METHODS
Bacterial growth conditions.
E. coli was grown in LC medium (similar to Luria-Bertani medium but with 5 g/liter NaCl) (64). R. rubrum was grown in yeast extract-supplemented malate-NH4+ (SMN) rich medium (21, 57). For derepression of nitrogenase activity, SMN cultures of R. rubrum were inoculated into malate-glutamate (MG) medium (45). The whole-cell nitrogenase activity assay and darkness/NH4Cl treatments have been described previously (75). Antibiotics were used at the following concentrations (mg/liter): for R. rubrum, streptomycin (Sm), 100; kanamycin (Km), 12.5; tetracycline (Tc), 1; gentamicin (Gm), 10; and for E. coli, ampicillin (Ap), 100; Km, 25; Gm, 5; Cm, 25; and Tc, 12.5.
Construction of R. rubrum glnD mutants.
About 3 kb of a BamHI-HindIII fragment containing R. rubrum glnD was cloned into pUX19 (48), yielding pUX1000. A QuikChange method (Stratagene, La Jolla, CA) was used according to the manufacturer's instructions to generate localized random mutagenesis of the G110 or D123 residue, using pUX1000 as a template and synthetic oligonucleotides with a randomized codon at G110 or D123 as a pair of primers. After PCR, the DNA fragment was digested with BamHI and HindIII and then cloned into pUX19. After transformation with E. coli DH5α, plasmids were isolated from transformants and sequenced to identify the mutation. Plasmids with mutated glnD were transferred into E. coli S17-1 (70) and then transferred into an R. rubrum ΔglnD mutant (UR1325) by mating as described previously (47). Smr Gmr Kmr R. rubrum colonies were selected such that these plasmids were integrated into the chromosome of UR1325 at the 5′ or 3′ end of glnD, yielding strains UR1551 to UR1564.
Different primers were designed to construct deletions of the ACT region in R. rubrum GlnD by PCR, using pUX1236 (80) as a template. One plasmid (pUX1614) has a second ACT domain and the C terminus deleted (ΔT839-A936), and another (pUX1615) has both ACT domains and the C terminus deleted (ΔS723-A936). Similarly, two deletions in the central HD domain were also constructed: one plasmid (pUX1617) has an in-frame deletion of the central region from an NruI site to the first ACT domain (ΔE394-A721), and another one (pUX1620) has a 66-base pair (bp) deletion in the HD domain (ΔA530-V551). These plasmids were transformed into E. coli S17-1 and then conjugated into an R. rubrum ΔglnD mutant (UR1325). Smr Gmr Kmr R. rubrum colonies were selected such that these plasmids were integrated into the chromosome of UR1325, yielding strains UR1446, UR1447, UR1448, UR1449, and UR1658.
Overexpression and purification of R. rubrum GlnD variants.
To overexpress R. rubrum GlnD in E. coli, the wild-type (wt) and truncated glnD were amplified by PCR with a set of primers with NdeI and EcoRI sites at each end and then cloned into pJAL503 (67) at the NdeI and EcoRI sites, yielding pUX1815, pUX1816, pUX1817, pUX1818, and pUX1819. Together with the vector pJAL503, these plasmids were transferred into E. coli strain BD (glnB glnD mutant) (39), yielding strains UQ4091 to UQ4096.
We also constructed an R. rubrum wt GlnD-MalE fusion protein by subcloning wt glnD into pMAL-C2 (New England Biolabs, Inc., Beverly, MA), yielding strain pUX1655. This MalE-GlnD fusion protein was purified using an amylose resin column (New England Biolabs, Inc., Beverly, MA) according to the manufacturer's instructions and then desalted using a G-25 column.
Construction, overexpression and purification of E. coli GlnD variants.
E. coli GlnD variants with substitutions at the G93, G94, D107, H514, and D515 residues and HD or ACT deletions were constructed in the same way as the R. rubrum GlnD variants except using different numbers for these conserved residues. Two truncated GlnD variants with the N terminus only or without the N terminus were also constructed. These mutated glnD genes were cloned into pET-15b (Novagen/EMD Chemicals, Gibbstown, NJ), and His-tagged GlnD variants were purified with a His-Bind resin column (Novagen) and then desalted with G-25 column. Most proteins were at least 90% pure.
UTase and UR activity assay.
The UTase activity of E. coli GlnD was assayed under two different conditions: in the presence of Mg2+ only or in the presence of both Mn2+ and Mg2+, as described previously (34). For Mg2+-UTase activity, the reaction mixture contained 100 mM Tris (pH 7.5), 100 mM KCl, 25 mM MgCl2, 1 mM dithiothreitol (DTT), 0.3 mg/ml bovine serum albumin (BSA), 1 mM ATP, 10 mM α-KG, 60 μM E. coli GlnB, and various concentrations of GlnD as indicated below, in a volume of 50 μl. The reaction mixture was preincubated at 30°C for 2 min, and the reaction was started by adding 1 μl of 50 mM UTP. After incubation at 30°C for 60 min, the reaction was stopped by adding 2.5 μl of 0.5 M EDTA. For Mg2+-Mn2+-UTase activity, the reaction conditions were the same as described above except that 1 mM MnCl2 was added.
The UR activity of E. coli GlnD was also assayed under two different conditions: in the presence of Mn2+ or in the presence of Mg2+, as described previously. For Mg2+-UR activity, the reaction mixture contained 100 mM Tris (pH 7.5), 100 mM KCl, 50 mM MgCl2, 1 mM DTT, 0.3 mg/ml BSA, 0.5 mM ATP, 0.5 mM α-KG, 2.5 mM glutamine, 40 μM E. coli GlnB-UMP, and various concentrations of GlnD as indicated below, in a volume of 50 μl. The reaction was carried out at 30°C for 60 min, and was stopped by adding 2.5 μl of 0.5 M EDTA. For Mn2+-UR activity, the reaction conditions were the same as described above except that 1 mM MnCl2 was added, the glutamine concentration was increased from 2.5 mM to 10 mM, and MgCl2, ATP, and α-KG were omitted.
For the assay of R. rubrum GlnD activity, UTase activity was monitored under the same conditions as described above for E. coli GlnD in the presence of both Mn2+ and Mg2+. However, the UR activity was monitored using different conditions than those described above for E. coli GlnD, as suggested previously (38). The reaction mixture of 50 μl contained 100 mM Tris (pH 7.5), 100 mM KCl, 25 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 0.3 mg/ml BSA, 2 mM ATP, and 10 mM glutamine. R. rubrum or E. coli GlnB-UMP (20 to 60 μM) was used as a substrate. Different levels of α-KG (0 to 1 mM) were added to the mixture.
Quantitation of the modification of GlnB on native gels.
Samples from the UTase or UR assay were mixed with nondenaturing sample buffer, and the degree of uridylylation of GlnB was detected using nondenaturing polyacrylamide gel electrophoresis (PAGE) analysis, as described previously (8). All 4 forms of GlnB (with 0 to 3 UMP groups attached) were well separated. The bands in the gel were visualized using an Alpha Imager (Alpha Innotech Co., San Leandro, CA) and quantified using ImageQuant software (GE Healthcare, Piscataway, NJ).
The modified states of GlnB were determined using the following formula according to the amount of modified and unmodified forms of the protein: [GlnB-UMP + 2 × GlnB-(UMP)2 + 3 × GlnB-(UMP)3] ÷ [GlnB + GlnB-UMP + GlnB-(UMP)2 + GlnB-(UMP)3]. GlnB represents the unmodified form, while GlnB-UMP, GlnB-(UMP)2, and GlnB-(UMP)3 represent forms with one, two, and three modified subunits, respectively.
SDS-PAGE and immunoblotting of R. rubrum GlnB.
A trichloroacetic acid precipitation method was used to quickly extract protein (76), and a low-cross-linker (ratio of acrylamide to bisacrylamide, 172/1) tricine gel was used to separate modified and unmodified GlnB, as described previously (66). Proteins were electrophoretically transferred onto a nitrocellulose membrane, immunoblotted with polyclonal antibody against R. rubrum GlnB, and visualized with horseradish peroxidase color detection reagents (Bio-Rad, Richmond, CA).
RESULTS
In vivo and in vitro studies of R. rubrum GlnD variants.
Previous mutagenesis and complementation studies showed that the N-terminal region of R. rubrum GlnD is responsible for UTase activity (80). GlnD has an NT domain in the N terminus with several conserved glycine and aspartate residues (Fig. 1). To determine the role of these residues in UTase activity, we performed random mutagenesis on two of the conserved residues in the NT domain, G110 and D123, and monitored nitrogenase activity, which indirectly measures UTase activity. In R. rubrum, GlnD modifies GlnB under nitrogen-limiting conditions, and GlnB-UMP then activates NifA, which leads to the synthesis of nitrogenase (78, 80, 81). Little nitrogenase activity is detected in a ΔglnD mutant, since GlnB remains in the unmodified form regardless of nitrogen status (80). As shown in Table 1, most substitutions at residues G110 and D123 eliminated nitrogenase activity, suggesting the absence of UTase activity. Only the G110A substitution supported normal nitrogenase activity, implying substantial UTase activity. This is surprising since this glycine (G110) and the adjacent one (G109) are highly conserved in GlnD. As we reported previously, the activation of NifA requires lower UTase activity than does the activation of other receptors, such as NtrC (80). Nevertheless, these data suggest that the NT motif is important for UTase activity.
TABLE 1.
Nitrogenase activity in R. rubrum glnD mutants
| Strains | Chromosomal genotype with gene integration | Nitrogenase activitya |
|---|---|---|
| UR2 | wt | 750 |
| UR1446 | ΔglnD mutant with wt glnD | 650 |
| UR1556 | ΔglnD mutant with glnD encoding GlnD-G110L | <20 |
| UR1557 | ΔglnD mutant with glnD encoding GlnD-G110D | <20 |
| UR1558 | ΔglnD mutant with glnD encoding GlnD-G110R | <20 |
| UR1559 | ΔglnD mutant with glnD encoding GlnD-G110I | <20 |
| UR1560 | ΔglnD mutant with glnD encoding GlnD-G110A | 660 |
| UR1561 | ΔglnD mutant with glnD encoding GlnD-G110V | <20 |
| UR1562 | ΔglnD mutant with glnD encoding GlnD-G110W | <20 |
| UR1563 | ΔglnD mutant with glnD encoding GlnD-G110N | 30 |
| UR1564 | ΔglnD mutant with glnD encoding GlnD-G110Q | <20 |
| UR1551 | ΔglnD mutant with glnD encoding GlnD-D123A | <20 |
| UR1552 | ΔglnD mutant with glnD encoding GlnD-D123G | <20 |
| UR1553 | ΔglnD mutant with glnD encoding GlnD-D123V | <20 |
| UR1554 | ΔglnD mutant with glnD encoding GlnD-D123L | <20 |
| UR1555 | ΔglnD mutant with glnD encoding GlnD-D123Y | <20 |
| UR1447 | ΔglnD mutant with glnD-ΔACT2 (ΔT839-A936) | 680 |
| UR1448 | ΔglnD mutant with glnD-ΔACT1&2 (ΔS723-A936) | 700 |
| UR1449 | ΔglnD mutant with glnD-Δcentral region (ΔE394-A721) | 450 |
| UR1658 | ΔglnD mutant with glnD-ΔHD-22aa (ΔA530-V551) | 650 |
Each unit of nitrogenase activity is expressed as nmol of ethylene produced per h per ml of cells at an optical density of 1 at 600 nm. Each activity value is from at least five replicate assays from different individually grown cultures. The standard deviations were between 5 and 15%.
GlnD also has a central HD domain and two ACT domains at the C terminus, but the roles of these domains in UTase/UR are unknown. To study the roles of the HD and ACT domains, we constructed four R. rubrum glnD mutants with a deletion of the second ACT domain (GlnD-ΔACT2, strain UR1447), a deletion of both ACT domains (GlnD-ΔACT1&2, strain UR1448), a deletion of the central region including the entire HD domain (Δcentral, strain UR1449), or a deletion of 22 amino acids including the core HD residues (ΔHD-22aa, strain UR1658). As shown in Table 1, these deletions in R. rubrum glnD showed no significant effects on the activation of NifA, indicating that these HD and ACT domains are not essential for UTase activity. We also monitored two other regulatory systems that have previously been shown to be affected by GlnD activity (80), the DRAT/DRAG regulatory system, by monitoring nitrogenase activity in response to dark/light shifts or ammonium addition, and the NtrB-NtrC regulatory system, by monitoring the accumulation of GlnJ, whose expression is regulated by NtrC. Overall, the regulation of nitrogenase activity, reflecting DRAT/DRAG regulation, was normal in these mutants (data not shown). GlnJ levels were slightly decreased in strains UR1448 (GlnD-ΔACT1&2) and UR1658 (GlnD-ΔHD-22aa), but very low in UR1449 (GlnD-Δcentral) (data not shown). This implies that there are different levels of unmodified PII in these mutants, which would interact with NtrB and thereby inactivate NtrC. These results reflect alteration of UTase and/or UR activities in these mutants.
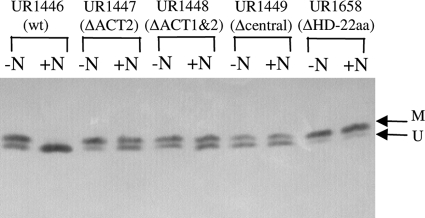
We also examined in vivo GlnD activity more directly by monitoring the modification of GlnB in response to the addition of NH4+. We monitored the modification of GlnB because its expression in R. rubrum is not tightly regulated by nitrogen status, unlike that of glnJ (37). In the presence of wt GlnD (UR1446), GlnB was partially modified under nitrogen-limiting conditions and became completely unmodified after the addition of NH4+ (Fig. 2). This is caused by the stimulation of UR activity and the inhibition of UTase of GlnD by the NH4+ signal (i.e., the glutamine pool). In GlnD-ΔACT2 (UR1447) and GlnD-ΔACT1&2 (UR1448), GlnB was also partially modified before NH4+ treatment, but only a fraction of GlnB became unmodified after the addition of NH4+, implying that these mutants have lower UR activities than does the wild type. Under nitrogen-limiting conditions, GlnB was also partially modified in the GlnD-Δcentral variant (UR1448) and almost completely modified in the GlnD-ΔHD-22aa variant (UR1658), but these two variants showed no significant changes in GlnB modification in response to NH4+, suggesting that these deletion mutants lacking the HD domain might have lost UR activity.
FIG. 2.
Modification of GlnB in R. rubrum strains UR1446 (wt GlnD), UR1447 (GlnD-ΔACT2), UR1448 (GlnD-ΔACT1&2), UR1449 (GlnD-Δcentral), and UR1658 (GlnD-ΔHD-22aa) in response to NH4+ treatment. R. rubrum strains were grown in MG medium for 2 days, and protein samples were collected before (−N) or 60 min after (+N) the addition of 20 mM NH4Cl by trichloroacetic acid precipitation, loaded on low-cross-linker tricine SDS-PAGE gels, and immunoblotted with antibody against R. rubrum GlnB. The positions of the modified (M) and unmodified GlnB subunits (U) are indicated by arrows.
However, it is difficult to interpret these data obtained from in vivo experiments. Poor deuridylylation of GlnB in response to NH4+ might be due to either low UR activity or inappropriately high (unregulated) UTase activity, which in turn could compete with the UR activity. It is impossible to separate these two GlnD activities in vivo, so we attempted to examine UTase and UR activities separately in vitro.
We overexpressed R. rubrum wt and four truncated GlnD proteins with deletions of ACT or HD domains in E. coli, using the heat-inducible pJAL503 (67). Unlike wt GlnD, most of the truncated R. rubrum GlnD proteins were insoluble. However, GlnD variants with deletions of ACT2, ACT1&2, central region, and HD-22aa had substantial amounts of UTase activity in crude extracts, but no UR activity was detected in any GlnD variants, including wt GlnD (data not shown). To rule out the possibility of an inhibitor of UR activity in crude extracts, we purified wild-type GlnD; it showed high UTase activity but no UR activity (data not shown). We also constructed and purified a MalE fusion to wt GlnD, which again showed high UTase activity but no UR activity (data not shown). We note that Jonsson and Nordlund did report UR activity in R. rubrum GlnD expressed with a glutathione S-transferase fusion that was later removed, but the activity seems to have been rather low and detected only with a very sensitive [α-32P]UMP assay using excess GlnD (38). Our inability to assay in vitro UR activity of R. rubrum GlnD forced us to consider another homolog, E. coli GlnD.
Construction, expression, and purification of E. coli glnD variants.
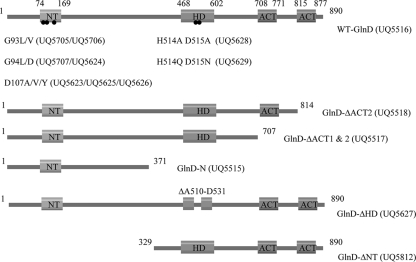
As shown in Fig. 3, we constructed a number of mutants of E coli GlnD, with substitutions at the G93, G94, and D107 residues (corresponding to the G109, G110, and D123 residues in R. rubrum GlnD), double substitutions at residues H514 and D515 (HD-AA and HD-QN) in the HD domain, deletions of the entire central and C-terminal region (GlnD-N), ΔACT1&2, ΔACT2, ΔHD (22 amino acids), and deletion of the N-terminal NT domain (GlnD-ΔNT). All of these GlnD proteins were His tagged and expressed in E. coli, purified using a His-bind resin column, and desalted using a G-25 column.
FIG. 3.
Substitutions and deletions of E. coli GlnD mutants. The location of each domain is indicated by the amino acid residue numbers. The positions of the replaced conserved residues in the NT and HD domains are indicated by the dots shown on the top line of the figure. E. coli strains are indicated in parenthesis.
The central HD domain of GlnD encodes UR activity.
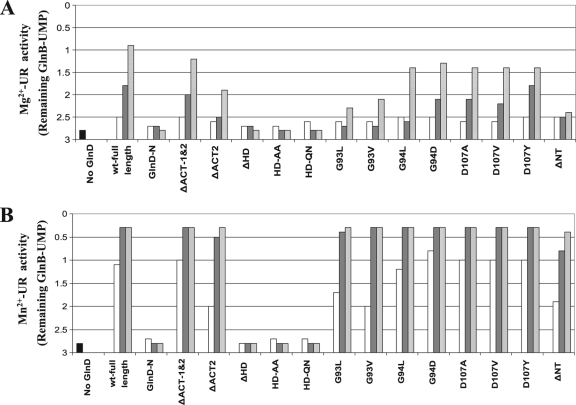
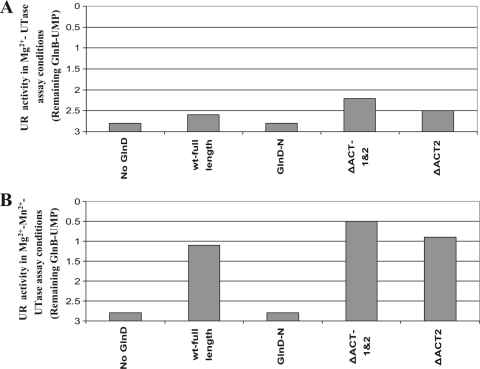
To compare UR activity, two assay conditions were used, as described in Materials and Methods; one is for Mg2+-UR activity, and the other is for Mn2+-UR activity. Three different concentrations of GlnD were used in the assays: 0.1, 0.5, and 2 μM. Similar to previous reports (34), E. coli wt GlnD showed more than 5-fold higher Mn2+-UR activity than Mg2+-UR activity (Fig. 4). The majority of the mutant proteins showed UR activity that was comparable to or only modestly lower than that of wt GlnD under both assay conditions. The NT deletion mutant (GlnD-ΔNT) and the G93L and G93V substitution mutants displayed Mn2+-UR activities similar to that of wt GlnD but had low Mg2+-UR activity. Only the GlnD-N, ΔHD, and two HD substitutions lacked UR activity under both conditions. These results indicate that the central HD domain is essential for UR activity and likely represents the active site of UR. The N-terminal NT domain is not essential for UR activity but might be involved in Mg2+ binding to support Mg2+-UR activity, based on the result with GlnD-ΔNT.
FIG. 4.
Mg2+-UR activities (A) and Mn2+-UR activities (B) in E. coli wt GlnD (full-length) and mutant variants. The starting substrate was fully uridylylated GlnB, and UR activity was measured by the decrease in the average number of UMP groups per GlnB trimer. Each variant had three concentrations of GlnD in the assay: 0.1 (open bars), 0.5 (dark gray bars), and 2 (light gray bars) μM. The results for the negative control (without GlnD) are shown by the first (black) bars. After the reaction, protein samples were separated on nondenaturing gels, and the degree of uridylylation of GlnB was determined as described in Materials and Methods.
The N-terminal NT domain is essential for UTase activity but not UR activity.
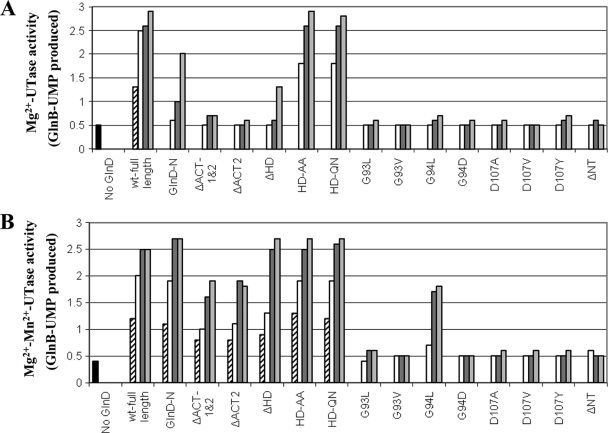
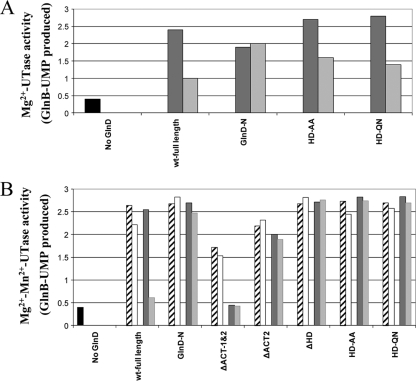
To compare UTase activities, two assay conditions were also used for the UTase assay, as described in Materials and Methods: Mg2+-UTase activity and Mg2+-Mn2+-UTase activity. wt GlnD had slightly higher Mg2+-UTase activity than Mg2+-Mn2+-UTase activity (Fig. 5), but the difference between these two activities was less than that seen previously (34). As expected, GlnD-N showed substantial UTase activity in both conditions, although it is much lower than wt GlnD in the presence of Mg2+; GlnD-ΔNT lacked UTase activity. These data are consistent with the hypothesis that the NT domain is the active site for UTase. All substitutions at G93, G94, and D107, except G94L, also caused almost complete loss of UTase activity under both conditions. These data confirm an important role for these NT domain residues in UTase activity, as predicted by the structural analysis of this domain from other family members (17, 18, 58, 62, 65).
FIG. 5.
Mg2+-UTase activities (A) and Mg2+-Mn2+-UTase activities (B) in E. coli wt GlnD and mutant variants. UTase activity was measured by monitoring the appearance of GlnB-UMP with unmodified GlnB as the substrate. Each variant had three or four concentration of GlnD in the assay: 0.025 (striped bars), 0.1 (open bars), 0.5 (dark gray bars) and 2 (light gray bars) μM. The results for the negative control (without GlnD) are shown by the first (black) bars.
Importantly, the HD substitution mutants (HD-AA and HD-QN) shown above to lack UR activity (Fig. 4) had UTase activities similar to that seen in wt GlnD in both assays (Fig. 5), indicating that these variants are not simply dead GlnD proteins. This argues that UTase and UR activities do not share the same active site. Both the GlnD-ΔACT1&2 and the GlnD-ΔACT2 variant appear to have lower UTase activities than that seen in wt GlnD in both assays, though to different degrees, suggesting that ACT might play some role in the regulation of UTase activity. However, as described below, the apparent reduced Mg2+-Mn2+-UTase activities in these ACT deletion mutants might actually be due to the altered regulation of UR activity, which then competes with UTase activity to decrease the modification of GlnB.
Competition of UTase and UR activities in in vitro assays.
In a UR activity assay, UTase does not interfere with UR activity, since there is no UTP in the assay for GlnD to use for PII modification. In contrast, under UTase assay conditions, inappropriately high UR activity in a mutant protein could compete with the UTase activity by deuridylylating GlnB. This would have the appearance of altered UTase activity, though the direct effect would actually be on UR activity. The reduced UTase activity in ACT mutants could therefore be caused by unregulated UR activity. To test this, we measured UR activity under the Mg2+-Mn2+-UTase assay conditions in these variants and in wt GlnD. Under these assay conditions and in the absence of glutamine, GlnD-ΔACT1&2 and GlnD-ΔACT2 showed substantial UR activity—even higher than that of wt GlnD (Fig. 6B). This result strongly suggests that there is competition of UTase and UR activities under Mg2+-Mn2+-UTase assay conditions in these ACT mutants and, probably, in the wild type as well, resulting in a futile cycle of the uridylylation and deuridylylation of GlnB. We also measured UR activity under Mg2+-UTase assay conditions, and the ACT mutants and wt GlnD showed low UR activities (Fig. 6A). Unlike Mg2+-Mn2+-UTase, the poor Mg2+-UTase activities in these ACT variants (Fig. 5A) do not seem to be due to the competition of UR activity, for the following reasons: (i) the Mg2+-UR activity in these variants is much lower than the Mn2+-UR activity (Fig. 4); (ii) very low UR activity was also seen under Mg2+-UTase assay conditions (Fig. 6B); and (iii) low Mg2+-UTase activity was also seen in GlnD-N and GlnD-ΔHD, which have no UR activity (Fig. 5A). In summary, there are substantial UTase and UR activities in the presence of Mn2+, causing a futile cycle of the uridylylation and deuridylylation of GlnB. The ACT domains might be important for the regulation of UR activity under these conditions to avoid this futile cycle.
FIG. 6.
UR activities in E. coli wt GlnD and mutant variants in the absence of glutamine. The assay was done in the presence of 1 mM ATP, 1 mM α-KG, and 50 mM MgCl2 (Mg2+-UTase assay conditions) (A) or in the presence of 1 mM ATP, 10 mM α-KG, 50 mM MgCl2, and 1 mM MnCl2 (Mg2+-Mn2+-UTase assay conditions) (B). A concentration of 0.5 μM GlnD was used for the assay. The results for the negative control (without GlnD) are shown by the first bars.
Glutamine stimulates UR activity in wt GlnD but not in the ACT deletion mutants.
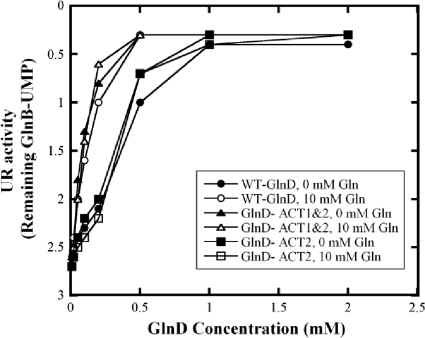
Previous studies showed that glutamine stimulates the UR activity of E. coli GlnD (20, 34). However, as seen in the results described above, GlnD-ΔACT1&2 and GlnD-ΔACT2 showed high UR activities in the absence of glutamine. Because of the potential role of the ACT domains in the binding of glutamine, we further investigated the effect of glutamine on UR activity in wt GlnD and ACT deletion variants in the presence of Mn2+, since all strains have very similar Mn2+-UR activities. As shown in Fig. 7, glutamine stimulates the Mn2+-UR activity of wt GlnD but has no significant effect on the Mn2+-UR activities of ACT deletion variants. This suggests that these ACT domains might play some regulatory role in the regulation of GlnD activity in response to glutamine.
FIG. 7.
Effects of glutamine on Mn2+-UR activities in E. coli wt GlnD and ACT deletion variants. Each variant had 8 different concentrations of GlnD in the assay: 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, and 2 μM. Glutamine concentrations were either 0 or 10 mM.
Inhibition of UTase activity by glutamine.
Previous studies have shown that glutamine inhibits the UTase activity of E. coli GlnD (1, 20, 34, 39) but not that of R. rubrum GlnD (38). However, this is complicated by the glutamine stimulation of UR activity noted above. A decrease in PII modification by glutamine, suggestive of the inhibition of UTase activity, could therefore be caused wholly or in part by the elevation of UR activity. Indeed, as shown in the results described above, this futile cycle of the uridylylation/deuridylylation of PII clearly occurs under our in vitro assay conditions. To avoid the interference in UTase activity by UR, we investigated the effects of glutamine on UTase activities in the mutants lacking UR activity. As shown in Fig. 8B, glutamine inhibits the uridylylation of GlnB by wt GlnD under Mg2+-Mn2+-UTase assay conditions. However, the glutamine effect was more striking at a high protein concentration (2 μM) than at a low concentration (0.5 μM). This is difficult to explain by the “UTase inhibition” model but is consistent with glutamine stimulating UR activity. With our GlnD variants lacking UR activity (the GlnD-N and HD deletion or substitution mutants), there was little effect of glutamine on the Mg2+-Mn2+-UTase activity. Mutants with ACT deletions also showed no significant effect of glutamine, although they have lower Mg2+-Mn2+-UTase activities than do other variants, due to the high and unregulated UR activity under these assay conditions (Fig. 6B). These results suggest that under the Mg2+-Mn2+ assay conditions, the effect of glutamine is solely on the stimulation of UR activity and that the ACT domains are important for this glutamine effect.
FIG. 8.
Effects of glutamine on Mg2+-UTase activities (A) and Mg2+-Mn2+-UTase activities (B) in E. coli wt GlnD and variants. The assay had each variant of GlnD present in one or two concentrations, as well as the presence or absence of glutamine: 0.5 μM GlnD without glutamine (striped bars), 0.5 μM GlnD with 10 mM glutamine (open bars), 2 μM GlnD without glutamine (dark gray bars), and 2 μM GlnD with 10 mM glutamine (light gray bars). The results for the negative control (without GlnD) are shown by the first (black) bars.
In contrast, in the absence of Mn2+, glutamine showed partial inhibition of the Mg2+-UTase in wt GlnD and GlnD-HD-AA or QN substitution mutants but had no effect on Mg2+-UTase in GlnD-N (Fig. 8A). Clearly this glutamine effect cannot be on the stimulation of UR activity, since these mutants lack that. Instead, this effect must reflect inhibition of UTase activity by glutamine under these conditions. The GlnD-N variant lacks this effect, which might mean either that glutamine inhibition requires a C terminus and/or a central region of GlnD or that this variant has an altered structure that affects regulation. Unfortunately, we are unable to study the roles of ACT in this inhibition, since ACT deletion mutants have very low Mg2+-UTase activity (Fig. 5A). In summary, these results suggest that glutamine probably inhibits Mg2+-UTase activity but not Mg2+-Mn2+-UTase activity.
DISCUSSION
Role of the N-terminal NT domain.
GlnD is a bifunctional uridylyltransferase/uridylyl-removing enzyme (UTase/UR), and it is clear that the UTase active site is located at the N terminus of the protein, based on mutational studies (56, 72, 80) and domain analysis (4, 27). The results of in vivo analysis of R. rubrum glnD mutants and the in vitro studies of E. coli GlnD variants reported here confirm that the N-terminal NT domain is critical for UTase activity. Most substitutions for conserved glycine and aspartate residues in this domain completely abolish its UTase activity but have no significant effect on UR activity.
Role of the central HD domain.
Although sequence comparisons suggested that the HD domain might be the active site for UR activity, previous mutational and kinetic studies implied that UR and UTase of E. coli GlnD might share an active site (34, 56). Our results clearly indicate that HD is critical for UR activity. Two substitution mutations of HD residues in the central HD domain, as well as the deletion of the HD domain, completely eliminate UR activity but have little effect on UTase activity. Furthermore, a truncated GlnD lacking the N-terminal NT domain showed high Mn2+-UR activity. These data indicate that the UR and UTase activities do not share an active site. Given the fact that this HD domain is found in many proteins with phosphohydrolase activity, this HD domain is likely the active site for the UR reaction.
Role of ACT domains.
Based on sequence comparisons and previous studies of proteins containing ACT domains that bind amino acids or small effectors, it is likely that this domain in GlnD is involved in the binding of glutamine. The results of previous studies indicate that glutamine stimulates UR activity and inhibits the UTase activity of GlnD, and it is believed that this is the mechanism for nitrogen signal transduction in the cell (1, 20, 34, 39). Our results show that the deletion of ACT domains eliminates this effect of glutamine on UR activity, strongly supporting this model. Presumably, the ACT domain causes some inhibitory effect on UR activity, which is eliminated when it binds glutamine.
However, in vivo studies of R. rubrum ACT mutants indicated that these mutants could still sense a nitrogen signal even in the absence of the ACT domains of GlnD. As shown in Fig. 2, R. rubrum GlnD variants with deletions of the ACT domains were able to sense NH4+, resulting in the deuridylylation of some portion of GlnB after the addition of NH4+, although the deuridylylation of GlnB in these mutants is much less complete than that seen in the wild type. Previously, Tøndervik et al. reported that an E. coli glnD mutant lacking the ACT domains was still able to sense nitrogen status (72). These authors suggested that either the N terminus of GlnD senses glutamine or that there is another metabolite or mechanism for nitrogen sensing. Given the present results, we favor a model in which cells also sense the nitrogen signal through PII. PII binds α-KG (40, 54), and a change in the glutamine level would affect the α-KG pool, which would change the PII structure and its modification state. This should be sufficient to explain the results of Tøndervik et al. (72) and our own results with the R. rubrum ACT mutants.
We have shown that a futile cycle of the uridylylation and deuridylylation of GlnB occurs under in vitro assay conditions. The ACT domains play important roles for the regulation of UR activity under these conditions to avoid this futile cycle. It would be interesting to know if this futile cycle occurs in vivo as well. We noticed that the addition of glutamine has significant effects on both UTase and UR activities, but the inhibition of UTase activity by glutamine is not very strong, at least under our in vitro assay conditions. If the regulation of GlnD is solely dependent on the glutamine level in the cell, it is likely that the futile cycles of the uridylylation/deuridylylation of PII occur in vivo as well.
The role of metal ions.
It is well known that Mn2+ and Mg2+ have significant effects on both the UTase and the UR activity of E. coli GlnD (1, 22, 34), though the precise role of these metals in the regulation of GlnD activity is still unknown. Mg2+ is believed to be the physiologically important metal ion effector (34), but it is impossible at present to know the effective concentrations of these metal ions in vivo. Thus, while GlnD appears to have two metal-binding sites, and our results indicate that the deletion of one site has a significant effect on the activity located at the different site, conjectures about the relative impacts of different ions at these sites would be pure speculation.
The structures of kanamycin nucleotidyltransferase (Kan-NT), DNA polymerase β, and other family members show that the NT domain can be directly involved in binding metal ions (18, 62, 65), so it is reasonable to suppose that the NT domain in GlnD is also involved in metal binding for UTase activity. However, variants altered in the NT domain showed robust Mn2+-UR activity but low Mg2+-UR activity, indicating that there are other metal-binding site(s) for Mn2+ to support Mn2-UR activity, likely in the HD domain.
In summary, we confirm that several conserved residues in the N-terminal NT domain of GlnD are critical for UTase activity. Our results indicate that the UR active site is located in the central HD domain. The ACT domains at the C terminus appear to play regulatory roles in GlnD activity, probably through the binding of glutamine.
Acknowledgments
This work was supported by NIGMS grant GM65891 to G.P.R.
We thank Alex Ninfa for generously providing the E. coli glnB glnD mutant and the GlnB and GlnD overexpression strains. We also thank Stefan Nordlund for providing information about in vitro UTase and UR activity assays for R. rubrum GlnD.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Adler, S. P., D. Purich, and E. R. Stadtman. 1975. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl-removing enzyme. J. Biol. Chem. 250:6264-6272. [PubMed] [Google Scholar]
- 2.Araújo, L. M., L. F. Huergo, A. L. Invitti, C. I. Gimenes, A. C. Bonatto, R. A. Monteiro, E. M. Souza, F. O. Pedrosa, and L. S. Chubatsu. 2008. Different responses of the GlnB and GlnZ proteins upon in vitro uridylylation by the Azospirillum brasilense GlnD protein. Braz. J. Med. Biol. Res. 41:289-294. [DOI] [PubMed] [Google Scholar]
- 3.Araujo, M. S., V. A. Baura, E. M. Souza, E. M. Benelli, L. U. Rigo, M. B. Steffens, F. O. Pedrosa, and L. S. Chubatsu. 2004. In vitro uridylylation of the Azospirillum brasilense N-signal transducing GlnZ protein. Protein Expr. Purif. 33:19-24. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., and E. V. Koonin. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind, L., and E. V. Koonin. 1999. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 287:1023-1040. [DOI] [PubMed] [Google Scholar]
- 6.Aravind, L., and E. V. Koonin. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 7.Arcondéguy, T., R. Jack, and M. Merrick. 2001. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 65:80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson, M. R., E. S. Kamberov, R. L. Weiss, and A. J. Ninfa. 1994. Reversible uridylylation of the Escherichia coli PII signal transduction protein regulates its ability to stimulate the dephosphorylation of the transcription factor nitrogen regulator I (NRI or NtrC). J. Biol. Chem. 269:28288-28293. [PubMed] [Google Scholar]
- 9.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benelli, E. M., M. Buck, E. M. de Souza, M. G. Yates, and F. O. Pedrosa. 2001. Uridylylation of the PII protein from Herbaspirillum seropedicae. Can. J. Microbiol. 47:309-314. [DOI] [PubMed] [Google Scholar]
- 11.Blauwkamp, T. A., and A. J. Ninfa. 2003. Antagonism of PII signalling by the AmtB protein of Escherichia coli. Mol. Microbiol. 48:1017-1028. [DOI] [PubMed] [Google Scholar]
- 12.Blondal, T., S. Hjorleifsdottir, A. Aevarsson, O. H. Fridjonsson, S. Skirnisdottir, J. O. Wheat, A. G. Hermannsdottir, G. O. Hreggvidsson, A. V. Smith, and J. K. Kristjansson. 2005. Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J. Biol. Chem. 280:5188-5194. [DOI] [PubMed] [Google Scholar]
- 13.Bonatto, A. C., G. H. Couto, E. M. Souza, L. M. Araújo, F. O. Pedrosa, L. Noindorf, and E. M. Benelli. 2007. Purification and characterization of the bifunctional uridylyltransferase and the signal transducing proteins GlnB and GlnK from Herbaspirillum seropedicae. Protein Expr. Purif. 55:293-299. [DOI] [PubMed] [Google Scholar]
- 14.Brown, M. S., A. Segal, and E. R. Stadtman. 1971. Modulation of glutamine synthetase adenylylation and deadenylylation is mediated by metabolic transformation of the PII-regulatory protein. Proc. Natl. Acad. Sci. U. S. A. 68:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chipman, D. M., and B. Shaanan. 2001. The ACT domain family. Curr. Opin. Struct. Biol. 11:694-700. [DOI] [PubMed] [Google Scholar]
- 16.Coutts, G., G. Thomas, D. Blakey, and M. Merrick. 2002. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 21:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies, J. F., II, R. J. Almassy, Z. Hostomska, R. A. Ferre, and Z. Hostomsky. 1994. 2.3 Å crystal structure of the catalytic domain of DNA polymerase β. Cell 76:1123-1133. [DOI] [PubMed] [Google Scholar]
- 18.Deng, J., N. L. Ernst, S. Turley, K. D. Stuart, and W. G. Hol. 2005. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 24:4007-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devedjiev, Y., Y. Surendranath, U. Derewenda, A. Gabrys, D. R. Cooper, R. G. Zhang, L. Lezondra, A. Joachimiak, and Z. S. Derewenda. 2004. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J. Mol. Biol. 343:395-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engleman, E. G., and S. H. Francis. 1978. Cascade control of E. coli glutamine synthetase. II. Metabolite regulation of the enzymes in the cascade. Arch. Biochem. Biophys. 191:602-612. [DOI] [PubMed] [Google Scholar]
- 21.Fitzmaurice, W. P., L. L. Saari, R. G. Lowery, P. W. Ludden, and G. P. Roberts. 1989. Genes coding for the reversible ADP-ribosylation system of dinitrogenase reductase from Rhodospirillum rubrum. Mol. Gen. Genet. 218:340-347. [DOI] [PubMed] [Google Scholar]
- 22.Francis, S. H., and E. G. Engleman. 1978. Cascade control of E. coli glutamine synthetase. I. Studies on the uridylyl transferase and uridylyl removing enzyme(s) from E. coli. Arch. Biochem. Biophys. 191:590-601. [DOI] [PubMed] [Google Scholar]
- 23.Gallagher, D. T., G. L. Gilliland, G. Xiao, J. Zondlo, K. E. Fisher, D. Chinchilla, and E. Eisenstein. 1998. Structure and control of pyridoxal phosphate dependent allosteric threonine deaminase. Structure 6:465-475. [DOI] [PubMed] [Google Scholar]
- 24.Garcia, E., and S. G. Rhee. 1983. Cascade control of Escherichia coli glutamine synthetase. Purification and properties of PII uridylyltransferase and uridylyl-removing enzyme. J. Biol. Chem. 258:2246-2253. [PubMed] [Google Scholar]
- 25.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19:1373-1384. [DOI] [PubMed] [Google Scholar]
- 26.Hogg, T., U. Mechold, H. Malke, M. Cashel, and R. Hilgenfeld. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57-68. [DOI] [PubMed] [Google Scholar]
- 27.Holm, L., and C. Sander. 1995. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20:345-347. [DOI] [PubMed] [Google Scholar]
- 28.Huai, Q., Y. Liu, S. H. Francis, J. D. Corbin, and H. Ke. 2004. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J. Biol. Chem. 279:13095-13101. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda, T. P., A. E. Shauger, and S. Kustu. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259:589-607. [DOI] [PubMed] [Google Scholar]
- 30.Jaggi, R., W. C. van Heeswijk, H. V. Westerhoff, D. L. Ollis, and S. G. Vasudevan. 1997. The two opposing activities of adenylyl transferase reside in distinct homologous domains, with intramolecular signal transduction. EMBO J. 16:5562-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javelle, A., E. Severi, J. Thornton, and M. Merrick. 2004. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 279:8530-8538. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, P., A. E. Mayo, and A. J. Ninfa. 2007. Escherichia coli glutamine synthetase adenylyltransferase (ATase, EC 2.7.7.49): kinetic characterization of regulation by PII, PII-UMP, glutamine, and α-ketoglutarate. Biochemistry 46:4133-4146. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, P., and A. J. Ninfa. 1999. Regulation of autophosphorylation of Escherichia coli nitrogen regulator II by the PII signal transduction protein. J. Bacteriol. 181:1906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Enzymological characterization of the signal-transducing uridylyltransferase/uridylyl-removing enzyme (EC 2.7.7.59) of Escherichia coli and its interaction with the PII protein. Biochemistry 37:12782-12794. [DOI] [PubMed] [Google Scholar]
- 35.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. Reconstitution of the signal-transduction bicyclic cascade responsible for the regulation of Ntr gene transcription in Escherichia coli. Biochemistry 37:12795-12801. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, P., J. A. Peliska, and A. J. Ninfa. 1998. The regulation of Escherichia coli glutamine synthetase revisited: role of 2-ketoglutarate in the regulation of glutamine synthetase adenylylation state. Biochemistry 37:12802-12810. [DOI] [PubMed] [Google Scholar]
- 37.Johansson, M., and S. Nordlund. 1996. Transcription of the glnB and glnA genes in the photosynthetic bacterium Rhodospirillum rubrum. Microbiology 142:1265-1272. [DOI] [PubMed] [Google Scholar]
- 38.Jonsson, A., and S. Nordlund. 2007. In vitro studies of the uridylylation of the three PII protein paralogs from Rhodospirillum rubrum: the transferase activity of R. rubrum GlnD is regulated by α-ketoglutarate and divalent cations but not by glutamine. J. Bacteriol. 189:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamberov, E. S., M. R. Atkinson, J. Feng, P. Chandran, and A. J. Ninfa. 1994. Sensory components controlling bacterial nitrogen assimilation. Cell. Mol. Biol. Res. 40:175-191. [PubMed] [Google Scholar]
- 40.Kamberov, E. S., M. R. Atkinson, and A. J. Ninfa. 1995. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 270:17797-17807. [DOI] [PubMed] [Google Scholar]
- 41.Kaplun, A., M. Vyazmensky, Y. Zherdev, I. Belenky, A. Slutzker, S. Mendel, Z. Barak, D. M. Chipman, and B. Shaanan. 2006. Structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from Escherichia coli. J. Mol. Biol. 357:951-963. [DOI] [PubMed] [Google Scholar]
- 42.Kobe, B., I. G. Jennings, C. M. House, B. J. Michell, K. E. Goodwill, B. D. Santarsiero, R. C. Stevens, R. G. Cotton, and B. E. Kemp. 1999. Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6:442-448. [DOI] [PubMed] [Google Scholar]
- 43.Kondo, N., S. Kuramitsu, and R. Masui. 2004. Biochemical characterization of TT1383 from Thermus thermophilus identifies a novel dNTP triphosphohydrolase activity stimulated by dATP and dTTP. J. Biochem. 136:221-231. [DOI] [PubMed] [Google Scholar]
- 44.Kondo, N., N. Nakagawa, A. Ebihara, L. Chen, Z. J. Liu, B. C. Wang, S. Yokoyama, S. Kuramitsu, and R. Masui. 2007. Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis. Acta Crystallogr. D 63:230-239. [DOI] [PubMed] [Google Scholar]
- 45.Lehman, L. J., and G. P. Roberts. 1991. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J. Bacteriol. 173:5705-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leigh, J. A., and J. A. Dodsworth. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349-377. [DOI] [PubMed] [Google Scholar]
- 47.Liang, J. H., G. M. Nielsen, D. P. Lies, R. H. Burris, G. P. Roberts, and P. W. Ludden. 1991. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J. Bacteriol. 173:6903-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lies, D. P. 1994. Genetic manipulation and the overexpression analysis of posttranslational nitrogen fixation regulation in Rhodospirillum rubrum. Ph.D. thesis. University of Wisconsin—Madison, Madison, WI.
- 49.Mangum, J. H., G. Magni, and E. R. Stadtman. 1973. Regulation of glutamine synthetase adenylylation and deadenylylation by the enzymatic uridylylation and deuridylylation of the PII regulatory protein. Arch. Biochem. Biophys. 158:514-525. [DOI] [PubMed] [Google Scholar]
- 50.Martin, G., W. Keller, and S. Doublie. 2000. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19:4193-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mas-Droux, C., G. Curien, M. Robert-Genthon, M. Laurencin, J. L. Ferrer, and R. Dumas. 2006. A novel organization of ACT domains in allosteric enzymes revealed by the crystal structure of Arabidopsis aspartate kinase. Plant Cell 18:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray, K. D., and H. Bremer. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 259:41-57. [DOI] [PubMed] [Google Scholar]
- 53.Nagata, M., C. Kaito, and K. Sekimizu. 2008. Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J. Biol. Chem. 283:2176-2184. [DOI] [PubMed] [Google Scholar]
- 54.Ninfa, A. J., and M. R. Atkinson. 2000. PII signal transduction proteins. Trends Microbiol. 8:172-179. [DOI] [PubMed] [Google Scholar]
- 55.Ninfa, A. J., M. R. Atkinson, E. S. Kamberov, J. Feng, and E. G. Ninfa. 1995. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria, p. 147-158. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 56.Ninfa, A. J., P. Jiang, M. R. Atkinson, and J. A. Peliska. 2000. Integration of antagonistic signals in the regulation of nitrogen assimilation in Escherichia coli. Curr. Top. Cell. Regul. 36:31-75. [DOI] [PubMed] [Google Scholar]
- 57.Ormerod, J. G., K. S. Ormerod, and H. Gest. 1961. Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch. Biochem. Biophys. 94:449-463. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen, L. C., M. M. Benning, and H. M. Holden. 1995. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry 34:13305-13311. [DOI] [PubMed] [Google Scholar]
- 59.Pelletier, H., M. R. Sawaya, A. Kumar, S. H. Wilson, and J. Kraut. 1994. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science 264:1891-1903. [PubMed] [Google Scholar]
- 60.Proudfoot, M., E. Kuznetsova, G. Brown, N. N. Rao, M. Kitagawa, H. Mori, A. Savchenko, and A. F. Yakunin. 2004. General enzymatic screens identify three new nucleotidases in Escherichia coli. Biochemical characterization of SurE, YfbR, and YjjG. J. Biol. Chem. 279:54687-54694. [DOI] [PubMed] [Google Scholar]
- 61.Quirk, S., and M. J. Bessman. 1991. dGTP triphosphohydrolase, a unique enzyme confined to members of the family Enterobacteriaceae. J. Bacteriol. 173:6665-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakon, J., H. H. Liao, A. M. Kanikula, M. M. Benning, I. Rayment, and H. M. Holden. 1993. Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-Å resolution. Biochemistry 32:11977-11984. [DOI] [PubMed] [Google Scholar]
- 63.Salgado-Pabón, W., S. Jain, N. Turner, C. van der Does, and J. P. Dillard. 2007. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol. Microbiol. 66:930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 65.Sawaya, M. R., R. Prasad, S. H. Wilson, J. Kraut, and H. Pelletier. 1997. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36:11205-11215. [DOI] [PubMed] [Google Scholar]
- 66.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 67.Schauder, B., H. Blöcker, R. Frank, and J. E. McCarthy. 1987. Inducible expression vectors incorporating the Escherichia coli atpE translational initiation region. Gene 52:279-283. [DOI] [PubMed] [Google Scholar]
- 68.Schuller, D. J., G. A. Grant, and L. J. Banaszak. 1995. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat. Struct. Biol. 2:69-76. [DOI] [PubMed] [Google Scholar]
- 69.Seto, D., S. K. Bhatnagar, and M. J. Bessman. 1988. The purification and properties of deoxyguanosine triphosphate triphosphohydrolase from Escherichia coli. J. Biol. Chem. 263:1494-1499. [PubMed] [Google Scholar]
- 70.Simon, R., U. B. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 71.Thompson, J. R., J. K. Bell, J. Bratt, G. A. Grant, and L. J. Banaszak. 2005. Vmax regulation through domain and subunit changes. The active form of phosphoglycerate dehydrogenase. Biochemistry 44:5763-5773. [DOI] [PubMed] [Google Scholar]
- 72.Tøndervik, A., H. R. Torgersen, H. K. Botnmark, and A. R. Strøm. 2006. Transposon mutations in the 5′ end of glnD, the gene for a nitrogen regulatory sensor, that suppress the osmosensitive phenotype caused by otsBA lesions in Escherichia coli. J. Bacteriol. 188:4218-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu, Y., R. Zhang, A. Joachimiak, P. D. Carr, T. Huber, S. G. Vasudevan, and D. L. Ollis. 2004. Structure of the N-terminal domain of Escherichia coli glutamine synthetase adenylyltransferase. Structure 12:861-869. [DOI] [PubMed] [Google Scholar]
- 74.Yakunin, A. F., M. Proudfoot, E. Kuznetsova, A. Savchenko, G. Brown, C. H. Arrowsmith, and A. M. Edwards. 2004. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J. Biol. Chem. 279:36819-36827. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1995. Comparison studies of dinitrogenase reductase ADP-ribosyl transferase/dinitrogenase reductase activating glycohydrolase regulatory systems in Rhodospirillum rubrum and Azospirillum brasilense. J. Bacteriol. 177:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Y., R. H. Burris, P. W. Ludden, and G. P. Roberts. 1993. Posttranslational regulation of nitrogenase activity by anaerobiosis and ammonium in Azospirillum brasilense. J. Bacteriol. 175:6781-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2001. Functional characterization of three GlnB homologs in the photosynthetic bacterium Rhodospirillum rubrum: roles in sensing ammonium and energy status. J. Bacteriol. 183:6159-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2000. Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum. J. Bacteriol. 182:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Y., E. L. Pohlmann, P. W. Ludden, and G. P. Roberts. 2003. Regulation of nitrogen fixation by multiple PII homologs in the photosynthetic bacterium Rhodospirillum rubrum. Symbiosis 35:85-100. [Google Scholar]
- 80.Zhang, Y., E. L. Pohlmann, and G. P. Roberts. 2005. GlnD is essential for NifA activation, NtrB/NtrC-regulated gene expression, and posttranslational regulation of nitrogenase activity in the photosynthetic, nitrogen-fixing bacterium Rhodospirillum rubrum. J. Bacteriol. 187:1254-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu, Y., M. C. Conrad, Y. Zhang, and G. P. Roberts. 2006. Identification of Rhodospirillum rubrum GlnB variants that are altered in their ability to interact with different targets in response to nitrogen-status signals. J. Bacteriol. 188:1866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimmerman, M. D., M. Proudfoot, A. Yakunin, and W. Minor. 2008. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 378:215-226. [DOI] [PMC free article] [PubMed] [Google Scholar]