Abstract
The conjugative coupling protein TrwB is responsible for connecting the relaxosome to the type IV secretion system during conjugative DNA transfer of plasmid R388. It is directly involved in transport of the relaxase TrwC, and it displays an ATPase activity probably involved in DNA pumping. We designed a conjugation assay in which the frequency of DNA transfer is directly proportional to the amount of TrwB. A collection of point mutants was constructed in the TrwB cytoplasmic domain on the basis of the crystal structure of TrwBΔN70, targeting the nucleotide triphosphate (NTP)-binding region, the cytoplasmic surface, or the internal channel in the hexamer. An additional set of transfer-deficient mutants was obtained by random mutagenesis. Most mutants were impaired in both DNA and protein transport. We found that the integrity of the nucleotide binding domain is absolutely required for TrwB function, which is also involved in monomer-monomer interactions. Polar residues surrounding the entrance and inside the internal channel were important for TrwB function and may be involved in interactions with the relaxosomal components. Finally, the N-terminal transmembrane domain of TrwB was subjected to random mutagenesis followed by a two-hybrid screen for mutants showing enhanced protein-protein interactions with the related TrwE protein of Bartonella tribocorum. Several point mutants were obtained with mutations in the transmembranal helices: specifically, one proline from each protein may be the key residue involved in the interaction of the coupling protein with the type IV secretion apparatus.
Bacterial conjugation can be viewed mechanistically as a rolling-circle replication system linked to a type IV secretion process. The two processes come into contact through the activity of a protein that couples the plasmid replication machinery to the export system in the membrane, allowing horizontal dissemination of the replicating DNA molecule (35). This key protein is called “coupling protein” (here “T4CP” for “type IV CP”). It is present in all conjugative systems as well as in many type IV secretion systems (T4SS) involved in bacterial virulence (16). The secreted substrate in bacterial conjugation is the relaxase or pilot protein, attached to the DNA strand. The shoot-and-pump model for bacterial conjugation proposes that, after secretion of the protein through the T4SS, the T4CP works as a motor for export of the rest of the DNA molecule (36). In addition to its presumed role as a DNA transporter, TrwB is also required for transport of relaxase TrwC in the absence of DNA transfer (15).
In accordance with its proposed coupling activity, early genetic experiments made patent that the function of conjugative T4CPs depended on interactions with both the cytoplasmic substrate complex (the relaxosome) and the T4SS (6, 7). Thus, T4CP interactions with other conjugation proteins are a key aspect of their function. There have been several reports of interactions between T4CPs from conjugative plasmids and either relaxosomal components—such as F-TraD with TraM (14, 38), RP4-TraG with TraI (49), and pCF10-PcfC with PcfF and PcfG (11)—or T4SS components such as R27-TraG with TrhB (17). T4CP-T4SS interactions have also been reported for the VirB/D4 T4SS involved in DNA transfer from Agrobacterium tumefaciens to plant cells (1, 9). Both sets of interactions have only been concurrently shown for TrwB, the T4CP of plasmid R388. TrwB interacts with proteins TrwA and TrwC, which form the R388 relaxosome, and with the R388 T4SS component TrwE (37). While the interaction with the relaxosome is highly specific for its cognate system (24, 37, 48), the interaction between the T4CP and the T4SS is less specific: a single T4CP can interact functionally with several conjugative T4SS. Interestingly, a correlation was observed between the strength of the T4CP-TrwE-like interaction and the efficiency of DNA transfer (37). T4CPs also interact with TrwE-like components of T4SS involved in virulence (13). In the case of the highly related Trw T4SS systems of plasmid R388 and the human pathogen Bartonella, it was further demonstrated that R388 TrwE could be functionally replaced by the Bartonella tribocorum TrwE homolog, TrwEBt (13).
T4CPs are integral membrane proteins anchored to the inner membrane by an N-terminal transmembrane domain (TMD). The soluble cytoplasmic domain of TrwB (TrwBΔN70), lacking this TMD, has been biochemically and structurally analyzed in detail. It retains the ability to bind NTPs and to unspecifically bind DNA (42). The characterization of its DNA-dependent ATPase activity (53) strengthened the possibility that T4CPs work as DNA motors. This activity is also stimulated by the oriT-binding protein TrwA (52).
The determination of the three-dimensional (3D) structure of TrwBΔN70 indicated a quaternary structure consisting of hexamers that form an almost spherical, orange-shaped structure with a 20-Å inner channel (ICH) (18, 19). Each monomer is composed of two main structural domains: the nucleotide-binding domain (NBD) and the all-alpha domain (AAD). The NBD has α/β topology and is reminiscent of RecA and DNA ring helicases. The AAD is facing the cytoplasmic side and bears significant structural similarity to the N-terminal domain of site-specific recombinase XerD and also to a 40-residue segment of the DNA binding domain of protein TraM, the component of the relaxosome of F-like plasmids that interacts with its cognate T4CP, TraD. The structure of the hexamer as a whole resembles that of the F1-ATPase, raising interesting perspectives into the possible way of action of coupling proteins as molecular motors in conjugation (5).
There have been several attempts to functionally dissect T4CPs. In F-TraD, it was determined that its C terminus is essential for relaxosomal specificity, probably through an interaction with TraM (4, 39, 48). The cytoplasmic domain of the related TraD protein of plasmid R1 stimulates both transesterase and helicase activities of its cognate relaxase, TraI (41, 51). A series of random mutations were shown to affect TraD oligomerization (23). In VirD4, the T4CP of the VirB T4SS of A. tumefaciens, both the periplasmic domain plus key residues of the NBD are required for its location at the cell poles (31); its interaction with the T4SS protein substrate VirE2 does not require the N-terminal TMD (2). Mutational analysis of R27 TraG showed that the periplasmic residues are essential for interaction with the T4SS (22). An N-terminal deletion variant of PcfC, the T4CP of the Enterococcus plasmid pCF10, loses its membrane localization but retains its ability to bind relaxosomal components (11). Biochemical analysis of full-length R388 TrwB showed that the N-terminal TMD stabilizes the protein, aids oligomerization, and affects nucleotide selection (25-27). This region is essential for T4SS interaction, but TrwBΔN70 retains the ability to interact with the relaxosomal components TrwA and TrwC (37). Taken together, these analyses suggested that the N-terminal TMD of the T4CPs is necessary for T4SS interaction, oligomerization, and cellular location and that the C-terminal cytoplasmic domain is necessary for relaxosomal interactions and ATPase activity associated with DNA transport.
In this study, we set up different assays to search for mutants affecting TrwB function in DNA and protein transfer. We constructed a series of TrwB point mutants based on the 3D structure of TrwBΔN70. Most selected residues were essential for TrwB function in conjugation, especially under conditions where TrwB was in limiting quantities. We analyzed the in vivo properties of selected mutants with a battery of in vivo assays to map functional domains. Also, random mutants in the TMD were screened for improved interactions with the T4SS, which allowed mapping of the TrwB-TrwE interaction domain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain DH5α (21) was used in all cloning procedures. Strain DY380 (32) was used to express the Red recombination system. Most plasmids were maintained in the lacIq strain D1210 (44). For conjugation experiments, the donor and recipient strains were D1210, DH5α, UB1637 (12), and HMS174 (8), as indicated. The cya-deficient strain DHM1 (28) was used for the bacterial two-hybrid assay. The plasmids used are described in Tables 1 and 2.
TABLE 1.
Published plasmids used in this work
| Plasmid | Description | Phenotype | Reference |
|---|---|---|---|
| pET29:trwAC | pET29c::PtrwA-trwA-trwC | Km TrwA TrwC | 15 |
| pHP100 | pSU24::trwE (Bartonella) | Cm TrwE (Bartonella) | 13 |
| pHP102 | pSU24::trwE | Cm TrwE | 13 |
| pKM101Δmob | pKM101 SmaI deletion | Ap IncN Tra-T4SS+ | 15 |
| pMTX502 | pT25::trwBΔN75 | Cm T25::TrwBΔN75 | 37 |
| pMTX503 | pT18::trwBΔN75 | Ap TrwBΔN75::T18 | 37 |
| pMTX513 | pUT18C::trwB | Ap T18::TrwB | 37 |
| pMTX514 | pT25::trwB | Cm T25::TrwB | 37 |
| pMTX611 | pSU4633::trwB(W216A) | Cm TrwA TrwB(W216A) | 53 |
| pMTX631 | pUT18C::trwE | Ap T18::TrwE | 37 |
| pMTX632 | pT25::trwE | Cm T25::TrwE | 37 |
| pMTX697 | pUT18C::trwE (Bartonella) | Ap T18::TrwE (Bartonella) | 13 |
| pMTX698 | pT25::trwE (Bartonella) | Cm T25::TrwE (Bartonella) | 13 |
| pSU19 | Cloning vector | Cm Rep (p15A) | 3 |
| pSU1443 | R388::Tn5tac1 in trwB | Tp Km Tra-IncW | 34 |
| pSU1445 | R388::Tn5tac1 in trwC | Tp Km Tra-IncW | 34 |
| pSU2007 | R388 Kmr | All Traw proteins | 40 |
| pSU4134 | R388::Tn5tac1 in trwE | Tp Km Tra-IncW | 37 |
| pSU4622 | pSU24::PtrwA-trwA-trwB | Cm TrwA TrwB | 42 |
| pSU4623 | pSU4622::trwB-KpnI | Cm TrwA TrwB-KpnI | 42 |
| pSU4632 | pSU4622::trwB(K136T) | Cm TrwA TrwB(K136T) | 42 |
| pSU4633 | pSU4622::trwB-KpnI/NdeI | Cm TrwA TrwB-KpnI/NdeI | 42 |
| pT25 | 2-hybrid cloning vector | Cm T25 | 29 |
| pT25zip | 2-hybrid positive control | Cm T25-leucine zipper | 29 |
| pUT18 | 2-hybrid cloning vector | Ap T18 | 30 |
| pUT18C | 2-hybrid cloning vector | Ap T18 | 30 |
| pUT18zip | 2-hybrid positive control | Ap T18-leucine zipper | 30 |
TABLE 2.
Bacterial plasmids constructed in this work
| Plasmid | Description | Constructiona |
||
|---|---|---|---|---|
| Vector | Insert | Digestion/oligonucleotides (5′→3′) | ||
| Plasmids expressing PtrwA-trwA-trwB | ||||
| pDEL010 | trwB(K275A) | pSU4633 | PCR on pSU4633 | TGAAGCGAGCGCGGCGCTGAC |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCTGGATCCGATGCATCCAGACGATCA | ||||
| pMTX520 | trwB(D158A) | pSU4633 | PCR on pSU4633 | CCGCATGGTAATTGTGGCGCCGAATGGCGATATG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX521 | trwB(D356A) | pSU4633 | PCR on pSU4633 | GGCTGTTCATCGCCGAGCTCGCTTCGCTGGAAAAG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX524 | trwB(Q386A) | pSU4633 | PCR on pSU4633 | GTGGCGGGCCTGGCGTCGACCTCGCAG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX525 | trwB(Q390A) | pSU4633 | PCR on pSU4633 | CTGCAATCGACCTCAGCGCTTGATGACGTGT |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX527 | trwB(R240A E241A) | pSU4633 | PCR on pSU4633 | ACGCCTTCCATGGCAGCGCTGTTCCACTGGA |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX528 | trwB(H244A W245A) | pSU4633 | PCR on pSU4633 | ATGCGCGAATTGTTCGCCGCGACAACGATCGCCAC |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX530 | trwB(R375A) | pSU4633 | PCR on pSU4633 | GCACTCACCAAAGGCGCCAAGGCAGGGCTTCGG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX533 | trwB(D252A D253A) | pSU4633 | PCR on pSU4633 | TCGCCACGTTTGCAGCGCTGCGGGGGTTTC |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX546b | trwB(D252R D253R E259R) | pSU4633 | PCR on pSU1443 | CATGCGCGAATTGTTCCACTGGACAACCATCGCCACGTTTCGTCGACCCCGGATGAATGTCA |
| CCAGCAAACAAAGATTCGGCCAAAGTTCCTCGCAGAAACCCCCGCAGTCGACGGATTTGCACTGCC | ||||
| pMTX549 | trwB(N271D) | pSU4633 | PCR on pSU4633 | TGCTGGGTCGGATGAAGCGAG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCTGGATCCGATGCATCCAGACGATCA | ||||
| pMTX552 | trwB(K421A D425A) | pSU4633 | PCR on pSU4633 | CCGACCCGGCAACCAATGAGGCCATGAGTTTGA |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCTGGATCCGATGCATCCAGACGATCA | ||||
| pMTX553c | trwB(K398A) | pSU4633 | PCR on pSU4633 | ACGGCGTGGCAGAGGCGCAG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCTGGATCCGATGCATCCAGACGATCA | ||||
| pMTX554 | trwB(Δ17C) | pSU4633 | PCR on pSU4633 | CCAGGATCCTCGGACAAGGCGAATTTG |
| CCAGAATTCAAAGCGGAACCTTGGCAA | ||||
| pMTX558 | trwB(R124A) | pSU4633 | PCR on pMTX518 | CCACATATGAATAGCGTCGGACAAGG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| pMTX559 | trwB(E357A) | pSU4633 | PCR on pSU4633 | CTGTTCATCGACGCGTTGGCTTCGCTG |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX560 | trwB(Δ12C) | pSU4633 | PCR on pSU4633 | CCAGAATTCAAAACTGCTTGATTTCAAG |
| CCAGGATCCTCGGACAAGGCGAATTTG | ||||
| pMTX590 | trwB(S270P) | pSU4633 | None | None (random mutagenesis) |
| pMTX593 | trwB(R318H) | pSU4633 | None | None (random mutagenesis) |
| Plasmids expressing trwB under control of lactose promoter | ||||
| pDEL003 | pSU19::trwA-trwB | pHP139 | PCR on pSU4622 | TGACATATGGCACTAGGCGAC |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| pDEL009 | pSU19::trwB(K275A) | pHP139 | pDEL010 | KpnI + EcoRI |
| pHP138d | pET29c::oriT-trwA-trwC | pET29:trwAC | PCR on pSU2007 | TTACTCTAGACTCATTTTCTGCATCATTGT |
| TTACTCTAGATTGTAGTGGCATAACACTA | ||||
| pHP139 | pSU19::trwB | pSU19 | PCR on pMTX601 | TTACAAGCTTAGGAGGATCCATATGCATCCAGACGATCAAA |
| ATGCGGCATCAGAGCAGATTG | ||||
| pHP140 | pSU19::trwB(Q390A) | pHP139 | pMTX525 | KpnI + EcoRI |
| pHP141 | pSU19::trwB(R240A E241A) | pHP139 | pMTX527 | KpnI + EcoRI |
| pHP142 | pSU19::trwB(H244A W245A) | pHP139 | pMTX528 | KpnI + EcoRI |
| pHP143 | pSU19::trwB(D252A D253A) | pHP139 | pMTX533 | KpnI + EcoRI |
| pHP145 | pSU19::trwB(N271D) | pHP139 | pMTX549 | KpnI + EcoRI |
| pHP149 | pSU19::trwB(V74I) | pHP139 | pHP106 | BamHI + KpnI |
| pHP150 | pSU19::trwB(P18S S95N) | pHP139 | pHP107 | BamHI + KpnI |
| pHP169 | pSU19::trwB(K421A D425A) | pHP139 | pMTX552 | KpnI + EcoRI |
| pHP170 | pSU19::trwB(K398A) | pHP139 | pMTX553 | KpnI + EcoRI |
| Constructs in vectors for bacterial 2-hybrid assay | ||||
| pHP106 | pUT18C::trwB(V74I) | pMTX601 | PCRe on pMTX601 | GAAGTTCTCGCCGGATGT |
| BamHI + KpnI | ATGCGGCATCAGAGCAGATTG | |||
| pHP107 | pUT18C::trwB(P18S S95N) | pMTX601 | PCRe on pMTX601 | GAAGTTCTCGCCGGATGT |
| BamHI + KpnI | ATGCGGCATCAGAGCAGATTG | |||
| pHP108 | pUT18C::trwB(P18S) | pMTX601 | PCRe on pMTX601 | GAAGTTCTCGCCGGATGT |
| BamHI + KpnI | ATGCGGCATCAGAGCAGATTG | |||
| pHP119 | pUT18C::trwEBt(T107S) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP120 | pUT18C::trwEBt(P57A) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP121 | pUT18C::trwEBt(P57L T107A) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP122 | pUT18C::trwEBt(P57S) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP123 | pUT18C::trwEBt(V54L T126A) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP124 | pUT18C::trwEBt(V52L) | pMTX697 | PCRe on pMTX697 | GAAGTTCTCGCCGGATGT |
| BamHI + NdeI | ATGCGGCATCAGAGCAGATTG | |||
| pHP126 | pT25::trwB(V74I) | pT25 | pHP106 | BamHI + EcoRI |
| pHP127 | pT25::trwB(P18S) | pT25 | pHP108 | BamHI + EcoRI |
| pMTX544 | pT25::trwB(K136T) | pT25 | PCR on pSU4632 | CCTGGATCCGATGCATCCAGACGATCA |
| CCAGGATCCTAGATAGTCCCCTCAACAA | ||||
| pMTX545 | pUT18C::trwB(K136T) | pUT18C | PCR on pSU4632 | CCTGGATCCGATGCATCCAGACGATCA |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| pMTX601 | pUT18C::trwB with KpnI | pUT18C | PCR on pSU4623 | CCTGGATCCGATGCATCCAGACGATCA |
| CCAGAATTCTAGATAGTCCCCTCAACAA | ||||
| pMTX615 | pUT18C::trwB(Δ12C) | pMTX513 | pMTX560 | BsmI + EcoRI |
| pMTX616 | pUT18C::trwB(R375A) | pMTX513 | pMTX530 | BsmI + StyI |
| pMTX617 | pUT18C::trwB(D356A) | pMTX513 | pMTX521 | BsmI + StyI |
| pMTX625 | pUT18C::trwB(R318H) | pMTX513 | pMTX593 | BsmI + EcoRI |
| pMTX627 | pT25::trwB(R375A) | pT25 | pMTX616 | BamHI + KpnI |
| pMTX628 | pT25::trwB(D356A) | pT25 | pMTX617 | BamHI + KpnI |
| pMTX629 | pUT18C::trwB(Q386A) | pMTX601 | pMTX524 | BsmI + StyI |
| pMTX630 | pT25::trwB(R318H) | pT25 | pMTX625 | BamHI |
| pMTX633 | pUT18C::trwB(D158A) | pMTX601 | pMTX520 | KpnI + StyI |
| pMTX645 | pUT18C::trwB(E357A) | pMTX601 | pMTX559 | KpnI + StyI |
| pMTX647 | pT25::trwB(Δ12C) | pT25 | pMTX615 | BamHI + EcoRI |
| pMTX648 | pT25::trwB(D158A) | pT25 | pMTX633 | BamHI + EcoRI |
| pMTX652 | pT25::trwB(Q386A) | pT25 | pMTX629 | BamHI + EcoRI |
| pMTX656 | pT25::trwB(E357A) | pT25 | pMTX645 | BamHI + EcoRI |
For construction, the first column lists the vector plasmids, the second column lists the plasmids from which the inserts were obtained and the method, and the third column indicates either the restriction enzymes used for cloning or the oligonucleotides used for PCR amplification of the desired fragment, with the restriction sites underlined. When three oligonucleotides are shown, the mutant was generated by the megaprimer method. The first two oligonucleotides were used for the first PCR (the first oligonucleotide codes for the mutation, and if a restriction site was introduced it is underlined), and this PCR was used as a megaprimer together with the third oligonucleotide in a second PCR. The final PCR products were cloned either as a KpnI-EcoRI fragment in pSU4633 (except in pMTX558, which was digested with NdeI plus EcoRI).
This plasmid was obtained by Red mutagenesis as described in Materials and Methods.
This mutant was found to contain the additional mutation R417S, which will be omitted for clarity.
The oriT fragment from plasmid pSU2007 was PCR amplified and inserted into the XbaI site of plasmid pET29c::trwAC, lying 5′ to the start of trwA; the orientation was selected to reconstruct the same oriT-trwA arrangement as in R388.
PCRs were performed with the GeneMorph system to introduce random mutations (see Materials and Methods). The oligonucleotides flank the inserts in vector pUT18C. After the mutagenic PCR, the fragment with the desired mutated region was obtained with the indicated restriction endonucleases.
Plasmid constructions.
Plasmids were constructed by standard methodology (45). A brief description of each plasmid is included in Table 2. TrwB mutants were constructed in plasmid pSU4633, which codes for trwA-trwB under the control of its own PtrwA promoter, for assay of in vivo functions (complementation of a trwB mutant R388 with a dominant-negative effect on wild-type R388). To limit the amount of TrwB, selected mutants were transferred to the pSU19 derivative pHP139, where trwB expression is under the control of the lactose promoter. trwA was added to pHP139 to express trwA-trwB from the lac promoter in plasmid pDEL003. For the two-hybrid assay, we used the cloning vectors pUT18C and pT25 (to create fusions to the T18 and T25 domains of adenylate cyclase).
Mutagenesis procedures.
Site-directed mutagenesis of selected TrwB residues was performed by the megaprimer method (47). The triple mutation in pMTX546 was not obtained by this method after several attempts, so the triple mutant was constructed by a Red recombination-based technique, as described previously (32); briefly, plasmid pSU4633 was introduced into E. coli strain DY380, which carries a defective λ prophage harboring the recombination genes exo, bet, and gam under the control of temperature-sensitive cI repressor. The Red recombination system was induced by 15-min incubation at 42°C, and cells were made competent for electroporation. The mutation to be introduced (a kanamycin resistance gene from transposon Tn5tac1, flanked by SalI unique restriction sites) was PCR amplified with oligonucleotides coding for the desired mutation plus 40-nucleotide (nt) regions of homology to the regions flanking the site of mutation. This PCR fragment was electroporated into the competent cells, and recombinants were selected by the antibiotic resistance marker and subsequently confirmed by PCR and restriction analysis. In a second step, the antibiotic resistance gene was deleted by SalI restriction and religation.
Random mutagenesis of trwB was performed by introducing the plasmid to be mutagenized into mutator strain XL1-Red (Stratagene) following the manufacturer′s recommendations. Plasmid pSU4633 was introduced into competent XL1-Red cells and grown overnight in 5 ml LB plus chloramphenicol (Cm) overnight in the dark. A 10−5 dilution continued to grow under the same conditions. Succesive dilutions were 10−4, 10−3, 10−2, and 10−1, since the mutagenic strain grows slower after several generations. After the 5 cycles of mutagenic growth, DNA was extracted and reintroduced into fresh competent XL1-Red cells for a second round of mutagenesis. After four complete rounds of mutagenesis, DNA was transformed into D1210(pSU1443) cells. In order to check for conjugation ability, transformants were replicated on LB plates with a lawn of DH5α recipient strain, left for 2 h at 37°C, and replicated again on plates supplemented with nalidixic acid (Nx) plus kanamycin (Km) (for transconjugants) and Cm plus Km (for donors). Each plate included positive (pSU4633) and negative (pSU4632) controls. Transfer-deficient mutants were selected for DNA extraction, confirmation of the Tra phenotype, and DNA sequencing to confirm the nature of the mutation.
Random mutagenesis of N-terminal regions of both TrwB and Bartonella tribocorum TrwE (TrwEBt) was performed by mutagenic PCR using the GeneMorph II random mutagenesis kit (Pharmacia) following the manufacturer′s recommendations. We adjusted the reactions to obtain an average of 1 mutation per 400 bp. The template DNAs were pMTX601 for mutagenesis of trwB and pMTX697 for mutagenesis of trwEBt. The oligonucleotides used in both cases were pUT18C-F (GAAGTTCTCGCCGGATGT) and pUT18C-R (ATGCGGCATCAGAGCAGATTG), which flank the cloning site in vector pUT18C. The PCR-amplified products were digested with enzymes BamHI and KpnI (for trwB) or BamHI and NdeI (for trwEBt); the fragment corresponding to the ca. first 400 bp of the genes was gel extracted and cloned into the corresponding template vector DNAs digested with the same enzymes. Ligations were electroporated into competent DHM1 cells containing the appropriate pT25 derivative (pMTX698 for screening of TrwB mutants, pMTX514 for TrwEBt mutants), and transformants plated in plates containing Cm plus ampicillin (Ap) plus X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) for screening of blue colonies representing mutants with stronger TrwB-TrwEBt interactions. DHMI cells harboring the plasmids with the wild-type genes (pMTX601 plus pMTX698, and pMTX514 plus pMTX697) were also plated as a control of the color of the TrwB-TrwEBt interaction.
Bacterial conjugation.
Standard matings were performed as described previously (15). The dominant-negative effect of TrwB mutants was checked in the presence of a wild-type R388 derivative (pSU2007). pSU4622-derived or pHP139-derived TrwB mutants were checked either for complementation of a trwB-deficient R388 derivative (pSU1443) or for mobilization of a plasmid containing oriT plus TrwA and TrwC (pHP138) through the T4SS of plasmid pKM101 (present in plasmid pKM101Δmob). Matings were carried out from the lacIq strain D1210 in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside) (repressed conditions), in the presence of 0.5 mM IPTG in the agar plate during the 1-h mating period (mild induction), or with an additional induction by previous 1/20 dilution of donor strains and growth for 2 h in the presence of 0.5 mM IPTG (strong induction). Results are shown as frequency of transconjugants per donor and are the mean of 2 to 5 independent experiments. Since standard deviations in mating assays can be as high as 50%, only differences of at least 1 log have been considered as mutant phenotypes.
Relaxase transport assay.
Triparental matings were performed to check for TrwC transport in the absence of DNA, as described previously (15). Plasmids containing TrwB or mutant derivatives were introduced in donor cells (D1210 or DH5α, as indicated) coding for the T4SS of pKM101 (plasmid pKM101Δmob) and TrwA plus TrwC in a nonmobilizable plasmid (pET29::trwAC). None of the three plasmids present in the donor cell contains an oriT gene, so there is no conjugative DNA transport from the donor. Donor cells were mated with a second strain (DH5α or UB1637, as appropriate) containing a trwC-deficient R388 derivative (plasmid pSU1445). TrwC transport into this second strain was detected by complementation of the trwC mutation and subsequent mobilization of pSU1445 into a third recipient strain (HMS174).
Two-hybrid assay.
In vivo protein-protein interactions were tested by the bacterial two-hybrid assay as described previously (13). In this assay, the strength of the interactions is reflected in the expression of lacZ. Strain DHM1 was grown at 30°C and cotransformed with plasmids bearing a T25 fusion and a T18 fusion. Transformants were grown overnight and spread on sectors of X-Gal-containing plates to observe and compare the blue color.
Western blots.
The amount of TrwB and TrwA in the cells was estimated by Western blotting of total protein extracts. E. coli D1210 cells containing the indicated plasmids were grown to an optical density (OD) of 0.6, and then IPTG was added as indicated. Cells were collected, centrifuged, resuspended in 1/10 volume of 2× SDS-gel loading buffer (45), and frozen at −20°C. Twenty-microliter samples were boiled for 5 min and applied to SDS-PAGE gels. After the run, gels were transferred to nitrocellulose filters. Filters were stained with 0.1% Coomassie brilliant blue R250 in 50% methanol to estimate protein transfer. Anti-TrwB and anti-TrwA antibodies from rabbits immunized with purified TrwBΔN70 or TrwA-His protein were a generous gift of Gabriel Moncalián (University of Cantabria) and were used at a 1:5,000 dilution. Secondary antibody (peroxidase-conjugated anti-rabbit IgG from ICN) was used at a 1:10,000 dilution. Detection was performed with the Supersignal kit (Pierce), and bands were analyzed on a Bio-Rad ChemiDoc apparatus. Control proteins TrwBΔN70 and TrwA-His were purified as described previously (52).
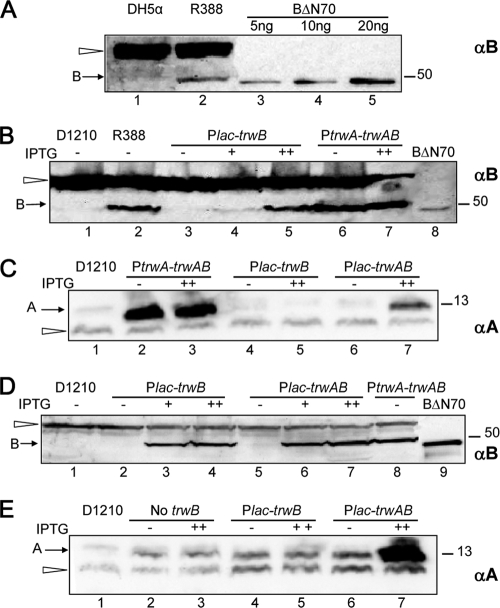
To determine the number of TrwB molecules in R388-containing cells, total extracts were blotted and compared to TrwBΔN70 dilution standards, as shown in Fig. 1A. It was estimated that an optical density at 600 nm (OD600) of 0.5 corresponded to a concentration of 108 bacteria/ml. Quantification of bands was done using the Quantity One software of Bio-Rad.
FIG. 1.
Western blots showing amounts of TrwB (A, B, and D) or TrwA (C and E) under different expression conditions. Total extracts from E. coli cells containing the trwB-expressing plasmid indicated at the top were processed as described in Materials and Methods, samples were run on SDS-PAGE, and proteins were detected with anti-TrwB (αB) or anti-TrwA (αA) serum. (A to C) SDS-PAGE with samples from cells harboring the trwB-expressing plasmids alone. (D and E) SDS-PAGE with samples from D1210 cells containing additional plasmids pKM101Δmob and pHP138, as in conjugation assay types DEF (Fig. 2). The position of TrwA and TrwB is indicated to the left. The white arrowhead points to unspecific bands visible in the strain with no plasmids. Molecular mass markers are indicated to the right (in kDa). BΔN70, purified TrwBΔN70 protein, shown in panel A for TrwB quantification and in panels B and D as a size marker. The induction conditions (IPTG for 0, 1, or 3 h) are indicated as −, +, or ++, respectively.
RESULTS
Mating assays under TrwB limiting conditions.
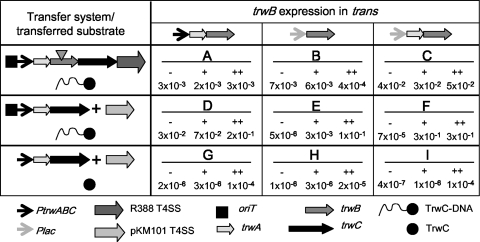
The amount of TrwB produced from plasmid R388 under normal liquid growth conditions was determined by comparison with known quantities of purified TrwBΔN70 protein in a Western blot (Fig. 1A). The different bands shown were used to make a calibration curve. The amount of TrwB in the R388 sample, corresponding to 3 × 107 bacteria, is 8.3 ng. Thus, there are 3,350 TrwB monomers per cell. This is a surprisingly large amount of protein compared to the RP4 conjugative system, for which a similar study determined that there are less than 5 molecules of the T4CP TraG per cell (20). This suggests that the R388 PtrwA promoter, driving the expression of the trwABC operon, produces TrwB in excess. If this were the case, mutations affecting the activity of TrwB only partially might not affect the conjugation frequency. In order to construct a system in which the amount of TrwB has a direct effect on the conjugation frequency, we designed a set of conjugation assays in which we modified trwB expression conditions and/or we used a heterologous T4SS for DNA transfer. The different transfer assay conditions are depicted in Fig. 2.
FIG. 2.
Scheme of the different conjugation assays used in this work and the transfer frequencies obtained. The elements of the transfer machinery present in each assay type (A to I) are represented as indicated in the lower part of the figure. The transposon insertion inactivating trwB in the R388 derivative is represented by a triangle. The different plasmids (separated by “+” symbols) present in the donor strains are the sum of those in the lane and column matching for each assay type; lanes also indicate the transferred product (either TrwC alone or with covalently attached DNA). Standard (A to F) or triparental (G to I) matings were performed as described in Materials and Methods. IPTG was added to the mating plate only (+) or was preceded by an additional 2-h incubation in liquid (++), as explained in Materials and Methods. Frequencies (as number of transconjugants per donor) are the mean of 3 to 5 independent experiments. Under these assay conditions, background transfer in the absence of a mobilizable plasmid is <10−7 transconjugants/donor and R388 transfer frequency is 10−1 transconjugants/donor.
We constructed a plasmid (pHP139) in which the expression of trwB was under the control of the lactose promoter. This plasmid was carried in the lacIq strain D1210 in order to keep trwB expression repressed. For induction of its expression, we assayed two different lac induction conditions: (i) mild overexpression by addition of IPTG to the mating plate, and (ii) strong overexpression by an additional 2-h incubation with IPTG prior to the mating. The amount of TrwB present in each condition was checked by Western blotting (Fig. 1B). The results confirmed that TrwB was not produced at detectable levels from pHP139 in the absence of induction, while upon addition of IPTG there was an increase in TrwB levels. The amount of TrwB under strong induction conditions was comparable to the TrwB levels in wild-type R388 (Fig. 1B, compare lanes 2 and 5). For comparison, TrwB levels were monitored in plasmid pSU4622 (Table 2), where trwB is expressed from the PtrwA promoter and an additional Plac promoter originating from the vector; the TrwB levels observed were higher than those in R388 and increased upon IPTG induction (Fig. 1B, compare lanes 2, 6, and 7).
Both trwB-expressing plasmids were tested for complementation of the R388 trwB mutant pSU1443 (Fig. 2, assays A and B) under different induction conditions. The results showed that even in the absence of induction, the amount of TrwB produced from the lactose promoter was high enough to complement pSU1443 mobilization to the same levels as with the plasmid expressing PtrwA-trwA-trwB (compare assays A and B). After several hours of overexpression of trwB from the lactose promoter in pHP139, the transfer efficiency dropped 1 log (Fig. 2), which could be due to an adverse effect of an excess of TrwB. However, the steady-state levels of TrwB were not higher than in assay A (Fig. 1B, compare lanes 5 and 6). A reason could be that in assay A, additional TrwA is produced from pSU4622, as checked by Western blotting (Fig. 1C, lanes 2 and 3). We constructed another plasmid (pDEL003) (Table 2) by adding trwA 5′ to trwB in plasmid pHP139; TrwA production increased with IPTG induction as expected (Fig. 1C, lanes 6 and 7). We used this plasmid to complement the R388 trwB mutant (assay type C) (Fig. 2). The results showed a 1-log increase in the conjugation frequency with respect to the Plac-trwB construct, and the frequency was not lowered after 3 h of induction with IPTG, suggesting that TrwA is counteracting the deleterious effect of an excess of TrwB.
We also used the T4SS of plasmid pKM101 for DNA mobilization. The conjugative machinery of this plasmid is related to that of R388, and it was shown that the T4SSs of both plasmids are functionally exchangeable (37). Plasmids coding for trwB under different expression conditions were tested for conjugation in the presence of compatible plasmids coding for the T4SS of pKM101 (pKM101Δmob) and the relaxosomal components of R388, oriT-trwA-trwC (Fig. 2, assay types D, E, and F). TrwB levels were checked by Western blots of donor cells from assays D, E, and F (Fig. 1D). The results of the conjugation assays are shown in Fig. 2; the higher frequencies obtained when the T4SS of pKM101 was used are probably due to the higher copy number of the mobilizable plasmid used in this case (a pMB1-derived replicon, versus the R388 derivative pSU1443). In the absence of induction, there was no detectable TrwB protein produced from the lactose promoter, correlating with the very low conjugation frequencies observed (Fig. 1D and 2). Upon addition of IPTG, we observed that conjugation efficiency became dependent on the amount of TrwB, especially when trwB expression was controlled from the lactose promoter only (assays E and F) (Fig. 2). In summary, the combination of controlled expression of trwB and the use of a heterologous T4SS provided an assay in which the conjugation frequency is dependent on the amount of TrwB.
There was a significant increase in the transfer frequencies when the plasmid expressing Plac-trwAB was used in place of Plac-trwB (compare assays E and F). This was not due to a different amount of TrwB, as judged by Western blotting (Fig. 1D), but could be due to the different amount of TrwA (Fig. 1E). Thus, the results could suggest that expression of trwA is itself another limiting factor or that it is important for TrwB function in conjugation. To pursue our study of TrwB function, we used assay type E, in which the only limiting factor is TrwB.
TrwB is required for TrwC transport into recipient cells, even in the absence of DNA transfer (15). Using triparental matings as described previously (15), transport of TrwC was detected by complementation of a trwC-deficient R388 in the recipient cell. Wild-type TrwB was assayed for TrwC transport under different induction conditions (Fig. 2) (assay types G, H, and I). To avoid entry exclusion, the T4SS present in the donor was that of pKM101 in all cases. The transfer frequencies obtained (Fig. 2) were much lower than those in DNA transfer assays, as described previously (15). It can be observed that frequencies were also increased by induction of trwB expression and that TrwC transport was 10 times more efficient when trwA expression was increased from Plac-trwAB.
Characterization of TrwBΔN70 mutants.
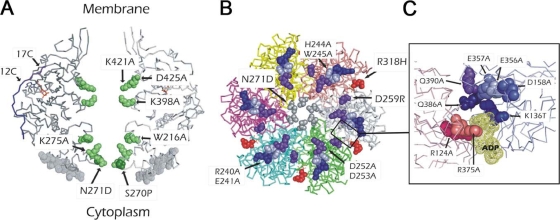
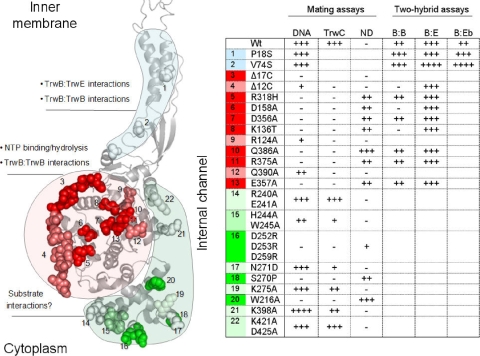
Based on the known 3D structure of protein TrwBΔN70, which contains the cytoplasmic domain of TrwB, we selected the residues highlighted in Fig. 3, which were more likely to have a key role in TrwB function. We performed site-directed mutagenesis of each one as detailed in Materials and Methods. We also performed a random mutagenesis procedure by continuous growth of a trwB-containing plasmid in the E. coli mutator strain XL1-Red and selection of transfer-deficient mutants, as detailed in Materials and Methods. Two transfer-deficient mutants were obtained: the S270P and R318H mutants.
FIG. 3.
Representations of the 3D structure of TrwBΔN70 bound to ADP (PDB accession no. 1GKI), where the mutated residues are highlighted in the space-filled format. (A) Side view of two opposing monomers, showing in green the mutated residues which protrude into the ICH of the hexamer. The extent of the 12 and 17 C-terminal residues of the protein is indicated with arrows (12C and 17C, respectively). Mutated AAD residues are also indicated (space filled in gray). (B) TrwBΔN70 hexamer seen from the cytoplasmic side, with the selected residues from the AAD domain shown in the space-filled format. Residue R318, located in the interface between monomers, is also highlighted (in red). The region included in the quadrangle is amplified in panel C for better resolution of the mutated residues in the proximity of the nucleotide molecule.
Mutations were initially introduced in plasmid pSU4622, which carries genes trwA and trwB under the control of PtrwA. In order to assess the steady-state levels of the transfer-deficient mutant TrwB proteins, Western blot analysis was carried out from extracts of cells containing the pSU4622 derivatives after IPTG induction. All mutants showed the same protein levels as wild-type TrwB, except when indicated. Plasmids containing TrwB mutations were checked for complementation of the transfer-deficient R388 trwB mutant pSU1443 and for the dominant-negative phenotype in the presence of wild-type TrwB. Results are shown in Table 3. Most NBD mutants, a triple-AAD mutant, and C-terminal deletions knocked down TrwB function in conjugation. TrwB mutations that did not have an effect in the standard complementation assay (type A in Fig. 2) were transferred to plasmid pHP139 and tested for TrwB function under the TrwB limiting conditions defined above. The results are shown in Table 4. Additional mutants were found to be impaired in conjugation under these mating conditions. These results are next analyzed in detail by mutant categories.
TABLE 3.
Transfer frequencies in the presence of different TrwB mutantsa
| TrwB type | Plasmid | Complementation with pSU1443b |
Negative dominancec |
||
|---|---|---|---|---|---|
| Frequency | % | Frequency | % | ||
| Wild type | pSU4622 | 2 × 10−3 | 100 | 7 × 10−2 | 100 |
| NBD mutation | |||||
| R124A | pMTX558 | 4 × 10−5† | 2† | 2 × 10−2 | 30 |
| K136T | pSU4632 | <10−7‡ | <0.01‡ | 3 × 10−4† | 0.44† |
| D158A | pMTX520 | <10−7‡ | <0.01‡ | 1 × 10−4† | 0.18† |
| D356A | pMTX521 | <10−7‡ | <0.01‡ | 6 × 10−5† | 0.09† |
| E357A | pMTX559 | <10−7‡ | <0.01‡ | 1 × 10−4† | 0.20† |
| R375A | pMTX530 | <10−7‡ | <0.01‡ | 8 × 10−5† | 0.10† |
| Q386A | pMTX524 | <10−7‡ | <0.01‡ | 1 × 10−6‡ | 0.002‡ |
| Q390A | pMTX525 | 1 × 10−3 | 61 | 7 × 10−3 | 11 |
| AAD mutations | |||||
| R240A E241A | pMTX527 | 4 × 10−3 | 200 | 2 × 10−2 | 32 |
| H244A W245A | pMTX528 | 3 × 10−3 | 150 | 9 × 10−3 | 13 |
| D252A D253A | pMTX533 | 3 × 10−3 | 150 | 1 × 10−2 | 19 |
| D252R D253R D259R | pMTX546 | <10−7‡ | <0.01‡ | 6 × 10−3 | 9 |
| ICH mutation(s) | |||||
| W216A | pMTX611 | <10−7‡ | <0.01‡ | 2 × 10−7‡ | 0.0003‡ |
| N271D | pMTX549 | 8 × 10−4 | 40 | 2 × 10−2 | 28 |
| K275A | pDEL010 | 4 × 10−3 | 200 | 9 × 10−2 | 129 |
| K398A | pMTX553 | 3 × 10−3 | 150 | 1 × 10−2 | 19 |
| K421A D425A | pMTX552 | 4 × 10−4 | 20 | 9 × 10−3 | 13 |
| C-terminal deletion | |||||
| Δ17C | pMTX554 | 2 × 10−7‡ | 0.01‡ | 1 × 10−2 | 18 |
| Δ12C | pMTX560 | 5 × 10−6† | 0.25† | 1 × 10−2 | 20 |
| Random mutation | |||||
| S270P | pMTX590 | <10−7‡ | <0.01‡ | 2 × 10−4† | 0.30† |
| R318H | pMTX593 | <10−7‡ | <0.01‡ | 1 × 10−4† | 0.10† |
The donor and recipient strains were D1210 and DH5α, respectively. Frequencies are given as transconjugants per donor and are the mean of 2 to 9 independent assays. Reduced frequencies are noted as follows: †, mild effects; ‡, stronger phenotypes.
Transfer of pSU1443 (Table 1), a transfer-deficient R388 derivative with a transposon insertion in trwB. It was complemented by pSU4633 derivatives carrying the indicated trwB mutations.
Donor strains carried plasmid pSU2007 (a Kmr R388 derivative) and pSU4622 or derivatives carrying the indicated mutations in trwB.
TABLE 4.
Mating assays with trwB mutants under different expression conditions
| TrwB type (domain) | Result for assay showna |
|||||
|---|---|---|---|---|---|---|
| A (IPTG −) | B (IPTG −) | E |
G (IPTG −) | |||
| IPTG − | IPTG + | IPTG ++ | ||||
| Wild type | 2 × 10−3 | 6 × 10−3 | 5 × 10−6 | 2 × 10−3 | 5 × 10−2 | 4 × 10−3 |
| Q390A mutant (NBD) | 1 × 10−3 | 4 × 10−4† | 5 × 10−8‡ | 2 × 10−7‡ | 4 × 10−4‡ | <7 × 10−7‡ |
| R240A E241A mutant (AAD) | 4 × 10−3 | 2 × 10−3 | 3 × 10−7 | 3 × 10−4 | 2 × 10−2 | 6 × 10−4 |
| H244A W245A mutant (AAD) | 3 × 10−3 | 6 × 10−4† | 1 × 10−8‡ | 7 × 10−5† | 2 × 10−2 | 1 × 10−6‡ |
| D252A D253A mutant (AAD) | 3 × 10−3 | 2 × 10−3 | 6 × 10−7 | 3 × 10−4 | 2 × 10−1 | 8 × 10−4 |
| N271D mutant (AAD/ICH) | 8 × 10−4 | 4 × 10−3 | 5 × 10−7 | 4 × 10−4 | 4 × 10−2 | 4 × 10−6‡ |
| K275A mutant (ICH) | 4 × 10−3 | 4 × 10−3 | 2 × 10−5 | 1 × 10−2 | 4 × 10−2 | 2 × 10−4† |
| K398A mutant (ICH) | 3 × 10−3 | 4 × 10−3 | 1 × 10−5 | 2 × 10−2 | 2 × 10−1 | 7 × 10−5† |
| K421A D425A mutant (ICH) | 1 × 10−3 | 7 × 10−2 | 1 × 10−7 | 5 × 10−2 | 1 × 10−1 | 4 × 10−3 |
Assays A through E test the effect of trwB mutants on DNA transfer and were performed as in Table 3. Assay G tests the effect on TrwC transport in the absence of DNA transfer and was performed as explained in Materials and Methods, using DH5α as the donor strain. The numbers shown are the mean of 2 to 10 independent assays. Reduced frequencies are noted as follows: †, mild effects (1- to 2-log drop); ‡, stronger phenotypes (more than 3-log drop). Frequencies higher than wild type are shown in boldface. Assays were performed as described in the legend to Fig. 2. For the assays shown, the trwB promoters used were PtrwA (assays A and G) and Plac (assays B and E). Assays A and G had trwA coexpression, while assays B and E did not. Assays A and B were performed with R388 T4SS, and assays E and G included plasmid pKM101 T4SS. For IPTG induction, −, +, and ++, respectively, represent 0, 1, and 3 h of IPTG incubation.
(i) Mutations in the NBD.
The NBD contains the NTP binding pocket. Also, monomer-monomer interactions run mainly along this domain. According to the structure, there are several residues in direct contact with the NTP belonging to two adjacent monomers (Fig. 3C). Residues K136, D158, D356, E357, Q386, and Q390 contact the NTP from one TrwB monomer, while residues R124 and R375 contact the NTP from the adjacent TrwB monomer. All of them were changed to alanine, except for the previously characterized K136T mutant (42, 53). In addition, the R318H mutant was obtained by random mutagenesis; residue R318 lies at the interface between monomers (Fig. 3B, shown in red). Standard complementation assays (Table 3) showed that almost all designed mutations in contact with the bound NTP were completely transfer deficient: the R124A mutant retained 2% transfer efficiency, and mutation Q390A (which lies slightly separated from the nucleotide molecule) did not affect transfer significantly in the standard mating assay, but the mutant showed a strong phenotype when trwB expression or the T4SS used was altered (Table 4) (assays B and E).
All transfer-deficient NBD mutations showed a dominant-negative phenotype (Table 3). The strongest effect (5 logs) occurred in the Q386A mutant. The mutants with a strong dominant-negative effect (2 to 3 logs) were the K136T mutant (previously reported in reference 53); the D158A, D356A, E357A, and R375A mutants; and the randomly obtained R318H mutant. Mutations R124A and Q390A showed an effect of less than 1 log.
(ii) Mutations in the AAD.
The AAD is located at the cytoplasmic side of the TrwB hexamer, and it surrounds the entrance to the ICH. Thus, it is a candidate for interactions with the components of the relaxosomal substrate: TrwA, TrwC, and the DNA to be transferred. We searched for residues which were polar and surface exposed and also residues that were conserved in different T4CPs, and we constructed the following double mutants (Fig. 3A and B): the H244A W245A mutant, based on the conservation of these two residues in the T4CP family; and the R240A E241A and D252A D253A mutants, based on the charge and proximity of the residues to the cytoplasmic entrance to the ICH. We also constructed a more aggressive D252R D253R D259R mutant (named the triple-AAD mutant), in which the three charged residues were mutated to residues with the opposite charge. Neither of the three double mutants affected transfer (Table 3). The D252R D253R D259R triple mutant, however, knocked down protein function completely. The H244A W245A double mutant showed an effect on conjugation under TrwB limiting conditions (1- to -log drop in transfer frequency) (Table 4).
(iii) Mutations in the ICH.
According to the current model, the ICH is the candidate region for DNA interaction, since transferred DNA is expected to travel through the center of the hexamer. Positively charged residues could have a role in contacting the negatively charged DNA backbone. Therefore, the three lysines protruding into the ICH (K275, K398, and K421) (Fig. 3A) were mutated to alanine. The K421A mutant contained the additional mutation D425A, also mapping in the ICH. In addition, a conserved tryptophan (W216) was previously mutated to alanine and characterized (53). Residue N271 blocks the entrance to the ICH and was mutated to Asp. The constructed mutants did not significantly affect transfer (Table 3), except for the already described W216A mutant. Mutation N271D did not affect transfer even under limiting conditions (Table 4), in contrast with the randomly obtained transfer-deficient S270P mutant, lying adjacent to N271 (Fig. 3A, shown in dark green). Mutations in the three lysines protruding into the ICH did not affect transfer frequencies, or even showed 10-times higher conjugation frequencies than the wild type in some cases (the K421A mutant in assay B and the K398A mutant in assay E) (Table 4).
The W216A mutant, as previously reported, showed a severe dominant-negative phenotype (5-log drop in the transfer efficiency of the coresident wild-type plasmid). The S270P mutant showed a dominant-negative effect of about 2 logs.
(iv) C-terminal deletions.
Both Δ12C and Δ17C deletions were constructed. As judged by the 3D structure (Fig. 3A), the C-terminal 12 residues of the TrwB monomer protrude away from the structure as a random coil, and residues 13 to 17 from the C terminus lie on top of the bound NTP molecule. Both deletions resulted in severely reduced protein functionality (3- and 4-log decreases, respectively, in the standard conjugation assay) (Table 3). The amount of protein present, as judged by Western blot analysis, was reduced: roughly, the Δ12C deletion showed half the amount of protein, and the Δ17C deletion showed approximately 1/5 of wild-type levels (data not shown), suggesting that this deletion affected the stability of TrwB. However, considering that under these conditions the amount of TrwB is not a limiting factor, it can be speculated that the C terminus of TrwB plays a role in its activity.
(v) Effect of TrwBΔN70 mutations in relaxase transport.
In order to determine if any of the TrwB mutations affected specifically protein transport through the T4SS, triparental matings were performed (Fig. 2) (assay G). Since the transfer frequency in this assay is much lower than that used to test DNA transfer, the assays were performed from donor strain DH5α, which allows expression from the lactose promoter even in the absence of IPTG. Using this donor strain, transfer frequencies without IPTG were higher than those with IPTG-induced D1210 donor cells.
All transfer-deficient or severely affected mutants (K136T, R124A, D158A, D356A, E357A, R375A, Q386A, W216A, S270P, R318H, triple-AAD, and Δ12C mutants) were deficient for TrwC transport in the absence of DNA: <4 × 10−7 transconjugants per donor, compared to a frequency of 4 × 10−3 in the presence of wild-type TrwB (data not shown). The assays with the mutants shown to affect TrwB function only under certain assay conditions are shown in the last column of Table 4. Mutants affected in DNA transfer (the Q390A mutant and, to a lesser extent, the H244A W245A double mutant) were proportionally affected in TrwC transfer. Interestingly, the K398A, K275A, and especially N271D mutants, inside and closing the entrance of the ICH respectively, were not affected in DNA transfer but showed a significant decrease in TrwC transport efficiency (decreases of 57, 20, and 1,000 times, respectively).
(vi) Effect of TrwBΔN70 mutations in oligomerization.
We used the adenylate cyclase-based bacterial two-hybrid assay (29) to study protein-protein interactions. Previous results using this assay showed that TrwB interacts with itself in this system, while deletion of the N-terminal 75 residues of TrwB severely affects the interaction (37). We transferred selected TrwB mutations, lying close to the monomer-monomer interface, to plasmids producing TrwB fusions to both adenylate cyclase domains (Table 2) and introduced them in the DHM1 test strain, as explained in Materials and Methods. The results are shown in the left columns in Table 5. NBD mutations K136T and D158A and the C-terminal deletion Δ12C strongly affected the interaction, suggesting these residues were implicated in TrwB hexamer formation. The rest of mutants tested did not significantly affect the TrwB-TrwB interaction, as judged by the blue colonies formed on selective plates.
TABLE 5.
Bacterial two-hybrid assays showing the effect of TrwB NBD mutations in protein-protein interactions
| TrwB type | Protein interaction testeda: |
||
|---|---|---|---|
| With itself | With TrwB | With TrwE | |
| Wild type | ++ | ++ | +++ |
| ΔN75 mutant | − | − | − |
| K136T mutant | − | + | +++ |
| D158A mutant | − | + | +++ |
| R318H mutant | ++ | ++ | +++ |
| D356A mutant | ++ | ++ | +++ |
| E357A mutant | ++ | ++ | +++ |
| R375A mutant | ++ | ++ | +++ |
| Q386A mutant | ++ | ++ | +++ |
| Δ12C mutant | − | + | +++ |
DHM1 cells containing pairs of plasmids coding for T18 or T25 fusions with the indicated TrwB mutants were grown in plates supplemented with X-Gal. The symbols represent the intensity of the blue color observed: −, white (no interaction), as in the negative control (pUT18 plus pT25zip); +, pale blue; ++, blue; +++, dark blue, as in the positive control (pUT18zip plus pT25zip).
Using this two-hybrid assay, TrwB has also been shown to interact strongly with the T4SS component TrwE (37). We tested the same TrwB mutants for their interaction with TrwE. None of them was affected in the interaction (Table 5, right column); thus, the NBD region involved in TrwB-TrwB interactions is not involved in the TrwB-TrwE interaction.
TrwB mutants improving the interaction with the T4SS.
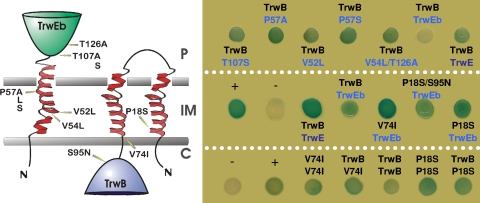
The TMD of TrwB was shown to be essential for the interaction with the T4SS component TrwE, as judged by two-hybrid analysis. In turn, the N-terminal 64 residues of TrwE, which also include a transmembranal segment, were essential for the interaction (37). This suggests that the TrwB-TrwE interaction occurs mainly at the inner membrane. We used the bacterial two-hybrid assay as a screening method to look for TrwB mutations that affected this interaction. Since mutants that lose the interaction are less informative, we decided to address the weaker interaction between TrwB and the TrwE homologue of the highly related Trw T4SS of Bartonella tribocorum, TrwEBt (13); the TrwB-TrwEBt interaction renders pale blue colonies in this assay, allowing screening for mutants with improved TrwB-TrwEBt interactions by selecting dark blue colonies on X-Gal-containing selective medium.
We performed random mutagenesis on the 5′ region of both genes, as explained in Materials and Methods, and selected mutants forming dark blue colonies for further analysis. Several colonies were obtained in both cases: 8 out of about 2,000 colonies when screening for TrwB mutations (plus 18 white colonies that were not further characterized) and 7 out of about 4,000 colonies when screening for TrwEBt mutations (plus 20 white colonies). The TrwB and TrwEBt mutants obtained are listed in Table 6 and illustrated in Fig. 4. Mutations altering the TrwB-TrwEBt interaction were located mostly in the transmembranal segments of both proteins. Six out of eight TrwB mutants carried the P18S mutation, alone or together with additional silent mutations; only one of them was selected for further analysis.
TABLE 6.
TrwB-TrwEBt mutants obtained by two-hybrid screen
| Plasmid | Protein | Mutation |
Complementationa |
||
|---|---|---|---|---|---|
| DNA | Residue | pSU1443 (R388 trwB) | pSU4134 (R388 trwE) | ||
| pUT18C | None | <10−7 | |||
| pMTX601 | TrwB | None | None | 4.8 × 10−3 | |
| pHP106 | TrwB | GTC→ATC | Val74→Ile | 4.9 × 10−3 | |
| pHP107 | TrwB | CCG→TCG | Pro18→Ser | 3.4 × 10−3 | |
| AGT→AAT | Ser95→Asn | ||||
| pHP108 | TrwB | CCG→TCG | Pro18→Ser | 3.8 × 10−3 | |
| pUT18C | None | <10−7 | |||
| pMTX631 | TrwE | None | None | 1.9 × 10−3 | |
| pMTX697 | TrwEBt | None | None | 1.3 × 10−6 | |
| pHP119 | TrwEBt | ACA→TCA | Thr107→Ser | 1.5 × 10−6 | |
| pHP120 | TrwEBt | CCA→GCA | Pro57→Ala | 2.0 × 10−6 | |
| pHP121 | TrwEBt | CCA→CTA | Pro57→Leu | 2.4 × 10−7 | |
| ACA→GCA | Thr107→Ala | ||||
| pHP122 | TrwEBt | CCA→TCA | Pro57→Ser | 2.9 × 10−6 | |
| pHP123 | TrwEBt | GTA→TTA | Val54→Leu | 1.0 × 10−6 | |
| ACA→GCA | Thr126→Ala | ||||
| pHP124 | TrwEBt | GTC→CTC | Val52→Leu | 1.8 × 10−6 | |
For complementation, the indicated pairs of plasmids introduced into strain DH5α were mated with strain D1210. pSU1443 and pSU4134 (Table 1) are transfer-deficient R388 derivatives with transposon insertions in trwB and trwE, respectively. The values shown represent the number of transconjugants per donor and are the means of 2 to 4 independent experiments.
FIG. 4.
Mutations affecting the TrwB-TrwE interaction. (Left) Scheme of the transmembranal regions of TrwEBt and TrwB. Predicted transmembranal segments are represented by helices embedded in the inner membrane (IM). C, cytoplasm; P, periplasm. N, N terminus of the proteins. The locations of the residues shown to be involved in the interaction are indicated with arrows. (Right) Protein-protein interactions tested by the two-hybrid assay. For each spot, the pairs of proteins produced as T25 and T18 fusions are indicated. The top panel shows the TrwEBt mutants tested against TrwB, the middle panel shows TrwB mutants tested against TrwEBt, and the bottom panel shows TrwB mutants tested against TrwB or themselves.
Mutants were checked for their functionality in conjugation by complementation assays of either an R388 TrwB mutant or an R388 TrwE mutant, since TrwEBt was previously shown to complement an R388 trwE mutation, albeit with 100-fold lower efficiency (13). The results of the complementation tests are shown in Table 6. The TrwEBt mutants maintained their ability to complement an R388 TrwE mutant as efficiently as wild-type TrwEBt; the only mutant that complemented about 10 times lower was the P57L T107A double mutant, while mutations affecting either residue independently did not affect complementation. Similarly, TrwB mutants were not altered in their function in R388 conjugation, showing the same complementation levels as wild-type TrwB (Table 6). We transferred TrwB mutations to pHP139 in order to assay them under restrictive conditions (assay type E). Both the V74I and P18S S95N mutants behaved as wild-type TrwB in this assay (data not shown).
All mutants were also tested for their interaction with R388 TrwE, and no significant differences from wild-type TrwB and TrwEBt were observed (data not shown). TrwB mutants were additionally tested for self-interaction and interaction with wild-type TrwB. The results (Fig. 4, right, lower panel) showed an increase in TrwB-TrwB interaction, especially for the P18S mutant. Thus, the mutants selected in the N-terminal region increased both TrwB-TrwEBt and TrwB-TrwB interactions.
DISCUSSION
Coupling proteins are key elements of the conjugative transfer machineries; their pivotal role as connectors between the relaxosome and the T4SS implies the existence of protein-protein interactions on both sides and thus an essential role in substrate recruitment. In addition, evidence is accumulating that T4CPs may also act as DNA pumping motors. Consequently, these proteins have to accomplish a number of functions, such as interactions with the T4SS and with the relaxosome, DNA binding, and ATPase activity. In this work, we addressed a mutagenesis analysis of the T4CP TrwB which allowed us to discern key residues of the protein involved in its different functional roles. In order to characterize these mutants, we designed a series of conjugation assays under different trwB expression conditions.
TrwB function under different assay conditions.
We determined that there are several thousand TrwB molecules per cell when expressed from its natural promoter in plasmid R388. Thus, we suspected the amount of TrwB was probably in excess under standard assay conditions, a situation which would hamper the search for transfer-deficient trwB mutants. Confirming this assumption, we set up a conjugation assay controlling trwB expression from a lactose promoter and using the T4SS of the related plasmid pKM101, where the amount of TrwB correlates directly with the frequency of transfer (Fig. 2). In the wild-type situation, the conjugation frequency was not dependent on the amount of TrwB. A plausible explanation is that the interaction of TrwB with the T4SS of pKM101 is less efficient, but this loss is compensated by an increased amount of TrwB.
The use of the TrwB limiting system allowed us to detect TrwB mutations affecting transfer efficiency, which were not apparent in the standard conjugation assay. Nevertheless, caution has to be exerted when interpreting data from TrwB mutants which show an impaired phenotype only in the limiting system. It has to be taken into account that a heterologous T4SS is being used. In the absence of any further phenotype shown by in vitro assays, it remains possible that the effect observed is solely due to a defective interaction of the T4CP with the T4SS of pKM101 and that the mutations would not affect the interaction of TrwB with its cognate T4SS. However, considering the residues showing a phenotype under these conditions (located in the NBD, the AAD, and in the ICH), this seems unlikely. The more plausible explanation is that the inefficient interaction of TrwB with the T4SS of pKM101 cannot be compensated for by the impaired TrwB variants when TrwB is not in excess.
We also found that DNA transfer frequency was increased more than 1 log when trwA expression was induced (Fig. 2); in addition, an imbalance in the TrwA/TrwB ratio seemed to be deleterious to the cell, since there was a drop in the transfer frequency upon IPTG induction of Plac-trwB, but not Plac-trwAB (Fig. 2).
These data suggest that TrwA is important for TrwB function. TrwA has been shown to increase TrwB DNA-dependent ATPase activity (52). It is plausible that the formation of a TrwA-TrwB complex will work most efficiently for substrate recruitment and translocation.
Production of TrwA from its own promoter has been described to be autorepressed (43). Interestingly, we found that TrwA production from pHP138 (oriT-trwAC) was indeed repressed, since only a slight amount of TrwA was detected by Western blot (Fig. 1E, lanes 2 and 3); however, TrwA was produced from pSU4622 in large amounts (PtrwA-trwABC) (Fig. 1C, lanes 2 and 3). A plausible explanation is that, while pHP138 includes the R388 oriT and thus, 400 bp upstream of the start of trwA, in pSU4622, the PtrwA promoter sequence starts at nucleotide 270 from the R388 oriT sequence (33), including TrwA binding site A but not binding site B (43); thus, TrwA needs both binding sites A and B to repress its own promoter, which should lead to a redefinition of the trwABC promoter.
Functional dissection of TrwB.
We mutagenized the N-terminal region of TrwB and screened for mutants affected in the interaction with the T4SS. Point mutants in the cytoplasmic domain were designed based on the protein 3D structure addressing putative key residues for NTP binding/hydrolysis, oligomerization, and interaction with the substrate protein and DNA. This approach was complemented with a search for transfer-deficient mutants after random mutagenesis. Mutants were characterized using the above-described assays. A schematic summary of the results obtained is shown in Fig. 5. The TrwB domains proposed to be involved in specific TrwB functions are indicated, with the mutated residues highlighted in different shades to indicate the magnitude of their effect in transfer.
FIG. 5.
(Left) Representation of a TrwB protomer with the mutated residues shown in the space-filled format and their proposed function indicated in a color code. Within each proposed functional domain, the space-filled residues show brighter colors when their mutation affects the transfer efficiency. The 3D structure of the N-terminal TMD has been modeled as in reference 19. Each mutated residue is marked with a number. (Right) The table summarizes the results obtained from the in vivo assays for each mutant. Mating assay abbreviations: DNA transfer efficiency (DNA), TrwC transport efficiency (TrwC), and negative dominance (ND). Two-hybrid assay abbreviations: TrwB-TrwB interactions (B:B), TrwB-TrwE interactions (B:E), and TrwB-TrwEBt interactions (B:Eb).
(i) Mutations in nucleotide binding/hydrolysis and oligomerization.
Residue K136 is in Walker box A, while residues D356 and E357 form the DEXX box (Walker box B) (54). Residue D158 is within a putative Asp box. Residues R124 and R375 contribute to the NTP-binding pocket from the adjacent subunit (Fig. 3C). When the 3D structures of TrwBΔN70 resembling the substrate and product of NTP hydrolysis were compared (18), the regions that varied most were the P-loop (including K136), residues R124 and R375, and the region that includes Q386 and Q390. The data suggest that the selected residues have a key role in nucleotide binding and hydrolysis.
Six out of eight mutants in contact with the NTP (K136T, D158A, D356A, E357A, R375A, and Q386A) lost their function in conjugation. The R124A mutant retained 2% transfer ability, and the Q390A mutant was impaired in conjugation under TrwB limiting conditions (Tables 3 and 4). The results obtained underscore the essential role of NTP binding hydrolysis by TrwB in conjugation. All NBD mutants showed a proportional transfer-deficient phenotype when only conjugative TrwC transport was tested, suggesting that the ATPase activity is equally required for DNA and protein transport through the T4SS.
All transfer-deficient mutants in the NBD exerted negative dominance to different extents (Table 3). Negative dominance is typically an effect of mutant proteins that form part of complexes, so an explanation for this effect would be that these mutants can oligomerize with wild-type TrwB forming nonfunctional hetero-hexamers. However, a two-hybrid assay showed that mutations K136T and D158A strongly affected the TrwB-TrwB interaction (Table 5). It is possible that the mutants with decreased binding ability form unstable, less-functional multimers. But, considering that these mutants maintain intact their interaction with TrwE (Table 5), it seems more plausible that negative dominance is due to the interaction of the TrwB mutants with the T4SS components and subsequent “poisoning” of the TrwB-T4SS functional interaction required for DNA transfer.
The C-terminal deletion Δ12C also affected TrwB-TrwB interactions (Table 5). Thus, in addition to the N-terminal region, the C-terminal segment and the NBD region of TrwB were involved in TrwB-TrwB interactions. Similar results were obtained with the T4CPs TraD of the F plasmid and TcpA of plasmid pCW3: the TMD is important for oligomerization, together with another segment of the cytoplasmic domain (23, 50). It was suggested that the N-terminal domain of TraD is required for the interaction of the T4CP with the rest of the conjugal machinery, while the region of the soluble domain would only be required for self-interactions. In accordance with these results, we have shown that the residues implicated in the interaction of TrwB with the T4SS component TrwE are located in the N-terminal segment (Fig. 4), while cytoplasmic mutations severely affecting TrwB-TrwB interactions do not affect the interaction with TrwE (Table 5).
(ii) Putative mutants and relaxosomal interactions.
TrwB interacts with the relaxosomal proteins TrwA and TrwC, and binds nonspecifically both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) (37, 42). The TrwB ICH is expected to be the channel for DNA transfer. The AAD of TrwB is located in the cytoplasmic side of the hexamer and surrounds the entrance to the ICH. We constructed a series of mutants with mutations targeting the residues more likely to be involved in interactions with TrwA, TrwC, or the DNA (Fig. 3A and B): conserved or polar residues at the AAD surface and positively charged residues protruding into the ICH. The targeted AAD region is within the TrwB region from Y233 to G260, which is structurally related to protein TraM of the F plasmid, involved in relaxosomal specificity (18).
When these mutants were tested in the standard conjugation assay, the AAD region triple mutant and the previously characterized W216A mutant in the ICH were transfer deficient (Table 3). In addition, a random mutagenesis experiment created the mutation S270P, which lies close to the cytoplasmic entrance to the ICH. Thus, the AAD polar surface, the ICH, and the entrance to the ICH are essential regions for TrwB function, but under standard conjugation assays, aggressive mutations are required to obtain an impaired phenotype. When transferred to the TrwB limiting assays depicted in Fig. 2, additional mutants showed a decrease in conjugation efficiency (Table 4), especially the AAD mutant with mutation in two conserved residues, H244A and W245A, which was 2 logs less efficient than wild-type TrwB.
It should be noted that all mutants affected in DNA transfer were proportionally affected in TrwC transport (Table 4). No mutants were obtained affecting specifically DNA transfer, which would have mapped a DNA pumping activity of TrwB. However, several mutants were obtained which were not affected in DNA transfer but showed a significant decrease in efficiency for TrwC transport (Table 4). These discriminatory mutants open up the unlikely possibility that DNA is transported independently of TrwC; however, there are other more plausible explanations. While only one or a few TrwC-DNA complexes are expected to be transferred during conjugation, TrwC alone may be transported in large numbers in order to recircularize the transferred DNA in the recipient cell. Both substrates (TrwC and TrwC-DNA complex) could be recruited with different affinities; in fact, TrwB interacts with the relaxase domain of TrwC (37), so the presence of the covalently bound DNA could affect this interaction. Since TrwB binds DNA, the TrwC-DNA complex could likely be the preferred substrate. In the triparental matings performed to check for TrwC transport, complementation of the trwC mutant in the “second donor” strain would be largely dependent on the amount of TrwC molecules entering the cell from the “first donor” strain. Thus, a defect in TrwC transport, while maintaining TrwC-DNA transfer levels may diminish significantly the number of free TrwC molecules entering the “second donor” and thus the complementation efficiency, from which TrwC transport rates are estimated.
The residues affected specifically in TrwC transport were located inside or at the entrance of the ICH (mutations K398A, K275A, and especially N271D). This location suggests that they could have a role in recruitment of TrwC. TrwB residue K398 is embedded in the ICH; the effect of mutation K398A on TrwC transport could imply that TrwC is secreted through the TrwB ICH in an unfolded state. More likely, TrwB could interact with TrwC as a monomer, and after TrwC translocation through the T4SS, TrwB would hexamerize on the DNA, as suggested by the “shoot-and-pump” model for DNA transfer (36).
(iii) Mutations in the TMD.
Previous works showed that the soluble derivatives TrwBΔN75 and TrwEΔN64 lost the interaction with each other and with themselves (37). We mutagenized the region coding for the N-terminal 135 residues of TrwB and TrwEBt, selecting for mutants which showed stronger interactions with each other in the two-hybrid assay. In this way, we mapped the interacting domain of these proteins, according to the location of the residues shown to increase the interaction when mutated (Fig. 4 and Table 6). Most mutations lie in the transmembranal regions of both proteins, although mutation T107S in TrwEBt indicated that regions outside the inner membrane are also required for the interaction. Two proline residues, Pro18 in TrwB and Pro57 in TrwEBt, appeared in independently isolated mutants, highlighting their importance in the interaction and ensuring that the mutagenesis procedure was exhaustive. Structural motifs that contain proline residues in α-helices are key elements in mechanisms of signaling through conformational “switches,” since prolines affect the α-helix topology in the inner membrane by changing the direction of the helix (46). Their mutation probably induced a conformational change that affected the interaction with other proteins.
The interaction between the T4CP and the VirB10 homologue is of special relevance. It was shown that this interaction drives the efficiency of DNA transfer through heterologous T4SS (37). In this case, mutants with stronger interactions did not show corresponding higher DNA transfer efficiency, but it has to be taken into account that, while interactions between TrwB and TrwEBt are improved, interactions between TrwEBt and the rest of the R388 T4SS may still limit the efficiency of DNA transfer.
VirB10 has been proposed to act as an energy sensor which could couple inner membrane ATP consumption to substrate transfer (9). We have found that the same TrwB residues involved in interaction with TrwE were also involved in TrwB-TrwB interactions (Fig. 4, right, lower panel). It is tempting to speculate that the transmembranal helices from TrwB and TrwE intertwine in the inner membrane; in this way, the ICH of the TrwB hexamer would contact directly the transmembranal channel formed by TrwE, recently shown to cross both bacterial membranes (10); so the mating signal could be transmitted from the outside to the relaxosome via the T4CP-VirB10 interaction.
Acknowledgments
This work was supported by grants BIO2008-00133 from the Spanish Ministry of Science and Innovation and API 07/01 from the Fundación Marqués de Valdecilla to M.L. and grants LSHM-CT-2005_019023 from the European Commission, BFU2008-00995/BMC from the Spanish Ministry of Science and Innovation, and RETICS Research Network RD06/0008/1012, Instituto de Salud Carlos III, Spanish Ministry of Health, to F.C. H.P. and D.L. were the recipients of predoctoral fellowships from the University of Cantabria and JAE-predoc (CSIC), respectively.
We are grateful to Gabriel Moncalián for providing TrwA and TrwB antisera and Elena Cabezón and Ignacio Aréchaga for helpful discussions.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmakuri, K., Z. Ding, and P. J. Christie. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 4.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabezon, E., and F. de la Cruz. 2006. TrwB: an F(1)-ATPase-like molecular motor involved in DNA transport during bacterial conjugation. Res. Microbiol. 157:299-305. [DOI] [PubMed] [Google Scholar]
- 6.Cabezón, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabezón, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. U. S. A. 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. U. S. A. 101:17228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran, V., R. Fronzes, S. Duquerroy, N. Cronin, J. Navaza, and G. Waksman. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y., X. Zhang, D. Manias, H. J. Yeo, G. M. Dunny, and P. J. Christie. 2008. Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J. Bacteriol. 190:3632-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Cruz, F., and J. Grinsted. 1982. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J. Bacteriol. 151:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Paz, H. D., F. J. Sangari, S. Bolland, J. M. Garcia-Lobo, C. Dehio, F. de la Cruz, and M. Llosa. 2005. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology 151:3505-3516. [DOI] [PubMed] [Google Scholar]
- 14.Disque-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draper, O., C. E. César, C. Machón, F. de la Cruz, and M. Llosa. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc. Natl. Acad. Sci. U. S. A. 102:16385-16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcillan-Barcia, M. P., M. V. Francia, and F. de la Cruz. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 17.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 18.Gomis-Rüth, F. X., G. Moncalián, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277:7556-7566. [DOI] [PubMed] [Google Scholar]
- 19.Gomis-Rüth, F. X., G. Moncalián, R. Pérez-Luque, A. González, E. Cabezón, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 20.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunton, J. E., M. W. Gilmour, G. Alonso, and D. E. Taylor. 2005. Subcellular localization and functional domains of the coupling protein, TraG, from IncHI1 plasmid R27. Microbiology 151:3549-3561. [DOI] [PubMed] [Google Scholar]
- 23.Haft, R. J., E. G. Gachelet, T. Nguyen, L. Toussaint, D. Chivian, and B. Traxler. 2007. In vivo oligomerization of the F conjugative coupling protein TraD. J. Bacteriol. 189:6626-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hormaeche, I., I. Alkorta, F. Moro, J. M. Valpuesta, F. M. Goñi, and F. de la Cruz. 2002. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 277:46456-46462. [DOI] [PubMed] [Google Scholar]
- 26.Hormaeche, I., I. Iloro, J. L. Arrondo, F. M. Goni, F. de la Cruz, and I. Alkorta. 2004. Role of the transmembrane domain in the stability of TrwB, an integral protein involved in bacterial conjugation. J. Biol. Chem. 279:10955-10961. [DOI] [PubMed] [Google Scholar]
- 27.Hormaeche, I., R. L. Segura, A. J. Vecino, F. M. Goni, F. de la Cruz, and I. Alkorta. 2006. The transmembrane domain provides nucleotide binding specificity to the bacterial conjugation protein TrwB. FEBS Lett. 580:3075-3082. [DOI] [PubMed] [Google Scholar]
- 28.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. [PubMed] [Google Scholar]
- 31.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 32.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 33.Llosa, M., S. Bolland, and F. de la Cruz. 1991. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol. Gen. Genet. 226:473-483. [DOI] [PubMed] [Google Scholar]
- 34.Llosa, M., S. Bolland, G. Grandoso, and F. de la Cruz. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J. Bacteriol. 176:3210-3217. (Erratum, 176:6414.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llosa, M., and F. de la Cruz. 2005. Bacterial conjugation: a potential tool for genomic engineering. Res. Microbiol. 156:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Llosa, M., F.-X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. U. S. A. 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, J., and L. S. Frost. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J. Bacteriol. 187:4767-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, J., J. J. Wong, R. A. Edwards, J. Manchak, L. S. Frost, and J. N. Glover. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 70:89-99. [DOI] [PubMed] [Google Scholar]
- 40.Martínez, E., and F. de la Cruz. 1988. Transposon Tn21 encodes a RecA-independent site-specific integration system. Mol. Gen. Genet. 211:320-325. [DOI] [PubMed] [Google Scholar]
- 41.Mihajlovic, S., S. Lang, M. V. Sut, H. Strohmaier, C. J. Gruber, G. Koraimann, E. Cabezon, G. Moncalian, F. de la Cruz, and E. L. Zechner. 2009. Plasmid r1 conjugative DNA processing is regulated at the coupling protein interface. J. Bacteriol. 191:6877-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moncalián, G., E. Cabezón, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goñi, and F. de la Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 43.Moncalián, G., G. Grandoso, M. Llosa, and F. de la Cruz. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270:188-200. [DOI] [PubMed] [Google Scholar]
- 44.Sadler, J. R., M. Tecklenburg, and J. L. Betz. 1980. Plasmids containing many tandem copies of a synthetic lactose operator. Gene 8:279-300. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sansom, M. S., and H. Weinstein. 2000. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol. Sci. 21:445-451. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. Biotechniques 8:404-407. [PubMed] [Google Scholar]
- 48.Sastre, J. I., E. Cabezón, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steen, J. A., T. L. Bannam, W. L. Teng, R. J. Devenish, and J. I. Rood. 2009. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 191:2926-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sut, M. V., S. Mihajlovic, S. Lang, C. J. Gruber, and E. L. Zechner. 2009. Protein and DNA effectors control the TraI conjugative helicase of plasmid R1. J. Bacteriol. 191:6888-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezon. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282:25569-25576. [DOI] [PubMed] [Google Scholar]
- 53.Tato, I., S. Zunzunegui, F. de la Cruz, and E. Cabezón. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. U. S. A. 102:8156-8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]