Abstract
Enterotoxigenic Escherichia coli (ETEC) is the leading bacterial cause of diarrhea in the developing world, as well as the most common cause of traveler's diarrhea. The main hallmarks of this type of bacteria are the expression of one or more enterotoxins and fimbriae used for attachment to host intestinal cells. Longus is a pilus produced by ETEC. These bacteria grown in pleuropneumonia-like organism (PPLO) broth at 37°C and in 5% CO2 produced longus, showing that the assembly and expression of the pili depend on growth conditions and composition of the medium. To explore the role of longus in the adherence to epithelial cells, quantitative and qualitative analyses were done, and similar levels of adherence were observed, with values of 111.44 × 104 CFU/ml in HT-29, 101.33 × 104 CFU/ml in Caco-2, and 107.11 × 104 CFU/ml in T84 cells. In addition, the E9034AΔlngA strain showed a significant reduction in longus adherence of 32% in HT-29, 22.28% in Caco-2, and 21.68% in T84 cells compared to the wild-type strain. In experiments performed with nonintestinal cells (HeLa and HEp-2 cells), significant differences were not observed in adherence between E9034A and derivative strains. Interestingly, the E9034A and E9034AΔlngA(pLngA) strains were 30 to 35% more adherent in intestinal cells than in nonintestinal cells. Twitching motility experiments were performed, showing that ETEC strains E9034A and E9034AΔlngA(pLngA) had the capacity to form spreading zones while ETEC E9034AΔlngA does not. In addition, our data suggest that longus from ETEC participates in the colonization of human colonic cells.
Enterotoxigenic Escherichia coli (ETEC) is an important cause of infant diarrhea in developing countries, a leading cause of traveler's diarrhea, and a reemergent diarrheal pathogen in the United States (1, 25, 29, 33, 38, 40, 41, 44, 51, 52, 55). ETEC strains were first recognized as a cause of diarrheal disease in animals, especially in piglets and calves, where the disease continues to cause lethal infection in newborn animals (3, 37). Studies of ETEC in piglets first elucidated the mechanisms of disease, including the presence of two plasmid-encoded enterotoxins. In humans, the clinical appearance of ETEC infection is identical to that of cholera, with severe dehydrating illness not commonly seen in adults (38, 46). DuPont et al. (12) subsequently showed that ETEC strains were able to cause diarrhea in adult volunteers. ETEC strains cause watery diarrhea similar to that caused by Vibrio cholerae through the action of two enterotoxins, the cholera-like heat-labile and heat-stable enterotoxins (LT and ST, respectively) (38). These strains may express an LT only, an ST only, or both LT and ST. To cause diarrhea, ETEC strains must first adhere to small bowel enterocytes, an event mediated by a variety of surface fimbrial appendages called colonization factor antigens (CFAs), coli surface antigens (CSs), and putative colonization factors (PCF) (22, 33, 38). Transmission electron microscopy (TEM) of ETEC strains typically reveals many peritrichously arranged fimbriae around the bacterium; often, multiple fimbrial morphologies can be visualized on the same bacterium (6, 19, 31, 38). ETEC strains also express the K99 fimbriae, which are pathogenic for calves, lambs, and pigs, whereas K88-expressing organisms are able to cause disease only in pigs (8). Human ETEC strains possess their own array of colonization fimbriae, the CFAs usually encoded in plasmids (10). Currently, more than 20 CFAs known in human ETEC infections have been described (17). The CFAs can be subdivided based on their morphological characteristics. Three major morphological varieties exist: rigid rods (CFA I), bundle-forming flexible rods (CFA III), and thin, flexible, wiry structures (CFA II and CFA IV) (7, 8, 26, 30, 49, 53, 54).
A high proportion of human ETEC strains contain a plasmid-encoded type IV pilus (T4P) antigen (CS20) also called longus for its length (19, 21). Longus is a T4P composed of a repeating structural subunit called LngA of 22 kDa, and its N-terminal amino acid sequences shares similarities with the class B type IV pili. These pili include the CFA III pilin subunit CofA of ETEC, the toxin-coregulated pilin (TCP) of V. cholerae, and the bundle-forming pilin (BFP) found in enteropathogenic E. coli (EPEC) and in a small percentage in other Gram-negative pathogens (21, 23). The lngA gene, which encodes the longus pilus in ETEC strains, is widely distributed in different geographic regions such Bangladesh, Chile, Brazil, Egypt, and Mexico (23). Interestingly, the lngA gene has been observed in association with ETEC strain producers of LT and ST (23). Sequence analysis of the fimbrial genes provided insight into the evolutionary history of longus. It appears that the highly conserved nonstructural lngA genes evolved in a similar manner to that of housekeeping genes.
Recently, another important adherence factor called E. coli common pilus (ECP) has been identified; it is composed of a 21-kDa pilin subunit whose amino acid sequence corresponds to the product of the yagZ (renamed ecpA) gene present in all E. coli genomes sequenced to date (47). ECP production was demonstrated in strains representing intestinal (enterohemorrhagic E. coli [EHEC], EPEC, and ETEC) and extraintestinal pathogenic E. coli as well as normal-flora E. coli.
In this study we report that longus plays an important role in the adherence to colonic epithelial cells. In addition to mediating cell adherence, longus is also associated with other pathogenicity attributes exhibited by other Gram-negative pathogenic bacteria producing T4P, which can contribute in part to the virulence of ETEC.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
To verify and evaluate the production of longus from the ETEC E9034A strain, the bacteria was cultured in pleuropneumonia-like organism (PPLO) broth (BD Difco, NJ), TSAB (Trypticase soy agar supplemented with 5% blood), brucella broth, brain heart infusion (BHI) broth, MacConkey agar, Luria-Bertani broth (LB), and CFA agar (1% Casamino Acids, 0.15% yeast extract, 0.005% MgSO4, 0.0005% MnCl2, 2% agar [pH 7.4]) overnight at 37°C in a 5% CO2 atmosphere. Antibiotics, when necessary, were added at concentrations of 100 μg/ml (ampicillin) and 50 μg/ml (kanamycin). Strains and plasmids employed in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
Construction of the ETEC lngA mutant.
The lambda Red recombinase system was used to generate a nonpolar deletion in the lngA gene from the ETEC E9034A strain, as previously described (9). Primers J1 and J2 were employed to mutate the lngA gene (Table 2), using the lambda Red system to replace this gene with a kanamycin resistance cassette derived from the template plasmid pKD4 (Table 1) (9). l-(+)-Arabinose (Sigma) was used at a final concentration of 100 mM to induce expression of the lambda red system from plasmid pKD46. Primers flanking the lngA gene, as well as primers inside the kanamycin cassette (K1 and K2), were used to confirm the desired gene replacement by PCR. Colonies were selected using antibiotics required to check the mutations in the lngA gene.
TABLE 2.
List of primers used in this study
| Primer name | Sequence (5′-3′) | Function or description |
|---|---|---|
| J1 | ATGAGCCTGCTGGAAGTTATCATTGTTCTTG | lngA mutant |
| GCATTATCGGTACGATTGC | ||
| J2 | TTAACGGCTACCTAAAGTAATTGAGTTTACC | lngA mutant |
| TGAGCAGTACAGGTACTTA | ||
| J5 | ATGAGCCTGCTGGAAGTTAT CATTG | Amplification of the lngA gene |
| J6 | TTAACGGCTACCTAAAGTAATTGAGTT | Amplification of the lngA gene |
| K1 | GCCCAGTCATAGCCGAATAGCCT | Inside the kanamycin resistance gene |
| K2 | CGGTGCCCTGAATGAACTGCAGG | Inside the kanamycin resistance gene |
Cloning of the lngA gene from E9034A.
To complement the lngA mutation, the gene was amplified from ETEC E9034A using primers J5 and J6 and cloned into the SspI site of the low-copy-number plasmid pBR322.
Purification of longus.
ETEC E9034A was grown in PPLO agar (BD, Difco, NJ) at 37°C in 5% CO2 to induce longus production. The pili were detached by vigorous shaking from bacteria and were harvested and suspended in phosphate-buffered saline ([PBS], pH 7.4). Briefly, bacteria were separated twice by centrifugation at 10,000 × g for 30 min, and the supernatant was centrifuged at 18,000 × g for 30 min to remove flagella, outer membranes, and bacterial debris. The pilus-containing supernatant was centrifuged at 78,000 × g for 4 h, and the pellet was dissolved in 5 ml of phosphate buffer (100 mM; pH 7.4) and applied onto a cesium chloride-1% Sarkosyl gradient to obtain different bands of relatively pure pili. The purified of longus was subjected to denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 16% acrylamide gels and Coomassie blue staining (28, 35). The presence of longus was confirmed by TEM. The purification was performed to generate polyclonal antilongus antibodies.
Polyclonal antisera.
Antilongus and anti-ETEC E9034A antibodies were obtained from rabbit as described previously (19). ETEC strains (E9034A and E9034AΔlngA) grown in PPLO broth and CFA agar were used to remove nonspecific antibodies against longus (eight times).
Detection of LngA production.
Immunoblotting was used to detect LngA production using a 1:2,000 dilution of antilongus antibodies, followed by an anti-rabbit IgG horseradish peroxidase (HRP) conjugate (1:20,000 dilution), as previously described (56). Blots were developed with HyGLO chemiluminescent HRP antibody detection reagent substrate (Denville). Anti-DnaK was used as a control for protein loading.
TEM and immunogold labeling.
Longus was produced in PPLO broth overnight at 37°C in 5% CO2. After incubation of ETEC strains with HT-29 cells for 6 h, the supernatants were recovered and mounted on a Formvar- and carbon-coated grid for 5 min and negatively stained with 1% sodium phosphotungstic acid (pH 7.2) for 5 min. The samples were visualized under a Philips TEM, as previously described (56). For immunogold assays, the supernatants were recovered after incubation of ETEC E9034A and derivative strains with HT-29 cells for 6 h at 37°C in 5% CO2. Samples were immunogold labeled using polyclonal antilongus antibodies, followed by goat anti-rabbit IgG conjugated to 10-nm gold particles (Sigma Aldrich) and stained with 1% sodium phosphotungstic acid as previously described. The antibodies were diluted 1:10 in PBS containing 1% (wt/vol) bovine serum albumin (BSA).
Bacterial adherence to intestinal and nonintestinal cells.
The adherence assays were performed employing intestinal colonic epithelial cells (HT-29, T84, and Caco-2) and nonintestinal (HeLa and HEp-2) cells, as previously described (56). The cell monolayers, at 70 to 80% confluence, were cultured in Dulbecco's minimal Eagle medium ([DMEM] Invitrogen, Carlsbad, CA) at 37°C under 5% CO2 in 24-well polystyrene plates (Cellstar) containing glass coverslips. Cell monolayers were infected with 10 μl of bacteria with an optical density (OD) of 1.1 for 6 h at 37°C in 5% CO2 and were washed with PBS to remove unbound bacteria. For the qualitative analysis, the samples were fixed with 2% formalin-PBS for 20 min and stained with Giemsa for 30 min to be visualized by light microscopy. For quantitative assessment, the cell monolayers were treated with 1 ml of 0.1% Triton X-100 for 10 min and were removed to perform serial dilutions; data are expressed as the number of CFU. Quantitative analyses of adherence assays were performed in triplicate on three different days to obtain an average of the results.
Immunofluorescence.
Adherence assays were performed by incubating 10 μl of overnight bacterial cultures (ETEC E9034A strains) with intestinal cell (T84, Caco-2, and HT-29 cells) and nonintestinal cell (HeLa and HEp-2 cells) monolayers for 6 h at 37°C with 5% CO2. Cell monolayers were washed with PBS, fixed with 2% formalin overnight, and incubated with primary antilongus antibodies diluted 1:2,000 for 2 h. After being washed three times with PBS-Tween-20 (PBST), the samples were incubated with goat anti-rabbit IgG conjugated with Alexa Fluor 488 diluted 1:5,000 in PBS-horse serum (HS) for 2 h each, at room temperature. A 1:5,000 dilution of propidium iodide in PBS-HS was used for DNA staining, and the coverslips were mounted on glass slides with 1% p-phenylenediamine mounting medium. Samples were visualized using an Axio Imager 1.0 Zeiss microscope as previously described (20).
Flow cytometry.
ETEC strains (E9034A and derivative strains) were cultured in PPLO broth and CFA agar at 37°C with 5% CO2 to determine the production of longus by flow cytometry, as previously described (6). For the assays, 45 μl of a bacterial culture at an OD at 600 nm (OD600) of 1.1 and grown overnight was incubated with 40 μl of antilongus antibodies using a dilution of 1:2,000 for 2 h on ice. After three gentle washes with cold PBS, the bacteria were resuspended in 40 μl of a 1:10,000 dilution of goat anti-rabbit IgG(H+L)-Alexa Fluor conjugate (Invitrogen, California). After 2 h of incubation on ice, the bacteria were gently washed three times with PBS and resuspended in an 800-ml final volume of PBS. For the analysis, the bacteria were labeled with 5 μl of a propidium iodide solution (Sigma Aldrich). Samples were analyzed in a Becton Dickinson FACSCalibur. The experiments were performed three times in triplicates on separate days, and the data are expressed as the mean of the averages of the results obtained from the three experiments performed.
ELISAs.
Previously, E9034A and derivative strains were cultured in PPLO broth overnight at 37°C in 5% CO2 to trigger the production of longus. Polystyrene 96-well plates containing HT-29 cells were incubated with 3 μl of the bacteria at an OD600 of 1.1 for 6 h at 37°C in 5% CO2. The recovered supernatants (unbound bacteria) were centrifuged at 9,000 × g, and the pellets were resuspended in 200 μl of 0.05 M carbonate buffer, pH 9.6. Immediately, the recovered bacteria were used to coat quadruplicate in 96-well plates and incubated overnight at 4°C. In both cases, unbound bacteria and bacteria attached to HT-29 cell monolayers were identified with anti-ETEC antibodies and were measured in an enzyme-linked immunosorbent assay (ELISA) reader at 405 nm (Multiskan Ascent; Thermo Labsystems). The production of pili bound to the bacteria in contact with HT-29 cells and in the supernatants was identified with polyclonal antilongus antibodies. The experiments were performed in quadruplicate, and the wells were blocked with 1% dry milk in PBST for 2 h at 37°C, washed, and then incubated with antilongus antibodies for 2 h at room temperature. After three washes, secondary antibodies conjugated to anti-rabbit IgG-alkaline phosphatase were added for 2 h at room temperature. The presence of longus binding to bacteria was measured by adding the appropriate substrate and by ELISA reader at 405 nm (Multiskan Ascent; Thermo Labsystems).
Twitching motility assay.
The twitching motility assay was performed, and twitching motility was evidenced as described by Xicohtencatl et al. (57). ETEC E9034A and the mutant strains were cultured in PPLO broth with 1% agar grown for 24 h and stained with Coomassie blue for 1 h. EHEC EDL933 wild-type and EDL933ΔhcpA strains were inoculated in Minca minimal medium with 1% agar and used as positive and negative controls, respectively.
RESULTS
Optimal medium to trigger longus production.
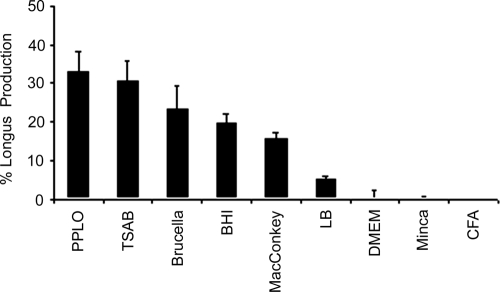
Longus is a type IV pilus produced by ETEC grown on PPLO agar at 37°C in 5% CO2 and is not produced on CFA agar or in DMEM (6). The production of longus using polyclonal antilongus antibodies was detected by flow cytometry. The antibodies against longus were absorbed eight times using the E9034A and E9034AΔlngA (not containing the lngA gene) strains grown on CFA agar and in PPLO broth overnight at 37°C. The E9034A strain cultured in PPLO broth, TSAB, brucella broth, BHI broth, MacConkey agar, and LB broth at 37°C in 5% CO2 produced longus in different percentages, with greater production of these pili in PPLO broth (Fig. 1). However, no production of longus was observed in the ETEC E9034A strain when it was cultured on CFA agar or Minca minimal medium agar or in DMEM and cultured overnight at 37°C in 5% CO2 (Fig. 1).
FIG. 1.
Percentages of longus production in ETEC E9034A strains grown in different media at 37°C in 5% CO2, showing the highest production in PPLO broth and TSAB and no production in CFA agar or in Minca minimal medium.
Longus production by ETEC E9034A and derivative strains.
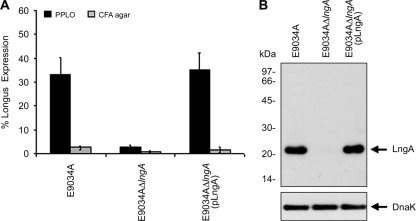
According to the data obtained by flow cytometry, PPLO broth and TSAB were the best media to express longus in the E9034A strain. Flow cytometry was used to evaluate the production of longus in the E9034A wild-type, E9034AΔlngA, and E9034AΔlngA(pLngA) strains. The results showed that 100% of the ETEC strains stained with propidium iodide produced 35% of longus when labeled with Alexa Fluor 488 (Fig. 2A). The mutation in the lngA gene from the ETEC E9034A strain grown in PPLO at 37°C in 5% CO2, as expected, showed a negative effect in the production of longus, confirming the lack of the lngA gene which was replaced with a kanamycin cassette (Fig. 2A). In addition, when the ETEC E9034A strain with a deletion of the lngA gene was complemented with the pLngA plasmid, the capacity to produce and assemble longus was restored at a level similar to that of the wild-type, i.e., 33 to 35%, respectively (Fig. 2A).
FIG. 2.
Percentages of longus production in ETEC E9034A and derivative strains. (A) ETEC E9034A and derivative strains were cultured in PPLO broth and CFA agar, and expression was measured by flow cytometry. No production of longus was observed in the lngA mutant. Longus production was restored in the ETEC E9034AΔlngA strain complemented with the pLngA plasmid. (B) Western blot of whole bacterial cell extracts analyzed in 16% SDS-PAGE gels using antilongus polyclonal antibodies. The ETEC wild-type strain and the lngA mutant complemented with pLngA showed the presence of a protein of 22 kDa that corresponds to the LngA pilin. The LngA pilin was not produced in the lngA mutant. Anti-DnaK antibody was used to detect the DnaK protein to ensure that equal amounts of antigen were tested.
Ultrastructural studies of longus.
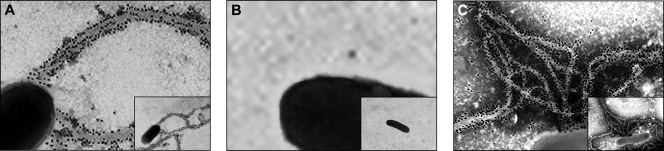
To verify the production of longus by ETEC E9034A and derivative strains, the bacteria were cultured in PPLO broth, labeled with anti-rabbit IgG conjugated to 10-nm gold particles, and negatively stained to be visualized by TEM (data not shown). The presence of longus was investigated and well characterized in the recovered supernatant from E9034A, E9034AΔlngA, and E9034AΔlngA(pLngA) strains after 6 h of incubation with HT-29 cells. Longus produced by ETEC strains was detected by immunogold labeling with antilongus antibodies to confirm the identity of longus. Using these growth conditions, we were able to observe the production of longus forming long bundles (>20 μm) associated to the bacteria in only the E9034A wild-type and E9034AΔlngA(pLngA) strains (Fig. 3A and C). Under the same growth conditions that produced longus, no gold particles were seen associated to the surface of the E9034AΔlngA strain, confirming the lack of the lngA gene and the purity and specificity of the antibody (Fig. 3B). Moreover, no gold particles and no assembly of longus were visualized in ETEC E9034A and derivative strains when they were cultured in CFA broth, and this suppression of the production of longus confirms that its expression and production depend on the composition of the growth medium (data not shown).
FIG. 3.
Immunogold labeling of longus produced by ETEC strains with antilongus antibody. (A) E9034A. (B) E9034AΔlngA. (C) E9034AΔlngA(pLngA). No gold is seen associated with the bacterial cell surface recognizing the antilongus antibody specific to the fimbriae. The electron micrographs were taken at a magnification of ×19,000. The insets in panels A and B show the whole bacterium producing longus covered with gold particles.
Production of LngA pilin.
To evaluate the production of longus, the wild-type ETEC and derivative strains were cultured under conditions where these pili are produced. Whole bacterial cell extracts were normalized and analyzed in 16% SDS-PAGE gels, and the proteins were transferred to a nitrocellulose membrane to be incubated with polyclonal rabbit antilongus antibodies, followed by a horseradish peroxidase conjugate. Results of Western blotting showed that the absence of the lngA gene had a negative effect on the production of the LngA protein (Fig. 2B). The ETEC E9034A wild-type strain and the lngA mutant complemented with pLngA showed the presence of a protein of 22 kDa that corresponds to the pilin, the structural subunit of longus. Anti-DnaK antibody was used to detect the DnaK protein to ensure that equal amounts of antigen were tested (Fig. 2B).
Role of longus in adherence to epithelial cells.
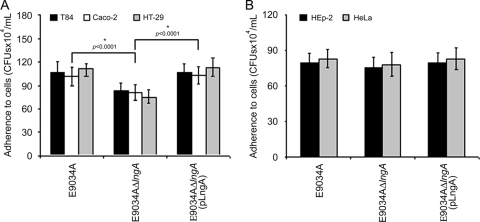
Assembly of longus of ETEC E9034A results from specific growth conditions. To trigger the production of longus and to determine its possible role, intestinal (HT-29, T84, and Caco-2 cells) and nonintestinal epithelial cells were incubated for 6 h with 10 μl of a bacterial suspension grown overnight in PPLO broth at 37°C in 5% CO2. Qualitative analysis from human colonic cells infected with ETEC 9034A showed similar levels of adherence, with values of 111.44 × 104 CFU/ml in HT-29, 101.33 × 104 CFU/ml in Caco-2, and 107.11 × 104 CFU/ml in T84 cells (Fig. 4A; see also Fig. 5). However, the E9034AΔlngA strain showed a significant reduction in adherence of 32% in HT-29, 22.28% in Caco-2, and 21.68% in T84 cells in comparison to the wild-type strain tested with the colonic cells described above (Fig. 4A). In addition, we observed a significant reduction in adherence of the bacteria to cultured intestinal cells (HT-29, T84, and Caco-2 cells) between the lngA mutant and the wild-type strain (P < 0.0001). Interestingly, the ETEC E9034AΔlngA(pLngA) strain restored 100% of the capacity to interact with the surface of human colonic cells, showing the same levels of adherence as those observed in the ETEC E9034A strain. Qualitative analysis of the different adherence assays with E9034A and derivative strains using HT-29 human colonic cells was directly correlated with the values of the quantitative analysis. The images processed by light microscopy confirmed the differences in adherence observed with the quantitative analyses between the wild-type and the E9034AΔlngA strains (Fig. 5A). Evidently the adherence levels were not completely abolished in the mutant, and this could be due to the presence of other adhesins that can act simultaneously or at distinct steps of the adherence process.
FIG. 4.
Qualitative comparison of adherence between ETEC E9034A and derivative mutant strains to epithelial cells. (A) Adherence assays performed with colonic human cells. In the three cellular lines HT-29, T84, and Caco-2 a significant reduction in adherence is observed in the mutant strain (E9034AΔlngA). (B) Adherence assays with nonintestinal cells where no reduction of adherence is observed. The monolayers of epithelial cells were obtained at 70 to 80% confluence and were incubated with ETEC strains for 6 h at 37°C in 5% CO2. The experiments were performed in triplicate on three different days. *, statistically significant (P < 0.0001) with respect to the values obtained from the wild type, the lngA mutant, and the lngA mutant complemented with the same gene.
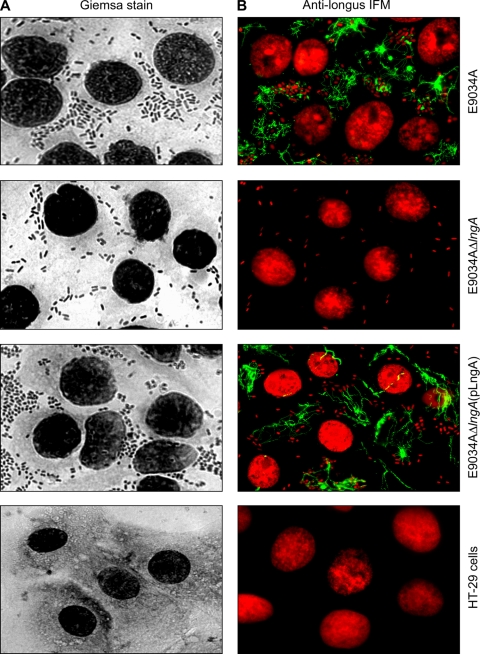
FIG. 5.
Qualitative analysis and immunofluorescence of wild-type ETEC E9034A and derivative mutants. (A) Images by light microscopy show differences in adherence between ETEC and derivative strains in contact with human colonic cells. The images were taken at a magnification of ×200 in a light microscope. (B) Images by IFM of ETEC E9034A and E9034AΔlngA(pLngA) strains showing long fluorescent fibers (green) extending several micrometers throughout the cell monolayer interconnecting the bacteria (red). No fluorescent fibers are seen in the E9034AΔlngA strain, confirming the lack of the lngA gene. DNA in bacteria and eukaryotic cells were stained red with propidium iodide. The images were processed at ×200. In both panels HT-29 cells without infection were used as controls.
In other studies, quantitative analysis of adherence assays using nonintestinal cells (HEp-2 and HeLa cells) did not show significant changes in the number of CFU among the wild-type and derivative strains (Fig. 4B). These data correlated with the images obtained by light microscopy, where no significant changes were observed in the levels of adherence (data not shown). All the strains tested [E9034AΔlngA with respect to the wild-type and E9034AΔlngA(pLngA) strains] in nonintestinal cells showed a minimal change of 5 to 10% in the values of adherence (Fig. 4B). Interestingly, significant changes were observed in the percentages of adherence among human colonic cells and nonintestinal cells (Fig. 4A). The results showed that when human colonic cells (HT-29) were used, a significant increase was observed (above 30%) in the levels of adherence of the E9034A wild-type and E9034AΔlngA(pLngA) strains compared with adherence in nonintestinal cells (Fig. 4). In sum, the reduction in adherence of 30 to 35% observed in the lngA mutant strain in contact with intestinal cells correlates with the values obtained for adherence in nonintestinal cells, showing similar levels for the E9034A, E9034AΔlngA, and E9034AΔlngA(pLngA) strains. Our data suggest the presence of a possible receptor localized in the surface of human colonic cells (HT-29, T84, and Caco-2) and absent in nonintestinal cells. The images from light microscopy (qualitative analysis) confirmed the result obtained by the quantitative analysis (Fig. 5A).
Correlation between longus production and adherence.
The presence of longus on bacteria adhering to cultured intestinal cells was investigated and analyzed by immunofluorescence microscopy (IFM). For the IFM assays, HT-29 cells in 1 ml of DMEM supplemented with 5% d-mannose were incubated with the E9034A, E9034AΔlngA, and E9034AΔlngA(pLngA) strains for 6 h at 37°C. Using rabbit antilongus antibodies and IFM, we saw that bacterial colonies produced abundant fluorescent structures forming large filaments that emerged from the E9034A and E9034AΔlngA(pLngA) strains, which is consistent with the presence of pili (Fig. 5B). Previously, the bacteria were cultured in PPLO broth at 37°C under 5% CO2 (conditions in which longus is produced) overnight, and the monolayer of epithelial cells (intestinal and nonintestinal) in 1 ml of DMEM were infected with 10 μl of a bacterial suspension at an optical density (OD600) of 1.1. The assays were performed for 6 h at 37°C in 5% CO2, and three washes with PBS were done to remove unbound bacteria. IFM showed that longus adhered to intestinal cells. It is important to indicate that longus is not produced in DMEM. The supernatants were used for the immunogold assays. The large filament structures appeared to be interconnecting bacteria mediating direct binding to the surface of human colonic cells (Fig. 5B). No fluorescence was visualized when the preimmune serum was used, confirming the specificity of the reaction (data not shown). Meanwhile, the images taken from the E9034AΔlngA strain showed no fluorescent structures, confirming the lack of the lngA gene in this strain (Fig. 5B).
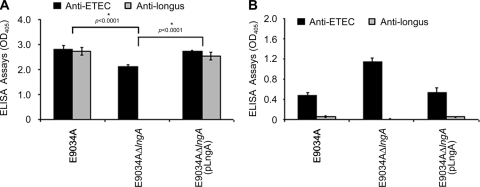
According to our preliminary data, HT-29 cells were used because they are intestinally derived cells with physiologically relevant implications in the study of intestinal pathogen adherence and because they withstand long incubation periods with the bacteria. ELISAs were performed four times to quantitatively evaluate the correlation between the levels of adherence and the production of longus. ELISAs showed different adherence levels of ETEC and the mutant strain, with a significant reduction of adherence in the E9034AΔlngA strain compared with the wild type (P < 0.0001) (Fig. 6A). These data confirmed and are consistent with the results obtained previously by plating serial dilutions and obtaining CFU counts (Fig. 4A). The ELISA plate containing the HT-29 cells and bacteria from ETEC strains was identified with anti-ETEC antibodies to quantitatively measure the levels of adherence. Previously, anti-ETEC was incubated with HT-29 cells to eliminate any antibody that could cross-react and interfere with the detection of the bacteria. In contrast, no signal was detected in HT-29 cells incubated with ETEC strains [wild-type E9034A, E9034AΔlngA, and E9034AΔlngA(pLngA)] when preimmune serum was used as a negative control (data not shown). Our data showed that the E9034A and E9034AΔlngA(pLngA) strains are attached to the surface of the HT-29 cells, with similar levels of adherence (Fig. 6A). However, when we tested the E9034AΔlngA strain, a significant reduction of 24.5% was observed in the levels of adherence in comparison to the wild-type E9034A (P < 0.0001) (Fig. 6A). Sample duplicates were used to determine the levels of longus production, with antilongus followed by an anti-rabbit IgG-alkaline phosphatase conjugate used for detection by ELISA. The result showed values of adherence for the E9034A and E9034AΔlngA(pLngA) strains of 2.73 and 2.53, respectively (Fig. 6A). As expected, no signal was detected in the E9034AΔlngA strain, confirming the lack of the lngA gene (Fig. 6A).
FIG. 6.
Immunofluorescence and quantification of bacterial adherence to HT-29 cells by ELISA. The strains were incubated with human colonic HT-29 cells for 6 h and were read by an ELISA reader. (A) After this incubation time, the nonbinding bacteria were removed, and the cells with attached bacteria were washed with PBS. The presence of the bacteria attached to the cells and the pili were read at 405 nm using antilongus and anti-ETEC antibodies, respectively. A significant reduction in the E9034ΔlngA was also observed, as described in the legend of Fig. 4. (B) We observed that not all the bacteria [E9034A, E9034AΔlngA(pLngA)] in the supernatants produced longus. Interestingly, E9034AΔlngA showed high levels of bacteria in the supernatants in comparison to E9034A and E9034AΔlngA(pLngA) strains. *, statistically significant (P < 0.0001) with respect to the values obtained without the presence of antibodies against longus.
Nonattached bacteria in HT-29 cells and contained in the supernatants were recovered and analyzed by ELISA. Bacteria not attached to human colonic cells and present in the supernatants were used for ELISAs in which the plates were coated completely with ETEC strains (E9034A and derivative strains). The number of bacteria measured with anti-ETEC antibodies by ELISA produced OD values of 0.4733 for the E9034A strain and 0.5333 for the E9034AΔlngA(pLngA) strain (Fig. 6B). Interestingly, we observed an increase in the ELISA readings in the E9034AΔlngA strain, with an OD value of 1.14 (Fig. 6B). Obviously, the increases in bacteria in the supernatants are due to colonies that do not adhere to the surface of human colonic cells. When the recovered supernatants reacted with antilongus antibodies, we observed low production of longus, with optical densities of 0.0571 and 0.0535 for strains E9034A and E9034AΔlngA(pLngA), respectively. The E9034AΔlngA strain did not react with antilongus antibodies, which confirms the presence of the mutation. These data suggest that longus-producing strains have the capacity to attach to the surface of cells.
Inhibition of adherence using antibodies against longus.
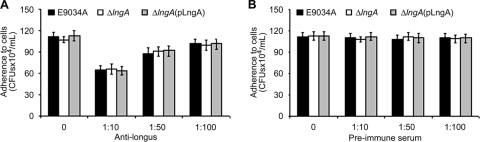
The inhibition assays with intestinal cells (HT-29) were performed using polyclonal antilongus antibodies at different dilutions (1:10, 1:50, and 1:100) against ETEC E9034A and derivative strains under conditions where longus is produced. The results obtained showed a striking dose-dependent inhibition in the E9034A and E9034AΔlngA(pLngA) strains, reaching more than 40% inhibition at a dilution of 1:10 (Fig. 7A). The inhibition levels were reduced in the E9034A and E9034AΔlngA(pLngA) strains when they were tested using a dilution of 1:100, and almost the same levels of adherence were observed as without the antilongus antibodies. However, the E9034AΔlngA strain did not show any changes at the three dilutions tested because this mutant did not assemble longus since it lacks the lngA gene (Fig. 7A). No change in the levels of inhibition was observed in the E9034A, E9034AΔlngA, and E9034AΔlngA(pLngA) strains when they were incubated with the preimmune serum (Fig. 7B). Our results indicated that differences observed at the three dilutions were due to the presence of longus antibodies in the serum employed. These data support the participation of longus in adherence of the bacteria to intestinal colonic epithelial cells.
FIG. 7.
Inhibition assays with antilongus antibodies. (A) HT-29 cells were used to perform this assay at 1:10, 1:50, and 1:100 dilutions. A dose-dependent inhibition can be seen in E9034A and E9034AΔlngA(pLngA) strains. Due to the inability of the E9034AΔlngA strain to assemble longus, no change was observed. (B) The bacteria were added to HT-29 cells in the absence (0) or the presence of three dilutions of the preimmune serum (1:10, 1:50, and 1:10). Error bars represent standard deviations.
Longus is involved in the twitching motility phenomenon.
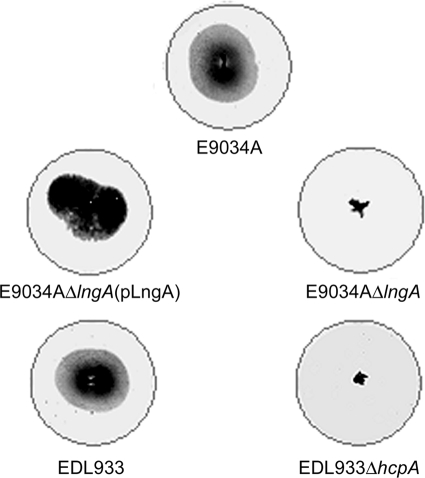
Twitching motility is a form of semisolid surface translocation capable of producing spreading zones in a wide range of bacteria, and this ability is associated with virulence and T4P production in many Gram-negative bacteria (31). The retraction and extension phenomenon is produced by the flexible pili of bacteria. Our results showed that longus produced by ETEC E9034A and E9034AΔlngA(pLngA) strains formed spreading zones. These longus-producing strains induce the formation of halos of different diameters, and show a twitching motility zone on the agar interstitial surface where the production of longus is triggered (Fig. 8). However, twitching motility was not observed in the ETEC E9034AΔlngA strain (Fig. 8). Hemorrhagic coli pili (HCP) produced by enterohemorrhagic E. coli and the hcpA mutant strain were used as positive and negative controls, respectively.
FIG. 8.
Twitching motility for ETEC E9034A and derivative strains. E9034A and E9034AΔlngA(pLngA) strains display spreading zones, showing the retraction and extension of these pili. This phenomenon is not observed in the ETEC E9034AΔlngA strain. HCP from enterohemorrhagic E. coli and the hcpA mutant were used as positive and negative controls, respectively.
DISCUSSION
Gram-negative bacteria are capable of producing distinct pilus types, which allow them to recognize different receptor moieties in the surface of epithelial cells (2). ETEC is an important cause of diarrhea among children in developing countries and among travelers to areas of ETEC endemicity. These bacteria produce a variety of surface appendages called colonization factor antigens (CFAs), coli surface antigens (CSs), and putative colonization factors (PCFs), which favor the interaction of the bacteria with the intestinal brush border cell (4, 14, 16). ETEC expresses one or both of two types of toxins, heat-stable and heat-labile enterotoxins (ST and LT, respectively), that are generally encoded or regulated by large virulence plasmids involved in diarrhea. Recent studies have described other pili produced by ETEC strains (6).
A considerable number of human isolates produce in addition to CFAs a plasmid-encoded, type IV pilus, termed longus due to its unusual length (>20 um long) (19). The longus structural gene lngA was found at frequencies similar to those reported for CFA I and CFA II (>38%) in some geographic regions. Some CFA-negative strains were also shown to have the lngA gene even though not all of the ETEC strains that produced these colonization factors produced longus (19, 21, 45). In this study, we report the production of longus from the ETEC E9034A strain in different culture media and showed that growth in PPLO broth or TSAB and contact with host cells are the best conditions for the expression of these pili. These results suggest that longus production in vivo could be modulated by the composition of the medium and other conditions such as nutrients, pH, and temperature and by the presence of CO2. Previous reports have indicated that the production of T4P in other pathogenic E. coli is under the control of environmental signals; therefore, the production of longus in ETEC E9034A is probably tightly regulated (14, 42). Some of the signaling factors involved in the induction of pilus expression have been studied for some pathogenic E. coli (13, 42). The environmental signals and molecular mechanisms involved in regulation of transcription of toxin-coregulated pilus (TCP) from V. cholerae, the bundle-forming pilus (BFP) of EPEC, and the 987P pili of ETEC have been recently studied (13, 15, 42, 48, 50). We believe that the environmental and nutritional signals that trigger longus production must resemble those found in vivo, perhaps during colonization of the small intestine in humans. Recently, studies showed that the EHEC O157:H7 strain produces HCP (T4P) after growth in Minca medium and in contact with host cells, suggesting that its expression is probably tightly regulated (24). These studies in vitro suggest that the assembly of HCP in the EHEC O157:H7 wild-type strain required and depends on environmental and culture medium conditions (56).
It is well established that ETEC strains colonize the small intestine mediated by appendices called colonization factors (39). CFA III from ETEC is a type IV pilus that has been found only in LT-producing strains of the O25:H16 and O25:H2 serotypes and has been associated with CS6 (21, 34). CFA III antigen is closely related to longus, a type IV pilus composed of a 22-kDa subunit which shares similarities at the level of N-terminal sequences. Many functions associated with pathogenicity in Gram-negative bacteria are attributed to T4P, including twitching motility, adherence to host cells, biofilm formation, DNA uptake, phage attachment, cell signaling, invasion, and evasion of the immune system (32, 56, 57).
In the present study, we explored whether longus produced by ETEC E9034A is involved in the phenomenon of adherence as a requirement for mediating the interaction between the bacteria and cultured epithelial cells (intestinal and nonintestinal cells). To achieve this aim, we analyzed in comparative and quantitative assays the ability of the wild-type strain, the lngA mutant, and the lngA mutant complemented with pLngA to adhere to various human intestinal colonic cell and nonintestinal cell lines. Our data provide evidence that clearly show that ETEC E9034A is able to assemble LngA, a type IV pilin, into bundles of pili called longus, forming a large structure when it is in contact with human intestinal cells. In addition, we observed that longus produced by ETEC strains is involved in adherence to human colonic cells (HT-29, T84, and Caco-2 cells). Other observations indicate that the lngA mutant exhibited a significant reduction (P < 0.0001) in the attachment to cultured intestinal cells, but the mutant could be restored to full adherence by complementation with the lngA gene. Interestingly, adherence assays with nonintestinal epithelial cells exhibited a significantly decreased attachment to the surface of epithelial cells in comparison with attachment to intestinal cells (P < 0.0001). However, the levels of adherence observed in the E9034A and derivative strains [E9034AΔlngA and E9034AΔlngA(pLngA)] did not show any difference. These data strongly suggest the presence of a receptor, possibly localized in the surface of the intestinal cells. The comparative studies between the percentages of adherence and the production of longus revealed that almost all bacteria attached to the intestinal cells produce these pili. Interestingly, bacteria unattached to intestinal cells and present in the supernatants showed low production of longus, confirming its role in the events of adherence to human colonic cells. According to the ultrastructural and IFM studies performed, these data strongly suggest that longus contributes in mediating the interaction between the bacteria and a particular receptor, which is apparently widely distributed among colonic intestinal cells. The production of longus by ETEC strains could act at early or late stages during the colonization process, but this requires further investigation. In this context, longus could be playing an accessory role in the colonization of the small intestine, supporting the hypothesis of the participation of these fimbriae in the first stages of the bacteria's adherence to intestinal cells. In fact, these data are a strong indication that longus participates in promoting the bacterial adherence to host intestinal epithelial cells, participating as a colonization factor in vitro.
Type IV pili in many pathogens are involved in diverse mechanisms related to colonization of host cells. Gram-negative bacteria like Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Neisseria meningitidis contribute to the formation of twitching motility, and their mutants defective in twitching motility show reduced virulence in vitro and in vivo (36, 43, 58). Recently, we reported that HCP, a type IV pilus from EHEC, is involved in the formation of twitching motility. BFP from EPEC is responsible for bacterial aggregation and microcolony formation on epithelial cells although it has not been reported to be involved in twitching motility (5, 11, 18, 27). Our data indicated that the production of longus by ETEC strains participates in the phenomenon of twitching motility, which may contribute to the pathogenicity of the bacteria. Still, it is necessary to perform other types of assays to verify the potential role of longus retraction in ETEC virulence.
Acknowledgments
We thank Maribel Martínez-Carrión for technical assistance in this work.
This work was supported by federal funds from the Hospital Infantil de México Federico Gómez (grant number HIM/2008/051).
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Acheson, D. W. 1992. Enterotoxins in acute infective diarrhoea. J. Infect. 24:225-245. [DOI] [PubMed] [Google Scholar]
- 2.Ahren, C. M., and A. M. Svennerholm. 1982. Synergistic protective effect of antibodies against Escherichia coli enterotoxin and colonization factor antigens. Infect. Immun. 38:74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, T. J. L. 1994. Neonatal diarrhoea in pigs, p. 151-170. In C. L. Gyles ed., Escherichia coli in domestic animals and humans. CAB International, Wallingford, United Kingdom.
- 4.Beachey, E. H. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J. Infect. Dis. 143:325-345. [DOI] [PubMed] [Google Scholar]
- 5.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, D., A. Husband, Z. Saldana, R. A. Nada, J. Klena, F. Qadri, and J. A. Giron. 2009. Distribution of the Escherichia coli common pilus among diverse strains of human enterotoxigenic E. coli. J. Clin. Microbiol. 47:1781-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhler, T., H. Hoschutzky, and K. Jann. 1991. Analysis of colonization factor antigen I, an adhesin of enterotoxigenic Escherichia coli O78:H11: fimbrial morphology and location of the receptor-binding site. Infect. Immun. 59:3876-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassels, F. J., and M. K. Wolf. 1995. Colonization factors of diarrheagenic E. coli and their intestinal receptors. J. Ind. Microbiol. 15:214-226. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Graaf, F. K., P. Klemm, and W. Gaastra. 1981. Purification, characterization, and partial covalent structure of Escherichia coli adhesive antigen K99. Infect. Immun. 33:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6:3427-3437. [DOI] [PubMed] [Google Scholar]
- 12.DuPont, H. L., S. B. Formal, R. B. Hornick, M. J. Snyder, J. P. Libonati, D. G. Sheahan, E. H. LaBrec, and J. P. Kalas. 1971. Pathogenesis of Escherichia coli diarrhea. N. Engl. J. Med. 285:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Ebel, F., C. Deibel, A. U. Kresse, C. A. Guzman, and T. Chakraborty. 1996. Temperature- and medium-dependent secretion of proteins by Shiga toxin-producing Escherichia coli. Infect. Immun. 64:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, R. A., and J. L. Puente. 1998. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 6:282-287. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, R. A., and D. M. Schifferli. 1997. Differential regulation of fasA and fasH expression of Escherichia coli 987P fimbriae by environmental cues. Mol. Microbiol. 25:797-809. [DOI] [PubMed] [Google Scholar]
- 16.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaastra, W., and A. M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 18.Girón, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 19.Girón, J. A., M. M. Levine, and J. B. Kaper. 1994. Longus: a long pilus ultrastructure produced by human enterotoxigenic Escherichia coli. Mol. Microbiol. 12:71-82. [DOI] [PubMed] [Google Scholar]
- 20.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 21.Girón, J. A., G. I. Viboud, V. Sperandio, O. G. Gomez-Duarte, D. R. Maneval, M. J. Albert, M. M. Levine, and J. B. Kaper. 1995. Prevalence and association of the longus pilus structural gene (lngA) with colonization factor antigens, enterotoxin types, and serotypes of enterotoxigenic Escherichia coli. Infect. Immun. 63:4195-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girón, J. A., J. G. Xu, C. R. Gonzalez, D. Hone, J. B. Kaper, and M. M. Levine. 1995. Simultaneous expression of CFA/I and CS3 colonization factor antigens of enterotoxigenic Escherichia coli by ΔaroC, ΔaroD Salmonella typhi vaccine strain CVD 908. Vaccine 13:939-946. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinee, P. A., W. H. Jansen, and C. M. Agterberg. 1976. Detection of the K99 antigen by means of agglutination and immunoelectrophoresis in Escherichia coli isolates from calves and its correlation with entertoxigenicity. Infect. Immun. 13:1369-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, Z. D., J. J. Mathewson, C. D. Ericsson, A. M. Svennerholm, C. Pulido, and H. L. DuPont. 2000. Characterization of enterotoxigenic Escherichia coli strains in patients with travelers' diarrhea acquired in Guadalajara, Mexico, 1992-1997. J. Infect. Dis. 181:779-782. [DOI] [PubMed] [Google Scholar]
- 26.Knutton, S., M. M. McConnell, B. Rowe, and A. S. McNeish. 1989. Adhesion and ultrastructural properties of human enterotoxigenic Escherichia coli producing colonization factor antigens III and IV. Infect. Immun. 57:3364-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutton, S., R. K. Shaw, R. P. Anantha, M. S. Donnenberg, and A. A. Zorgani. 1999. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33:499-509. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Levine, M. M., C. Ferreccio, V. Prado, M. Cayazzo, P. Abrego, J. Martinez, L. Maggi, M. M. Baldini, W. Martin, D. Maneval, et al. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 30.Levine, M. M., P. Ristaino, G. Marley, C. Smyth, S. Knutton, E. Boedeker, R. Black, C. Young, M. L. Clements, C. Cheney, et al. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect. Immun. 44:409-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. F., S. Poole, K. Nishio, K. Jang, F. Rasulova, A. McVeigh, S. J. Savarino, D. Xia, and E. Bullitt. 2009. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:10793-10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 33.McConnell, M. M., M. L. Hibberd, M. E. Penny, S. M. Scotland, T. Cheasty, and B. Rowe. 1991. Surveys of human enterotoxigenic Escherichia coli from three different geographical areas for possible colonization factors. Epidemiol. Infect. 106:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McConnell, M. M., and B. Rowe. 1989. Prevalence of the putative colonization factors CFA/III and PCFO159:H4 in enterotoxigenic Escherichia coli. J. Infect. Dis. 159:582-586. [DOI] [PubMed] [Google Scholar]
- 35.McMichael, J. C., and J. T. Ou. 1979. Structure of common pili from Escherichia coli. J. Bacteriol. 138:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, B., and P. Z. Fekete. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 295:443-454. [DOI] [PubMed] [Google Scholar]
- 38.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura, L. S., J. A. Giron, S. L. Nunes, and B. E. Guth. 2002. Prevalence of enterotoxigenic Escherichia coli strains harboring the longus pilus gene in Brazil. J. Clin. Microbiol. 40:2606-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paniagua, M., F. Espinoza, M. Ringman, E. Reizenstein, A. M. Svennerholm, and H. Hallander. 1997. Analysis of incidence of infection with enterotoxigenic Escherichia coli in a prospective cohort study of infant diarrhea in Nicaragua. J. Clin. Microbiol. 35:1404-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peruski, L. F., Jr., B. A. Kay, R. A. El-Yazeed, S. H. El-Etr, A. Cravioto, T. F. Wierzba, M. Rao, N. El-Ghorab, H. Shaheen, S. B. Khalil, K. Kamal, M. O. Wasfy, A. M. Svennerholm, J. D. Clemens, and S. J. Savarino. 1999. Phenotypic diversity of enterotoxigenic Escherichia coli strains from a community-based study of pediatric diarrhea in periurban Egypt. J. Clin. Microbiol. 37:2974-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 43.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A. 96:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qadri, F., S. K. Das, A. S. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack, and A. M. Svennerholm. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 38:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qadri, F., J. A. Giron, A. Helander, Y. A. Begum, M. Asaduzzaman, J. Xicohtencatl-Cortes, E. Negrete, and M. J. Albert. 2000. Human antibody response to longus type IV pilus and study of its prevalence among enterotoxigenic Escherichia coli in Bangladesh by using monoclonal antibodies. J. Infect. Dis. 181:2071-2074. [DOI] [PubMed] [Google Scholar]
- 46.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rendon, M. A., Z. Saldana, A. L. Erdem, V. Monteiro-Neto, A. Vazquez, J. B. Kaper, J. L. Puente, and J. A. Giron. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sjoberg, P. O., M. Lindahl, J. Porath, and T. Wadstrom. 1988. Purification and characterization of CS2, a sialic acid-specific haemagglutinin of enterotoxigenic Escherichia coli. Biochem. J. 255:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 51.Sommerfelt, H., H. Steinsland, H. M. Grewal, G. I. Viboud, N. Bhandari, W. Gaastra, A. M. Svennerholm, and M. K. Bhan. 1996. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J. Infect. Dis. 174:768-776. [DOI] [PubMed] [Google Scholar]
- 52.Svennerholm, A. M., J. Holmgren, and D. A. Sack. 1989. Development of oral vaccines against enterotoxinogenic Escherichia coli diarrhoea. Vaccine 7:196-198. [DOI] [PubMed] [Google Scholar]
- 53.Taniguchi, T., M. Arita, M. Sato, K. Yamamoto, T. Miwatani, and T. Honda. 1994. Evidence that the N-terminal amino acid sequence of pilus colonization factor antigen III produced by human enterotoxigenic Escherichia coli is similar to that of TcpA pilin of Vibrio cholerae. J. Infect. Dis. 170:1049-1050. [DOI] [PubMed] [Google Scholar]
- 54.Taniguchi, T., Y. Fujino, K. Yamamoto, T. Miwatani, and T. Honda. 1995. Sequencing of the gene encoding the major pilin of pilus colonization factor antigen III (CFA/III) of human enterotoxigenic Escherichia coli and evidence that CFA/III is related to type IV pili. Infect. Immun. 63:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viboud, G. I., M. J. Jouve, N. Binsztein, M. Vergara, M. Rivas, M. Quiroga, and A. M. Svennerholm. 1999. Prospective cohort study of enterotoxigenic Escherichia coli infections in Argentinean children. J. Clin. Microbiol. 37:2829-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xicohtencatl-Cortes, J., V. Monteiro-Neto, M. A. Ledesma, D. M. Jordan, O. Francetic, J. B. Kaper, J. L. Puente, and J. A. Giron. 2007. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J. Clin. Invest. 117:3519-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xicohtencatl-Cortes, J., V. Monteiro-Neto, Z. Saldana, M. A. Ledesma, J. L. Puente, and J. A. Giron. 2009. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 191:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zolfaghar, I., D. J. Evans, and S. M. Fleiszig. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71:5389-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]