Abstract
Primase, encoded by dnaG in bacteria, is a specialized DNA-dependent RNA polymerase that synthesizes RNA primers de novo for elongation by DNA polymerase. Genome sequence analysis has revealed two distantly related dnaG genes, TtdnaG and TtdnaG2, in the thermophilic bacterium Thermoanaerobacter tengcongensis. Both TtDnaG (600 amino acids) and TtDnaG2 (358 amino acids) exhibit primase activities in vitro at a wide range of temperatures. Interestingly, the template recognition specificities of these two primases are quite distinctive. When trinucleotide-specific templates were tested, TtDnaG initiated RNA primer synthesis efficiently only on templates containing the trinucleotide 5′-CCC-3′, not on the other 63 possible trinucleotides. When the 5′-CCC-3′ sequence was flanked by additional cytosines or guanines, the initiation efficiency of TtDnaG increased remarkably. Significantly, TtDnaG could specifically and efficiently initiate RNA primer synthesis on a limited set of tetranucleotides composed entirely of cytosines and guanines, indicating that TtDnaG initiated RNA primer synthesis more preferably on GC-containing tetranucleotides. In contrast, it seemed that TtDnaG2 had no specific initiation nucleotides, as it could efficiently initiate RNA primer synthesis on all templates tested. The DNA binding affinity of TtDnaG2 was usually 10-fold higher than that of TtDnaG, which might correlate with its high activity but low template specificity. These distinct priming activities and specificities of TtDnaG and TtDnaG2 might shed new light on the diversity in the structure and function of the primases.
Primase is the single-stranded-DNA (ssDNA)-dependent RNA polymerase that synthesizes RNA primers to be extended by DNA polymerase in DNA replication (14, 38). The primase functions at least once during leading-strand DNA synthesis and repeatedly for Okazaki fragment synthesis (17, 48). The bacterial primase gene, named dnaG, is usually colocated in a macromolecular synthesis (MMS) operon, which encodes proteins involved in the initiation of translation, DNA replication, and transcription (6, 11, 49, 50, 52). DnaG primase usually contains three functional domains (37, 46): the N-terminal zinc-binding domain (ZBD) that mediates interaction with the ssDNA template (24), the central TOPRIM catalytic domain responsible for primer synthesis (RPD) (1), and the C-terminal domain that interacts with DNA helicase (DnaB-ID) (30, 44, 46). High-resolution structures of these domains have been revealed for Geobacillus stearothermophilus and Escherichia coli (21, 31, 33, 36, 39).
At the initiation of bacterial DNA replication, primase is recruited to the replication fork by an association with the replicative helicase DnaB (9), and the priming activity of DnaG is usually regulated by DnaB (27, 45). In E. coli, DnaB helicase stimulates primer synthesis activity, decreases the primase initiation specificity, and prevents overlong primers from forming (3, 19). E. coli DnaB helicase is a ring-shaped hexameric molecule with a central channel (35). The interaction between DnaB and DnaG allows the helicase to serve as a mobile docking station, which increases the local concentration of ssDNA template relative to primase, and hence stimulates the activity of primase (9, 29). This helicase stimulation is cooperative at low helicase concentrations and inhibitory at high helicase concentrations in vitro (19, 29). As an important component of the replisome, DnaG plays key roles in coordination of leading- and lagging-strand DNA synthesis (53) and coupling of DNA replication to chromosome partitioning and DNA damage response (15, 28), and it is also indispensable in reactivation of blocked replication forks (16).
Primases from phages and bacteria that have been studied so far require specific nucleotide sequences to initiate synthesis of RNA primers (13). For phages, the T7 primase predominantly recognizes sequences containing 5′-(G/T)GGTC-3′ and less frequently recognizes sequences with 5′-(A/C)GGTC-3′ and 5′-NTGTC-3′ (41). T4 primase recognizes 5′-GTT-3′ and 5′-GCT-3′ (8), while SP6 primase specifically recognizes 5′-GCA-3′ (47). For bacteria, the E. coli primase synthesizes RNA primers predominantly on templates containing a 5′-CTG-3′ sequence (3, 55). Similarly, the primases from Staphylococcus aureus and G. stearothermophilus predominantly initiate RNA synthesis on 5′-CTA-3′ and 5′-TTA-3′ sequences (22, 43). Interestingly, the specific initiation sequences for the primase from the hyperthermophilic bacterium Aquifex aeolicus are comprised of only cytosines and/or guanines, with the preferred initiation trinucleotide sequence 5′-CCC-3′ and two other sequences, 5′-GCC-3′ and 5′-CGC-3′, to a much lesser degree (26).
Thermoanaerobacter tengcongensis is an anaerobic, rod-shaped, low-GC (37.6%), thermophilic bacterium which was isolated from a freshwater hot spring in China and grows optimally at 75°C (54). The genome sequence of T. tengcongensis has been determined, showing that 86.7% of its genes are carried on the leading strand of DNA replication (2). It is interesting that two dnaG genes, named T. tengcongensis dnaG (TtdnaG [TTE1756]) and TtdnaG2 (TTE2018), are annotated in the chromosome and likely encode two primases with different sizes. This phenomenon of more than one dnaG gene existing simultaneously in one bacterial chromosome appears to be common in the Firmicutes but has yet to be studied. In the present study, we report the distinct enzyme activities and template recognition specificities of TtDnaG and TtDnaG2, which may play different roles in T. tengcongensis.
MATERIALS AND METHODS
Bacteria, plasmids, and oligonucleotides.
E. coli DH5α served as the host for cloning experiments, while E. coli BL21(DE3) was used for overproduction of recombinant proteins. E. coli strains were cultured at 37°C in Luria-Bertani (LB) medium with shaking. Kanamycin (50 μg ml−1) was supplied when necessary. T. tengcongensis was grown in modified MB medium at 75°C without shaking (2, 54).
The expression vector pET28a was obtained from Novagen (Madison, WI). The oligodeoxyribonucleotides used as templates for TtDnaG and TtDnaG2 were synthesized by Invitrogen or Sangon (Beijing, China). Each template was purified by urea-PAGE and high-performance liquid chromatography (HPLC) and qualified by mass spectrometry. The 3′-OH termini were blocked with a C3 spacer (26) when necessary.
Sequence alignment and phylogenetic analysis of DnaG proteins.
Protein sequence homology analysis was performed using the BLAST service (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the GeneDoc program (http://www.nrbsc.org/gfx/genedoc/index.html). The phylogenetic tree for DnaG proteins was constructed using the neighbor-joining method (34) with MEGA4 (42). The topology of the phylogenetic tree obtained was evaluated by bootstrap analysis with 1,000 replications.
Construction of expression plasmids.
The open reading frames of TtdnaG and TtdnaG2 were amplified from T. tengcongensis with the primer pairs ATGGATCCGCTTATACGAGAGAGTCC/TCCTCGAGCTATGGGTTTCTCTCTTTC and ATGGATCCCACAGAGATTTGGACATT/TCCTCGAGTCACACAACTTCCTTTTG (BamHI and XhoI sites are underlined), respectively. The PCR products were digested with BamHI/XhoI and then cloned into the same sites of the vector pET28a, resulting in the expression plasmids p28TtDnaG and p28TtDnaG2, respectively. For all of these constructs, the PCR-amplified sequences were verified by DNA sequencing.
Protein expression and purification.
To overexpress and purify TtDnaG and TtDnaG2, E. coli BL21(DE3) cells harboring p28TtDnaG and p28TtDnaG2 were cultivated in LB medium containing kanamycin to reach an optical density at 600 nm (OD600) of 0.4 to 0.6, at which point IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM. The induced cultures were allowed to grow for an additional 3 h. The cells were harvested by centrifugation, resuspended in lysis buffer containing 0.3 M NaCl, 10 mM imidazole, and 50 mM sodium phosphate buffer (NaH2PO4/Na2HPO4, pH 8.0), and then lysed by ultrasonication. The lysates were centrifuged at 12,000 rpm for 30 min, and supernatants were heated at 70°C for 30 min prior to an additional centrifugation step. The His-tagged recombinant proteins were then purified by use of a Ni2+-nitrilotriacetatic acid-agarose column (Novagen), followed by gel filtration chromatography through a Superdex 200 10/300 GL column (GE Healthcare). Proteins were concentrated with Amicon Ultrafra-15 concentrators (Millipore) when necessary. The purified proteins were examined by SDS-PAGE, and the protein concentrations were determined by using a bicinchoninic acid (BCA) protein concentration assay kit (Pierce).
RNA primer synthesis assay.
The typical 25-μl reaction mixture contained 50 mM HEPES (pH 7.5), 100 mM potassium glutamate, 10 mM dithiothreitol, 5 mM magnesium acetate, 100 μM (each) ATP, CTP, and GTP in a mixture, 50 μM UTP, 0.5 μCi [α-32P]UTP (3,000 Ci/mmol), 250 ng M13mp18 ssDNA or 1 μM synthetic ssDNA template, and the specific primase (1 μM or the concentration indicated). Incubations were performed at 70°C (unless indicated otherwise) for 30 min. Reactions were stopped by the addition of 50 μl of 3 M sodium acetate (pH 5.2). The RNA products were then precipitated overnight at −70°C with 100% cold ethanol in the presence of 40 μg glycogen. The precipitates were washed with 75% cold ethanol and dissolved in 20 μl loading buffer (98% formamide, 0.025% xylene cyanol FF, 0.025% bromphenol blue, 10 mM EDTA, pH 8.0). Samples were heated at 95°C for 3 min, and 5 μl was then loaded onto a 20% denaturing polyacrylamide sequencing gel containing 7 M urea and 1× TBE (Tris-borate-EDTA) buffer. After electrophoresis, the gels were analyzed by autoradiography with X-ray film and/or by phosphorimaging with a Storm Phosphor-Imager and ImageQuant software (Amersham). All experiments were performed at least in triplicate.
SPR assay.
The binding affinities of TtDnaG and TtDnaG2 for ssDNA templates were detected by surface plasmon resonance (SPR) assays on a BIAcore 3000 system (BIAcore AB, Uppsala, Sweden) as previously described (32), with minor modifications. An SA streptavidin-coated sensor chip was used, which was conditioned 3 to 5 times with 1-min injections of 1 M NaCl and 50 mM NaOH (at 10 μl/min) until a stable baseline was observed. The cells were then washed twice with 0.05% SDS in buffer I (10 mM HEPES, pH 7.5, 200 mM potassium glutamate, 10 mM magnesium acetate, 0.005% BIAcore surfactant P20) for 3 min. Afterwards, specific biotinylated single-stranded oligonucleotides were diluted to 100 pM in buffer II (10 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA) and were immobilized until ∼150 resonance units (RU) were reached through manual injections with a flow rate of 5 μl/min. For detection of the interaction, TtDnaG or TtDnaG2 was diluted from 0 to 800 nM with buffer I containing 33 ng/μl poly(dI-dC) and injected with the K-inject command. Buffer I was used as the running buffer throughout all binding and dissociation analyses, at a flow rate of 30 μl/min. At the end of each cycle, 0.05% SDS was used as regeneration buffer to remove the bound protein. For data analysis, the BIAevaluation 3.0 program (BIAcore AB, Uppsala, Sweden) was used.
RESULTS
Sequence and phylogenetic analysis of TtDnaG and TtDnaG2.
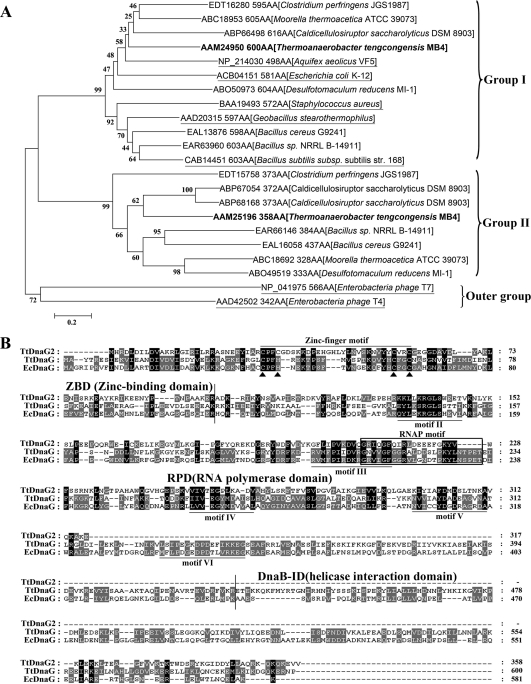
T. tengcongensis harbors two dnaG genes in the chromosome. The TtdnaG gene is located in an MMS operon with the organization X-dnaG-rpoD, as observed in many other bacteria (6, 11, 40, 49, 50, 52), while TtdnaG2 is located in an unconserved position. This phenomenon of one bacterium encoding more than one DnaG primase seems quite common in the phylum Firmicutes, to which T. tengcongensis belongs. Among the 184 bacteria in this phylum whose genomes have been sequenced, about 35% have at least two dnaG genes per chromosome (data not shown). Phylogenetic analysis of these multiple DnaGs from several selected bacteria along with some other known primases revealed two main DnaG groups (I and II). Both groups were only distantly related to the primases of phages T4 and T7 (Fig. 1A). The DnaGs in group I, including TtDnaG (600 amino acids [aa]), are likely the actual replicative primases. The bacterial DnaGs that are usually the only primase in the chromosome are all clustered in this group. In contrast, the DnaGs in group II, including TtDnaG2 (358 aa), are usually smaller, and none have been characterized. Therefore, comparative studies of the two DnaGs of T. tengcongensis might provide novel knowledge on the existence of diverse dnaG genes in a bacterium.
FIG. 1.
(A) Phylogenetic analysis of DnaG proteins from selected bacteria whose genomes encode more than one DnaG homolog, along with several well-known bacterial and phage primases (underlined). TtDnaG and TtDnaG2 from T. tengcongensis are shown in bold. The GenBank accession number and protein size (AA) of each DnaG are given, followed by the microorganism name in brackets. The tree was obtained by using the neighbor-joining algorithm of MEGA software, version 4.0 (34, 42). The numbers next to the nodes indicate the bootstrap values based on 1,000 replications (expressed as percentages). Bar = 0.2 substitution per site. (B) Protein sequence alignment of homologous DnaG proteins. EcDnaG, E. coli DnaG. The major domains and motifs are indicated with lines or a box, while the key cysteine and histidine residues in the zinc fingers are indicated by triangles. Identical amino acids are shown with a black background, while two similar amino acids are shown with a gray background.
Based on structural and functional information on E. coli DnaG (EcDnaG) (18, 21), the corresponding domains and motifs of TtDnaG and TtDnaG2 were deduced (Fig. 1B). The N-terminal zinc finger motifs, although showing distinct features, could be found in both proteins. As observed in EcDnaG, TtDnaG has a Cys3His1 zinc finger motif with a 17-residue spacer. In contrast, TtDnaG2 has a Cys4 zinc finger motif with a longer spacer of 22 residues, resembling those in phage (T4 or T7) primases. Five other important motifs, as well as the RNAP motif, were also predicted according to the EcDnaG sequence (18, 21) (Fig. 1B). However, a large C-terminal fragment, including motif VI and most of the deduced helicase interaction domain, was absent in TtDnaG2. These results indicate that TtDnaG is more conserved with the bacterial replicative primases than TtDnaG2.
Priming activities of TtDnaG and TtDnaG2.
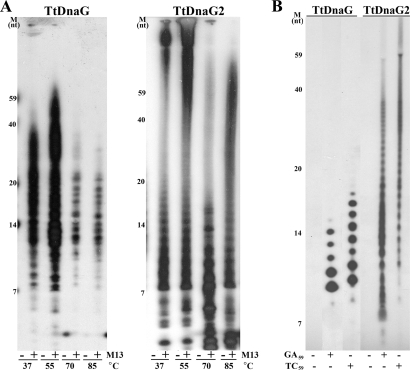
To detect and compare the priming activities of TtDnaG and TtDnaG2, the TtdnaG and TtdnaG2 genes were cloned into pET28a and overexpressed in E. coli. The purified recombinant TtDnaG and TtDnaG2 proteins were then subjected to RNA primer synthesis analysis at different temperatures ranging from 37 to 85°C, with M13mp18 ssDNA as the template. The results indicated that both TtDnaG and TtDnaG2 could synthesize RNA polymers on the M13mp18 ssDNA template at a wide range of temperatures (Fig. 2A). Under these in vitro conditions, TtDnaG synthesized RNA primers with lengths corresponding to 7 to 59 nucleotides (nt) (Fig. 2A, left panel), while a much wider range of RNA products were synthesized by TtDnaG2 (Fig. 2A, right panel). Since 70°C is close to the optimal growth temperature of T. tengcongensis, 75°C, it was used as the standard priming assay temperature in this study.
FIG. 2.
(A) RNA polymers synthesized ([α-32P]UTP was incorporated) by TtDnaG (left) and TtDnaG2 (right) at 37 to 85°C, with (+) or without (−) M13 template. (B) RNA primers synthesized ([α-32P]UTP was incorporated) by TtDnaG and TtDnaG2 at 70°C, with template GA59 or TC59. Note that there was no RNA synthesis in the control reactions without ssDNA template (A and B). Numbers at the left of each panel indicate the sizes, in nucleotides (nt), of the γ-32P-labeled ssDNA markers.
When several random templates were assayed, two 59-nt ssDNAs with complementary sequences, named GA59 and TC59 (Table 1), were found to support RNA primer syntheses by both TtDnaG and TtDnaG2 (Fig. 2B). Similar to the case with the M13mp18 ssDNA template (Fig. 2A), the patterns of the RNA primers generated by the two primases from these oligonucleotides were also different. The RNA products of TtDnaG were distributed from 10 to 16 nt, but those of TtDnaG2 were distributed over a much wider range, from smaller than 7 nt to longer than 59 nt, even longer than the lengths of the templates (59 nt) (Fig. 2B). When the templates were omitted in the reaction mixtures, no RNA products were generated by either TtDnaG or TtDnaG2 (Fig. 2), indicating that the RNA priming activities of both TtDnaG and TtDnaG2 were strictly template dependent.
TABLE 1.
Important templates used in this study
| Template | Sequence (5′ to 3′)a | Concentration |
|---|---|---|
| GA59 | GTTTTTTTAACTGGATAGCGGCAAAAAATTTTTATTTTATCCAGGTAATTGTGACTTTA | 1 μM |
| TC59 | TAAAGTCACAATTACCTGGATAAAATAAAAATTTTTTGCCGCTATCCAGTTAAAAAAAC | 1 μM |
| TC10 | ATTTTTTGCCGCTATCCAGT | 1 μM |
| TC10-9 | ATTTTTTGCCGC | 100 pM (SPR assay) |
| TC-M1 | ATTTTTTGTCGC | 100 pM (SPR assay) |
| TC-M2 | ATTTTTTGCCTC | 100 pM (SPR assay) |
The specific initiation sites for TtDnaG on the templates are underlined, while the possible nonspecific initiation sites are shown in italics. The mutated nucleotides in the initiation sites for TC-M1 and TC-M2 are shown in bold.
Specific initiation sites of TtDnaG on templates derived from GA59 and TC59.
Since template recognition specificity is a common feature of primases from bacteria and phages, so far as we know (8, 13, 22, 26, 41, 47, 55), it would be interesting to investigate whether TtDnaG and TtDnaG2 require specific recognition sequences to synthesize RNA primers de novo.
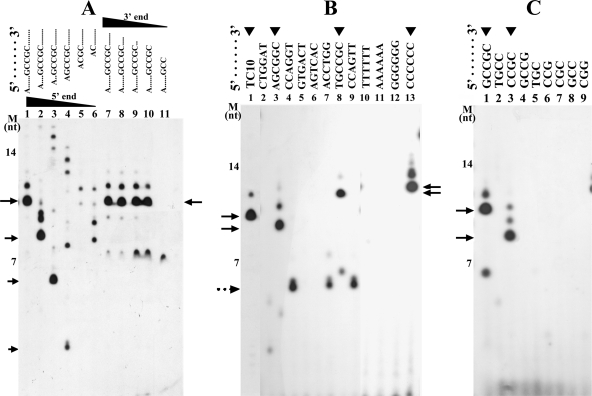
Since both GA59 and TC59 (Fig. 2B) could be utilized efficiently by TtDnaG and TtDnaG2, a series of ssDNA templates derived from both GA59 and TC59, with lengths of ∼20 nt, were selected as the test templates to search for possible specific initiation sequences of the two TtDnaGs. While mainly nonspecific RNA polymers were synthesized by TtDnaG2, fortunately a predominant and specific RNA primer product (∼10 nt) was generated by TtDnaG on the template TC10 (Table 1), which was derived from TC59 (Fig. 3A and B, lanes 1).
FIG. 3.
RNA primer syntheses by TtDnaG on templates with sequences derived from GA59 and TC59. The lanes with RNA primers synthesized are indicated by arrowheads. The main RNA products are indicated with arrows, and the assumed nonspecific or aborted RNA products (with unexpectedly short lengths) are indicated with a dotted arrow. (A) RNA primer syntheses by TtDnaG on TC10 (lane 1) and its derived templates with nucleotide deletions (2 nt deleted each time) from the 5′ or 3′ terminus (lanes 2 to 12). (B) RNA primer syntheses by TtDnaG on TC10 (lane1) and on templates with the sequence 5′-A6XA6-3′, in which the X represents a GC-rich region from GA59 or TC59 (lanes 2 to 9) or a homohexamer of each nucleotide (lanes 10 to 13), as indicated at the top of the figure. (C) RNA primer syntheses by TtDnaG on templates with the sequence 5′-AATA5XA6-3′ (lanes 1 to 9), with X representing specific sequences derived from 5′-TGCCGC-3′, as indicated at the top of the figure. M, ssDNA markers as in Fig. 2.
To identify the specific initiation sites of TtDnaG in TC10, additional templates derived from TC10 by sequence deletion (2 nt deleted each time) from its 5′ or 3′ terminus were investigated further. Deletions from the 5′ terminus of TC10 led to progressively shorter RNA products (Fig. 3A, lanes 2 to 4). Further deletions from the 5′ terminus up to the sequence 5′-(A)CGCTATCCAGT-3′ abolished the ability to support primer synthesis by TtDnaG (Fig. 3A, lanes 5 and 6). In contrast, when the sequences of TC10 were deleted from the 3′ terminus, the sizes of the RNA polymers did not change at first (Fig. 3A, lanes 7 to 10). When the 3′-terminal deletion occurred from 5′-ATTTTTTGCCGC-3′ to 5′-ATTTTTTGCC-3′, the specific and predominant RNA primer disappeared immediately (Fig. 3A, lane 11). These results suggest that the important recognition sequence of TtDnaG on TC10 lies within the sequence 5′-GCCGC-3′, the GC-rich region.
There were 8 GC-rich regions in total in GA59 and TC59 (Table 1), and we relocated these GC-rich sequences (indicated by “X”) into templates with the sequence 5′-A6XA6-3′ to test if there were additional initiation sequences for TtDnaG. We also tested 4 other templates in which X was a homohexamer of one kind of nucleotide (T, A, G, or C) (Fig. 3B). We selected dA6 for the flanking sequences because TtDnaG could not utilize oligo(dA)18 to synthesize any RNA primers (Fig. 3B, line 11), and thus we could neglect the effect of the flanking dA6 on the specific sequences tested. From the results shown in Fig. 3B, we concluded that TtDnaG could recognize the sequences 5′-AGCGGC-3′, 5′-TGCCGC-3′, and 5′-CCCCCC-3′ (Fig. 3B, lanes 3, 8, and 13). The 5′-AGCGGC-3′ sequence might be the initiation site for TtDnaG on GA59 (Fig. 2B; Table 1), while the 5′-TGCCGC-3′ sequence pointed to the initiation sites for TtDnaG on TC59 (Fig. 2B; Table 1) and TC10 (Fig. 3A; Table 1), and 5′-CCCCCC-3′ could be an additional initiation site for TtDnaG. Notably, several abortive RNA products with smaller-than-expected lengths were synthesized by TtDnaG from three other GC-rich sequences (Fig. 3B, lanes 4, 7, and 9), which are discussed later.
To further determine the specific initiation nucleotides of TtDnaG within the three identified sequences, we relocated the possible trinucleotides, tetranucleotides, and pentanucleotides from 5′-TGCCGC-3′ (with AGCGGC and CCCCCC to be investigated later) into the supporting template sequence 5′-AATA5XA6-3′, where X was TGC, CCG, CGC, GCC, CGG, TGCC, CCGC, GCCG, or GCCGC. Unexpectedly, none of the five templates containing a trinucleotide (TGC, GCC, CCG, CGC, or CGG) could support RNA primer synthesis (Fig. 3C, lanes 5 to 9). However, a template containing the tetranucleotide 5′-CCGC-3′ (Fig. 3C, lane 3), but not 5′-TGCC-3′ or 5′-GCCG-3′ (Fig. 3C, lanes 2 and 4), could support RNA primer synthesis. Consequently, a template containing 5′-GCCGC-3′ could also be used for initiation, as it contains the same specific tetranucleotide (Fig. 3C, lane 1). This is quite interesting, as unlike the specific recognition of trinucleotides reported so far for many other bacterial primases, TtDnaG recognizes a specific tetranucleotide (5′-CCGC-3′) composed entirely of cytosines and guanines.
Investigation of initiation specificity of TtDnaG by use of templates containing all 64 trinucleotides.
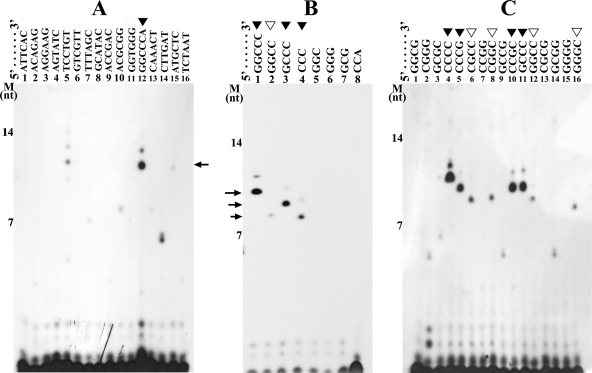
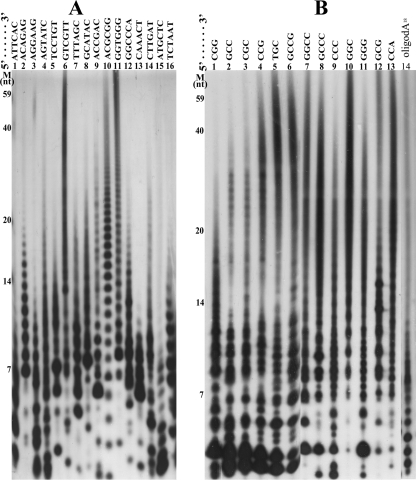
To determine whether TtDnaG could initiate RNA primer synthesis specifically from trinucleotides, we located all 64 potential trinucleotides into 16 different templates with the sequence 5′-A6XA6-3′. In each template, the X represents a carefully designed hexanucleotide sequence containing 4 different combinations of trinucleotides derived from TEOS1 and TEOS2 (26). With these 16 templates, we identified that the template containing 5′-GGCCCA-3′ could be used specifically for initiation by TtDnaG, as a predominant and specific RNA product with the expected length was synthesized (Fig. 4A, lane 12). Similarly, when the possible trinucleotides, tetranucleotides, and pentanucleotides in 5′-GGCCCA-3′ were investigated further, it was revealed that TtDnaG could initiate RNA primer synthesis on templates containing the trinucleotide 5′-CCC-3′ (Fig. 4B, lane 4) but not on templates containing any other trinucleotides (Fig. 4B, lanes 5 to 8). Interestingly, templates with 5′-GGCCC-3′ and 5′-GCCC-3′, which contain 5′-CCC-3′ plus additional guanines, were more efficient than 5′-CCC-3′ in supporting RNA primer synthesis of TtDnaG (Fig. 4B, lanes 1, 3, and 4).
FIG. 4.
RNA primer syntheses by TtDnaG on different templates. The lanes with RNA primers synthesized are indicated by closed arrowheads (more significant) or open arrowheads (less significant), and the main RNA products are indicated with arrows. (A) RNA primers synthesized by TtDnaG on 16 templates with the sequence 5′-A6XA6-3′ (3′-OH terminus blocked) (lanes 1 to 16), with the exact sequences of X shown at the top of the figure, containing all 64 potential trinucleotides in total. (B) RNA primer syntheses by TtDnaG on templates with the sequence 5′-AATA5XA6-3′ (lanes 1 to 8), with X representing specific sequences derived from 5′-GGCCCA-3′, as shown at the top of the figure. (C) RNA primer syntheses by TtDnaG on 16 templates with the sequence 5′-AATA5XA6-3′ (lanes 1 to 16), with the exact sequences of X shown at the top of the figure, containing all 16 tetranucleotides composed entirely of cytosines and/or guanines. M, ssDNA markers as in Fig. 2.
It is noteworthy that faint RNA products were also synthesized when a template containing 5′-TCCTGT-3′ was used (Fig. 4A, lane 5). Further investigation revealed that no trinucleotides, tetranucleotides, or pentanucleotides within this hexanucleotide were able to support RNA primer syntheses by TtDnaG (data not shown). We thus supposed that these faint RNA products might be due to nonspecific synthesis by TtDnaG. Therefore, detection of the initiation specificity of TtDnaG with all 64 trinucleotides showed that only the trinucleotide 5′-CCC-3′, comprised entirely of cytosines, could be used efficiently for initiation by TtDnaG.
Investigation of initiation specificity of TtDnaG by use of templates containing all 16 GC-only tetranucleotides.
Since the specific initiation sequences of TtDnaG revealed thus far, i.e., the trinucleotide 5′-CCC-3′ (Fig. 4B, lane 4) and the tetranucleotide 5′-CCGC-3′ (Fig. 3C, lane 3), are comprised entirely of cytosines and/or guanines, it would be interesting to investigate whether other GC-only tetranucleotides could support RNA primer syntheses by TtDnaG. For this purpose, templates containing all 16 corresponding tetranucleotides were investigated. In addition to the two previously identified tetranucleotides, 5′-CCGC-3′ and 5′-GCCC-3′ (Fig. 4C, lanes 10 and 11), the tetranucleotides 5′-CCCC-3′ and 5′-CCCG-3′, containing 5′-CCC-3′, could also be used efficiently by TtDnaG (Fig. 4C, lanes 4 and 5). Four other tetranucleotides, 5′-CGCC-3′, 5′-CGGC-3′, 5′-GGGC-3′ (Fig. 4C, lanes 6, 8, and 16), and 5′-GGCC-3′ (Fig. 4C, lane 12, and B, lane 2), were found to be used for initiation by TtDnaG, to a much lesser degree. Obviously, the specific initiation nucleotides in 5′-AGCGGC-3′ and 5′-CCCCCC-3′ (Fig. 3B, lanes 3 and 13) were the tetranucleotide 5′-CGGC-3′ (Fig. 4C, lane 8) and the trinucleotide 5′-CCC-3′ (Fig. 4B, lane 4), respectively. Similarly, templates (5′-AGCGGC-3′ and 5′-CCCCCC-3′) with guanines or cytosines flanking the initiation nucleotides 5′-CGGC-3′ or 5′-CCC-3′ could be used more efficiently.
Therefore, the specific initiation sequences of TtDnaG are mainly several tetranucleotides (Fig. 4C) and only one trinucleotide sequence (5′-CCC-3′) (Fig. 4B), and these sequences are comprised entirely of cytosines and/or guanines. This is quite novel and might be a thermoadaptation of this primase from a thermophilic bacterium.
No specific initiation sequence for TtDnaG2.
At the same time as the above analyses, all templates tested above were also subjected to RNA primer synthesis analysis with TtDnaG2. Several typical results for selected templates, containing all 64 trinucleotide sequences or GC-rich trinucleotides or tetranucleotides, as well as oligo(dA)18, are shown in Fig. 5. All templates tested could efficiently support RNA polymer synthesis by TtDnaG2. Thus, unusually, TtDnaG2 has no apparent template specificity for RNA primer synthesis.
FIG. 5.
RNA primer syntheses by TtDnaG2 on different templates. (A) RNA primers synthesized by TtDnaG2 on 16 templates with the sequence dA6XA6 (3′-OH terminus blocked) (lanes 1 to 16), which were the same as those in Fig. 4A. (B) RNA primers synthesized by TtDnaG2 on templates with the sequence 5′-AATA5XA6-3′ (lanes 1 to 13), with the exact sequences of X, including 3 tetranucleotides and 10 trinucleotides, shown at the top of the figure (lanes 1 to 13). Oligo(dA)18 was also included (lane 14). M, ssDNA markers as in Fig. 2.
Notably, apart from the short RNA products, TtDnaG2 tended to synthesize RNA polymers that were even longer than the templates (Fig. 2B and Fig. 5). These longer products were not likely extended from the 3′-OH termini of the DNA templates, as the longer RNA primers (which could be degraded by RNase) were still generated by TtDnaG2 when the 3′-OH termini of the templates were blocked with a C3 spacer (Fig. 5A). The exact reason remains to be investigated and is discussed further below.
Comparison of template binding affinities of TtDnaG and TtDnaG2.
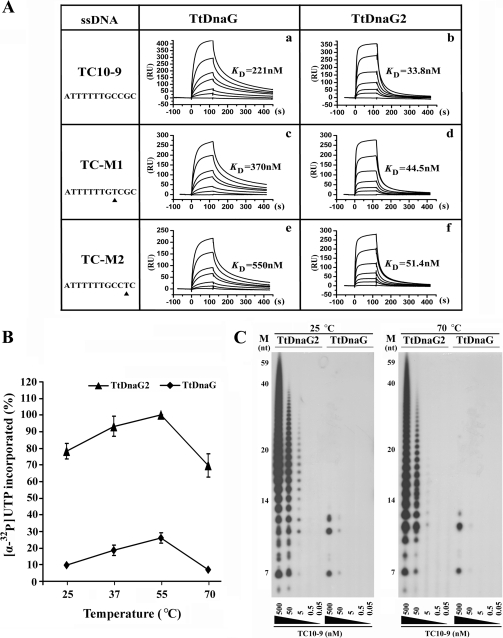
To understand the possible reasons that TtDnaG and TtDnaG2 exhibited distinct priming activities and template initiation specificities, the template binding affinities of these two proteins were compared by SPR assays with the templates TC10-9, TC-M1, and TC-M2 (Fig. 6A). The template TC10-9 contained the 5′-CCGC-3′ recognition sequence of TtDnaG and supported efficient primer synthesis by TtDnaG (Fig. 3A, lane 10). Templates TC-M1 and TC-M2 did not support primer syntheses by TtDnaG due to a single-base mutation. All three templates could serve for RNA primer syntheses by TtDnaG2 (data not shown). As shown in Fig. 6A, template TC10-9 was bound by TtDnaG with a KD (equilibrium dissociation constant) of 221 nM (Fig. 6A, panel a), whereas the affinities for templates TC-M1 and TC-M2 were lower, with KD values of 370 nM and 550 nM, respectively (Fig. 6A, panels c and e). These results demonstrated that the binding affinity of TtDnaG for a template with a specific initiation sequence was higher than those for templates with mutant sequences at room temperature. Significantly, TtDnaG2 bound to the three templates with KD values of 33.1 nM, 44.5 nM, and 51.4 nM, respectively (Fig. 6A, panels b, d, and f). Thus, the binding affinities of TtDnaG2 were almost 10-fold higher than those of TtDnaG and were less different among the three tested templates.
FIG. 6.
(A) Binding affinities of TtDnaG and TtDnaG2 for templates TC10-9, TC-M1, and TC-M2, detected by SPR assays. The binding affinities are shown in arbitrary resonance units (RU; where 1,000 RU corresponds to a surface density of 1 ng/mm2) and as KD values. Binding curves for TtDnaG were obtained with protein concentrations (bottom to top) of 0, 50, 100, 200, 300, 600, and 800 nM (a, c, and e). Binding curves for TtDnaG2 were obtained with protein concentrations (bottom to top) of 0, 6.25, 12.5, 25, 37.5, 50, and 100 nM (b, d, and f). (B) RNA primer syntheses (measured by the amount of [α-32P]UTP incorporated) by TtDnaG (⧫) and TtDnaG2 (▴) (0.5 μM [each]) with the TC10 template (3′-OH terminus blocked; 5 μM) at temperatures from 25°C to 70°C. The RNA amounts were quantified by phosphorimaging. The relative value of the products of TtDnaG2 at 55°C was assumed to be 100%. Assays were performed in triplicate. (C) RNA primer syntheses by TtDnaG and TtDnaG2 (0.5 μM [each]) with decreasing concentrations (from 500 nM to 0.05 nM) of template TC10-9 (3′-OH terminus blocked). Assays were performed at both 25°C (left) and 70°C (right). M, ssDNA markers as in Fig. 2.
To understand the relationship between the template binding activities detected by SPR assay at room temperature (25°C) and the RNA priming activities detected at 70°C, the enzyme activities of TtDnaG and TtDnaG2 were further compared at different temperatures as well as different template concentrations. First, when assays were performed at temperatures ranging from 25°C to 70°C, with a constant template concentration (5 μM TC10), it was obvious that the RNA primer syntheses (measured by the amount of [α-32P]UTP incorporated) of TtDnaG2 were always more efficient than those of TtDnaG, while the general trends of enzyme activities in this temperature range were similar, with the highest activity at 55°C for both proteins (Fig. 6B). Second, when template concentration dependences of these two enzymes were compared, it was clear that TtDnaG2 could synthesize RNA primers at template concentrations as low as 5 nM at 25°C, while TtDnaG could only weakly synthesize RNA primers even at a 10-fold-higher template concentration of 50 nM (Fig. 6C, left panel). This is quite consistent with the SPR experiment results showing that the binding affinities of TtDnaG2 were almost 10-fold higher than those of TtDnaG at room temperature (Fig. 6A). Importantly, similar results were also observed at 70°C (Fig. 6C, right panel), although the RNA priming activities of both enzymes at this temperature were a little lower than those at 25°C. Since the minimum template concentration for TtDnaG2 was also about 10-fold lower than that for TtDnaG at 70°C (Fig. 6C, right panel), it could be speculated that the template binding affinity of TtDnaG2 might also be approximately 10-fold higher than that of TtDnaG at such a high temperature. Thus, we presumed that the high template binding affinity of TtDnaG2 might be an important reason for its high priming activity as well as low template specificity.
DISCUSSION
Two distantly homologous dnaG genes were annotated in the genome of the thermophilic bacterium T. tengcongensis. This phenomenon of more than one dnaG gene per chromosome is widespread in the phylum Firmicutes, to which T. tengcongensis belongs. Phylogenetic analysis revealed that these DnaGs are divided into two groups, which we labeled groups I and II. TtDnaG and TtDnaG2 from T. tengcongensis are representatives of these two groups, respectively (Fig. 1A). Thus, comparative study of TtDnaG and TtDnaG2 would provide new information on the diversity in DnaG proteins, which is still poorly understood.
TtDnaG as a thermophilic bacterial replicative primase with a novel GC-only recognition sequence.
Taking into account all the results of protein sequence alignment, phylogenetic analysis, gene distribution pattern on the chromosome, and sequence recognition specificity, it becomes clear that TtDnaG is the ordinary replicative primase in T. tengcongensis. TtDnaG is homologous to and shares a similar molecular size (∼600 aa) with the previously identified bacterial primases, e.g., the DnaGs from E. coli, A. aeolicus, and S. aureus (Fig. 1). Moreover, like the replicative primases from bacteria and phages (8, 13, 22, 26, 41, 55), TtDnaG requires specific recognition sequences to synthesize RNA primers de novo.
The specific recognition sequences of TtDnaG included not only the trinucleotide 5′-CCC-3′ but also 8 tetranucleotides comprised entirely of cytosines and/or guanines (Fig. 4). In addition, the tetranucleotides 5′-CCCC-3′, 5′-GCCC-3′, and 5′-CCCG-3′, with additional cytosines or guanines flanking 5′-CCC-3′, could be used more efficiently by TtDnaG (Fig. 4B and C). These GC-only recognition sequences of TtDnaG are reminiscent of those observed in the hyperthermophilic organism A. aeolicus, whose primase preferentially recognizes 5′-CCC-3′ as well as 5′-GCC-3′ or 5′-CGC-3′, to a much lesser degree (26). Interestingly, by examining the supporting sequences, 5′-CAGA(CA)5XYZ(CA)3-3′, that were used to test the possible initiation nucleotides in A. aeolicus, we found that there is always a cytosine (C) flanking the predicted initiation trinucleotides (26). Thus, it might be possible that this flanking cytosine assists the trinucleotides in being used for initiation by A. aeolicus primase. If that is so, then the exact recognition sequences of the A. aeolicus primase might be the trinucleotide 5′-CCC-3′ and the tetranucleotides 5′-GCCC-3′ and 5′-CGCC-3′, which are all in the list of the recognition sequences of TtDnaG. Since both A. aeolicus and T. tengcongensis are low-GC-content thermophilic bacteria, these results suggest that the GC composition in the initiation sequences is important for maintaining the primase-nucleoside triphosphate (NTP)-ssDNA complex during the initiation step at high temperature (26), as the strong base-pairing strength of G-C versus A-T would provide greater thermostability of this complex.
Preference to GC-only nucleotides (especially tetranucleotides) in priming initiation seemed a novel characteristic of thermophilic primases. It is noteworthy that three GC-rich regions derived from GA59 and TC59 could generate abortive RNA products (Fig. 3B, lanes 4, 7, and 9). These GC-rich regions contain the tetranucleotide 5′-CCAG-3′ or 5′-CCTG-3′ (Table 1), just resembling the specific initiation tetranucleotide 5′-CCCG-3′. These sequences might occasionally support RNA synthesis by TtDnaG. However, since an A-T base pair is less stable than a G-C base pair, these initiation sites would make the primase-NTP-ssDNA complex unstable at high temperatures and tend to release abortive RNA products with shorter lengths (Fig. 3B, lanes 4, 7, and 9). This observation further supported the hypothesis that the GC-only recognition sequences might represent an adaptation of TtDnaG to high-temperature environments.
TtDnaG2 as a novel primase with unknown but possibly important physiological functions.
Although TtDnaG2 (358 aa) is much smaller than TtDnaG, it also exhibited the ability to synthesize RNA polymers, dependent on ssDNA, at a broad temperature range (Fig. 2A). However, unlike all known replicative primases, TtDnaG2 did not exhibit apparent template specificity, as it efficiently initiated RNA primer syntheses on all templates tested. In addition, the RNA synthesized by TtDnaG2 was usually very long, even longer than the templates, in vitro (Fig. 2B and 5). These unusual characteristics have made TtDnaG2 more doubtable as an ordinary replicative primase.
The ability of TtDnaG2 to synthesize primers that are longer than the template could be explained by a number of mechanisms. First, the primase might slip repeatedly during synthesis, thereby generating extended products. Second, a primer generated in one round of synthesis might anneal to a second template and be extended by the primase (25). The high DNA binding affinity and priming activity of TtDnaG2 provide support for these possibilities. Another reason was also conceivable, i.e., that the primase might possess a terminal nucleotidyltransferase-like activity, which adds nucleotides to the 3′ end of a DNA molecule in a template-independent manner (25). However, 3′-terminal nucleotidyltransferase activity was investigated but was not detectable for TtDnaG2 (data not shown).
Previous proteomic analysis revealed that TtDnaG2 is actively expressed in T. tengcongensis under optimal growth conditions (51). Although there is limited experimental support, as an in vivo gene manipulation system for T. tengcongensis is still not established, we presume that TtDnaG2 might also play important but unknown roles in this bacterium. It might function in some emergency situations, such as DNA replication restarting after severe DNA damage. The low template recognition specificity of TtDnaG2 provides it with an advantage to function in these situations. Moreover, there is usually a bacterial DnaG orthologue existing in archaea, except for the archaeal two-subunit replicative primase. This DnaG orthologue in archaea has been found tightly associated with some other proteins to form an exosome-like protein complex, which is essential for many pathways of RNA processing and degradation (12). It would be interesting to investigate whether TtDnaG2 is also associated with other proteins in vivo and plays similar or novel physiological roles.
Implications of structure comparison of TtDnaG and TtDnaG2.
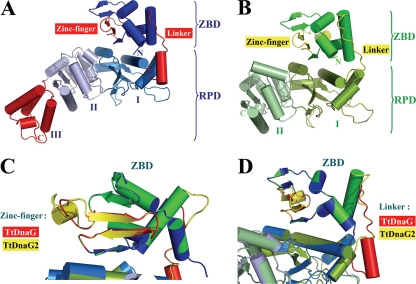
Its lack of apparent template specificity (Fig. 5), much higher ssDNA binding affinity (Fig. 6), and higher RNA primer synthesis activity (Fig. 2 and 6B) make TtDnaG2 distinct from TtDnaG. To gain information on the relevant implications of these differences by structure comparison, the three-dimensional (3D) structures of these two proteins were modeled by using the web-based server Phyre, according to the crystal structure of A. aeolicus primase. Unlike TtDnaG, which has all three complete domains (ZBD, RPD, and DnaB-ID) found in conserved bacterial DnaG primases (Fig. 1B and 7A), TtDnaG2 has only the N-terminal ZBD and the first two subdomains of RPD (Fig. 7B), resembling the T7 primase (20).
FIG. 7.
Comparison of modeled 3D structures of TtDnaG and TtDnaG2. Protein 3D structures were modeled by the web-based server Phyre (http://www.sbg.bio.ic.ac.uk/phyre/). The 3D models of TtDnaG and TtDnaG2 were superimposed by using Coot and were viewed from the same orientation. (A and B) Predicted 3D models of two domains (ZBD and RPD) of TtDnaG (A) and TtDnaG2 (B). The two main different structures (zinc finger motif in ZBD and linker region between ZBD and RPD) for these two proteins are shown in red for TtDnaG (A) and yellow for TtDnaG2 (B). The subdomains (I, II, and III) of RPD are also indicated. (C) Superimposed comparison of zinc finger motif regions in the ZBD of TtDnaG (red) and TtDnaG2 (yellow). The model is rotated 90° around the vertical axis relative to those in panels A and B. (D) Superimposed comparison of linker regions between ZBD and RPD of TtDnaG (red) and TtDnaG2 (yellow). The model is rotated 30° around the vertical axis relative to those in panels A and B. The figures were prepared by using PYMOL (http://www.pymol.org).
There are several structural implications for the biochemical characteristics of TtDnaG2. First, in the zinc finger motif, TtDnaG2 has a longer connecting region between the second and third β sheets (Fig. 7C) and more basic/hydrophobic residues (Fig. 1B) than TtDnaG. The longer and rigid connecting region with a partial α helix of TtDnaG2 might form a cleft more easily with the β sheets and may generate a tighter protein-DNA interaction, as observed in replication protein A (RPA) and some other proteins (4, 5, 7). The basic and hydrophobic residues located on the surfaces of the β sheets of ZBD are considered important for ssDNA binding and specific sequence recognition (23, 31), so the larger number of basic/hydrophobic residues in TtDnaG2 might also contribute to its higher DNA binding affinity and lower sequence recognition specificity. Second, at the linker region between the ZBD and RPD, TtDnaG2 has an extended polypeptide loop, like T7 primase (20), while TtDnaG has a longer but more rigid linker with an α helix (Fig. 7D). The extended loop in TtDnaG2 might make it more flexible than the rigid loop of TtDnaG, hence providing a more efficient switch in bringing together the ZBD and RPD for primer synthesis. In addition, different residues located in other important regions, such as the nonspecific capture and tracking locus site for DNA templates in the N-terminal subdomain of RPD (10), the predicted potential DNA binding site, and the active catalytic site in the TOPRIM subdomain (33), might also contribute to the different DNA binding abilities and RNA priming activities of TtDnaG and TtDnaG2. Indeed, many similarities to phage primases were found in TtDnaG2, implying that it possibly originated from lysogenic phages and evolved different biochemical and physiological functions in T. tengcongensis.
In summary, the two DnaGs in T. tengcongensis have evolved into two distinct primases with significant differences in both structure and function. TtDnaG seems to be more conserved as an ordinary replicative primase and has been well adapted for function at high temperature, while TtDnaG2 is likely involved in other unknown processes. Although several possible reasons for the distinctions between TtDnaG and TtDnaG2 have been implied from the modeled 3D structures, it would be interesting to determine their crystal structures, as well as those of the protein-ssDNA complexes, to gain a more precise and comprehensive view on the structure and function of these DnaG proteins.
Acknowledgments
We greatly appreciate Shixuan Liu for assistance with protein purification and 3D structure modeling and Zheng Fan for assistance with the SPR assay.
This work was supported by grants from the National Natural Science Foundation of China (NSFC) (grant 30621005). H. Xiang is a Distinguished Young Investigator of the NSFC (grant 30925001).
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, Q., Y. Tian, W. Li, Z. Xu, Z. Xuan, S. Hu, W. Dong, J. Yang, Y. Chen, Y. Xue, Y. Xu, X. Lai, L. Huang, X. Dong, Y. Ma, L. Ling, H. Tan, R. Chen, J. Wang, J. Yu, and H. Yang. 2002. A complete sequence of the T. tengcongensis genome. Genome Res. 12:689-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya, S., and M. A. Griep. 2000. DnaB helicase affects the initiation specificity of Escherichia coli primase on single-stranded DNA templates. Biochemistry 39:745-752. [DOI] [PubMed] [Google Scholar]
- 4.Bochkarev, A., R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176-181. [DOI] [PubMed] [Google Scholar]
- 5.Bogden, C. E., D. Fass, N. Bergman, M. D. Nichols, and J. M. Berger. 1999. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell 3:487-493. [DOI] [PubMed] [Google Scholar]
- 6.Burton, Z. F., C. A. Gross, K. K. Watanabe, and R. R. Burgess. 1983. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell 32:335-349. [DOI] [PubMed] [Google Scholar]
- 7.Cavarelli, J., B. Rees, M. Ruff, J. C. Thierry, and D. Moras. 1993. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature 362:181-184. [DOI] [PubMed] [Google Scholar]
- 8.Cha, T. A., and B. M. Alberts. 1986. Studies of the DNA helicase-RNA primase unit from bacteriophage T4. A trinucleotide sequence on the DNA template starts RNA primer synthesis. J. Biol. Chem. 261:7001-7010. [PubMed] [Google Scholar]
- 9.Corn, J. E., and J. M. Berger. 2006. Regulation of bacterial priming and daughter strand synthesis through helicase-primase interactions. Nucleic Acids Res. 34:4082-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corn, J. E., J. G. Pelton, and J. M. Berger. 2008. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat. Struct. Mol. Biol. 15:163-169. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, B. D., Z. F. Burton, K. K. Watanabe, and R. R. Burgess. 1985. Nucleotide sequence of the rpsU-dnaG-rpoD operon from Salmonella typhimurium and a comparison of this sequence with the homologous operon of Escherichia coli. Gene 40:67-78. [DOI] [PubMed] [Google Scholar]
- 12.Evguenieva-Hackenberg, E., P. Walter, E. Hochleitner, F. Lottspeich, and G. Klug. 2003. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 4:889-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frick, D. N., and C. C. Richardson. 2001. DNA primases. Annu. Rev. Biochem. 70:39-80. [DOI] [PubMed] [Google Scholar]
- 14.Griep, M. A. 1995. Primase structure and function. Indian J. Biochem. Biophys. 32:171-178. [PubMed] [Google Scholar]
- 15.Grompe, M., J. Versalovic, T. Koeuth, and J. R. Lupski. 1991. Mutations in the Escherichia coli dnaG gene suggest coupling between DNA replication and chromosome partitioning. J. Bacteriol. 173:1268-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller, R. C., and K. J. Marians. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439:557-562. [DOI] [PubMed] [Google Scholar]
- 17.Hiasa, H., and K. J. Marians. 1994. Primase couples leading- and lagging-strand DNA synthesis from oriC. J. Biol. Chem. 269:6058-6063. [PubMed] [Google Scholar]
- 18.Ilyina, T. V., A. E. Gorbalenya, and E. V. Koonin. 1992. Organization and evolution of bacterial and bacteriophage primase-helicase systems. J. Mol. Evol. 34:351-357. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, S. K., S. Bhattacharyya, and M. A. Griep. 2000. DnaB helicase stimulates primer synthesis activity on short oligonucleotide templates. Biochemistry 39:736-744. [DOI] [PubMed] [Google Scholar]
- 20.Kato, M., T. Ito, G. Wagner, C. C. Richardson, and T. Ellenberger. 2003. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell 11:1349-1360. [DOI] [PubMed] [Google Scholar]
- 21.Keck, J. L., D. D. Roche, A. S. Lynch, and J. M. Berger. 2000. Structure of the RNA polymerase domain of E. coli primase. Science 287:2482-2486. [DOI] [PubMed] [Google Scholar]
- 22.Koepsell, S. A., M. A. Larson, M. A. Griep, and S. H. Hinrichs. 2006. Staphylococcus aureus helicase but not Escherichia coli helicase stimulates S. aureus primase activity and maintains initiation specificity. J. Bacteriol. 188:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusakabe, T., A. V. Hine, S. G. Hyberts, and C. C. Richardson. 1999. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl. Acad. Sci. U. S. A. 96:4295-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusakabe, T., and C. C. Richardson. 1996. The role of the zinc motif in sequence recognition by DNA primases. J. Biol. Chem. 271:19563-19570. [DOI] [PubMed] [Google Scholar]
- 25.Lao-Sirieix, S. H., and S. D. Bell. 2004. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344:1251-1263. [DOI] [PubMed] [Google Scholar]
- 26.Larson, M. A., R. Bressani, K. Sayood, J. E. Corn, J. M. Berger, M. A. Griep, and S. H. Hinrichs. 2008. Hyperthermophilic Aquifex aeolicus initiates primer synthesis on a limited set of trinucleotides comprised of cytosines and guanines. Nucleic Acids Res. 36:5260-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, Y. B., P. V. Ratnakar, B. K. Mohanty, and D. Bastia. 1996. Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc. Natl. Acad. Sci. U. S. A. 93:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marini, F., A. Pellicioli, V. Paciotti, G. Lucchini, P. Plevani, D. F. Stern, and M. Foiani. 1997. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 16:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitkova, A. V., S. M. Khopde, and S. B. Biswas. 2003. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J. Biol. Chem. 278:52253-52261. [DOI] [PubMed] [Google Scholar]
- 30.Oakley, A. J., K. V. Loscha, P. M. Schaeffer, E. Liepinsh, G. Pintacuda, M. C. Wilce, G. Otting, and N. E. Dixon. 2005. Crystal and solution structures of the helicase-binding domain of Escherichia coli primase. J. Biol. Chem. 280:11495-11504. [DOI] [PubMed] [Google Scholar]
- 31.Pan, H., and D. B. Wigley. 2000. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure 8:231-239. [DOI] [PubMed] [Google Scholar]
- 32.Pei, H., J. Liu, J. Li, A. Guo, J. Zhou, and H. Xiang. 2007. Mechanism for the TtDnaA-Tt-oriC cooperative interaction at high temperature and duplex opening at an unusual AT-rich region in Thermoanaerobacter tengcongensis. Nucleic Acids Res. 35:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podobnik, M., P. McInerney, M. O'Donnell, and J. Kuriyan. 2000. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 300:353-362. [DOI] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.San Martin, M. C., N. P. Stamford, N. Dammerova, N. E. Dixon, and J. M. Carazo. 1995. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J. Struct. Biol. 114:167-176. [DOI] [PubMed] [Google Scholar]
- 36.Su, X. C., P. M. Schaeffer, K. V. Loscha, P. H. Gan, N. E. Dixon, and G. Otting. 2006. Monomeric solution structure of the helicase-binding domain of Escherichia coli DnaG primase. FEBS J. 273:4997-5009. [DOI] [PubMed] [Google Scholar]
- 37.Sun, W., J. Tormo, T. A. Steitz, and G. N. Godson. 1994. Domains of Escherichia coli primase: functional activity of a 47-kDa N-terminal proteolytic fragment. Proc. Natl. Acad. Sci. U. S. A. 91:11462-11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swart, J. R., and M. A. Griep. 1993. Primase from Escherichia coli primes single-stranded templates in the absence of single-stranded DNA-binding protein or other auxiliary proteins. Template sequence requirements based on the bacteriophage G4 complementary strand origin and Okazaki fragment initiation sites. J. Biol. Chem. 268:12970-12976. [PubMed] [Google Scholar]
- 39.Syson, K., J. Thirlway, A. M. Hounslow, P. Soultanas, and J. P. Waltho. 2005. Solution structure of the helicase-interaction domain of the primase DnaG: a model for helicase activation. Structure 13:609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szafranski, P., C. L. Smith, and C. R. Cantor. 1997. Cloning and analysis of the dnaG gene encoding Pseudomonas putida DNA primase. Biochim. Biophys. Acta 1352:243-248. [DOI] [PubMed] [Google Scholar]
- 41.Tabor, S., and C. C. Richardson. 1981. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A. 78:205-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 43.Thirlway, J., and P. Soultanas. 2006. In the Bacillus stearothermophilus DnaB-DnaG complex, the activities of the two proteins are modulated by distinct but overlapping networks of residues. J. Bacteriol. 188:1534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tougu, K., and K. J. Marians. 1996. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J. Biol. Chem. 271:21391-21397. [DOI] [PubMed] [Google Scholar]
- 45.Tougu, K., and K. J. Marians. 1996. The interaction between helicase and primase sets the replication fork clock. J. Biol. Chem. 271:21398-21405. [DOI] [PubMed] [Google Scholar]
- 46.Tougu, K., H. Peng, and K. J. Marians. 1994. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J. Biol. Chem. 269:4675-4682. [PubMed] [Google Scholar]
- 47.Tseng, T. Y., D. N. Frick, and C. C. Richardson. 2000. Characterization of a novel DNA primase from the Salmonella typhimurium bacteriophage SP6. Biochemistry 39:1643-1654. [DOI] [PubMed] [Google Scholar]
- 48.van der Ende, A., T. A. Baker, T. Ogawa, and A. Kornberg. 1985. Initiation of enzymatic replication at the origin of the Escherichia coli chromosome: primase as the sole priming enzyme. Proc. Natl. Acad. Sci. U. S. A. 82:3954-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic, J., T. Koeuth, R. Britton, K. Geszvain, and J. R. Lupski. 1993. Conservation and evolution of the rpsU-dnaG-rpoD macromolecular synthesis operon in bacteria. Mol. Microbiol. 8:343-355. [DOI] [PubMed] [Google Scholar]
- 50.Versalovic, J., and J. R. Lupski. 1993. The Haemophilus influenzae dnaG sequence and conserved bacterial primase motifs. Gene 136:281-286. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J., Y. Xue, X. Feng, X. Li, H. Wang, W. Li, C. Zhao, X. Cheng, Y. Ma, P. Zhou, J. Yin, A. Bhatnagar, R. Wang, and S. Liu. 2004. An analysis of the proteomic profile for Thermoanaerobacter tengcongensis under optimal culture conditions. Proteomics 4:136-150. [DOI] [PubMed] [Google Scholar]
- 52.Wang, L. F., and R. H. Doi. 1986. Nucleotide sequence and organization of Bacillus subtilis RNA polymerase major sigma (sigma 43) operon. Nucleic Acids Res. 14:4293-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C. A., E. L. Zechner, J. A. Reems, C. S. McHenry, and K. J. Marians. 1992. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis.J. Biol. Chem. 267:4074-4083. [PubMed] [Google Scholar]
- 54.Xue, Y., Y. Xu, Y. Liu, Y. Ma, and P. Zhou. 2001. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int. J. Syst. Evol. Microbiol. 51:1335-1341. [DOI] [PubMed] [Google Scholar]
- 55.Yoda, K., and T. Okazaki. 1991. Specificity of recognition sequence for Escherichia coli primase. Mol. Gen. Genet. 227:1-8. [DOI] [PubMed] [Google Scholar]