Abstract
Archaeal transcriptional machinery is similar to that of eukaryotes. We studied the fates of various components of the Sulfolobus solfataricus transcriptional apparatus under different stresses and found that in cells incubated at 90°C for 1 h, transcription factor E (TFE) is selectively depleted, but its mRNA levels are increased. We discuss the implications of these findings.
The archaeal transcriptional apparatus closely resembles the eukaryotic RNA polymerase II system (11). Biochemical studies have shown that the core components of the Sulfolobus transcriptional machinery are comprised of a TATA-binding protein (TBP), transcription factor B-1 (TFB1), and a 13-subunit RNA polymerase and that all three components are essential for accurate and efficient transcription from a variety of promoters (2, 9, 16, 20, 25, 28, 35). TBP and TFB1 bind sequence specifically to the A box and BRE, respectively, and the resulting ternary complex recruits RNA polymerase (1, 3, 4, 29). The Sulfolobus solfataricus genome also encodes other proteins implicated in transcription, such as transcription factor E (TFE), TBP-interacting protein 49 (TIP-49), and two TFB1 paralogs, TFB2 and TFB3 (6), but their respective roles in this process are less well understood.
In eukaryotes, TFIIE serves as a general transcription factor (GTF), composed of α and β subunits, that is essential for recruiting TFIIH (23). Deletion analysis of TFIIEα has demonstrated that its 20-kDa N-terminal region is required for it to maintain function (21) and that this region is conserved in archaeal TFE (2, 15). Archaeal TFE contains a winged helix-turn-helix structure in its N-terminal region, which, along with other surfaces that mediate protein-protein interactions, is conserved in TFIIEα (24). Like TFIIEα, archaeal TFE also interacts with TBP and RNA polymerase (2, 37). Additionally, there appears to be functional interdependence between archaeal TFE and TFB, since mutations in one can be complemented by the other (34). Biochemical studies have found that TFE associates with the RNA polymerase, stimulates transcription from some promoters, and is a part of both initiation and elongation complexes (2, 14, 34). Taken together, these findings suggest that, like eukaryotic TFIIE, archaeal TFE is also a GTF that is essential for gene transcription. No homolog of any of the TFIIH subunits has been found in archaea.
The heat shock response is widespread in organisms belonging to all three domains of life and allows cells to cope with thermal stress (8). In the euryarchaeote Pyrococcus furiosus, heat shock is sensed directly by the negatively acting transcription factor Phr, which dissociates from the DNA at elevated temperatures, allowing heat shock genes to be transcribed (33). While the mechanism through which heat shock is sensed in crenarchaeotes remains unknown, transient exposure of Sulfolobus shibatae to temperatures between 85 and 90°C rapidly culminates in the accumulation of heat shock proteins TF55α and -β, which, together with TF55γ, assemble into rosettasomes (18, 19). Both TF55α and -β are highly conserved in S. solfataricus and Sulfolobus acidocaldarius and in other crenarchaeotes. Several studies have employed DNA microarray-based methods to identify sets of genes whose expression is impacted by heat shock (7, 10, 27, 30-32). In S. solfataricus, heat shock affects expression of about one-third of the genes, some of which are transcribed 30 min after heat shock (32). Discrete sets of genes whose expression is influenced by UV-induced DNA and hydrogen peroxide-mediated oxidative stress have also been identified (12, 36). While data generated from such studies are extremely useful, they are restricted to the RNA level. To gauge the impact that DNA damage, oxidative stress, and cold and heat shock have on the levels of different components of the core transcriptional machinery, we generated highly specific antibodies against TBP, TFB, RpoB, TFE, and TIP49. Here, we report that DNA damage, oxidative stress, and cold shock do not affect the levels of any of the five proteins but that TFE is selectively depleted by heat shock.
MATERIALS AND METHODS
S. solfataricus growth and extract preparation.
S. solfataricus P2 was grown aerobically at 76°C in Brock's medium (13), and cell extracts were prepared according to the method of Hüdepohl et al. (17). The protein concentration was determined by the Bradford assay (Bio-Rad). A 200-ml mid-log-phase Sulfolobus culture (optical density at 595 nm [OD595], ∼0.3) growing at 76°C was dispensed in 1-ml aliquots in 1.5-ml plastic microcentrifuge tubes and quickly transferred to ice water, 76°C, 80°C, 85°C, and 90°C for 0 and 60 min. After treatment, the cells were rapidly cooled in ice water and centrifuged at 4°C at 12,000 rpm for 2 min and directly lysed in 200 μl 2× SDS sample loading buffer. Sulfolobus cells growing at 76°C were also incubated with 5 μg/ml mitomycin C (5) and 30 μM hydrogen peroxide (22, 36) for 1 h, and the impacts of DNA damage and oxidative stress on the expression of TBP, TFB1, TFE, RpoB, and TIF49 were examined by Western blotting. For temperature shift and recovery experiments, a 5-ml culture (OD595, ∼0.3) growing at 76°C in a 15-ml plastic Falcon tube was incubated at 90°C for 60 min and then shifted to 76°C for various lengths of time.
DNA cloning, expression, and purification of recombinant proteins.
Full-length open reading frames (ORFs) of S. solfataricus TFB1, TFE, TIP49, and TBP were PCR amplified using the following oligonucleotide sets: TFB1-F (GATCGGATCCTTGTATTTGTCTGAAGAAAATAAA) and TFB1-R (GATCAAGCTTTATTGAGTAGGTATTGATAT), TFE-F (GATCGGATCCGTTAACGCAGAGGATCTGTTT) and TFE-R (GATCAAGCTTCTAATTTAGGTTCTATTCCTAG), TIP49-F (GATCGGATCCTGTCATAATATAAAACCTCAC) and TIP-His6-R (ATGATGATGATGATGATGTTTCAATAATAGATTCTCATAC), and TBP-F (GATCGGATCCGCAACAGTTACGTTAGAGCAG) and TBP-R (GATCAAGCTTTTAGAGCTCTAACTCTTCCTC), respectively. The TFB1, TFE, and TIP49 PCR products were cloned into BamHI- and SmaI-digested pGEX4T1 (GE Healthcare Bio-Science), while TBP and TFE were cloned into BamHI- and HindIII-digested pQE30-Xa (Qiagen). Recombinant pGEX4T1 and pQE30-Xa plasmids were introduced into BL21-DE3-pLysS (Novogen) and M15-pREP4 (Qiagen) cells, respectively. After induction with IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma-Aldrich), cells were harvested, lysed with BPER (Pierce), and centrifuged. Glutathione S-transferase (GST)-fused TFE and TFB1 were purified from the crude extracts using 100 μl washed glutathione-Sepharose beads (GE Healthcare Bio-Science). His-tagged TIP49 was purified under denaturing conditions, whereas TBP and TFE were purified under nondenaturing conditions according to the manufacturer's instructions (Qiagen).
Antibody generation.
Gel-purified GST-fused TFE, TFB1, and TIP49 were mixed with Freund's complete adjuvant (Sigma-Aldrich) and injected intraperitoneally into BALB/c mice, followed by 4 additional injections containing Freund's incomplete adjuvant at 10-day intervals. The animals were euthanized 10 days after the last injection, and blood was collected. Antibodies against S. solfataricus TBP and RpoB were generated in rabbits as reported previously (28).
RNA isolation and qPCR.
RNA was isolated from 10 ml S. solfataricus P2 culture (OD595, ∼0.3) using TRI reagent (Sigma-Aldrich). The purified RNA was treated with DNase I (Takara Bio Inc.) and converted into cDNA using a mixture of random hexamers and oligo(dT) with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega). cDNA (200 ng) was used for amplification with the following sets of forward and reverse TBP, TFB1, and TFE oligonucleotides: TBPQ-F (ACTAATTTTCAGAATGGATGATCC) and TBPQ-R (CTAATTCTGCTAACTTATCAAATATC), TFB1Q-F (CCTGATAAGATTATCTTTGATGCAG) and TFB1Q-R (CAACTCTGCTTCTCTTTTCTTTCT), and TFEQ-F (AATCAGTTGAATATAAAAGTTAATG) and TFEQ-R (CTTTTCCTATTTAACAGTATTTC). Quantitative PCR (qPCR) was carried out on a Chromo-4 cycler (Bio-Rad) with Maxima SYBR green Master Mix (Fermentas) using the following thermocycling conditions: 94°C for 5 min, followed by 45 cycles of 94°C for 30 s, 51°C for 30 s, and 72°C for 30 s. The conditions for TFB1 and TFE were similar, except that annealing was carried out at 54°C and 46°C, respectively. Data were collected and analyzed using Opticon Monitor 3 software (Bio-Rad). TBP, TFB1, and TFE qPCRs were carried out in triplicate, and standard errors were calculated. No-template and no-reverse-transcriptase negative controls were carried out in triplicate and duplicate, respectively, and showed no DNA amplification.
Western blotting.
Proteins were separated on 12% SDS-PAGE, and the gel contents were electrotransferred onto nitrocellulose membranes (GE Healthcare Bio-Science). The blots were blocked, probed with anti-TFE (α-TFE) and α-TBP (1:1,000 each), α-TFB1 (1:2,000), α-TIP-49 (1:5,000), and α-RpoB (1:20,000) antibodies, and washed. After incubation with either anti-mouse or anti-rabbit horseradish peroxidase (HRP)-conjugated IgG antibodies, the blots were washed and then developed with the ECL-plus reagent (GE Healthcare Bio-Science). Images were captured on X-ray film (Fuji).
Protein stability assay.
Five hundred nanograms of purified bacterially expressed His6-TBP, His6-TFE, and bovine serum albumin (BSA) (Sigma-Aldrich) in 20 μl transcription buffer (20 mM HEPES, pH 7.9, 5 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, and 1 mM dithiothreitol (DTT) were incubated at 37°C, 76°C, and 90°C for 1 hour, and the products were analyzed on 12% SDS-PAGE. Proteins were visualized by Coomassie brilliant blue staining.
RESULTS
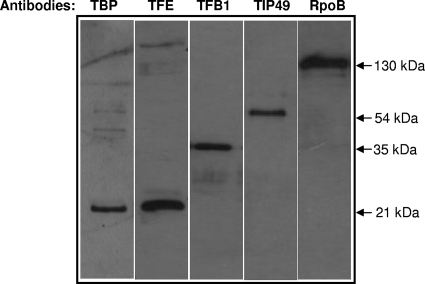
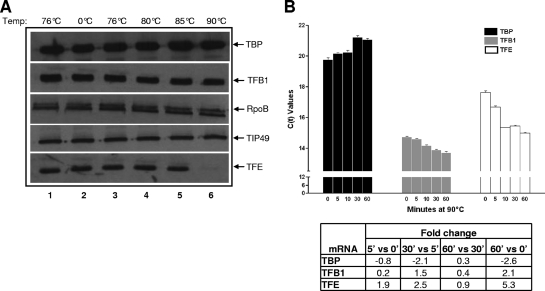
Several studies have employed DNA microarrays to study changes in mRNA patterns under various stress conditions (7, 10, 27, 30-32). As mRNA levels do not necessarily mirror protein levels, we studied the fates and expression levels of various components of the S. solfataricus core transcriptional machinery under different stress conditions. To that end, we generated antibodies against TBP, TFB, TFE, TIP49, and RpoB that could detect approximately 1 ng of its purified recombinant antigen on Western blots (data not shown) and were able to specifically bind their respective antigens in a crude extract (Fig. 1). This antibody set was used to assess whether oxidative stress, DNA damage, or temperature shock influenced the levels of their respective antigens. Western blots carried out with crude extracts prepared from hydrogen peroxide (oxidative stress)- or mitomycin C (DNA damage)-treated cells for 1 h failed to show any change in the level of any of the 5 proteins (data not shown). Similarly, cells which had been cold shocked at 0°C for 1 h also failed to show any significant change in the levels of any of the factors (Fig. 2A, lane 2). However, incubation of S. solfataricus cells at 90°C (heat shock) for 1 h reduced TFE to an undetectable level (lane 6) but did not affect levels of TBP, TFB1, RpoB, or TIP49 (Fig. 2A, lanes 2 to 5). No changes in the amounts of any of five proteins were observed in cells incubated for 1 h at 76°C, 80°C, or 85°C (lanes 2 to 5). To ascertain that transferring a 1-ml aliquot from a large culture to a tube by itself did not influence protein levels, extracts were also prepared from cells growing at 76°C (lane 1). To determine whether the observed heat shock-induced decline of TFE occurs at the protein or RNA level, real-time PCR was employed to estimate the quantities of TFE, TBP, and TFB1 mRNAs in cells that had been heat shocked for various lengths of time. This experiment clearly showed that TFE mRNA levels increased 5-fold in cells that had been heat shocked for 1 h. TFE mRNA levels in these cells rose dramatically in the first 30 min of heat shock but also continued to increase after that (Fig. 2B). While heat shock also increased TFB1 mRNA levels 2-fold over 60 min, it reduced the amount of TBP mRNA by 2.6-fold during the same period.
FIG. 1.
Antibody specificity. Western blots containing ∼5 μg of S. solfataricus cell extract per lane were probed with antibodies against TBP, TFE, TFB1, TIP49, and RpoB. Anti-TBP and -TFE antisera were diluted 1:1,000, while anti-TFB1, -TIP49, and -RpoB were diluted 1: 2,000, 1:5,000, and 1:20,000, respectively.
FIG. 2.
TFE is selectively depleted under heat shock conditions, but transcription is not affected. (A) Sulfolobus cells growing at 76°C (lane1) were incubated at 0°C, 76°C, 80°C, 85°C, and 90°C (lanes 2 to 6, respectively) for 1 h. The cells were harvested and suspended directly in SDS-PAGE loading buffer. Western blots were probed with the indicated antibodies. (B) qPCR analysis of TFE, TFB1, and TBP mRNAs. Total RNA was isolated from 10 ml of cells incubated at 90°C for 0, 5, 10, 30, and 60 min and converted into cDNA. PCR amplification of TFE, TFB1, and TBP cDNAs was carried out on a Chromo-4 cycler with Maxima SYBR green Master Mix. DNA amplification was monitored in real time, and threshold cycle (Ct) values were obtained for each sample. TBP, TFB1, and TFE qPCRs were carried out in triplicate, and the standard error for each sample (error bars) was calculated. No-template and no-reverse-transcriptase negative controls were carried out in triplicate and duplicate, respectively, and showed no DNA amplification. Ct values were converted into fold change in TBP, TFB1, and TFE mRNA levels at different times and are shown in the table.
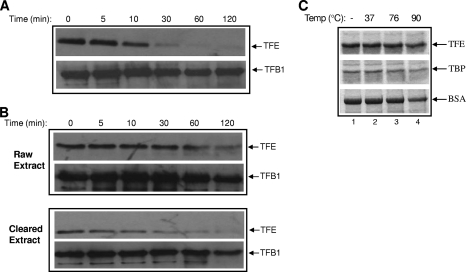
Subsequently, the kinetics of TFE depletion was studied in vivo by incubating cells at 90°C for different lengths of time. This experiment revealed that TFE levels were reduced by approximately 50% after 10 min and became nearly undetectable after 30 min (Fig. 3A). To determine whether a rate of TFE decline similar to that found in live cells is also observed in lysed cells, both raw (i.e., membrane-containing) and cleared (i.e., membrane-devoid) extracts were incubated at 90°C for various times and subjected to Western blotting. TFE declines in the raw and cleared extracts were equivalent but significantly less than that observed in intact cells (Fig. 3B). To ascertain that TFE does not spontaneously decay at elevated temperatures, recombinant His6-tagged TBP and TFE, along with BSA, were incubated at 37°C, 76°C, and 90°C for 1 h, and the products were analyzed by SDS-PAGE. No significant decay of TFE was found to occur at any of the three temperatures (Fig. 3C).
FIG. 3.
Kinetics of TFE depletion. (A) Sulfolobus cells were incubated at 90°C for 0, 5, 10, 30, 60, and 120 min. After being harvested, the cells were lysed directly in SDS-PAGE sample buffer. (B) Extracts were prepared from cells growing at 76°C, and aliquots of both crude and cleared extracts were incubated at 90°C for various periods. The Western blots in panels A and B were probed with anti-TFE and -TFB1 antibodies. (C) TFE does not decay spontaneously at 90°C. Approximately 500 ng recombinant His6-TFE and His6-TBP, along with BSA (66 kDa), was incubated at 37°C (lane 2), 76°C (lane 3), and 90°C (lane 4) for 60 min and subsequently mixed with sample buffer, heated, and analyzed on 12% SDS-PAGE, which was stained with Coomassie. Lane 1 represents input.
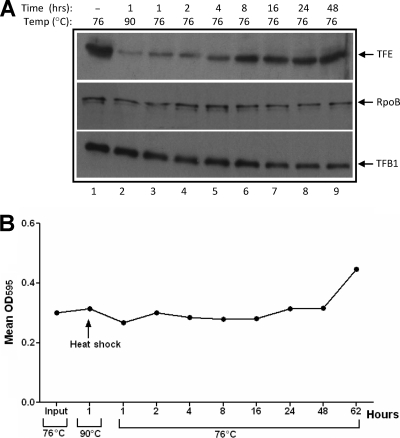
Lastly, temperature shift experiments were carried out to determine the length of time it takes for TFE levels to return to normal after heat shock and to assess whether there is any association between the amount of TFE and cell growth. For this, Sulfolobus cells (OD595, ∼0.3) growing at 76°C were heat shocked for 1 h at 90°C, and the culture was subsequently shifted to 76°C. Aliquots from the culture were taken after 1, 2, 4, 8, 16, 24, and 48 h, and the lysate was subjected to Western blotting with anti-TFE antisera. This exercise revealed that TFE levels in heat-shocked cells increased gradually over time and recovered fully after 48 h (Fig. 4A). The growth of cells that had been heat shocked for 1 h was also monitored by absorbance at 595 nm over 65 h. This experiment showed that no cell proliferation occurred until after 48 h (Fig. 4B).
FIG. 4.
Restoration of TFE levels at 76°C after heat shock and their association with cell growth. (A) Sulfolobus cells growing at 76°C (lane1) were heat shocked at 90°C for 1 h, and the culture was shifted back to 76°C for 1, 2, 4, 8, 16, 24, and 48 h (lanes 3 to 9, respectively). The levels of TFE, RpoB, and TFB1 in the various extracts were assessed by Western blotting. (B) Growth of cells at different times pre- and post-heat shock was monitored by absorbance at 595 nm. Shown is a plot of mean absorbance versus time.
DISCUSSION
In the study, we employed highly specific antibodies against TFE, TFB1, TBP, TIP49, and RpoB to show that TFE is selectively depleted under heat shock conditions whereas other factors are spared. Oxidative stress, DNA damage, and cold shock do not influence the levels of any of the five factors. Strikingly, cells that had experienced heat shock for 1 h were left with little to no TFE but contained 5-fold more TFE mRNA than naïve cells. We also found that heat-shocked cells nearly devoid of TFE did not begin to proliferate until their TFE levels were fully restored.
The fact that TFE does not decay spontaneously at 90°C suggests that it is targeted for destruction. This may occur in several ways. TFE could be marked for degradation under heat shock conditions. To determine whether TFE is posttranslationally modified by heat shock, we probed Western blots containing heat-shocked extracts with various phosphoserine, phosphotyrosine, and methyl-lysine antibodies but were unsuccessful (unpublished observations). Our inability to observe any phosphorylated, acetylated, or methylated forms of TFE may be due to the fact that modified TFE is degraded rapidly or that Sulfolobus TFE undergoes a different type of chemical modification, one that is not recognized by any of the antibodies that were employed. It is equally plausible that TFE is targeted by a specific protease that is activated by heat shock. Finally, heat shock-mediated translational arrest of TFE mRNA could also lead to depletion of TFE.
Our results show that under heat shock conditions, the amounts of TBP, TFB1, RpoB, and TIP49 remain constant and that transcription from the TFE and TFB1 promoters continues to occur. These findings are consistent with a previous report that showed that in S. solfataricus cells, TFE and TFB1 mRNA levels increase 5-fold and 2-fold, respectively, and that mRNA synthesis continues to occur after 30 min of heat shock at 90°C (32). Since TFE in these cells is nearly depleted after 30 min, we infer that transcription of housekeeping genes, TFE and TFB1 genes, and the 18 genes that are upregulated 30 min after heat shock (32) is not dependent on TFE. In light of these observations, and considering that TFE associates with RNA polymerase, it is tempting to speculate that TFE negatively regulates the transcription of those genes that are induced late in heat shock. Support for this notion stems from a recent study that showed that TFB3, which is highly upregulated in response to DNA damage (12), associates with RpoK and stimulates transcription from a number of promoters (26). The presence or absence of TFE, or other RNA polymerase subunits or associated factors, therefore has the potential to modulate targeting of RNA polymerase to different genomic loci under various stress or environmental conditions. We have shown previously that Sulfolobus RNA polymerase by itself is capable of initiating transcription accurately and efficiently from the TFB minor promoter (28). Alternatively, since TFE has been found to stimulate transcription from 5S and 16S rRNA gene promoters (2), it is possible that its absence during prolonged heat shock culminates in reduced mRNA levels of only those genes that are required for growth. Interestingly, we note that after heat shock Sulfolobus cells do not appear to resume growth until TFE levels have recovered. This observation suggests that TFE may be one of several factors that are required for outgrowth following heat shock.
Although in vitro biochemical studies have indicated that TFE stabilizes initiation complexes (34), is part of elongation complexes (14), and enhances A-box recognition by TBP (2), it is unclear how transcription in vivo under heat shock conditions may proceed in its absence. Whatever the precise role of TFE might turn out to be, the results presented in this study can be used to infer that in Sulfolobus TFE does not appear to serve the role of a general transcription factor. Further studies are required to understand the in vivo role of TFE in transcription, as well as to decipher the mechanism by which TFE is depleted under heat shock conditions and how it impacts gene expression patterns to confer thermoprotection.
Acknowledgments
We are extremely grateful to Romena Qazi (Department of Pathology and Microbiology, AKU) for technical help and valuable suggestions.
This work was supported by grants from the Aga Khan University Research Council (J.I.) and the Pakistan Science Foundation (S.A.Q.). J.I. is supported by a predoctoral fellowship awarded by AKU.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Bartlett, M. S. 2005. Determinants of transcription initiation by archaeal RNA polymerase. Curr. Opin. Microbiol. 8:677-684. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., A. B. Brinkman, J. van der Oost, and S. P. Jackson. 2001. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, S. D., and S. P. Jackson. 2000. The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem. 275:12934-12940. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. D., P. L. Kosa, P. B. Sigler, and S. P. Jackson. 1999. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. U. S. A. 96:13662-13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 1998. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180:3237-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, L., K. Brugger, M. Skovgaard, P. Redder, Q. She, E. Torarinsson, B. Greve, M. Awayez, A. Zibat, H. P. Klenk, and R. A. Garrett. 2005. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coker, J. A., P. DasSarma, J. Kumar, J. A. Muller, and S. DasSarma. 2007. Transcriptional profiling of the model archaeon Halobacterium sp. NRC-1: responses to changes in salinity and temperature. Saline Systems 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conway de Macario, E., and A. J. Macario. 1994. Heat-shock response in Archaea. Trends Biotechnol. 12:512-518. [DOI] [PubMed] [Google Scholar]
- 9.Darcy, T. J., W. Hausner, D. E. Awery, A. M. Edwards, M. Thomm, and J. N. Reeve. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181:4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frols, S., P. M. Gordon, M. A. Panlilio, I. G. Duggin, S. D. Bell, C. W. Sensen, and C. Schleper. 2007. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189:8708-8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiduschek, E. P., and M. Ouhammouch. 2005. Archaeal transcription and its regulators. Mol. Microbiol. 56:1397-1407. [DOI] [PubMed] [Google Scholar]
- 12.Gotz, D., S. Paytubi, S. Munro, M. Lundgren, R. Bernander, and M. F. White. 2007. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 8:R220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grogan, D. W. 1989. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J. Bacteriol. 171:6710-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grunberg, S., M. S. Bartlett, S. Naji, and M. Thomm. 2007. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 282:35482-35490. [DOI] [PubMed] [Google Scholar]
- 15.Hanzelka, B. L., T. J. Darcy, and J. N. Reeve. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEalpha. J. Bacteriol. 183:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hethke, C., A. C. Geerling, W. Hausner, W. M. de Vos, and M. Thomm. 1996. A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res. 24:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudepohl, U., W. D. Reiter, and W. Zillig. 1990. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc. Natl. Acad. Sci. U. S. A. 87:5851-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagawa, H. K., J. Osipiuk, N. Maltsev, R. Overbeek, E. Quaite-Randall, A. Joachimiak, and J. D. Trent. 1995. The 60 kDa heat shock proteins in the hyperthermophilic archaeon Sulfolobus shibatae. J. Mol. Biol. 253:712-725. [DOI] [PubMed] [Google Scholar]
- 19.Kagawa, H. K., T. Yaoi, L. Brocchieri, R. A. McMillan, T. Alton, and J. D. Trent. 2003. The composition, structure and stability of a group II chaperonin are temperature regulated in a hyperthermophilic archaeon. Mol. Microbiol. 48:143-156. [DOI] [PubMed] [Google Scholar]
- 20.Korkhin, Y., U. M. Unligil, O. Littlefield, P. J. Nelson, D. I. Stuart, P. B. Sigler, S. D. Bell, and N. G. Abrescia. 2009. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 7:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuldell, N. H., and S. Buratowski. 1997. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol. Cell. Biol. 17:5288-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maaty, W. S., B. Wiedenheft, P. Tarlykov, N. Schaff, J. Heinemann, J. Robison-Cox, J. Valenzuela, A. Dougherty, P. Blum, C. M. Lawrence, T. Douglas, M. J. Young, and B. Bothner. 2009. Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PLoS One 4:e6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxon, M. E., J. A. Goodrich, and R. Tjian. 1994. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 8:515-524. [DOI] [PubMed] [Google Scholar]
- 24.Meinhart, A., J. Blobel, and P. Cramer. 2003. An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 278:48267-48274. [DOI] [PubMed] [Google Scholar]
- 25.Ouhammouch, M., F. Werner, R. O. Weinzierl, and E. P. Geiduschek. 2004. A fully recombinant system for activator-dependent archaeal transcription. J. Biol. Chem. 279:51719-51721. [DOI] [PubMed] [Google Scholar]
- 26.Paytubi, S., and M. F. White. 2009. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol. Microbiol. 72:1487-1499. [DOI] [PubMed] [Google Scholar]
- 27.Pysz, M. A., D. E. Ward, K. R. Shockley, C. I. Montero, S. B. Conners, M. R. Johnson, and R. M. Kelly. 2004. Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8:209-217. [DOI] [PubMed] [Google Scholar]
- 28.Qureshi, S. A., S. D. Bell, and S. P. Jackson. 1997. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 16:2927-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qureshi, S. A., and S. P. Jackson. 1998. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol. Cell 1:389-400. [DOI] [PubMed] [Google Scholar]
- 30.Rohlin, L., J. D. Trent, K. Salmon, U. Kim, R. P. Gunsalus, and J. C. Liao. 2005. Heat shock response of Archaeoglobus fulgidus. J. Bacteriol. 187:6046-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shockley, K. R., D. E. Ward, S. R. Chhabra, S. B. Conners, C. I. Montero, and R. M. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tachdjian, S., and R. M. Kelly. 2006. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 188:4553-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vierke, G., A. Engelmann, C. Hebbeln, and M. Thomm. 2003. A novel archaeal transcriptional regulator of heat shock response. J. Biol. Chem. 278:18-26. [DOI] [PubMed] [Google Scholar]
- 34.Werner, F., and R. O. Weinzierl. 2005. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell. Biol. 25:8344-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner, F., and R. O. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10:635-646. [DOI] [PubMed] [Google Scholar]
- 36.Wiedenheft, B., J. Mosolf, D. Willits, M. Yeager, K. A. Dryden, M. Young, and T. Douglas. 2005. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. U. S. A. 102:10551-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokomori, K., C. P. Verrijzer, and R. Tjian. 1998. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc. Natl. Acad. Sci. U. S. A. 95:6722-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]