Abstract
Two-component systems are widespread prokaryotic signal transduction devices which allow the regulation of cellular functions in response to changing environmental conditions. The two-component system DccRS (Cj1223c-Cj1222c) of Campylobacter jejuni is important for the colonization of chickens. Here, we dissect the DccRS system in more detail and provide evidence that the sensor DccS selectively phosphorylates the cognate effector, DccR. Microarray expression profiling, real-time reverse transcription-PCR (RT-PCR), electrophoretic mobility shift assay, and primer extension analyses revealed that the DccRS regulon of strain 81116 consists of five promoter elements, all containing the consensus direct repeat sequence WTTCAC-N6-TTCACW covering the putative −35 promoter regions. One of these promoters is located in front of an operon encoding a putative macrolide efflux pump while the others are in front of genes coding for putative periplasmic or membrane proteins. The DccRS-regulated genes in C. jejuni strain 81116 are needed to enhance early in vivo growth of C. jejuni in 7-day-old chickens. The DccRS system is activated in the late stationary bacterial growth phase, probably by released metabolic products. Whole-genome mRNA profiling and real-time RT-PCR analysis under these conditions demonstrated that the system has no influence on the transcription of genes outside the DccRS regulon.
Eubacterial species typically harbor several dozen two-component systems, each responding to distinct cues and controlling the expression of discrete sets of genes. Usually they are composed of a histidine sensor protein that perceives environmental stimuli via its N-terminal input domain and a cognate response regulator. In the presence of the appropriate stimulus, the sensor protein autophosphorylates at a highly conserved histidine residue in the transmitter domain (13). Subsequently, the kinase activity of the histidine sensor transfers the phosphoryl group to an aspartic acid residue in the N-terminal receiver domain of the response regulator, resulting in a conformational change and activation of its C-terminal output domain, which frequently has DNA-binding properties. This ultimately leads to a bacterial phenotype that is optimally adapted to its ecological niche.
Campylobacter jejuni is the leading causative agent of bacterial enteritis worldwide (1). It is widespread throughout nature and can be isolated from many sources, including birds, domestic animals, and water. However, its most favored habitat appears to be the intestine of avian species, in which up to 109 CFU per gram of feces can be found (9). Ingestion of as few as 100 Campylobacter bacteria is sufficient to colonize the avian gut (12, 24). Little information regarding the molecular mechanisms regulating the expression of Campylobacter genes needed for optimal colonization of the host is available.
The relatively small genome (between 1,650 to 1,800 genes) of C. jejuni is regulated by only 37 putative transcription factors, of which 10 are response regulators (20). Therefore, many regulatory networks found in other bacterial species are missing in C. jejuni. Due to the lack of global stationary-phase regulatory elements, the physiological response to the conditions existing in stationary-phase cultures is restricted, and the transcription factors that might regulate this adaptation are largely unknown. One system shown to be important for the C. jejuni colonization of 1-day-old chickens is the response regulator DccR (8). This protein directly activates two hypothetical genes, Cj0200c and Cj1356c, but may regulate additional genes, as hypothesized from the presence of conserved DNA motifs (8). The role of the DccR/DccS system in chicken colonization as well as the signal that activates the system remains to be discovered.
In this study, we analyzed the DccR two-component system in more detail, and we provide evidence of the following in C. jejuni strain 81116: (i) DccR is the cognate response regulator for the sensor DccS, (ii) the two-component system is constitutively expressed under the conditions employed and not subject to autoregulation, (iii) the system upregulates seven genes through interaction of DccR with the promoter region, (iv) strong activation of the genes regulated by the DccRS system occurs in the late stationary phase, and (v) the system is not required for but accelerates colonization in the chicken.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. C. jejuni 81116 and derivatives were routinely maintained at 37°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) either on blood agar base II medium (Oxoid Ltd., London, United Kingdom) containing 5% horse blood lysed with 0.5% saponin (Sigma, St. Louis, MO) or in heart infusion (HI) broth (Oxoid). Escherichia coli was grown in Luria-Bertani medium at 37°C. When appropriate, growth media were supplemented with ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic(s) | Source or reference |
|---|---|---|
| C. jejuni strains | ||
| 81116 | Wild type | 10 |
| 81116 dccR::Cm mutant | 81116 derivative dccR::Cm | This study |
| E. coli strains | ||
| PC2955 | relA1 φ80dlacZΔM15 phoA8 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 luxS glnV44 | NCCB |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen Inc. |
| Plasmids | ||
| PGEM-T Easy | PCR cloning vector; Ampr | Promega Corp., Madison, WI |
| pAV35 | pBluescript II SK containing Campylobacter coli Cmr cassette | 16 |
| PGEMdccR | pGEM-T Easy containing dccR gene on a 2,009-bp fragment | This study |
| PGEMdccR::Cm | pGEMdccR with Cmr inserted in dccR | This study |
| pMA1 | C. jejuni-E. coli shuttle conjugative expression vector; Kmr | 15 |
| pMA1-1223 | pMA1 containing DccRS operon | This study |
| pT7.7 | Expression vector; Ampr | 14 |
| pT7-1223-H6C | pT7.7 containing Cj1223c gene with C-terminal His tag | This study |
| pT7-PhosR-H6N | pT7.7 containing Cj0890 gene with N-terminal His tag | 19 |
| pT7-1491-H6N | pT7.7 containing Cj1491 gene with N-terminal His tag | This study |
| pT7-1222-H6N | pT7.7 containing Cj1222c gene with N-terminal His tag | This study |
| pT7-RacR-H6N | pT7.7 containing racR gene with N-terminal His tag | This study |
Construction of recombinant protein overexpression plasmids.
To overexpress the complete response regulators DccR (Cj1223c), RacR (Cj1261), and Cj1491 and the cytoplasmic domain of sensor protein DccS (Cj1222c), chromosomal DNA of C. jejuni 81116 was amplified by PCR using the primer combinations Cj1223start/Cj1223Hisstop, RacRPstI/RacRNHISNdeI, HisCj1491start/Cj1491stop, and HisCj1222start/Cj1222stop, respectively (Table 2). The resulting PCR products of 683, 670, 680, and 678 bp were digested with NdeI and PstI and cloned into NdeI and PstI sites of plasmid pT7.7 to form pT7-DccR-H6C, pT7-RacR-H6N, pT7-1491-H6N, and pT7-DccSc-H6N (where H6 is an N-terminal [H6N]or C-terminal [H6C] His6 tag and where DccSc is the cytoplasmic domain of DccS), respectively. The nucleotide sequence of the cloned PCR products was verified by sequencing using an ABI Prism 310 genetic analyzer (Applied Biosystems Inc., Foster City, CA) and an ABI dye terminator cycle sequencing kit.
TABLE 2.
Primers used in this study
| Primer function and name | Sequence (5′-3′) |
|---|---|
| Mobility shift assay | |
| Cj200RDig | GTTTTAGACTATCTGCAAAA |
| CJ201F | TTTCATCTTCAATATACTCTAA |
| Cj605FDig | ATGTGATGATGGCTTTAGAATT |
| CJ606R2 | GAGTTAAATAGCTGATTTTTT |
| Cj1003cRDig | GATATTGTAATAATCATAATCATAAGAAA |
| Cj1004F | TTTAAAGATAAGCTTAAAATAGTAA |
| Cj1223FDig | ATGATCTCACTCAAGCTCAA |
| Cj1223CR | TGAAAGGATTATAGATAAAA |
| Cj1356RDig | AGAACAAACTAAGATAGATAT |
| Cj1357F | AGCTTGCTCCTTATAATTAT |
| Cj1626RDig | ATCTTTAGCTAAAAGCACCG |
| CJ1627F | AAGATGGAAAAGAAATAAAATTT |
| Cj1723RDig | AGAAATTGTATTTGCACTTG |
| CJ1724F | CGGAAATGTACACACAGTCA |
| Real-time RT-PCR | |
| Cj0200Ftaq | CAACAAAACCTTGCTTATTGACTCTATT |
| Cj0200Rtaq | CACAATCATCCATTTCTATTTCGGTAT |
| Cj0606Ftaq | ATGATTTTTTGCGTATTGGTATGAGT |
| Cj0606Rtaq | TTAATTGCGTATTGGTATGAGT |
| Cj0607Ftaq | CATACTTGCACACAAACTTCGTTCT |
| Cj0607Rtaq | GCCTAATCCTAAAGCTACAACACAA |
| Cj0608Ftaq | CAGATGTGAGATCTAAACTCAATAGCTTAAA |
| Cj0608Rtaq | TCCCACTTAATGCTCCACCTAAA |
| Cj1004Ftaq | TCCTTCCAAGCTCCATCTATATCAA |
| Cj1004Rtaq | CGTGCAAAAAGACAAAAATTCCT |
| Cj1356Rtaq | TCTTTACTTGGAGATATGATTATGCTTCAT |
| Cj1356Ftaq | CAAACAAAGTAATCATAAGCCCTGAAA |
| Cj1626Rtaq | TTCCTTGCATATCTGTATAAACTTCCA |
| Cj1626Ftaq | GCTTTGCCACATACTATCTTACCTAATG |
| Cj1222Ftaq | AGCCTTGTTTTAAATCATTTGTTGTG |
| Cj1222Rtaq | AGAATTAAGAAAAAGCGGAAAGCA |
| Protein expression | |
| Cj1223HisPstI | TTTACTGCAGTTAATGGTGATGGTGATGGTGGCCATAGCAATATCCCCTG |
| Cj1223NdeI | TATACATATGGCTGCTAAAATTTTACTTTT |
| Cj1222HisNdeI | ATTCATATGCACCATCACCATCACCATAAAATTGCTCTAAAGCCTTTG |
| Cj1222PstI | TGACTGCAGTTACCAATTTAATTTAAAAAC |
| RacRPstI | TTCTGCAGTCATCCTATCAGTTTATAT |
| RacRNHISNdeI | TACATATGCATCACCATCACCATCACATTAATGTGTTGATGATAGA |
| Cj1492HisC | CTGCAGCTAATGGTGATGGTGATGGTGCTCTACCCTCTCTTTTAGTTTTA |
| Cj1492EcoRI | AAGAATTCTTAGCGATATAGCACATCAATGG |
| Primer extension | |
| Cj1223prex | ATGATCTCACTCAAGCTCAA |
| Cj0200R | GTTTTAGACTATCTGCAAAA |
| Cj1356R | AGAACAAACTAAGATAGATAT |
| Cj1626R | ATCTTTAGCTAAAAGCACCG |
| prex1004 | CAGAAATTATTATATCAGCA |
| prex0606 | AATAAGCCCCAACACTTCCA |
| Cloning | |
| Cj1223CF | AATATCTGCTCTAACCTCTT |
| Cj1223R | TGAAAGGATTATAGATAAAA |
| Cj1223RamHI | TCAGGATCCAAGCTTTGCTTAAAAGAGCT |
| Cj1223FamHI | GCAGGATCCTAACCCTGATAGATAACTCG |
Purification of recombinant proteins.
Histidine-tagged DccS, DccR, PhosR, Cj1491, RacR, and FlgR proteins were expressed in E. coli BL21(DE3) containing either plasmid pT7-DccS-H6C, pT7-DccR-H6N, pT7-PhosR-H6N, pT7-1491-H6N, pT7-RacR-H6N, or pT7-FlgR-H6N.Protein expression and purification were performed as described previously (21). Protein concentrations were determined by the Bradford method (3).
Phosphorylation assays.
Recombinant DccSc-H6C (5.2 pmol) and DccR-H6N (0.85 pmol) proteins were incubated at room temperature for 15 min, with 10 μCi of [γ-32P]ATP (MP Biomedicals Netherlands) in 15 μl of phosphorylation buffer (50 mM Tris-HCl, 75 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol [DTT], pH 8.3). Phosphorylation of 0.85 pmol of DccR-H6 was achieved by the addition of 5.2 pmol of autophosphorylated DccSc-H6 in 15 μl of phosphorylation buffer. The reaction was stopped with SDS loading buffer after 0.5, 2, 4, 8, 16, or 32 min. Cross-phosphorylation experiments were performed by incubating 0.85 pmol of DccR-H6N, 0.8 pmol of PhosR-H6N, 1.5 pmol of Cj1491-H6N, 1.0 pmol of RacR-H6N, or 0.5 pmol of FlgR-H6N protein with 5.2 pmol of phosphorylated DccSc-H6C protein for 15 min at room temperature in 15 μl of phosphorylation buffer. The reaction was stopped with loading buffer. Samples were run on 12.5% SDS-polyacrylamide gels. After electrophoresis, the gel was dried and autoradiographed.
Construction of the dccR mutant.
A 1,990-bp DNA fragment containing the complete dccS and dccR genes was amplified from the C. jejuni 81116 chromosome using the primers Cj1223CF and Cj1223R (Table 2). The product was cloned into pGEM-T Easy, resulting in plasmid pGem1223. This plasmid was subsequently amplified with primers Cj1223RBamHI and Cj1223FBamHI. The resulting PCR product was digested with BamHI and ligated to a 0.7-kb BamHI fragment containing a chloramphenicol resistance gene (Cmr) of pAV35, resulting in the knockout construct pGEMdccR::Cm. Natural transformation was used to introduce the knockout constructs in C. jejuni 81116 (18). Homologous recombination resulting in double-crossover events was demonstrated by PCR.
Construction of a DccR complementation plasmid.
To complement the dccR::Cm mutant, the complete DccRS operon located on a 2.1-kb SphI-SacI fragment of plasmid pGem1223 was cloned into the E. coli-C. jejuni shuttle conjugative plasmid pMA1 (15). In the resulting complementation plasmid, pMA1-1223, the dccR and dccS genes are regulated by their own promoters. Finally, pMA1-1223 was introduced in the dccR::Cm mutant via conjugation (7).
RNA isolation.
Precultures of C. jejuni were grown at 32°C, 37°C, or 42°C in HI broth, diluted to an A550 of 0.02 in 5 ml of HI broth, and incubated on a gyratory shaker (150 rpm) under microaerobic conditions at 32°C, 37°C, or 42°C, respectively.
Total RNA was extracted from cultures of early logarithmic (EL), late logarithmic (LL), and late stationary (LS) phases (A550 of 0.1 to 0.3, 0.7 to 0.9, and 1.0 to 1.4, respectively) with an RNA-Bee kit (Tel-Test, Inc.) according to the manufacturer's specifications.
RNA was also isolated from early logarithmic cell cultures in which the medium had been replaced at 2 h before RNA extraction by fresh HI broth, phosphate-buffered saline (PBS), or spent filtrated (0.4 μm) stationary-phase culture medium.
Primer extension.
Localization of the 5′ mRNA start site of DccR-regulated promoters was performed by primer extension analysis using RNA extracted from wild-type strain 81116 and the dccR::Cm mutant grown to late logarithmic phase at 42°C and the primers Cj0200prex, Cj0606prex, Cj1004prex, Cj1356prex, and Cj1626prex (Table 2). cDNA synthesis was performed in a 25-μl volume with 20 μg of total RNA, 2 pmol of primer labeled with T4 polynucleotide kinase and 50 μCi [γ-32P]ATP, and 50 units of SuperScriptII RNase H− reverse transcriptase (Life Technologies, Inc.) according to the manufacturer's instructions. Extension products were analyzed by electrophoresis on a 6% polyacrylamide-7.5 M urea gel and compared with sequence ladders initiated with primer extension primers.
Real-time RT-PCR.
Real-time reverse transcription-PCR (RT-PCR) analysis was performed as previously described (21). The calculated threshold cycle (CT) for each gene amplification was normalized to the CT value for the gyrA gene amplified from the corresponding sample before calculating fold change using the arithmetic formula 2−ΔΔCT, where ΔΔCT = [(CT target − CT gyrA) phasex − (CT target − CT gyrA)phasey] or [(CT target − CT gyrA)mutant − (CT target − CT gyrA)wild type)] (11) and where target is the gene of interest, x is EL, LL, or LS, and y is EL. Each sample was examined in four replicates and was repeated with at least two independent preparations of RNA. Standard deviations were calculated and are displayed as error bars.
Microarray hybridization and analysis.
Microarray hybridization and analysis were performed as previous described (19) but with the use of total RNA extracted from wild-type and dccR::Cm mutant bacteria grown at late stationary phase.
Gel mobility shift assays.
The promoter regions upstream of the genes Cj0200c, Cj0606, Cj1004, dccR, Cj1356c, Cj1626c, and Cj1723c were amplified by PCR using C. jejuni 81116 chromosomal DNA as a template while the region upstream of Cj1723c was amplified using Cj11168 chromosomal DNA. Primers Cj1003cRDIG, Cj605FDIG, Cj200RDIG, Cj1626RDIG, Cj1723RDIG, and Cj1356RDIG were digoxigenin (DIG) labeled (Isogen). The Cj1004 (104 bp), Cj605 (240 bp), Cj200c (250 bp), Cj1626c (200 bp), Cj1723c (300 bp), and Cj1356c (232 bp) promoter regions were amplified using primer sets Cj1003cRDIG/Cj1004R, Cj605FDIG/Cj606R, Cj200RDIG/Cj201F, Cj1626RDIG/Cj1627F, Cj1723RDIG/Cj1724F, and Cj1356RDIG/Cj1357F, respectively (Table 2). Approximately 35 pmol of labeled DNA and 0, 7.5, 37.5, or 187.5 pmol of DccR-H6 protein in a 15-μl volume were incubated at room temperature for 15 min. The binding buffer used for protein-DNA incubations contained 20 mM Tris, 5 mM MgCl2, 50 mM KCl, 50 μg of bovine serum albumin, 1.0 μg of poly(dI-dC), and 5% glycerol, pH 7.4. Samples (20 μl) were run on a 4% nondenaturing Tris-glycine polyacrylamide gel at 4°C. Following electrophoresis, DNA was transferred to nylon membranes (Amersham) and UV cross-linked. DIG-labeled DNA was detected with anti-DIG antibodies according to the manual of the manufacturer (Roche).
Chicken experiments.
Challenge experiments were performed with Ross 308 broiler chickens housed in isolators as described previously (5). At day 7 after hatching, two groups of 25 animals were inoculated with an overnight HI broth culture of 105 CFU of either wild-type Campylobacter or dccR::Cm mutant bacteria. At days 2, 4, 7, and 14 after inoculation, five animals from each group were sacrificed, and C. jejuni was reisolated from cecal contents. The cecal contents were serially diluted and plated onto charcoal cefoperazone deoxycholate agar plates ([CCDA] item CM0739; Oxoid, Basingstoke, United Kingdom).
Microarray data accession number.
Microarray data were deposited in the Gene Expression Omnibus (GEO) database under accession number GSE19803.
RESULTS
Identification of the DccS/DccR regulon.
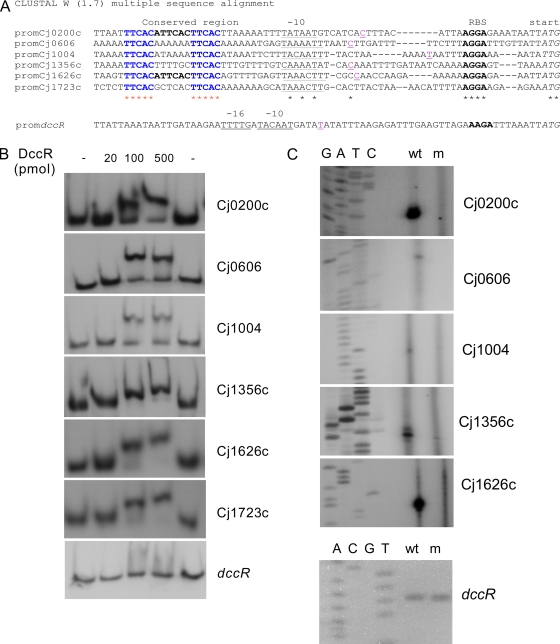
In C. jejuni strain 81-176, the DccRS system directly regulates the transcription of at least two genes encoding putative periplasmic and membrane proteins. These genes share a conserved repeat sequence (TTCAC) in their promoter regions (8). Analysis of all 14 C. jejuni genome sequences available in the public databases revealed that they contain either five or six promoter elements with the TTCAC motif that might be directly regulated by the DccR/DccS system (Fig. 1A). These elements were located upstream of the genes Cj0200c, Cj0606, Cj1004, Cj1356c, Cj1626c, and Cj1723c in strain 11168, which are annotated as putative periplasmic or membrane proteins. To obtain experimental evidence that these promoter elements are directly regulated by the DccR/DccS system, we performed electrophoretic mobility shift assays (EMSAs) (Fig. 1B). The DccR response regulator protein was isolated as a His-tagged recombinant protein (DccR-H6N) and incubated with DNA fragments containing the promoter regions of Cj0200c, Cj0606, Cj1004, Cj1356c, Cj1626c, and Cj1723c and of the dccR (Cj1223c) gene itself. DNA mobility shifts were seen for the Cj0200c, Cj0606, Cj1004, Cj1356c, Cj1626c, and Cj1723c promoter fragments, indicating that all of these promoters bind DccR. The dccR (Cj1223c) promoter fragment, used as a negative control, did not shift.
FIG. 1.
Identification of the DccS/DccR regulon. (A) Alignment of promoter DNA sequences containing a TTCAC repeat sequence which might be directly regulated by the DccRS system. Start codon (italic), ribosomal binding sequence (bold), putative −10 and −16 regions (underlined), transcription start points (pink underlined), and direct TTCAC repeats (blue) are indicated. (B) EMSAs with the DccR-H6N protein and the promoter regions in front of gene Cj0200c, Cj0606, Cj1004, Cj1356c, Cj1626c, Cj1723c, or dccR. The concentration of DccR-H6N in each reaction mixture is indicated at the top of each lane. (C) Mapping of the transcription start site of dccR promoter and DccR-regulated promoters by primer extension. Primer extension products were generated after reverse transcription of total RNA isolated from late-logarithmic phase wild-type (wt) strain or dccR mutant (m) grown in HI broth at 37°C. The primer extension product was run on a 6% sequencing gel against dideoxy sequencing reactions primed with the same primer.
To determine that the identified promoter regions are indeed regulated by the DccR/DccS system in vivo, we performed primer extension experiments. Total RNA was isolated from cultures of wild-type 81116 grown overnight and from a constructed dccR mutant. We did not perform primer extension on gene Cj1723c as it is not present in C. jejuni strain 81116. Primer extension assays yielded a single cDNA product for the genes Cj0200c, Cj0606, Cj1004, Cj1356c, Cj1626c, and dccR when wild-type RNA was used as a template (Fig. 1C). However, when total RNA of the dccR mutant was used, a cDNA product was detected for only the dccR gene. As the identified DccR-dependent transcription start sites were all located just downstream of the putative −10 region (Fig. 1A), these results clearly show that the transcription of the Cj0200c, Cj0606, Cj1004, Cj1356c, and Cj1626c genes is dependent on a functional DccR protein and, in addition, that the dccR gene is not autoregulated. These results are in agreement with the results of EMSAs.
Selective phosphorylation of DccR by DccS.
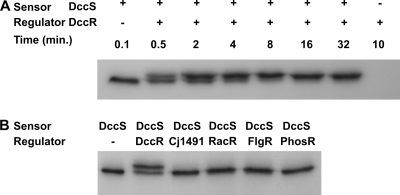
Components of two-component systems communicate with each other via phosphate transfer, but phosphate transfer may also occur between different two-component systems, thus expanding the DccS regulon. To investigate whether the sensor DccS is able to autophosphorylate and to transmit the phosphate group to the DccR response regulator, phosphorylation experiments were carried out. The DccR protein and the cytoplasmic domain of DccS were isolated as His-tagged labeled recombinant proteins and incubated with [γ-32P]ATP. The cytoplasmic domain of the sensor DccS showed rapid autophosphorylation in the presence of radioactive ATP (Fig. 2A). A rapid dephosphorylation of the sensor was seen when recombinant regulator DccR was added. After 16 min all the phosphate from the sensor was transferred to the regulator. In the absence of the sensor, phosphorylation of the response regulator was not observed.
FIG. 2.
Phosphorylation assays of the cytoplasmic domain of the sensor DccS and C. jejuni response regulators in vitro. Autophosphorylation of the truncated sensor DccSc-H6C was accomplished by incubation of DccSc-H6C with [γ-32P]ATP for 15 min at room temperature. (A) Time course of phosphotransfer from 32P-labeled DccSc-H6C to DccR-H6N. The DccR protein was incubated for 20 min with [γ-32P]ATP. (B) Phosphotransfer specificity of the sensor DccS. In this assay 32P-labeled DccSc-H6C was incubated with His-tagged recombinant DccR-H6N, Cj890-H6N, RacR-H6N, Cj1491-H6N, or l FlgR-H6N protein in vitro. Samples were run on a 12% SDS-polyacrylamide gel. Autophosphorylation and phosphotransfer were visualized by autoradiography.
To investigate whether DccS cross-phosphorylates other response regulators and thus regulates more regulons than DccR, we incubated phosphorylated DccS with other purified C. jejuni response regulators, e.g., FlgR, PhosR, RacR, and Cj1491 (Fig. 2B). DccS was able to phosphorylate only DccR. Conversely, the sensors FlgS, PhosS, RacS, and Cj1492 were not able to phosphorylate DccR (data not shown). These results indicate that the interaction of DccS and DccR is highly specific and that no cross-communication exists (at least at the level of phosphorylation) between the DccS/DccR, PhosS/PhosR, RacR/RacS, FlgS/FlgR, and Cj1491/Cj1492 two-component systems.
Environmental regulation of the activity of the DccRS system.
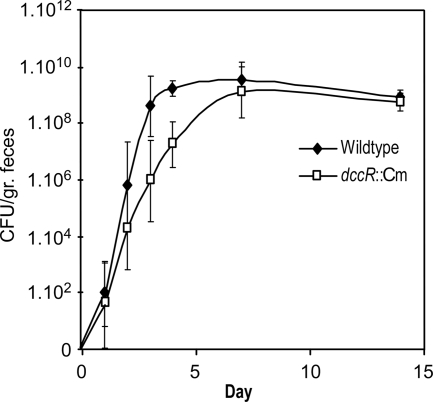
The DccRS system of strain 81-176 is required for the optimal colonization of 1-day-old chicks (8). We monitored, over time, the colonization of our 81116 dccR knockout strain in 7-day-old chickens (Fig. 3). Similar final colonization levels in the cecum were observed for the wild-type and mutant strains. However, the colonization was delayed for the mutant strain: maximum colonization levels were achieved at day 3 for the wild type but only at day 7 for the DccR mutant. This may indicate that the DccRS system is particularly relevant in the initial adaptation to the gastrointestinal milieu rather than for persistent colonization of the cecum.
FIG. 3.
Kinetic study of dccR mutant recovered from the cecum following oral inoculation of chickens. Ross 308 broiler chickens were inoculated with the wild type or dccR::Cm at a dose of approximately 1 × 105 CFU per chick. At various times postinoculation up to 14 days, five chickens per strain were sacrificed, and the number of CFU per gram of cecum content was determined. Each symbol represents the average value from five animals.
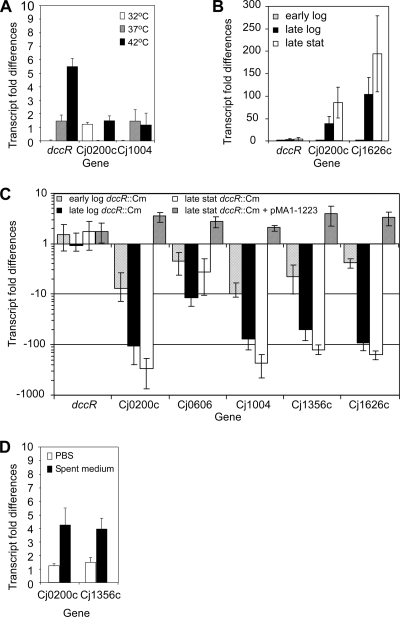
In the search for environmental cues that activate the DccRS system, we tested whether temperature influenced the transcription of the DccR regulon. Real-time RT-PCR on RNA isolated from cultures grown at 32°C, 37°C, and 42°C showed that transcript levels for the DccR-regulated genes Cj0200c and Cj1004 were independent of the growth temperature while dccR mRNA levels were almost 6-fold higher when bacteria were grown at 32°C than when they were grown at 42°C (Fig. 4A). These data indicate that the relatively high ambient body temperature of chickens of 42°C is not likely the cause of the DccRS activation.
FIG. 4.
Activation of the DccRS two-component system. (A) Real-time RT-PCR data showing the transcript fold difference of the dccR, Cj0200c, and Cj1004 mRNAs in wild-type C. jejuni grown at 32°C, 37°C, or 42°C. Fold change relative to the transcription levels was calculated using the arithmetic formula 2−ΔΔCT, where ΔΔCT = [(CT target − CT gyrA)temperature x − (CT target − CT gyrA)temperature y], where x is 42, 37, or 32°C and y is 32 or 37°C. (B) Growth-phase-dependent transcription of the dccR, Cj0200c, and Cj1656 genes as estimated for wild-type bacteria by real-time RT-PCR. Total RNA was extracted from cultures of early logarithmic, late logarithmic, and stationary phases (A550 of 0.1 to 0.3, 0.7 to 0.9, and 1.0 to 1.4, respectively) (C). Real-time RT-PCR data showing the transcript fold difference of the DccRS-regulated genes (Cj0200c, Cj0606, Cj1004, Cj1356c, and Cj1626c) in wild-type C. jejuni compared to the dccR mutant or to the dccR mutant complemented with plasmid pMA1-1223 at different phases of growth at 32°C. (D) Change in the transcript levels of the Cj0200c and Cj1004 genes in early-logarithmic-phase wild-type bacteria in response to the replacement of the medium by PBS or spent filtrated stationary-phase medium relative to the addition of fresh medium, as estimated by real-time RT-PCR. Data represent the mean values and standard deviations of four independent experiments with two independent preparations of RNA.
The putative DccRS homolog in Helicobacter pylori, the CrdRS two-component system, is required for copper-mediated induction of the copper resistance determinant CrdA (17). Growth curves of the C. jejuni 81116 wild-type strain and the dccR mutant in the presence of different concentrations of copper (0 to 0.5 μM) were identical. Real-time RT-PCR analysis confirmed that the transcript levels for the DccRS-dependent genes were not regulated by copper (data not shown).
Next, we investigated whether the bacterial growth phase influenced the transcription of the DccR regulon. Comparison of mRNA levels at different stages of bacterial growth revealed a clear increase in Cj0200c and Cj1656 mRNA levels after prolonged growth while dccR mRNA levels remained constant (Fig. 4B). To investigate whether the increase in mRNA for the DccR-regulated genes was due to the activation of the DccRS system, we performed real-time RT-PCR on the DccRS-regulated genes and the dccR gene itself by using total RNA of the wild-type strain, the dccR mutant, and the dccR mutant containing complementation plasmid pMA1-1223 isolated at different growth phases. No difference in dccR mRNA levels was observed, confirming the primer extension results that the dccR gene is not autoregulated (Fig. 4C). The differences in mRNA levels of different DccRS-upregulated genes were most prominent at the late stationary phase. This effect was less pronounced for Cj0606. Plasmid pMA1-1223 was able to complement the defect in the dccR gene as no transcript differences were observed between the wild-type and the complemented dccR mutant strain. Taken together, these results indicate that prolonged C. jejuni growth generates a signal that activates the DccRS system.
To determine if depletion of the medium or the accumulation of factors in the medium is the signal that activates the DccRS system at late stationary phase, we replaced the medium of wild-type bacteria grown to logarithmic phase with either PBS (no nutrients), spent filtrated stationary-phase culture medium (lacking nutrients and containing accumulated secreted factors), or fresh medium. Two hours after the medium replacement, RNA was extracted, and transcript levels were determined by real-time RT-PCR. After a 2-h incubation, the optical density levels at 600 nm (OD600s) of the PBS and spent filtrated medium were equal, indicating that C. jejuni was unable to grow in both media. Transcript levels for the DccR-regulated genes Cj0200c or Cj1004 were 5-fold higher for bacteria kept in spent medium than in for bacteria in fresh medium while no difference in their mRNA levels was detected for bacteria kept in PBS (Fig. 4D). These results suggest that the DccRS system may be activated by a factor that accumulates in the stationary phase rather than by a lack of nutrients.
Identification of indirectly DccR-regulated genes.
Knowledge of the growth phase condition that activates the DccRS system provided the opportunity to test whether additional genes/transcription factors are indirectly regulated by this system. To identify all DccR-dependent genes of strain 81116, mRNA profiles were compared between the parent and dccR mutant by microarray analysis. Total RNA isolated from stationary-phase cells of the parent or dccR mutant was used. Microarray analysis indicated that no genes were ≥2-fold upregulated in the dccR mutant compared to the wild type. Six of the seven (Cj0200c, Cj0606, Cj0607, Cj1004, Cj1356c, and Cj1626c) directly DccR-regulated genes in strain 81116 were found among the eight genes which were more than 2-fold downregulated in the dccR mutant compared to the wild type (Table 3). The maximum change in transcription of indirectly DccR-regulated genes was 2.6-fold while that of directly regulated genes was more than 99-fold. None of the changes in indirectly DccR-regulated gene transcripts identified by microarray analysis could be confirmed by real-time RT-PCR (Table 3). These results suggest that mutation of DccR affects only directly regulated genes and thus that the complete regulon of the DccR has been elucidated.
TABLE 3.
Gene expression data given as transcript fold reduction for the C. jejuni 81116 dccR mutant
| Gene | Transcript fold reduction in stationary-phase cells (mean ± SD) |
|
|---|---|---|
| Microarray | Real-time RT-PCR | |
| Cj0200c | 99.8 ± 9.7 | 601.0 ± 221.3 |
| Cj0606 | 2.4 ± 0.38 | 7.4 ± 3.7 |
| Cj0607 | 2.4 ± 0.25 | 7.2 ± 3.2 |
| Cj0608 | 1.7 ± 0.31 | 7.2 ± 2.5 |
| Cj1004 | 5.0 ± 1.5 | 289.2 ± 155.3 |
| Cj1356c | 11.6 ± 1.1 | 301.7 ± 151.6 |
| Cj1626c | 18.4 ± 2.9 | 244.3 ± 120.3 |
| Cj1080c | 2.2 ± 1.2 | 1.2 ± 0.2 |
| Cj0988c | 2.6 ± 1.4 | 1.0 ± 0.2 |
DISCUSSION
In the present study, we provide experimental evidence that the C. jejuni proteins DccS and DccR form a classical two-component system that is activated in the late stationary growth phase. The system is not autoregulated but, upon activation, upregulates the activity of five or six Campylobacter promoters (depending on the C. jejuni strain) through direct binding of a specific DNA sequence located in front of these promoters. The DccS/DccR system neither communicates with other two-component systems nor influences transcription of genes outside the DccR regulon. The system was found to facilitate rapid colonization of 7-day-old chickens but was not essential for colonization of the cecum.
Previous genome analysis, DNA binding studies, and comparable phenotypes of Cj1223c and Cj1222c mutants indicated that DccS and DccR proteins likely form a two-component system (8). Our findings for the first time demonstrate that the DccS protein, indeed, autophosphorylates in the presence of ATP and that the phosphate is rapidly transferred to the cognate effector, DccR. This event was shown to be highly specific as DccS was unable to transfer its phosphoryl group to other effector molecules. Similarly, the sensor proteins FlgS, PhosS, RacS, and Cj1492 were unable to transfer the phosphoryl group to the DccR protein (data not shown). This specificity indicates that the cross talk between different two-component systems in C. jejuni may be limited. In other bacterial pathogens, promiscuity between two-component systems has been observed (23), allowing bacteria to combine signals to achieve beneficial cross-regulation among pathways (2). The lack of cross talk between DccS/DccR and other two-component systems indicates that the DccR-regulated genes are important only in response to the signal that activates the sensor DccS.
Our microarray results, real-time RT-PCR, and primer extension analysis revealed that the C. jejuni strain 81116 response regulator, DccR, activates five promoter elements located upstream of the genes Cj0200c, Cj0606, Cj1004, Cj1356c, and Cj1626c. These putative σ70-dependent promoters are all preceded by a WTTCAC-N6-TTCACW direct repeat located around the −35 region. Searching all available finished Campylobacter genome sequences for the single TTCAC sequence or direct repeat revealed that the gene Cj1723c in C. jejuni strain NCTC 11168 (and its homolog in other C. jejuni strains) is the only other gene that might be directly regulated by the DccRS system. EMSA confirmed binding of the DccR protein to all repeat-containing fragments. This suggests that the complete regulon of the DccRS has been identified. It should be noted that the pattern of transcription activation of the Cj0606 gene did not entirely follow that of other genes directly regulated by DccRS. This may indicate that the promoter of this gene is controlled by not only DccR but also other regulatory-mechanism factors. The absence of genes indirectly regulated by DccR confirms the observed lack of cross talk between DccS/DccR and other two-component systems and provides further evidence that this system is dedicated to a specific function.
Thus far, all C. jejuni two-component regulator mutants tested have been involved in the optimum colonization of chickens (4, 6, 8, 21) although the specific role of these systems has not been determined. MacKichan et al. (8) noted that a dccR mutant of strain 81-176 poorly colonized the cecum of 1-day-old chickens. Our in vivo challenge experiments, in which we monitored over time the colonization with the dccR mutant of strain 81116 in 7-day-old chickens, indicated that the two-component system is not essential but, rather, accelerates the colonization of the chicken cecum. The reason for this different result is unclear but may be related to the age of the animals, the experimental setup, and/or the use of different C. jejuni strains. Our data suggest that the genes activated by the DccR system may serve in the primary adaptation of C. jejuni to the chicken environment rather than in conferring bacterial persistence in the chicken. Identification of the specific signals that activate this system may explain the behavior of the bacterium under different environmental conditions and the specific functions associated with the genes present in DccR regulon.
We were able to eliminate several potential signals that might activate the DccRS system. Chickens have a relatively high body temperature of 42°C, yet we showed that temperature is not the signal that actives the DccRS system. Based on amino acid sequence homology, the DccRS system appears most closely related to the copper-induced CrdRS two-component system of H. pylori (17). Like temperature, the availability of copper did not influence the transcription of the DccR-regulated genes. However, our results show that the genes that are directly regulated by the DccRS system are upregulated in the late stationary phase. This indicates that the DccRS system is activated by a specific signal that is present in the late stationary phase. C. jejuni undergoes numerous gene expression changes and substrate switching when it enters the stationary phase (22). Obvious stationary-phase signals, like pH, oxygen or nitrogen radicals (data not shown), starvation, or depletion of ions, however, did not influence the activation of the DccRS system. Based on the findings that the DccRS-regulated genes are activated in the stationary phase, together with the reduced initial chicken colonization of the dccR mutant, we speculate that the signals that activate the two-component system in the stationary phase are also present in the upper intestinal tract rather than in the cecum.
Acknowledgments
This work was supported by a fellowship of the Royal Netherlands Academy of Arts and Science, NWO-VIDI grant 917.66.330 to M.M.S.M. Wösten, by a USDA ARS award to C.T. Parker, and by funds from USDA CRIS project 5325-42000-045-00D.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourret, R. B. 2008. Signal transduction meets systems biology: deciphering specificity determinants for protein-protein interactions. Mol. Microbiol. 69:1336-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bras, A. M., S. Chatterjee, B. W. Wren, D. G. Newell, and J. M. Ketley. 1999. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J. Bacteriol. 181:3298-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 6.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 7.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacKichan, J. K., E. C. Gaynor, C. Chang, S. Cawthraw, D. G. Newell, J. F. Miller, and S. Falkow. 2004. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol. Microbiol. 54:1269-1286. [DOI] [PubMed] [Google Scholar]
- 9.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer, S. R., P. R. Gully, J. M. White, A. D. Pearson, W. G. Suckling, D. M. Jones, J. C. Rawes, and J. L. Penner. 1983. Water-borne outbreak of Campylobacter gastroenteritis. Lancet 1:287-290. [DOI] [PubMed] [Google Scholar]
- 11.Schmittgen, T. D. 2001. Real-time quantitative PCR. Methods 25:383-385. [DOI] [PubMed] [Google Scholar]
- 12.Shreeve, J. E., M. Toszeghy, M. Pattison, and D. G. Newell. 2000. Sequential spread of Campylobacter infection in a multipen broiler house. Avian Dis. 44:983-988. [PubMed] [Google Scholar]
- 13.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 14.Tabor, S., and C. C. Richardson. 1987. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 84:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Mourik, A., N. M. Bleumink-Pluym, L. van Dijk, J. P. van Putten, and M. M. Wösten. 2008. Functional analysis of a Campylobacter jejuni alkaline phosphatase secreted via the Tat export machinery. Microbiology 154:584-592. [DOI] [PubMed] [Google Scholar]
- 16.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waidner, B., K. Melchers, F. N. Stahler, M. Kist, and S. Bereswill. 2005. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J. Bacteriol. 187:4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 19.Wösten, M. M., C. T. Parker, A. van Mourik, M. R. Guilhabert, L. van Dijk, and J. P. van Putten. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278-291. [DOI] [PubMed] [Google Scholar]
- 20.Wösten, M. M., A. van Mourik, and J. P. Van Putten. 2008. Regulation of genes in Campylobacter, p. 611-624. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 21.Wösten, M. M., J. A. Wagenaar, and J. P. Van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 22.Wright, J. A., A. J. Grant, D. Hurd, M. Harrison, E. J. Guccione, D. J. Kelly, and D. J. Maskell. 2009. Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155:80-94. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto, K., K. Hirao, T. Oshima, H. Aiba, R. Utsumi, and A. Ishihama. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448-1456. [DOI] [PubMed] [Google Scholar]
- 24.Young, C. R., R. L. Ziprin, M. E. Hume, and L. H. Stanker. 1999. Dose response and organ invasion of day-of-hatch Leghorn chicks by different isolates of Campylobacter jejuni. Avian Dis. 43:763-767. [PubMed] [Google Scholar]