Abstract
Prokaryotic ubiquitin-like protein (Pup) is a posttranslational modifier that targets proteins for degradation by the mycobacterial proteasome. We show that the disordered amino terminus of Pup is required for degradation, while the helical carboxyl terminus mediates its attachment to proteins. Thus, Pup has distinct regions that either interact with pupylation enzymes or initiate proteasomal degradation.
Proteasomes are compartmentalized ATP-dependent proteases found in all eukaryotes and archaea, as well as in bacteria of the order Actinomycetales (reviewed in reference 2). Proteins destined for proteasomal degradation in eukaryotes are usually tagged with chains of the small protein ubiquitin (Ub) (3, 11, 13). Ub is processed from larger polypeptides to expose a diglycine (Gly-Gly) sequence, which is subjected to a series of transthioesterification reactions that ultimately attach Ub via an isopeptide bond to a lysine (Lys) on a target substrate (reviewed in reference 12). Additional Ub molecules are successively attached to the first Ub, forming polyubiquitin chains that can be recognized by the regulatory complex of the proteasome, leading to the destruction of the protein.
In eukaryotes, a two-part degron, including a proteasome binding signal and a disordered degradation initiation site, has been shown to be required for optimal degradation by the proteasome (reviewed in reference 18). Ub is a highly structured protein, with a characteristic “β-grasp” fold (20), and is thought to provide the binding signal for the degron. Disordered regions on the doomed proteins, which are not uncommon to eukaryotic proteins, serve as the degradation initiation sites to complete the two-part degron.
In Mycobacteria, prokaryotic ubiquitin-like protein (Pup) serves an analogous function to Ub by forming isopeptide bonds to Lys of proteins doomed for bacterial proteasomal degradation (1, 5, 17), although the enzymology of pupylation appears to be highly different from that of ubiquitylation (5). The only common features between Ub and Pup appear to be their small size and a diglycine motif at or near the carboxyl (C) terminus. Nuclear magnetic resonance (NMR) studies revealed that Pup is largely unstructured, unlike Ub (4, 14, 19). Additionally, NMR studies showed that the C-terminal helical part of Pup interacts directly with the proteasome-associated ATPase Mpa, an observation that confirmed earlier genetic analysis (17). Pup interacts with the amino (N)-terminal coiled-coil domain of Mpa (19) at a ratio of 1:6 (4, 19), supporting the hypothesis that Pup is required for targeting proteins to the proteasome. Because the N-terminal half of Pup is disordered, which is an uncommon characteristic of prokaryotic proteins, we hypothesized that this portion of Pup provides a signal to initiate degradation by the bacterial proteasome.
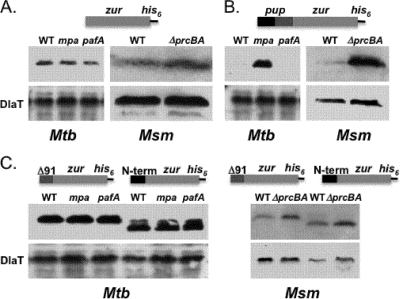
To test this hypothesis, we constructed a linear fusion of Pup to a protein that is not a robust proteasome substrate. Linear fusions of Ub have provided powerful tools for studying the Ub-proteasome system in eukaryotes (16). We cloned pup upstream of zur (zinc uptake regulator; Rv2359), with a his6 sequence at the 3′ end of zur in vector pSYMP (Fig. 1; see also Table S1 in the supplemental material). Zur is a transcriptional regulator that was studied in our lab, but it was not a robust proteasome substrate in either Mycobacterium tuberculosis or Mycobacterium smegmatis (Fig. 1A; see also Table S1 in the supplemental material). Pup-Zur-His6 was barely detectable in wild-type M. tuberculosis H37Rv but dramatically accumulated protein in the mpa mutant (Fig. 1, left). Pup-Zur-His6 also accumulated in the M. smegmatis proteasome-deleted (ΔprcBA) strain (Fig. 1B). These observations suggest that Pup targeted Zur-His6 to the M. tuberculosis proteasome in an Mpa-dependent manner. Pup-Zur-His6 was less detectable in the M. tuberculosis pafA (Pup ligase) mutant than in the wild-type strain (Fig. 1B); we originally anticipated that the steady-state levels of the fusion protein would be similar between wild-type and pafA strains; however, it is possible that the fusion protein was more efficiently degraded in the pafA mutant because there were no other pupylated proteins in the bacterium to compete for access to the proteasome. Importantly, our data show that PafA is itself not directly required for proteolysis. Taken together, the fusion of Pup to Zur resulted in the synthesis of a recombinant protein dependent on the proteasome for turnover.
FIG. 1.
Fusion of Pup to a nonproteasome substrate targets it for proteasomal degradation in an Mpa- and proteasome-dependent manner. Representations of linear fusions expressed in M. tuberculosis (Mtb) and M. smegmatis (Msm) are shown above each panel. (A) M. tuberculosis H37Rv Zur-His6 (Rv2359) is similarly stable at steady state in all M. tuberculosis and M. smegmatis strains tested. (B) Pup-Zur-His6 accumulates in an mpa mutant but not in wild-type or pafA M. tuberculosis strains (left). Pup-Zur-His6 accumulates in ΔprcBA but not wild-type M. smegmatis. The entire pup gene was fused to zur. The terminal Pup Gln codon was changed to Glu. In three independent experiments, less fusion protein was detected in the pafA mutant than in wild-type M. tuberculosis. (C) Deletion of nucleotides 4 to 90 (Δ91; deletion of amino acids 2 to 30) or 92 to 195 (N-term; deletion of amino acids 31 to 64) resulted in fusion genes that were equally stable in wild-type and degradation-defective strains (right). Experiments are representative of two independent replicate samples for each strain. Dihydrolipoamide acyltransferase (DlaT) is the loading control. Mutants are described elsewhere (1, 6). All cultures were processed as described elsewhere (7). For immunoblot analysis, cell lysates were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed with monoclonal antibodies to His5 (Qiagen) or polyclonal antibodies to DlaT (a gift from R. Bryk and C. Nathan).
We next tested the hypothesis that the N-terminal disordered half of Pup is required for degradation. We deleted nucleotides 4 to 90 to create “pup91-zur-his6” in the same plasmid used to examine full-length Pup-Zur-His6 (see Table S1 in the supplemental material). Unlike full-length Pup, the N-terminal truncated Pup-Zur fusion protein had the same steady-state levels in wild-type and degradation-defective M. tuberculosis and similar steady-state levels in wild-type and proteasome-deleted M. smegmatis (Fig. 1C). These results show a robust difference in the stability of full-length Pup-Zur-His6 compared to that of Pup91-Zur-His6 in wild-type M. tuberculosis and M. smegmatis. We also made a plasmid with only the first 30 amino acids of Pup fused to the N terminus of Zur-His6 (“pup-nterm-zur-his6”). The first 30 residues of Pup were not sufficient to target the recombinant protein to the proteasome (Fig. 1C). Thus, each half of Pup is necessary but not sufficient to target a protein to the mycobacterial proteasome.
NMR analysis determined that Mpa interacts with the C-terminal half of Pup but not with the disordered N terminus (4, 14, 19). In addition to providing an unstructured degron for proteasome substrates, we hypothesized that the N terminus of Pup interacts with enzymes of the pupylation pathway. To test this, we deleted nucleotides 4 to 90 from a strong expression plasmid used to purify the pupylome of M. tuberculosis, pUV15-his6-pup (see Table S1 in the supplemental material) (9). pUV15-his6-pup results in the synthesis of His6-Pup, which pupylates proteins in either M. tuberculosis or M. smegmatis. We purified the pupylome using either full-length His6-Pup or His6-Pup91 from wild-type M. smegmatis. To our surprise, the truncated Pup, consisting of only the C-terminal 34 amino acids, resulted in the modification of proteins (Fig. 2A and B). The entire pupylome appeared to shift to a lower molecular weight. We examined the pupylation pattern of a known proteasome substrate, Ino1 (inositol-1-phosphate synthase) (9). Using antibodies to Ino1, we observed the following two species of Ino1 in each purified pupylome sample: unpupylated Ino1 and either Pup∼Ino1 or Pup91∼Ino1 (Fig. 2C). Ino1 forms tetramers (15), but not all monomers within a tetramer are necessarily pupylated; unpupylated Ino1 copurifies with pupylated Ino1 (K. E. Burns and K. H. Darwin, unpublished observations). Ino1 was robustly modified by either Pup or Pup91, demonstrating that the N-terminal disordered region is not required for pupylation.
FIG. 2.
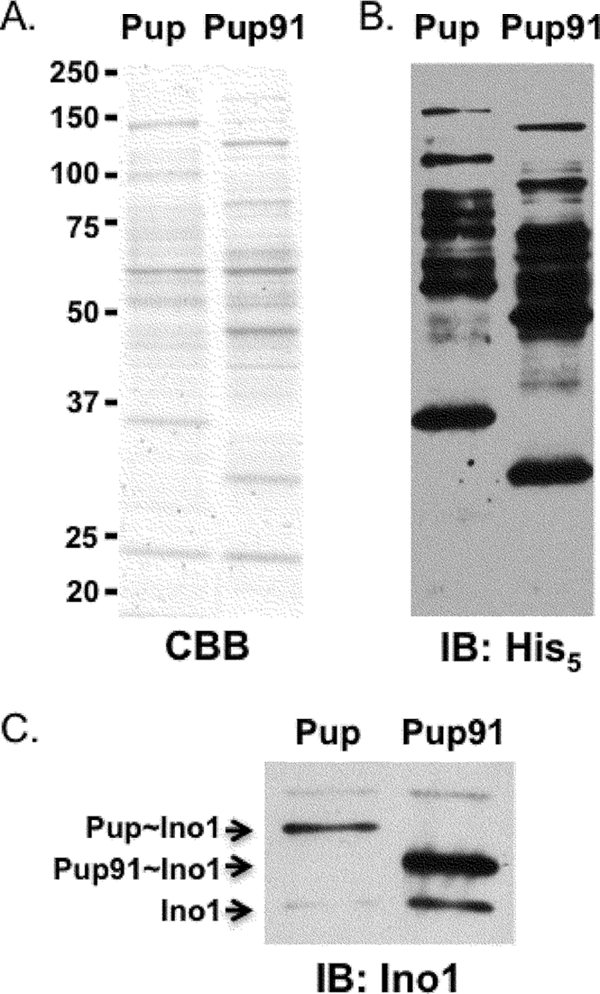
The Pup C terminus is sufficient for pupylation. Ectopic expression of his6-pup or his6-pup91 (ΔN-term-Pup) and subsequent Ni-nitrilotriacetic acid (NTA) purification of pupylated proteins from M. smegmatis. (A) Coomassie brilliant blue (CBB)-stained SDS-PAGE gel of Ni-NTA purified pupylomes. (B) Immunoblot (IB) analysis of purified proteins using antibodies to His5. (C) Anti-Ino1 immunoblot shows Ino1 is pupylated with either full-length or truncated Pup. The pupylomes were purified from wild-type M. smegmatis cultures harvested at an optical density at 580 nm (OD580) of 1. The amount of protein loaded was normalized based on protein concentration of the eluted proteins, as determined by A280 using a NanoDrop spectrophotometer.
Our data suggest that the truncated Pup91 can modify proteins (Fig. 2); however, based on the Pup-Zur-His6 linear fusion experiments, it cannot target them for proteasomal degradation (Fig. 1). We reasoned that strong overproduction of Pup91 in mycobacteria would compete with wild-type Pup to modify degradation substrates like Ino1, ultimately resulting in reduced proteolysis of the substrate. To test this, we examined endogenous Ino1 steady-state levels in lysates containing pUV15+ (vector control), pUV15-His6-pup, or pUV15-His6-pup91 using antibodies to Ino1. pUV15+ encodes one of the strongest known mycobacterial promoters (8). We detected endogenous Ino1 in all samples; however, there was less Ino1 in wild-type M. smegmatis overproducing full-length His6-Pup than in all other samples (Fig. 3, second lane); this suggested that overproduction of His6-Pup could increase the degradation of endogenous Ino1 in wild-type M. smegmatis. In contrast, overproduction of His6-Pup91 resulted in a similar accumulation of Pup91-Ino1 in both wild-type and proteasome-deleted M. smegmatis (Fig. 3); this supports the hypothesis that the N-terminal half of Pup is not needed for pupylation but is required for proteasome-dependent degradation.
FIG. 3.
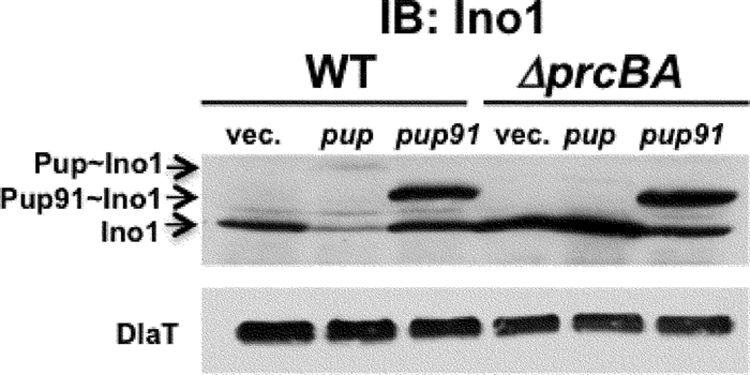
Expression of pup91 abrogates Ino1 degradation. Ectopic expression of his6-pup91 abrogates degradation of endogenous Ino1. Equivalent cell numbers were harvested, lysed by bead beating, and clarified by microcentrifugation for 10 min at 16,000 × g. Antibodies to Ino1 are described elsewhere (9). DlaT is the loading control.
Interestingly, although unpupylated Ino1 accumulated in the ΔprcBA strain as expected, full-length Pup-Ino1 did not (Fig. 3, compare lanes 2 and 5). We speculate that this is due to a possible feedback inhibitory mechanism by which pupylation is inhibited in the ΔprcBA strain. This may be unlikely, as the truncated Pup91 efficiently pupylates substrates in the ΔprcBA mutant (Fig. 3C, last lane) and the pupylome can be purified from the ΔprcBA mutant (Burns and Darwin, unpublished observations). Another hypothesis is that the absence of proteasome function results in the increased “depupylation” of Ino1. Nonetheless, our data show that the removal of the N-terminal half of Pup prevents proteasomal degradation of Ino1 and most likely other substrates.
In this study, we tested the model that the N-terminal half of Pup is critical to promote the degradation of a protein. Both linear fusions in mycobacterium mutants and overproduction of Pup lacking its N-terminal region strongly support this hypothesis. We found that the C-terminal half of Pup, in addition to the proteasomal ATPase Mpa, is required to interact with pupylation enzymes (4, 14, 17, 19). Taken together, we propose a model where (i) the C-terminal half of Pup interacts with enzymes to facilitate pupylation, (ii) the same region interacts with Mpa to target the protein to the proteasome, and (iii) the N-terminal half of Pup initiates degradation by the proteasome. It is important to point out that although it is likely that Pup is itself degraded when translationally fused to a protein like Zur, we do not know if Pup is degraded when conjugated by an isopeptide bond to a substrate. Thus, future work will determine if Pup can be removed and recycled like ubiquitin in eukaryotes (10).
Supplementary Material
Acknowledgments
We thank Ricky Festa for critical review of the manuscript. We are grateful to Randy Hampton and Kylie Walters for helpful discussions.
This work was supported by the National Institutes of Health (grant HL092774). K.H.D. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
Published ahead of print on 16 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Burns, K. E., W. T. Liu, H. I. Boshoff, P. C. Dorrestein, and C. E. Barry III. 2009. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J. Biol. Chem. 284:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerda-Maira, F., and K. H. Darwin. 2009. The Mycobacterium tuberculosis proteasome: more than just a barrel-shaped protease. Microbes Infect. 11:1150-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., W. C. Solomon, Y. Kang, F. Cerda-Maira, K. H. Darwin, and K. J. Walters. 2009. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J. Mol. Biol. 392:208-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin, K. H. 2009. Prokaryotic ubiquitin-like protein, proteasomes, and pathogenesis. Nat. Rev. Microbiol. 7:485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darwin, K. H., S. Ehrt, N. Weich, J.-C. Gutierrez-Ramos, and C. F. Nathan. 2003. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302:1963-1966. [DOI] [PubMed] [Google Scholar]
- 7.Darwin, K. H., G. Lin, Z. Chen, H. Li, and C. F. Nathan. 2005. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 55:561-571. [DOI] [PubMed] [Google Scholar]
- 8.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Festa, R. A., F. McAllister, M. J. Pearce, J. Mintseris, K. E. Burns, S. P. Gygi, and K. H. Darwin. 2010. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One 5:e8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finley, D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 12.Hochstrasser, M. 2009. Origin and function of ubiquitin-like proteins. Nature 458:422-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hough, R., and M. Rechsteiner. 1986. Ubiquitin-lysozyme conjugates. Purification and susceptibility to proteolysis. J. Biol. Chem. 261:2391-2399. [PubMed] [Google Scholar]
- 14.Liao, S., Q. Shang, X. Zhang, J. Zhang, C. Xu, and X. Tu. 2009. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem. J. 422:207-215. [DOI] [PubMed] [Google Scholar]
- 15.Norman, R. A., M. S. McAlister, J. Murray-Rust, F. Movahedzadeh, N. G. Stoker, and N. Q. McDonald. 2002. Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure 10:393-402. [DOI] [PubMed] [Google Scholar]
- 16.Papa, F. R., and M. Hochstrasser. 1993. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366:313-319. [DOI] [PubMed] [Google Scholar]
- 17.Pearce, M. J., J. Mintseris, J. Ferreyra, S. P. Gygi, and K. H. Darwin. 2008. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322:1104-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrader, E. K., K. G. Harstad, and A. Matouschek. 2009. Targeting proteins for degradation. Nat. Chem. Biol. 5:815-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutter, M., F. Striebel, F. F. Damberger, F. H. Allain, and E. Weber-Ban. 2009. A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 583:3151-3157. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar, S., C. E. Bugg, and W. J. Cook. 1987. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 194:531-544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.