Abstract
Toxin-antitoxin (TA) systems are plasmid- or chromosome-encoded protein complexes composed of a stable toxin and a short-lived inhibitor of the toxin. In cultures of Escherichia coli, transcription of toxin-antitoxin genes was induced in a nondividing subpopulation of bacteria that was tolerant to bactericidal antibiotics. Along with transcription of known toxin-antitoxin operons, transcription of mqsR and ygiT, two adjacent genes with multiple TA-like features, was induced in this cell population. Here we show that mqsR and ygiT encode a toxin-antitoxin system belonging to a completely new family which is represented in several groups of bacteria. The mqsR gene encodes a toxin, and ectopic expression of this gene inhibits growth and induces rapid shutdown of protein synthesis in vivo. ygiT encodes an antitoxin, which protects cells from the effects of MqsR. These two genes constitute a single operon which is transcriptionally repressed by the product of ygiT. We confirmed that transcription of this operon is induced in the ampicillin-tolerant fraction of a growing population of E. coli and in response to activation of the HipA toxin. Expression of the MqsR toxin does not kill bacteria but causes reversible growth inhibition and elongation of cells.
Bacterial toxin-antitoxin (TA) systems (for reviews, see references 16 and 54) are complexes consisting of a stable toxin component and a short-lived antitoxin. Toxins of TA systems are autotoxic; they target vital functions of the producing bacterium itself. TA complexes were discovered because they are plasmid-stabilizing entities. When a bacterium carries a plasmid which encodes a toxin and an antitoxin, both molecules are produced continuously and have no effect on the activities of the cell. When the plasmid is lost during cell division, the toxin is released and kills or inhibits the cell, because the unstable antitoxin is degraded faster. Chromosomal TA systems were found later (36), and comparative studies determined that they are widespread in free-living bacteria (35, 41).
There are two different types of bacterial TA systems, which depend on the nature of the antitoxin. In type I TA systems, the antitoxin is a small regulatory RNA (15, 18). In type II TA systems, which are relevant to this study, both the toxin and the antitoxin are proteins. Protein antitoxins neutralize toxins by direct interaction, forming catalytically inactive complexes. All known TA genes are organized as operons. The antitoxin is usually encoded by the first gene and always acts as a transcriptional autorepressor of the operon either alone or in a complex with the toxin molecule. Thus, antitoxins control toxin activity in two ways: through direct binding and through transcriptional regulation (17).
The toxins of type II systems attack essential functions of a bacterial cell, either protein synthesis through cleavage of free or ribosome-bound mRNA (e.g., RelE, MazF, HigB, and HicA) (10, 11, 25, 43) or the replication and integrity of DNA through interference with DNA gyrase (e.g., CcdB and ParE) (6, 24). TA pairs are grouped into 7 to 10 families (different authors propose different divisions) based on sequence similarities between the toxins (24, 54). The mechanisms of action and other characteristics of different TA families have been reviewed recently (16, 54) and are not discussed here. Targets of an increasing number of TA systems have been identified and their crystal structures have been described, but their role in the physiology of bacterial cells is unclear. Different authors have ascribed seemingly opposite functions to TA systems (for reviews, see references 34 and 56). Chromosome-encoded TA systems have been proposed to act as bacterial programmed cell death executioners, and in Escherichia coli, mazEF-mediated death under a variety of stressful conditions has been described by Hazan and colleagues (23). A homolog of MazF has recently been shown to have an essential role in cell death and lysis during Myxococcus xanthus fruiting body formation (39). Other workers have shown that TA toxins are activated in response to stress and starvation but have not reported any cell death (12). These workers proposed that TA systems have a protective role in survival under nutritional stress conditions and that toxins induce reversible growth arrest. Thus, TA toxins are actually not self-inflicted poisons but rather are global regulators of cell metabolism, growth, and division. Both explanations are challenged by experiments which have revealed no effect of deletions of the chromosomal TA operons on the survival and fitness of organisms (54). Whether the TA toxins are cytostatic or cytotoxic may depend on the strain background and experimental setup (29, 54). Growth arrest caused by a toxin typically can be reversed by controlled overexpression of the sequestering antitoxin (42). Proponents of the cell death hypothesis claim that reversal is possible only for a limited time. Thereafter, cells reach a “point of no return” and are unable to recover (3, 31).
Another open question is the role of TA systems in the formation of persisters. All bacterial populations contain individual cells that are not killed by antibiotics. These microbes are called persisters and are genetically identical to sensitive cells (33). A key to the nature of persisters seems to be their temporary nondividing state, since in most cases only proliferating bacteria are sensitive to the bactericidal effects of antibiotics (55). Different experiments have demonstrated that persisters are temporarily nonproliferating bacteria in a growing culture that, some time after removal of a drug, switch to the “normal,” proliferating state (5, 46). Understanding the mechanisms of persistence requires physical separation of the nondividing bacteria and analysis of their macromolecular content. This has been done by using two techniques: (i) killing and lysing the growing bacteria with ampicillin (27) and (ii) cell sorting of translationally inactive bacteria (on the basis of de novo green fluorescent protein [GFP] expression) (52). In both cases, transcription profiles of the dormant bacteria revealed strong and reproducible overexpression of chromosomal toxin-antitoxin operons (27, 52). As transcription of the TA operons is repressed by antitoxins, increases in transcription indicate that there are decreases in antitoxin levels and consequently activation of toxins.
An independent finding linking TA and persisters resulted from a targeted search for high-persistence mutants in E. coli (38). The point mutations that increased persister frequency several orders of magnitude were mapped to the hipA gene encoding a toxin of the hipBA TA module (31). Recently, the HipA toxin was demonstrated to have protein kinase activity and to phosphorylate elongation factor Tu (51). Also, deletion of hipAB affected survival in the stationary phase (27), and deletion of another TA toxin gene, yafQ, had a strong effect on the persister frequency in biofilms (22). Other data cast doubt on the role of TA systems in persister formation. For example, individual deletions of relBE, mazEF, or dinJ-yafQ had no effect on production of persisters by E. coli K-12 (27). A possible explanation for this is that there is functional redundancy of all or some of the TA systems. Also, the genome of E. coli may encode unknown TA systems with no sequence similarity to known TA pairs.
Among the genes that were induced in ampicillin-tolerant and quiescent E. coli subpopulations along with the known toxin-antitoxin operons are b3022 (ygiU, later renamed mqsR [19]) and b3021 (ygiT) (27, 52). These genes captured our attention because they have several characteristics of TA cassettes. mqsR and ygiT are located next to each other, and there is just 1 bp between the two open reading frames. Thus, these genes are probably cotranscribed. The lack of space for a ribosome-binding site between the two genes suggests that there is translational coupling, which is another characteristic of TA cassettes (47).
All protein antitoxins studied so far are transcriptional autorepressors. ygiT also encodes a putative DNA-binding protein consisting of 131 amino acids. The ygiT product contains a Cro/C1-type helix-turn-helix DNA-binding domain (http://www.uniprot.org/blast/?about=Q46864[74-127]) in its C-terminal region (amino acids 74 to 127) (49). It belongs to COG1396 (Clusters of Orthologous Groups of proteins; http://www.ncbi.nlm.nih.gov/COG), which also contains the known antitoxins HipB and HigA. The antitoxin gene candidate ygiT is the second gene, not the first gene, of the putative operon (see Fig. 3A); this organization is opposite to the widespread TA gene order but occurs in the higBA and hicAB toxin-antitoxin families (11, 25). mqsR encodes a 98-amino-acid protein with no similarity to known toxins and has previously been linked to biofilm formation (19, 45). Disruption of mqsR has been reported to abolish bacterial motility and reduce production of biofilms, particularly autoinducer 2-stimulated formation of biofilms (19). Moreover, the MqsR (YgiU)-YgiT family is among the new TA families recently predicted by bioinformatic analyses of microbial genome sequences (35).
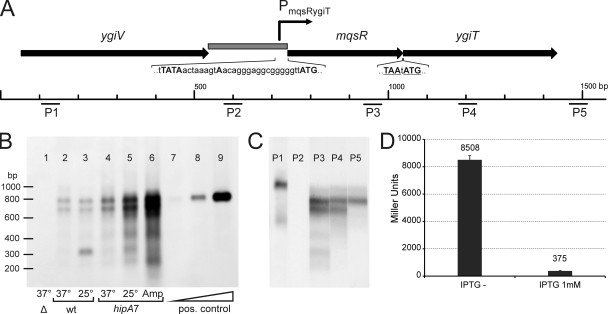
FIG. 3.
Organization and transcription of the mqsR-ygiT operon. (A) Map of the mqsR-ygiT region in the E. coli K-12 chromosome. The arrows represent the open reading frames. The stop codon of mqsR and the start codon of ygiT are underlined. The gray box represents the PmqsR-ygiT promoter region cloned in front of the lacZ reporter in the pPTX3 promoter probe plasmid. The Pribnow (−10) box, the 5′ end of the mqsR-ygiT transcript, and the start codon of mqsR are indicated by uppercase letters in the nucleotide sequence of PmqsR-ygiT. P1 to P5 indicate the locations of oligoprobes used in Northern hybridization. (B) Northern analysis of mqsR-ygiT transcription in different strains and under different conditions. Lane 1, HM21Δ [Δ(mqsR ygiT)]; lanes 2 and 3, HM21 (wt); lanes 4, 5, and 6, HM22 (hipA7); lanes 7, 8, and 9, 5-fold serial dilutions of the in vitro-synthesized mqsR-ygiT transcript. Bacteria were grown at 37°C for 2.5 h (to the exponential growth phase), and the cultures used for lanes 3 and 5 were transferred to 25°C and incubated for an additional 1.5 h. Ampicillin (100 μg ml−1) was added to the culture used for lane 6, and the culture was incubated at 37°C for additional 3 h to induce lysis of the sensitive bacteria and isolate the ampicillin-tolerant subpopulation. Total RNA was extracted from each sample, and 10-μg aliquots were subjected to electrophoresis, transferred to a membrane, and hybridized with fluorescein-labeled oligoprobe P3. Hybrids were detected using antifluorescein-AP conjugate and chemiluminescent detection. (C) Mapping of mqsR-ygiT transcripts by Northern oligoprobe hybridization. RNA was extracted from HM21 cells grown at 37°C for 2.5 h, and 10-μg portions were subjected to electrophoresis and transferred to a membrane. The membrane was cut into strips and hybridized with different fluorescein-labeled probes. P1 to P5 indicate the probes used for hybridization. (D) Repression of transcription from the PmqsR-ygiT promoter by YgiT. E. coli HM21Δ2 PTX lacZ carries a single copy of a Pmqs-RygiT::lacZ transcriptional fusion in the chromosome and was transformed with plasmid pAT3 for IPTG-inducible expression of YgiT. Overnight cultures were diluted 100-fold in fresh medium lacking IPTG (IPTG−) or containing 1 mM IPTG, and β-galactosidase levels were determined after 4 h. The values are the averages of results from two assays. The error bars indicate the standard errors.
Previously, we showed that overexpression of mqsR halts growth and induces tolerance to ofloxacin and cefotaxime, while a ygiT null mutant could not be obtained (52). In this work we determined experimentally that these two neighboring genes of E. coli K-12 encode a toxin-antitoxin system belonging to a completely new family.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains and plasmids used are listed in Table 1. Deletions Δ(mqsR ygiT), Δ(ygiV mqsR ygiT), ΔmazEF, and ΔlacZ were made using the method of Datsenko and Wanner (14) The PCR products for Red-mediated recombination were generated using knockout primers listed in Table 2. Deletions were verified with primers mqsR1, ygiT2, ygiV1, mazE1, and mazF2 (Table 2) in combination with primers k1, k2, and kt (14). For removal of the Kanr cassette from derivatives of HM21 and HM22, cells transformed with pCP20 were regrown nonselectively at 42°C because growth of these strains is hampered at 43°C. Single-copy integration of CRIM plasmid pPTX3 into the attλ site in HM21Δ2 was carried out, and integrants were verified as described by Haldimann and Wanner (21).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| MG1655 | F− λ−ilvG rfb-50 rph-1 | 7 |
| MG1655Δ2 | MG1655 Δ(mqsR ygiT) | This study |
| MG1655Δ3 | MG1655 Δ(ygiV mqsR ygiT) | This study |
| BW23473 | Δ(lacIZYA argF)U169 rph-1 rpoS396(Am) robA1 creC510 hsdR514 ΔendA9 uidA(ΔMluI)::pir(wt) recA1 | 21 |
| BW25113 | lacIqrrnB3 lacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | 21 |
| BW25113 ΔmotA | BW25113 ΔmotA::kan | 4 |
| BW25113 ΔfliC | BW25113 ΔfliC::kan | 4 |
| BW25113 ΔmotA | BW25113 ΔmotA::kan | 4 |
| BW25113 ΔmqsR | BW25113 ΔmqsR::kan | 4 |
| BW25113 ΔmqsR Kans | BW25113 ΔmqsR | This study |
| BW25113 ΔmqsR-ygiT | BW25113 Δ(mqsR ygiT) | This study |
| BW25113 ΔmazF | BW25113 ΔmazF::kan | 4 |
| BW25113 ΔmazEF | BW25113 ΔmazEF::kan | This study |
| BW25113 ΔrelE | BW25113 ΔrelE::kan | 4 |
| BW25113 ΔrelBE | BW25113 ΔrelBE | H. Luidalepp |
| HM21 | F+dapA zde-264::Tn10 tet | 38 |
| HM21Δ | HM21 Δ(mqsR ygiT)::kan | This study |
| HM21Δ2 | HM21 Δ(mqsR ygiT) ΔlacZ::kan | This study |
| HM21Δ2 PTXlacZ | ΗΜ21 Δ(mqsR ygiT) ΔlacZ attλ::pPTX3 | This study |
| HM22 | F+dapA zde-264::Tn10 hipA7 cs | 38 |
| Plasmids | ||
| pKD13 | bla FRT aph FRT PS1 PS2 oriR6K | 14 |
| pKD46 | bla PBAD-gam bet exo pSC101 oriTS | 14 |
| pCP20 | bla cat cI857 λPR-flp pSC101 oriTS | 14 |
| pBAD33 | cat araC PBAD pACYC184 ori | 20 |
| pBRlacItac | bla lacIq Ptac pBR322 ori | 40 |
| pAH125 | kan lacZ attλ oriR6K | 21 |
| pINT-ts | bla intλ cI857 λPR-flp pSC101 oriTS | 21 |
| pGEM9Zf | bla T7P SP6P orif1, pGEM ori | Promega |
| pTX3 | pBAD33-mqsR | This study |
| pAT3 | pBRlacItac-ygiT | This study |
| pPTX3 | pAH125-PmqsRygiT | This study |
| pTX4 | pGEM9Zf-mqsR | This study |
| pTX5 | pGEM9Zf-mqsRygiT | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a |
|---|---|
| Knockout primers | |
| mqsR11 | TTCCATTAATTAACGGATTTCATTCAATAGTTCTGGATGCTTATCCAGAAGTGTAGGCTGGAGCTGCTTC |
| ygiV11 | ACCGCTCCCGGGACGCGTTCCCGGGAATAATTTCGCAGGGAGGCAAAATGGTGTAGGCTGGAGCTGCTTC |
| ygiT24 | TTCATTGCTGTAATTAACCTTTTAGGTTATAACTAAAGTAACAGGGAGGCATTCCGGGGATCCGTCGACC |
| lacZ11 | ATTTTTGACACCAGACCAACTGGTAATGGTAGCGACCGGCGCTGAGCTGGGTGTAGGCTGGAGCTGCTTC |
| lacZ24 | CGGATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCATTCCGGGGATCCGTCGACC |
| mazE11 | AGATTGATATATACTGTATCTACATATGATAGCGGTTTGAGGAAAGGGTTGTGTAGGCTGGAGCTGCTTC |
| mazF24 | TCTGTCAGGTGGAAACCTGTGACCAGAATAGAAGTGAGTTAGTAACACTAATTCCGGGGATCCGTCGACC |
| Other PCR primers | |
| mqsR1 | TAGAGAGGAGCCGCACTTAC |
| ygiV1 | ATCCCTGCGTGACGACACCT |
| ygiT2 | AATGCGCCACTTTCTTATATGGTC |
| mazE1 | TAGCGACACCAAACAGCAAC |
| mazF2 | AGGTGGAAACCTGTGACCAG |
| mqsR-KpnUP | CGGGGTACCGGAGCCAACAATATGGAAAAACGCACACCACA |
| mqsR-SphDWN | ACATGCATGCTTACTTCTCCTTAAACGAGA |
| ygiT-KpnUP | CGGGGTACCGGAGCCAACAATATGAAATGTCCGGTTTGCCA |
| ygiT-EcoRDWN | CCGGAATTCTTAACGGATTTCATTCAATA |
| PmqsRSphUP | CCAACGCATGCGCCTGACTCCAGCTTCCCTT |
| PmqsRKpnDWN | CGGGGTACCAACCCCCGCCTCCCTGT |
| mqsRBglUP | CGGAGATCTGGAGCCAACAATATGGAAAAACGCACACCACA |
| mqsRBamDWN | ACGGATCCTTACTTCTCCTTAAACGAGA |
| race P1 | CGAGACGATCAGTACGTCATG |
| race P3 | GCCTGTAACAAGCCTGGGTC |
| C-anchor | GGCCACGCGTCGACTAGTACC15 |
| arb2 | GGCCACGCGTCGACTAGTAC |
| mqsR-seqDWN | AGCCTGGGTCTGTAAACATC |
| Hybridization probes | |
| P1 | TAGTTGAGCAATTCAGGGCTACAGCGGTGCGGCAACATCGCCACA |
| P2 | TAAGGAAAGTGATTGACCATATAAGAAAGTGGCGCATTAGTAGCG |
| P3 | GATAAACCTGGCCTGTAACAAGCCTGGGTCTGTAAACATCCTGCC |
| P4 | CGAAGCCCGAAATGCCTTTACTTGCGCCATGAAAGCATCTGACTC |
| P5 | GTTCGCTCAAACTTATCGCGAGTGATTTGGCTCACACTCCGGTAA |
Restriction sites in noncomplementary overhangs are underlined. Start and stop codons are indicated by bold type.
Plasmids were constructed as follows. To construct pAT3, ygiT was PCR amplified with primers ygiT-KpnUP and ygiT-EcoRDWN and inserted into pBRlacItac using KpnI and EcoRI. To construct pTX3, mqsR was amplified using primers mqsR-KpnUP and mqsR-SphDWN and inserted into pBAD33 using KpnI and SphI. pTX4; mqsR was amplified with primers mqsR-BglUP and mqsR-BamDWN, digested with BglII and BamHI, and inserted into BamHI-digested pGEM9Zf(+). To construct pTX5, DNA of the mqsR and ygiT region was amplified using primers mqsR-KpnUP and ygiT-EcoRDWN and inserted into the pGEM9Zf(+) T vector (Promega). To construct pPTX3, the intergenic region between ygiV and mqsR was amplified with primers PmqsR-SphUP and PmqsR-KpnDWN and inserted into pAH125 using the SphI and KpnI restriction sites. All PCR amplifications were done using the chromosome of HM21, and all primers used are listed in Table 2.
Luria-Bertani (LB) broth and LB agar plates were used for growth. To grow HM21 and its derivatives, media were supplemented with 75 μg ml−1 of diaminopimelic acid (DAP). Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 50 μg ml−1; kanamycin, 25 μg ml−1; and tetracycline, 15 μg ml−1.
Growth inhibition experiments and viability counts.
Overnight cultures were inoculated with bacteria obtained from thawed 8% dimethyl sulfoxide (DMSO) stocks (stored at −80°C) and grown for 18 h in media supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 0.2% glucose. Overnight cultures were diluted 1:1,000 in media supplemented with IPTG at the concentration indicated below and incubated at 37°C with shaking. l-Arabinose was added after 2.5 h. Samples were taken at different time points (see below), and the optical density at 600 nm was determined. To obtain viable counts, overnight cultures were diluted 1:1,000 in media supplemented with 50 μM IPTG and incubated at 37°C with shaking for 3.5 h. Then cultures were diluted 1:8 in media containing 1 mM l-arabinose. Samples were taken at different time points (see below) and serially diluted in sterile phosphate-buffered saline (PBS), and appropriate dilutions were plated on LB agar supplemented with 0.2% glucose and 1 mM IPTG. Colonies were counted after 24 h.
Flow cytometry.
Bacteria were counted using a bacterial counting kit from Invitrogen. Bacteria were grown as described above for the growth inhibition experiments, and 10-μl samples were taken, mixed with 490 μl sterile filtered PBS and 1 μl of a solution of the SYTO BC bacterial stain in dimethyl sulfoxide (DMSO) (Invitrogen), and stained at room temperature for 10 min. Samples were mixed with 500 μl sterile filtered PBS containing 30% glycerol and stored at −80°C. Before counting, samples were vortexed carefully and sonicated in an ultrasonic bath (Bandelin SONOREX Digital 10 P). The same number of sonicated 6.0-μm-diameter microspheres was added to all samples, and flow cytometry was performed using an LSRII and a high-throughput sampler (BD) with a laser beam maximum wavelength of 488 nm. All particles in a 100-μl sample were counted. The results were analyzed by using FlowJo 7.2.1 software (Treestar, Inc.).
Epifluorescence microscopy.
Bacteria were grown as described above for the growth inhibition experiments and collected by centrifugation. Cells from 1 ml of a culture were resuspended in 200 to 400 μl of nutrient-free M9 medium (37) and stained with the SYTO BC bacterial stain (Invitrogen) at room temperature for 10 min. Bacteria were spotted onto a solidified 1% agarose pad on a glass slide, the excess fluid was allowed to dry, the slide was covered with a coverslip, and the bacteria were observed immediately. Fluorescence microscopy was performed by using an Olympus BX51 microscope (magnification, ×1,000), images were captured by using a charge-coupled device (CCD) camera and Cell B software (Olympus), and the lengths of 100 cells per slide were measured using Image J software.
Northern blotting.
Overnight cultures were grown as described above for the growth inhibition experiments, diluted 250-fold in 20 ml of medium (250 ml for isolation of RNA from cells refractory to ampicillin lysis), and grown at 37°C for 2.5 h. To induce a cold shock, cultures were transferred to 25°C and incubated for 1.5 h. For isolation of RNA from bacteria refractory to ampicillin lysis, ampicillin (100 μg ml−1) was added and cultures were incubated at 37°C for 3 h to lyse the sensitive bacteria. RNA was isolated using Trizol reagent (Invitrogen). In vitro mqsR-ygiT mRNA was synthesized from plasmid pTX5 cut with NdeI using a T7 transcription kit (Fermentas). RNA samples (10 μg of total RNA and 5× serial dilutions of an in vitro transcription reaction mixture) were separated by electrophoresis in a denaturing 1.2% agarose gel and transferred onto positively charged nylon membranes (Roche) by capillary blotting. The membranes were prehybridized for 1.5 h and hybridized with a 5′,3′-fluorescein-labeled oligonucleotide DNA probe (1 pmol ml−1; Metabion) overnight at 42°C in modified Church buffer (53) in a rolling bottle in a hybridization oven or in bags or on a rocker-incubator. Prehybridization buffer was prepared according to the instructions in the DIG application manual for filter hybridization (Roche). Stringency washes were performed as described by Trayhurn et al. 53.
After the stringency washes, each membrane was rinsed in maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl; pH 7.5) supplemented with 0.3% Tween 20 for 3 min and blocked with a 1% solution of blocking reagent (Roche) in maleic acid buffer for 1 h at room temperature with gentle rocking. Fluorescein-labeled oligoprobe-mRNA hybrids were detected by chemiluminescence using antifluorescein-alkaline phosphatase conjugate (Fab fragments; Roche) and the CDP Star alkaline phosphatase substrate (Sigma-Aldrich). The membrane was incubated with antibody conjugate in blocking solution for 30 min, washed twice for 15 min in maleic acid buffer containing 0.3% Tween 20, and equilibrated in detection buffer (0.1 M Tris-HCl, 0.1 M NaCl; pH 9.5) for 5 min. AP detection was performed according to the recommendations in the CDP Star instruction manual, and the membrane was exposed to X-ray film.
5′ RACE.
Rapid amplification of cDNA ends (RACE) was used to map the 5′ end of the mqsR mRNA essentially as described by Sambrook and Russell (48). Total RNA from a log-phase culture of HM22 was isolated using Trizol reagent (Invitrogen) as described above for Northern blotting. Additional DNase treatment was performed, and the DNA-free RNA was purified using a Nucleospin RNA II kit (Macherey-Nagel). The cDNA was synthesized using primer race-P1 and 1.2 μg of total RNA as the template. All primers used in the experiment are listed in Table 2. Free nucleotides and primers were removed with an MSB Spin PCRapace kit (Invitek). cDNA was poly(dG) tailed using a terminal deoxyribonucleotidyl transferase (Fermentas) and dGTP. Free nucleotides and primers were removed as described previously. A chain complementary to the tailed cDNA was then synthesized using primer C-anchor and performing a single-cycle reaction with a PCR apparatus with the following program: 5 min at 95°C, increase the temperature from 55 to 65°C in 10 min, and 10 min at 72°C. Free nucleotides and primers were removed again as described above, and the DNA was amplified with primers arb2 and race-P3 using 30 cycles consisting of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C. The PCR product was then purified with the MSB Spin PCRapace kit (Invitek) and directly sequenced using primer mqsR-seqDWN.
β-Galactosidase assay.
β-Galactosidase levels were determined using the modified protocol of Miller (37). Overnight cultures were diluted 100-fold in 2 ml fresh medium containing 1 mM IPTG or no IPTG and grown at 37°C for 4 h, and the optical density at 600 nm (A600) of each culture was determined. Samples (50 μl) were mixed with 350 μl Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol; pH 7.0). Cells were permeabilized with 5 μl of chloroform and 10 μl 0.1% SDS, and 80 μl of a substrate solution (1 mg ml−1 o-nitrophenyl-d-galactoside in Z buffer) was added. When a sufficient amount of yellow color had developed, the reaction was stopped by adding 200 μl of 1 M Na2CO3, the cell debris was removed by centrifugation at 14,000 × g for 2 min, and the optical density at 420 nm (A420) of the supernatant was determined. β-Galactosidase levels were determined as follows: A420 × 1,000 × t−1 × V−1 × A600−1, where t and V are the time (in minutes) of the reaction and the volume (in milliliters) of the culture used in the assay, respectively.
Pulse-labeling.
Bacteria were grown as described above for the growth inhibition experiments. Optical densities at 600 nm of the cultures were determined at sampling time points. For measurement of translation, 100-μl portions of a culture were mixed with 10 μl of [35S]methionine (200 μCi ml−1; 1,175 Ci mmol−1) and incubated for 3 min at 37°C. For measurement of the transcription rate, 200-μl samples were mixed with 10 μl [5′-3H]uridine (5 mCi ml−1; 27 Ci mmol−1) and incubated for 6 min at 37°C. For measurement of DNA synthesis, 200-μl samples were mixed with 10 μl [methyl-3H]thymidine (400 μCi ml−1; 80Ci mmol−1) and incubated for 4 min at 37°C. Incorporation of the label was stopped by adding 1 ml of 5% trichloroacetic acid (TCA) supplemented with 2% Casamino Acids (CAA) and cooling the preparation on ice for 1 h. Before precipitation on ice, [35S]methionine-labeled samples were heated for 10 min at 95°C. Precipitates were collected on glass fiber filters prewetted with 5% TCA, washed with 5 ml 5% TCA and then with 96% ethanol, air dried for 15 min at 65°C, and counted using 5 ml of scintillation fluid (ScintiSafe 3; Fisher Scientific).
In vitro coupled transcription-translation assay.
In vitro coupled transcription-translation was performed using the E. coli T7 S30 extract system for circular DNA (Promega) according to the manufacturer's instructions. The reaction mixtures (25 μl) contained 1 μg of each template plasmid. For radioactive labeling of in vitro-synthesized proteins, 1 μl of [35S]methionine (10 mCi ml−1; 1,175 Ci mmol−1) was added. The reaction mixtures were incubated at 37°C for 1 h and frozen. For analysis of in vitro-synthesized proteins, 5-μl aliquots of a reaction mixture were mixed with 20 μl of acetone and incubated for 10 min on ice. Each precipitate was centrifuged for 5 min at 12,000 × g, dried under a vacuum for 15 min, dissolved in 20 μl of SDS-PAGE sample buffer, and heated at 95°C for 5 min. Aliquots (5 μl) were analyzed by SDS-PAGE using a 14% polyacrylamide gel.
Biofilm formation.
Biofilm formation was monitored in microtiter plates as described by Pratt and Kolter (44). Overnight cultures were diluted 100-fold in 150 μl LB media and grown in polystyrene 96-well plates for 48 h at 30°C without shaking. The optical densities at 600 nm of grown cultures were determined using a Tecan Sunrise microplate reader. Planktonic cells were removed by rinsing preparations with deionized water. Each biofilm was stained with 160 μl of a 1% crystal violet (CV) solution for 20 min, and excessive dye was washed off with deionized water. Plates were dried for 10 min, and the absorbed CV dye was dissolved in 170 μl of an ethanol-acetone (80:20) mixture. The adsorption at 590 nm of 125 μl of the solution in a microtiter plate was measured. The background value for the control sample containing no bacteria was subtracted. Three to five biological replicates (individual overnight cultures) and eight technical replicates (eight wells started from each overnight culture) of each strain were analyzed.
Identifying mqsR-ygiT gene pairs.
We used the MqsR protein sequence of E. coli (gi|16130918) as a query and performed a search against the complete microbial genome database at NCBI (Genomic Blast) by using BLASTP (2). The BLAST output was restricted by an E value of 1.
RESULTS
MqsR inhibits bacterial growth, and YgiT relieves the toxicity of MqsR.
To test if mqsR and ygiT encode a toxin-antitoxin pair, both genes were cloned into expression vectors. The ygiT sequence was inserted into the vector plasmid pBRlacItac (40) downstream of the IPTG-inducible Ptac promoter, resulting in plasmid pAT3. The mqsR gene was inserted into pBAD33 (20) under control of the arabinose-inducible PBAD promoter, resulting in pTX3. Because of the toxic effects of mqsR, cloning of this gene was possible only after introduction of the ygiT expression plasmid pAT3 and induction of the YgiT protein during growth of competent cells used for transformation of the ligation product. We found that HM21Δ, which contained both expression plasmids (pAT3 and pTX3), required constant low-level induction of ygiT for growth (Fig. 1A). This was probably because of the low levels of MqsR produced due to leakage of the PBAD promoter, which could not be stopped completely by addition of glucose to the growth medium. We tested the effects of expression of the candidate toxin and antitoxin on bacterial growth and found that MqsR halts growth and that expression of YgiT is able to suppress the inhibitory effect. Expression of YgiT alone did not influence bacterial growth (Fig. 1A). These observations show that mqsR encodes a toxic protein and that the product of ygiT can protect cells from the toxicity of MqsR.
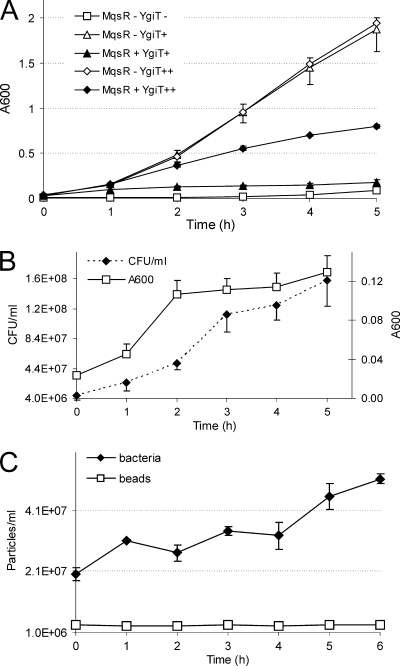
FIG. 1.
MqsR induces growth arrest which is reversible by expression of YgiT. HM21Δ contained plasmid pAT3 for IPTG-inducible ygiT expression and plasmid pTX3 for l-arabinose-inducible mqsR expression. (A) Effect on bacterial growth as measured by A600. Bacteria were grown without IPTG (squares) (YgiT−), in the presence of 50 μM IPTG (triangles) (YgiT+), or in the presence of 5 mM IPTG (diamonds) (YgiT++). At time zero, some cultures (filled symbols) (MqsR+) were supplemented with 1 mM l-arabinose. (B) Colony formation after induction of mqsR. Bacteria were grown in the presence of 50 μM IPTG, and mqsR was induced at time zero by addition of 1 mM l-arabinose. At the time points indicated, samples were taken, the A600 was measured, and appropriate dilutions were plated onto solid medium supplemented with 0.2% glucose and 50 μM IPTG. Colonies were counted after 24 h, and the number of CFU per ml of culture was calculated. (C) Counting of individual bacteria after induction of mqsR. Bacteria were grown, mqsR was induced, and samples were taken as described above for the plating experiments. Bacteria were stained with the SYTO BC dye and mixed with a constant number of standard microspheres. Particles were counted by flow cytometry. All results are averages of three independent experiments. The error bars indicate standard deviations.
MqsR causes reversible growth inhibition and elongation of bacterial cells.
To test if MqsR kills bacteria or causes growth inhibition, we induced MqsR production by addition of 1 mM l-arabinose to a growing culture, took samples at different time points, and plated them on solid media containing IPTG for ygiT induction. The number of colony-forming bacteria did not decrease during 5 h of continuous expression of MqsR, showing that later production of YgiT can rescue cells from the inhibiting effect of the toxin (Fig. 1B). Thus, we found that production of MqsR does not kill bacteria but causes a temporary, reversible growth arrest. Actually, both the optical density of the culture and the number of colonies continued to increase slowly after induction of the MqsR toxin (Fig. 1B). This could indicate that there was continuation of slow growth; alternatively, the increase in optical density might have been caused by cell filamentation and the increase in the number of CFU might have reflected an improved ability to form colonies. To test these hypotheses, we determined the absolute numbers of bacterial cells by flow cytometry (Fig. 1C; see Fig. S1 in the supplemental material) and, in parallel, examined the possible changes in cell shape and size using a microscope (Fig. 2). Fluorescence-activated cell sorting (FACS) counting showed that there were increases in the numbers of cells (Fig. 1C). Thus, expression of MqsR did not halt growth completely, and bacteria continued to divide slowly.
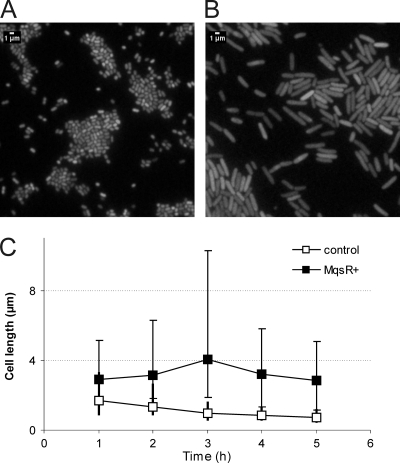
FIG. 2.
MqsR causes elongation of bacterial cells. Expression of mqsR was induced in the logarithmic growth phase at time zero by addition of 1 mM l-arabinose to a culture of HM21Δ cells bearing plasmids pAT3 (ygiT expression) and pTX3 (mqsR expression). An uninduced culture of the same strain was used as a control. Samples were taken at the times indicated, and bacteria were stained with the SYTO BC dye and examined by epifluorescence microscopy. Control bacteria (A) and the MqsR-arrested bacteria (B) were observed 4 h after induction of mqsR. (C) Average lengths of 100 bacterial cells. Filled squares, MqsR-expressing bacteria; open squares, uninduced control. The vertical bars indicate the limits of cell length (the longest and shortest bacteria in a set of cells).
Microscopy showed that there was elongation, but not filamentation, of bacteria in response to production of MqsR. Normally, exponentially growing cells of the HM21 strain and its Δ(mqsR ygiT) derivative used in our experiments were short rods resembling the stationary-phase cells of E. coli (Fig. 2A). Induction of mqsR increased the length of bacteria considerably (Fig. 2B). Both the average cell length and the variability of the cell length increased during the first 3 h after mqsR induction. After this, the average cell length and the variability of the cell length started to decrease (Fig. 2C), indicating that there was relaxation of the MqsR-induced block of cell division. The MqsR-arrested bacteria were also fatter; the average diameter increased gradually, and at 3 h after induction of mqsR it was 0.94 ± 0.09 μm, compared to 0.60 ± 0.08 μm for the control.
mqsR and ygiT form an operon.
To map the transcripts of the mqsR-ygiT region and to verify the expression array data from previous studies (27, 52), the mRNA of these two genes was analyzed by Northern hybridization. For validation of the results, serial dilutions of an in vitro-synthesized mqsR-ygiT transcript consisting of 806 bases were blotted as a positive control (Fig. 3B, lanes 7 to 9). Hybridization with a DNA oligoprobe complementary to mqsR revealed two major transcripts that were approximately 700 and 800 bases long (Fig. 3B). The Δ(mqsR ygiT) deletion mutant did not express either transcript (Fig. 3B lane 1), indicating that these two transcripts do correspond to the mqsR mRNA. Further mapping using probes complementary to ygiT and different regions near the mqsR-ygiT locus (Fig. 3C) indicated that mqsR and ygiT are cotranscribed and that the two major transcripts cover both genes. The 5′ ends of these transcripts are apparently identical and start at a promoter of the operon. Exact mapping of the 5′ end by the 5′ RACE method located the transcription start site 18 bp upstream of the start codon of mqsR (Fig. 3A). The 3′ end of the longer transcript extends farther downstream of the ygiT coding sequence, into the intergenic region, while the shorter transcript ends farther upstream and could not be detected with the P5 probe (Fig. 3C, lanes P3 to P5). Several minor, shorter transcripts map to the 5′ region of the operon (Fig. 3C, lane P3). The ygiV gene upstream of mqsR is transcribed separately (Fig. 3C, lane P1).
To reexamine expression of mqsR and ygiT in the nondividing subpopulation, we isolated RNA from cells of the HM22 high-persistence (hip) mutant refractory to lysis by ampicillin and analyzed it by Northern hybridization (Fig. 3B, lane 6). The increase in frequency of persisters due to the hipA7 mutation was considered to be a direct result of activation of the HipA toxin in a subpopulation of bacteria (38, 50). The same mutation also conferred cold sensitivity, indicating that there was HipA activation in all bacteria in the population at a lower temperature. Thus, we decided to look at mqsR and ygiT transcription in a cold-arrested HM22 culture as a model population consisting of artificially generated persisters. Our Northern blot data confirmed the previously described array results (27, 52); the level of expression of the mqsR-ygiT mRNA was considerably higher in a subpopulation of bacteria that were not sensitive to ampicillin lysis (Fig. 3B lane 6). An increase in mqsR-ygiT transcription in HM22 after transfer to a lower temperature (25°C) was observed as well (Fig. 3B, lane 5). The mqsR-ygiT transcripts, which were induced in the nondividing, HipA-arrested bacteria, were the same major transcripts that were observed in growing cells. At the same time, the level of expression of the major mqsR-ygiT transcripts was not higher in the growing cells of the hipA7 mutant than in the wild-type parent (HM21) (Fig. 3B, lanes 2 and 4) and was not induced in response to a decrease in the growth temperature in the wild-type strain (Fig. 3B, lanes 2 and 3). However, in the wild type, we observed induction of a small transcript corresponding to the 5′ portion of mqsR at 25°C (Fig. 3B, lane 3). Shorter transcripts were also detected in the the hipA7 mutant both in ampicillin-refractory bacteria and in response to the temperature downshift (Fig. 3B, lanes 5 and 6).
To summarize, the Northern blot results confirmed that the mqsR and ygiT genes constitute an operon which is transcriptionally induced both in response to HipA activation and in the nondividing subpopulation of a growing culture.
YgiT is a transcriptional repressor of the mqsR-ygiT operon.
We used the β-galactosidase assay to test the potential autoregulation of the mqsR-ygiT operon by YgiT antitoxin. The intergenic region between ygiV and mqsR was fused to the lacZ reporter and inserted as a single copy into the chromosome of an HM21 derivative lacking both the mqsR-ygiT operon and lacZ. Expression of YgiT from the pAT3 plasmid was induced with IPTG. After YgiT induction, the levels of β-galactosidase were repressed more than 20-fold (Fig. 3D). This confirmed the ability of YgiT to strongly repress transcription from the mqsR-ygiT promoter.
MqsR induces inhibition of protein synthesis in vivo.
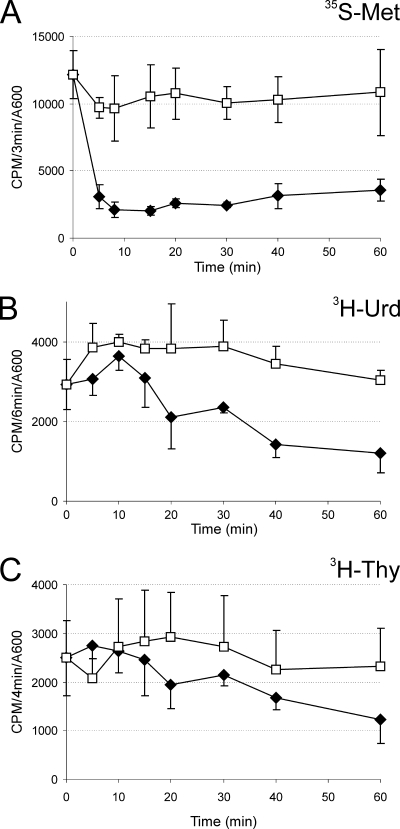
The molecular target of MqsR could not be predicted on the basis of its primary sequence. Therefore, we performed pulse-labeling experiments to measure the effect of the toxin on macromolecular synthesis in vivo. As shown in Fig. 4, induction of mqsR caused rapid inhibition of protein synthesis, whereas the decrease in RNA synthesis was slower and the effect on DNA synthesis was not clearly pronounced. The rate of protein synthesis dropped about 5-fold within 8 min and remained at that level.
FIG. 4.
Effect of mqsR expression on macromolecular synthesis in vivo. Pulse-labeling experiments showed that there was incorporation of radiolabeled precursors into protein (A), RNA (B), and DNA (C). Ectopic expression of mqsR (filled symbols) was induced at time zero by addition of l-arabinose (1 mM) to HM21Δ cultures bearing plasmids pAT3 (for ygiT expression) and pTX3 (for mqsR expression). Open symbols indicate the results for the negative control (no l-arabinose added). Samples were taken at the times indicated, radioactive precursors were added, and samples were incubated at 37°C for 3 min (A), 6 min (B), and 4 min (C) for optimal labeling. Incorporation of the label was stopped by adding trichloroacetic acid (TCA). The rates of synthesis are expressed as the number of cpm incorporated during a pulse-labeling reaction divided by the A600 of the bacterial culture. The values are the averages of three independent experiments; the error bars indicate the standard deviations.
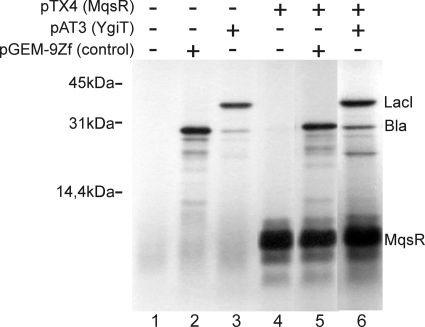
To determine if translation is the primary target of MqsR, we examined the effect of MqsR in a cell-free system. MqsR was expressed from the T7 promoter in an in vitro coupled transcription-translation reaction. As shown in Fig. 5, a polypeptide with the predicted molecular mass of MqsR (11.2 kDa) was produced in an S30 extract and did not inhibit the production of two heterologous proteins, β-lactamase (28 kDa) expressed from the pGEM-9Zf vector and LacI (38.6 kDa) expressed from pAT3. The product of ygiT with predicted molecular mass of 14.7 kDa could not be detected, probably due to the instability of this protein.
FIG. 5.
MqsR does not inhibit protein synthesis in vitro. Coupled transcription-translation was carried out using the E. coli T7 S30 extract system for circular DNA. Templates for expression of β-lactamase as a positive control (pGEM9Zf) (lanes 2 and 5), MqsR (pTX4) (lanes 4 to 6), and YgiT plus LacI (pAT3) (lanes 3 and 6) were added. In vitro-synthesized proteins were labeled with [35S]methionine and analyzed by SDS-PAGE using a 14% polyacrylamide gel. The positions of synthesized proteins are indicated on the right.
Phyletic distribution of mqsR-ygiT homologues.
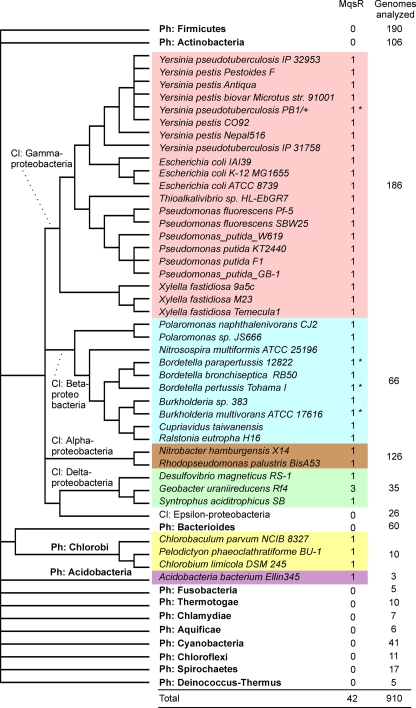
We searched for genes encoding homologues of the MqsR toxin in 914 fully sequenced bacterial genomes. A total of 42 genes coding for MqsR homologues were found in 40 genomes, and most of them were found in gamma- and betaproteobacteria (Fig. 6). We also found putative MqsR toxins in alpha- and deltaproteobacteria, in chlorobi, and in a species of Acidobacter, whereas phyla such as the Firmicutes and actinobacteria did not contain MqsR toxins. Most of the genomes contained a single mqsR gene; only Geobacter uraniireducens contained three mqsR genes. As a general rule, the presence of the mqsR gene in strains of the same species is variable. For example, only about one-half of the sequenced E. coli genomes contain this gene. Thirty-six homologous proteins were proteins whose sizes were similar to that of the E. coli MqsR used as the query and showed homology throughout the sequence, whereas five homologues were truncated and lacked either ∼40 (4 proteins) or 27 (Ralstonia eutropha) N-terminal amino acids. The MqsR of Pseudomonas fluorescens Pf-5 had a 43-amino-acid N-terminal extension (see Table S1 in the supplemental material).
FIG. 6.
Phyletic distribution of MqsR homologues on a cladistic tree of bacteria. Phyla for which only one or two genomes have been sequenced are not shown, and the total number of genomes analyzed was 914. Asterisks indicate MqsR variants with a ∼40-amino-acid N-terminal truncation. The cladistic tree was constructed by using a deeper branching order (phylum [Ph] and class [Cl] levels) than an NCBI microbial taxonomic tree (Genome Browser taxonomic tree). The more subtle branching order of genomes which contained toxin-antitoxin pairs is based on a 16S rRNA alignment retrieved from RDP II (13) and was computed by using neighbor-joining tools of MEGA4 (32).
All of the genes encoding MqsR homologues were followed by a downstream gene encoding a putative DNA-binding protein having a helix-turn-helix (HTH) motif, a gene encoding a putative antitoxin, a functional analogue of YgiT (see Table S1 in the supplemental material). Many of these open reading frames followed the mqsR gene at a distance of a few nucleotides or overlapped the mqsR gene, suggesting that there is translational coupling of the encoded proteins.
Effects of mqsR on biofilm formation and growth depend on the strain background.
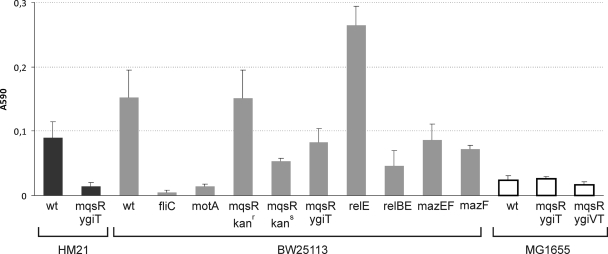
Disruption of mqsR has been reported to reduce biofilm formation (19). This phenotype was demonstrated previously in the MG1655 background for a mutant that had a Tn5 insertion in the mqsR open reading frame (26). Recently, a defect in biofilm formation was reported for an E. coli strain with deletions in five chromosomal TA operons, while single deletions of the corresponding toxin genes did not cause this phenotype. Multiple TA deletions decreased both bacterial attachment and dispersal (28). Here, we decided to reexamine the effect of mqsR using deletion mutants with different strain backgrounds. We tested three E. coli K-12 strains lacking either the toxin gene or the entire TA operon. In parallel, we examined similar mutants with mutations in the relBE and mazEF systems. Bacteria were grown for 48 h in LB medium in 96-well plates, and crystal violet staining was used for measurement of biofilms (Fig. 7). Since the densities in all of the wells were very similar, we did not normalize biofilm data using planktonic growth. Two known biofilm-defective mutants (fliC and motA mutants) were used as controls in the BW25113 background (44).
FIG. 7.
Biofilm formation by wild-type and mutant E. coli K-12 strains. Bacteria with the genotypes indicated were diluted from an overnight culture and grown in LB medium in 96-well polystyrene microtiter dishes at 30°C without shaking. Eight wells were inoculated using a single overnight culture. After 48 h, the microtiter plates were rinsed and stained with crystal violet. The surface-bound stain was dissolved and quantified by measuring the adsorption at 590 nm. The averages of three to five independent experiments are shown for each strain. The error bars indicate the standard errors. kanr, kanamycin resistant (contains resistance cassette acquired during disruption of mqsR); kans, kanamycin sensitive (resistance cassette eliminated). wt, wild type.
Our results show that the mqsR-ygiT system does affect biofilm formation, although this apparently depends on the strain background (Fig. 7). We observed a negative effect of the mqsR-ygiT deletion on biofilm formation in both HM21 and BW25113. The ability of the mqsR single mutant from the Keio collection (4) to form a biofilm, however, was not reduced compared to the ability of wild-type strain BW25113. Unexpectedly, elimination of the kanamycin resistance cassette from the chromosome of this mutant strain resulted in a decrease in biofilm formation. The production of biofilms by the BW25113 ΔrelBE and ΔmazEF deletion mutants was also reduced, while individual deletions of relE and mazF had the opposite effect. Under our assay conditions, E. coli MG1655, for which the biofilm-related phenotype of mqsR was originally reported, was a poor biofilm producer, and deletion of mqsR-ygiT had no effect on biofilm formation by this strain.
The study which reported that mqsR affected the biofilm phenotype also indicated that expression of this gene in trans could restore biofilm formation. This seemed to be incompatible with our observations; we had difficulty cloning mqsR when ygiT was not overexpressed. In another study by the same research group, the toxicity of MqsR was reported and deletion of the nearby ygiV gene was shown to reduce this toxicity in MG1655 (59). Therefore, we studied the effect of ectopic expression of MqsR in MG1655 and found that this strain is indeed less toxic than HM21 (see Fig. S2 in the supplemental material). MG1655Δ2 bearing pAT3 and pTX3 grows normally without addition of IPTG to the medium. However, we were not able to confirm the effect of ygiV on the toxicity of MqsR in the MG1655 background. No differences between the Δ(mqsR ygiT) and Δ(ygiV mqsR ygiT) mutants in either the ability to form a biofilm (Fig. 7) or inhibition of growth by MqsR were detected (see Fig. S2 in the supplemental material). In conclusion, the strain background had a considerable effect on MqsR activity.
DISCUSSION
The existence of the new MqsR (YgiU)-YgiT toxin-antitoxin family was predicted by sequence analysis (35) and our initial observation of the growth-inhibiting effect of MqsR (52). The present study confirmed experimentally that the mqsR-ygiT locus of E. coli K-12 encodes a toxin-antitoxin system. We showed that this locus shares essential properties of known TA loci: (i) mqsR encodes a toxic protein that inhibits bacterial growth and induces rapid inhibition of protein synthesis; (ii) the product of ygiT counteracts the toxicity of MqsR; (iii) these two genes are cotranscribed, forming an operon; and (iiii) YgiT is a transcriptional repressor of the promoter of this operon.
Growth inhibition is a vague parameter and must be interpreted with caution; overproduction of a single enzyme subunit, for example, may be detrimental to growth and can be compensated for by simultaneous production of the other, interacting subunit (1). The specialized nature and different half-lives of the subunits in a protein complex are definitive characteristics of a toxin-antitoxin pair. The only activity of known TA toxins is toxicity, and the only function of antitoxins is control of this toxicity both by complex formation and at the transcriptional level. The sequence data and experimental evidence indicate that YgiT is a specialized antitoxin that lacks any known function except control of MqsR. Primary sequence analysis of the MqsR protein revealed no similarity to any of the previously described toxins, which allowed us to classify MqsR and YgiT as members of a new TA family. MqsR homologues are encoded in the genomes of many bacteria (mostly members of different groups of proteobacteria), and the genes are always paired with a putative antitoxin-encoding open reading frame (ORF). However, the arrangement of the toxin and antitoxin genes in all gene pairs belonging to the mqsR-ygiT family is unusual; the toxin gene precedes the antitoxin gene. Previously, this gene order has been described for hicAB (25) and the higBA family (8). An important difference is that the higA gene has its own promoter located in higB (8), whereas we did not find a separate promoter in front of ygiT and careful mapping indicated that major transcripts of the operon cover both genes (Fig. 3C).
We were unable to identify the target of MqsR. Expression of the toxin caused rapid inhibition of translation in living cells but had no effect on protein synthesis in an S30 extract an in vitro coupled transcription-translation reaction. The MqsR protein itself was effectively produced in vitro. A similar contradiction has been reported previously for HipA (31). Another important feature of the TA systems is the instability of the antitoxin. The stability of YgiT remains to be determined.
In this study, we confirmed by Northern hybridization that transcription of the mqsR and ygiT genes is highly upregulated in cells refractory to ampicillin lysis (Fig. 3B). According to our previous analysis (46), this subpopulation contains persisters along with more numerous cells that remain quiescent after transfer into ampicillin-free medium. We do not know if these two groups of bacteria have the same transcription profile and if TA operons are overexpressed in both groups. It is noteworthy that transcription of mqsR and ygiT is also induced in response to activation of another toxin, HipA (Fig. 3B), given that the cold sensitivity of a hipA7 mutant is obviously due to activation of HipA at lower temperatures (50, 51). The same mutation causes an increase in the frequency of persisters. However, we cannot say if the ampicillin-tolerant cells of HM22 (hipA7), which were used for RNA isolation, were dormant due to the HipA activity. Actually, we do not know why these cells are not sensitive to ampicillin or even if this subpopulation is homogeneous or composed of physiologically diverse bacteria which entered into dormancy due to different causal mechanisms. Wild-type strains also produce a large fraction of ampicillin-tolerant cells (46; A. Jõers, H. Luidalepp, and T. Tenson, unpublished data), and transcription of the TA operons was similarly induced in dormant bacteria in wild-type cultures (52). Our observation that HipA activation leads to induction of the mqsR-ygiT system is not a unique example of sequential or simultaneous activation of different TA systems. Ectopic expression of VapC toxins originating from Salmonella and Shigella activated YoeB mRNA interferase in E. coli (57). Also, production of the Doc toxin activated RelE in E. coli (16), and amino acid starvation in E. coli activated both RelE and MazF (ChpA) (9, 10). Further studies are needed to shed light on the spread and mechanisms of this kind of serial TA activation.
Another interesting finding was the confirmation of the effect of the mqsR-ygiT system on biofilm formation and similar effects of the well-studied relBE and mazEF systems (Fig. 7). Deletion of five different TA systems in E. coli has been demonstrated to influence biofilm formation via YjgK (TabA) and fimbriae (28); however, the molecular mechanism(s) by which a toxin-antitoxin system affects this trait is not clear. The effect could be due to production of a fraction of lysing or dormant cells which somehow contribute to biofilm development or due to some limited, non-growth-inhibiting toxin activity in growing bacteria. The latter explanation would mean that TA systems not only are guardians of extreme stresses responding to catastrophic events but also may work as “normal” regulators of gene activity. Because of the complexity of biofilm formation and the strong effects of the strain background observed in this work (Fig. 7), further studies are required to identify the role of TA systems in different states in this process. Currently, it is not known why deletions of full toxin-antitoxin operons and single toxin genes have different effects. While this paper was being prepared, Kolodkin-Gal et al. described the effects of individual deletions of five TA operons on biofilm formation (30). These authors (i) demonstrated that there is a significant fraction of dead bacteria in an E. coli biofilm and (ii) showed that deletion of mazEF and deletion of dinJ-yafQ strongly reduced biofilm production, while ΔrelBE had a smaller effect. All of these results were obtained using the MC4100relA+ strain background, for which MazEF-dependent death has been reported by the same research group.
To test the hypotheses about the physiological role of TA systems, it is important to create model organisms devoid of all TA loci (54). Here we found a new TA system in addition to the previously characterized TA loci for eight families previously found in the chromosomes of sequenced E. coli isolates (25, 56). Finding all existing TA loci and deleting them from the chromosome of a TA-free model bacterium will be crucial for conducting experiments to test the role of TA systems in formation of persisters, as well as all other potential functions of these genes.
After the original submission of this paper, two research groups reported that mqsR and ygiT encode a TA system and showed that the MqsR toxin is an mRNA interferase that cleaves RNA independently of translation (12, 58). The same authors also mapped the promoter of the mqsR-ygiT operon and showed that it is transcriptionally repressed by YgiT. The transcription start site reported by Christensen-Dalsgaard and coworkers (12) is supported by our data.
Supplementary Material
Acknowledgments
We thank Ülo Maiväli, Arvi Jõers, and Rita Hõrak for valuable comments on the manuscript and Hannes Luidalepp for providing BW25113 ΔrelBE.
This work was supported by Estonian Science Foundation grant 6764 and by the European Regional Development Fund through the Center of Excellence in Chemical Biology.
Footnotes
Published ahead of print on 16 March 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, G. C., Jr., and A. Kornberg. 1991. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J. Biol. Chem. 266:22096-22101. [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Budde, P. P., B. M. Davis, J. Yuan, and M. K. Waldor. 2007. Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae. J. Bacteriol. 189:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 11.Christensen-Dalsgaard, M., and K. Gerdes. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397-411. [DOI] [PubMed] [Google Scholar]
- 12.Christensen-Dalsgaard, M., M. G. Jorgensen, and K. Gerdes. 2010. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75:333-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fozo, E. M., M. R. Hemm, and G. Storz. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 72:579-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Pino, A., M. Christensen-Dalsgaard, L. Wyns, M. Yarmolinsky, R. D. Magnuson, K. Gerdes, and R. Loris. 2008. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 283:30821-30827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes, K., and E. G. Wagner. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10:117-124. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez Barrios, A. F., R. Zuo, Y. Hashimoto, L. Yang, W. E. Bentley, and T. K. Wood. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison, J. J., W. D. Wade, S. Akierman, C. Vacchi-Suzzi, C. A. Stremick, R. J. Turner, and H. Ceri. 2009. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen, M. G., D. P. Pandey, M. Jaskolska, and K. Gerdes. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y., X. Wang, Q. Ma, X. S. Zhang, and T. K. Wood. 2009. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191:1258-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2008. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J. Bacteriol. 190:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodkin-Gal, I., R. Verdiger, A. Shlosberg-Fedida, and H. Engelberg-Kulka. 2009. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One 4:e6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 33.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 34.Magnuson, R. D. 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarova, K. S., Y. I. Wolf, and E. V. Koonin. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55-66. [DOI] [PubMed] [Google Scholar]
- 40.Ojangu, E. L., A. Tover, R. Teras, and M. Kivisaar. 2000. Effects of combination of different −10 hexamers and downstream sequences on stationary-phase-specific sigma factor σS-dependent transcription in Pseudomonas putida. J. Bacteriol. 182:6707-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 44.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 45.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 46.Roostalu, J., A. Joers, H. Luidalepp, N. Kaldalu, and T. Tenson. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol. 8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz-Echevarria, M. J., G. de la Cueva, and R. Diaz-Orejas. 1995. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol. Gen. Genet. 248:599-609. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sauer, R. T., R. R. Yocum, R. F. Doolittle, M. Lewis, and C. O. Pabo. 1982. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature 298:447-451. [DOI] [PubMed] [Google Scholar]
- 50.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schumacher, M. A., K. M. Piro, W. Xu, S. Hansen, K. Lewis, and R. G. Brennan. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah, D., Z. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trayhurn, P., J. S. Duncan, A. Nestor, M. E. Thomas, and D. V. Rayner. 1994. Chemiluminescent detection of mRNAs on Northern blots with digoxigenin end-labeled oligonucleotides. Anal. Biochem. 222:224-230. [DOI] [PubMed] [Google Scholar]
- 54.Tsilibaris, V., G. Maenhaut-Michel, N. Mine, and L. Van Melderen. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuomanen, E. 1986. Phenotypic tolerance: the search for beta-lactam antibiotics that kill nongrowing bacteria. Rev. Infect. Dis. 8(Suppl. 3):S279-S291. [DOI] [PubMed] [Google Scholar]
- 56.Van Melderen, L., and M. Saavedra De Bast. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winther, K. S., and K. Gerdes. 2009. Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 72:918-930. [DOI] [PubMed] [Google Scholar]
- 58.Yamaguchi, Y., J. H. Park, and M. Inouye. 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284:28746-28753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, X. S., R. Garcia-Contreras, and T. K. Wood. 2008. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA). ISME J. 2:615-631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.