Abstract
Lantibiotics are peptide-derived antibacterial substances produced by some Gram-positive bacteria and characterized by the presence of unusual amino acids, like lanthionines and dehydrated amino acids. Because lantibiotic producers may be attacked by self-produced lantibiotics, they express immunity proteins on the cytoplasmic membrane. An ATP-binding cassette (ABC) transport system mediated by the LanFEG protein complex is a major system in lantibiotic immunity. Multiple-sequence alignment analysis revealed that LanF proteins contain the E loop, a variant of the Q loop, which is a well-conserved motif in the nucleotide-binding domains (NBDs) of general ABC transporters. To elucidate E loop function, we introduced a mutation in the NukF protein, which is involved in the nukacin-ISK-1 immunity system. Amino acid replacement of glutamic acid in the E loop with glutamine (E85Q) resulted in slight decreases in the immunity level and transport activity. Additionally, the E85A mutation severely impaired the immunity level and transport activity. On the other hand, ATPase activities of purified E85Q and E85A mutants were almost similar to that of the wild type. These results suggested that the E loop found in ABC transporters involved in lantibiotic immunity plays a significant role in the function of these transporters, especially in the structural change of transmembrane domains.
Lantibiotics are antibacterial peptides produced by some Gram-positive bacteria and are characterized by the presence of unusual amino acids, such as lanthionine and dehydrated amino acid residues (4, 9, 20). The unusual amino acids are introduced after translation by a modification enzyme(s), and their subsequent processing and secretion are carried out by a leader peptidase and transporter, respectively. Since the secreted mature lantibiotics have the potential to attack producer cells, lantibiotic-producer cells express self-protection systems against their own lantibiotics. These self-protection systems have 2 major mechanisms: a lantibiotic transport mechanism mediated by an ATP-binding cassette (ABC) transporter (LanFEG) and a lantibiotic-binding mechanism mediated by a lipoprotein (LanI) or a membrane protein (LanH) (2, 8, 26, 33, 34).
Transport by LanFEG is a common and major mechanism in the lantibiotic immunity systems. LanFEG and LanI are needed for full immunity against nisin and subtilin, which are type A(I) lantibiotics (33, 34). However, the immunity level conferred by LanFEG is much higher than that conferred by LanI. LanFEG and LanH are expressed against nukacin ISK-1, which is a type A(II) lantibiotic produced by Staphylococcus warneri ISK-1 (2). As in the case of nisin and subtilin, LanFEG plays a major role in the level of immunity against nukacin ISK-1. Moreover, against lacticin 481, which is also a type A(II) lantibiotic, only LanFEG is expressed and it confers full immunity (9).
ABC transporters function as molecular pumps and transport various substrates, such as nutrients, lipids, and antibiotics coupled to ATP hydrolysis (10, 31). Bacterial ABC transporters consist of 2 transmembrane domains (TMDs) and 2 nucleotide-binding domains (NBDs). They utilize ATP hydrolysis as a source of energy for the transport. The NBD of an ABC transporter has several conserved motifs, such as Walker A, Walker B, Q loop, Signature, and H loop, in its amino acid sequence, and these motifs are involved in the functions of ABC transporters (31). Although the detailed substrate-binding mechanism is still unknown, the dimerization of NBDs concomitant with ATP binding leads to the conformational change of 2 TMDs, resulting in transport of the substrate (31). Sequence similarities and hydrophobicity profiles suggest that LanFEG consists of 2 heterodimeric subunits containing TMDs (LanEG) and 2 homodimeric subunits containing NBDs (LanF) (4, 27).
In general, ABC transporters that had been identified together with their substrates mediate the transport of the substrate across the membrane. An exception reported previously is the Lol system, which releases lipoproteins from the inner membrane to the outer membrane in Gram-negative bacteria (40). However, LanFEG proteins are believed to scavenge lantibiotics present on the membrane. This hypothesis is strongly supported by the mode of action of lantibiotics: many lantibiotics, especially type A(I) lantibiotics, show pore-forming activity against model membranes (4). Taken together, the transport mechanism of LanFEG seems to be different from that of general ABC transporters.
The immunity mechanism against nukacin ISK-1 mediated by NukFEG and NukH has been investigated before (2, 21-23, 39). On the basis of our analysis, we suggested that NukFEG transports both nukacin ISK-1 on the membrane and nukacin ISK-1 captured by NukH (2, 22). Since the transport reaction depended on the metabolic energy of the cells, we presumed that ATP hydrolysis by NukF is a driving force for the transport (22).
Using multiple sequence alignment analysis, we have found that all the LanF proteins have the E loop as a variant of the Q loop in general ABC transporters. Therefore, in this study, we investigated the function of the E loop existing in NukF by using site-directed mutagenesis. A bioassay using nukacin ISK-1 and recombinant Lactococcus lactis expressing nukF and its mutants showed that the E loop is important for immunity. Additionally, a transport assay with fluorescein isothiocyanate (FITC)-labeled nukacin ISK-1 indicated that the E loop is involved in transport activity. Since purified NukF and E loop mutants showed similar ATPase activity, we proposed that the E loop has an important role in the function of LanFEG, especially in coupled movement with the transmembrane subunit NukEG.
MATERIALS AND METHODS
Multiple sequence alignment.
The alignment of the LanF proteins and other ABC transporters was generated using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/). The following proteins were studied: NukF (RefSeq accession no. NP_940775; nukacin ISK-1 immunity system), LctF (GenBank accession no. AAC72253; lacticin 481 immunity system), SboF (GenBank accession no. ABI63642; salivaricin B immunity system), ScnF (GenBank accession no. AAB92604; streptococcin A-FF22 immunity system), McdF (GenBank accession no. ABI30231; macedocin immunity system), RumF (EMBL accession no. CAB93668; ruminococcin A immunity system), MrsF (EMBL accession no. CAB60255; mersacidin immunity system), NisF (EMBL accession no. CAA82547; nisin immunity system), SpaF (GenBank accession no. AAB91597; subtilin immunity system), EpiF (GenBank accession no. AAB61163; epidermin immunity system), GdmF (GenBank accession no. AAB61132; gallidermin immunity system), MutF (GenBank accession no. AAF99691; mutacin I immunity system), BcrA (GenBank accession no. AAD21213; bacitracin resistance system), DrrA (GenBank accession no. AAA74717; daunorubicin and doxorubicin resistance system), LmrA (RefSeq accession no. YP_001033128; multidrug resistance system), MalK (GenBank accession no. AAB59057; maltose transport system), BtuD (RefSeq accession no. NP_416224; vitamin B12 transport system), LolD (RefSeq accession no. AP_001743; lipoprotein release system), MsbA (RefSeq accession no. NP_752981; lipid transport system), HisP (RefSeq accession no. NP_416809; histidine transport system), FhuC (RefSeq accession no. NP_414693; ferrichrome transport system), GlcV (RefSeq accession no. NP_344170; glucose transport system), and MJ0796 (RefSeq accession no. NP_247785; unknown function).
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. warneri ISK-1 was grown in MRS broth (Oxoid, Hampshire, United Kingdom) at 37°C. L. lactis strains were grown in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) or GM17 agar at 30°C. Lactobacillus sakei subsp. sakei JCM 1137 was grown in MRS broth at 30°C. Escherichia coli JM109 and E. coli BL21(DE3) were grown in Luria-Bertani broth at 37°C. For the selection of transformants carrying pNZ8048 and its derivatives, chloramphenicol was used at a concentration of 10 μg/ml. For the selection of transformants carrying pBAD and its derivatives, ampicillin was used at a concentration of 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. warneri ISK-1 | Nukacin ISK-1 producer | 32 |
| L. lactis NZ9000 | MG1363 derivative; nisRK::pep | 7 |
| L. sakei subsp. sakei JCM 1157T | Indicator strain for bioassay | JCM |
| E. coli JM109 | Cloning host | Novagen |
| E. coli BL21(DE3) | Expression host; F−ompT hsdSB(rB−mB−) gal drm (DE3) | Novagen |
| Plasmids | ||
| pNZ8048 | Cmr; PnisA; L. lactis expression vector | 7 |
| pNZFHisEGH | pNZ8048 with hexahistidine sequence and nukFEGH | This study |
| pNZF(E85Q)HisEGH | E85Q mutant of pNZFHisEGH | This study |
| pNZF(E85A)HisEGH | E85A mutant of pNZFHisEGH | This study |
| pNZF(E158Q)HisEGH | E158Q mutant of pNZFHisEGH | This study |
| pNZF(H191A)HisEGH | H191A mutant of pNZFHisEGH | This study |
| pNZEGH | pNZ8048 with nukEGH | 22 |
| pBAD His/B | Ampr; ParaC; E. coli expression vector | Invitrogen |
| pBADFEGH | pBAD His/B with nukFEGH | This study |
| pBADmalK | pBAD His/B with malK | This study |
Site-directed mutagenesis of nukF.
First, a gene fragment containing nukFEGH was amplified with KOD plus DNA polymerase (Toyobo, Osaka, Japan) by using the primer sets pBADnukF_F and pBADnukH_R (the oligonucleotides used are listed in Table S1 in the supplemental material). After digestion of the amplified fragment by XhoI and EcoRI, the fragment was ligated with a similarly digested pBAD vector. After transformation into E. coli JM109, the resulting plasmid, pBADFEGH, was isolated. Then, the gene fragment containing nukFEGH and the His6 tag-encoding sequence located upstream of nukF were amplified with KOD plus DNA polymerase by using the primer sets pBADnukF_F2 and 8048nukH_R. After digestion with NcoI and PstI, the amplified fragment was ligated with the similarly digested pNZ8048 vector. After transformation into L. lactis NZ9000 according to the method developed by Holo and Nes (12), the resulting plasmid, pNZFHisEGH, was isolated. Site-directed mutagenesis of nukF in pNZFHisEGH was performed by inverse PCR according to the method described previously (21), using the following primer sets: E85Q_F and E85AQ_R (E85Q mutation), E85A_F and E85AQ_R (E85A mutation), E158Q_F and E158Q_R (E158Q mutation), and H191A_F and H191_R (H191A mutation). After transformation of the self-ligated fragments into L. lactis NZ9000, the plasmids pNZF(E85Q)HisEGH, pNZF(E85A)HisEGH, pNZF(E185Q)HisEGH, and pNZF(H191A)HisEGH were obtained.
Immunity assays.
For the expression of immunity genes, the nisin-controlled expression (NICE) system was used (6, 7). Overnight cultures of recombinant L. lactis were diluted 100-fold in GM17 broth, and after the cultures reached an optical density at 600 nm (OD600) of 0.8, nisin solution was added to achieve a final concentration of 10 ng/ml. Subsequently, gene expression was induced by overnight cultivation at 30°C. For the agar diffusion assay, 50 μl of the nisin-induced cultures was added to 5 ml of lactobacillus agar (Difco Laboratories, Detroit, MI) supplemented with nisin and chloramphenicol at concentrations of 10 ng/ml and 10 μg/ml, respectively, and overlaid on GM17 agar plates. Then, 40 μl solutions of serially diluted nukacin ISK-1, which had been purified as described previously (2), was added to the holes (diameter, 5 mm) made in the agar. After overnight culture at 30°C, we observed the growth inhibition zones. For measuring the optical density, 250 μl of the nisin-induced cultures was added to 5 ml GM17 broth supplemented with nisin and chloramphenicol at concentrations of 10 ng/ml and 10 μg/ml, respectively, and 135 μl of the cell suspension was added to wells in 96-microwell plates. After addition of 15 μl of nukacin ISK-1 solution to achieve the appropriate final concentrations, the growth of recombinant L. lactis strains was monitored at OD590 by using an Immuno Mini NJ-2300 plate reader (Biotec, Tokyo, Japan).
Transport assays.
The metabolic inhibition of recombinant L. lactis strains was induced using dinitrophenol as described previously (22). For the bioassay-based transport assay, nukacin ISK-1 was added to 1 ml of the cell suspensions (OD600 = 10) to achieve a final concentration of 2 μM and incubated for 30 min at 30°C. After one wash with 1 ml of 50 mM potassium phosphate buffer (pH 7.0) and resuspension of the cells with same buffer, 40 μl of each cell suspension was added to the hole on the MRS agar plate overlaid with 5 ml of lactobacillus agar and seeded with 50 μl of overnight-cultured L. sakei subsp. sakei JCM 1157T as an indicator strain. After overnight culture at 30°C, growth inhibition zones were observed. The transport reaction was monitored in real time according to the method described previously (22).
Preparation of inside-out membrane vesicles of L. lactis and Western blot analysis.
L. lactis strains were grown in GM17 broth to an OD600 of 0.8 and induced in the presence of nisin at a final concentration of 10 ng/ml for 3 h. Cells were harvested by centrifugation at 6,500 × g for 15 min, and the cell pellet was washed in 50 mM Tris-HCl buffer (pH 7.0) and incubated in the same buffer containing 2 mg/ml lysozyme (Seikagaku Co., Tokyo, Japan) and protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). After a 30-min incubation at 30°C, the cells were disrupted by passing them thrice through a French pressure cell at 20,000 lb/in2; unbroken cells were removed by centrifugation at 6,500 × g for 15 min. Inside-out membrane vesicles were collected from the supernatant obtained after ultracentrifugation at 176,000 × g for 60 min and stored in 50 mM Tris-HCl buffer (pH 7.4) containing 100 mM NaCl and 10% glycerol (buffer A) at −80°C. We determined the expression levels of NukF and its mutants in the membrane fractions by Western blot analysis for hexahistidine tags fused to the proteins, using a Penta·His horseradish peroxidase (HRP) conjugate kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol.
Purification of NukF and its mutants.
NukF and its mutants were solubilized from the membrane vesicles by incubation in buffer A containing 0.5% n-dodecyl-β-d-maltoside (DDM) for 1 h at 4°C. After ultracentrifugation of the mixture at 176,000 × g for 60 min, the supernatant was applied to a His-Trap FF column (GE Healthcare, Uppsala, Sweden) equilibrated with buffer A containing 0.05% DDM (buffer B). The column was washed with 10 volumes of buffer B and 5 volumes of buffer B containing 100 mM imidazole. The proteins were then eluted using 5 volumes of buffer B containing 300 mM imidazole. Imidazole and DDM in the samples were removed using HiTrap Desalting (GE Healthcare) equilibrated with buffer A; subsequently, the samples were concentrated using Amicon Ultra 10K (Millipore, Billerica, MA).
Expression and purification of E. coli MalK.
The gene fragment encoding MalK was amplified with the primer sets pBADmalK_F and pBADmalK_R by using the total genome of E. coli JM109 as a template. The amplified fragment was digested with XhoI and HindIII and ligated with a similarly digested pBAD vector, and the resulting plasmid, pBADmalK, was introduced into E. coli BL21(DE3). E. coli cells were grown to an OD600 of 0.5, and the expression of malK was induced by addition of 0.02% l-arabinose and further incubation for 3 h. The harvested cells were disrupted using the French pressure cell as mentioned above, and MalK was purified from the cytoplasmic fraction by using the His-Trap FF column. The purification protocol was the same as that for NukF and its mutants except that buffer A was used instead of buffer B.
Measurement of ATPase activity.
ATPase activities of purified NBDs were measured by assaying the amount of inorganic phosphate liberated from ATP. As a standard, NaH2PO4 solutions were used. Purified NBDs were added to achieve a final concentration of 60 μg/ml in 50 μl of 50 mM Tris-HCl (pH 7.4) containing 5 mM MgSO4 and 5 mM ATP, and then, the solution was incubated for 20 min at 30°C in 96-microwell plates. Subsequently, 100 μl of Biomol Green (Biomol Research Laboratories, Plymouth Meeting, PA) was added to the solution. After a 20-min incubation at room temperature, the OD620 was measured using the Immuno Mini NJ-2300 plate reader.
RESULTS
E loop, a novel conserved motif in LanF.
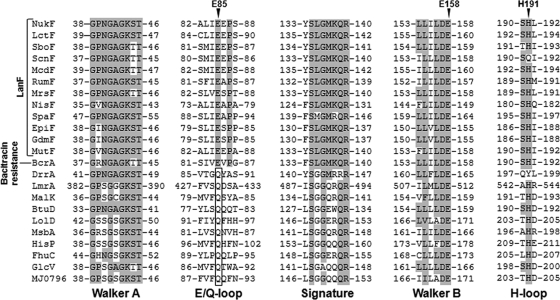
Multiple sequence alignment revealed that all LanF proteins have the E loop as a variant of the Q loop, which is well conserved in NBDs of general ABC transporters (Fig. 1). It should be noted that BcrA, which is an NBD of the bacitracin resistance ABC transporter, also contains the E loop. Bacitracin is a nonribosomally synthesized antibacterial peptide produced by Bacillus licheniformis, and this peptide contains unusual amino acids and the cyclic structure (17). Bacitracin inhibits the biosynthesis of the bacterial cell wall by inhibiting dephosphorylation of C55-isoprenyl pyrophosphate (35, 36). Like LanFEG, the bacitracin resistance ABC transporter was presumed to transport bacitracin from the membrane (28, 29). To investigate the function of the E loop, we constructed 2 E-loop mutants of NukF, namely, E85Q and E85A, corresponding to replacement with glutamine and alanine, respectively. Additionally, we introduced substitutions into the amino acids of 2 other conserved motifs, Walker B and H loop, in the NukF sequence, so that the E158Q and H191A mutants were obtained. In this study, we coexpressed NukF or its mutants with NukEGH to investigate their immunity against nukacin ISK-1. To compare the expression levels of NukF and its mutants in the cytoplasmic membrane, we expressed these mutants with hexahistidine tags (His6 tags) on their N termini. Western blot analysis using anti-His6 tag antibody and membrane fractions prepared from recombinant L. lactis showed that all proteins are localized in the membrane at similar expression levels (Fig. 2).
FIG. 1.
Multiple sequence alignment of LanF proteins and NBDs of other ABC transporters. The alignment of the LanF proteins and other ABC transporters was generated using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/). The residues involved in conserved motifs (Walker A, the E/Q loop, the Signature motif, Walker B, and the H loop) are shown, and glutamine residues conserved in the Q loop are boxed. The residues conserved in at least 50% of the studied proteins are shaded gray. The mutation points in NukF are shown above the sequence.
FIG. 2.

Expression levels of NukF and its mutants. Membrane vesicles were prepared from cells of recombinant L. lactis by using the French press; subsequently, 1 μg of membrane protein was subjected to Tricine SDS-PAGE, followed by Western blot analysis with anti-His6 tag antibody.
Effects of mutations on the immunity level.
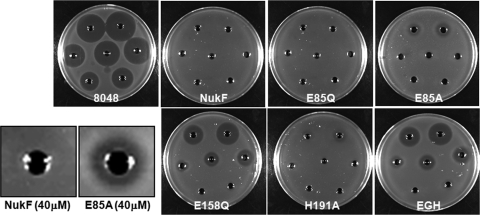
The level of immunity of each recombinant L. lactis against nukacin ISK-1 was analyzed by 2 bioassay methods. The agar diffusion assay, which defines the immunity level on the basis of the diameter of the growth inhibition zone formed on the agar, showed that E85Q-expressing cells and H191A-expressing cells had immunity levels comparable to that of nukF-expressing cells (Fig. 3). On the other hand, E85A-expressing cells showed a slightly decreased immunity level compared to that of nukF-expressing cells. The immunity level of E158Q-expressing cells was similar to that of nukEGH-expressing cells, suggesting that this mutation abolished the function of NukF completely (Fig. 3). Since the agar diffusion assay defines the immunity level of the strain after overnight culture, it is impossible to analyze the behavior of cells (e.g., time of lag phase, maximum cell density, and growth rate). Therefore, we investigated the immunity levels by monitoring the optical densities (590 nm) of the culture of each strain in the presence of nukacin ISK-1. In the absence of nukacin ISK-1, all recombinant L. lactis strains showed almost the same growth pattern (Fig. 4A). Even in the presence of 5 μM and 10 μM nukacin ISK-1 as final concentrations, the growth level of the nukF-expressing strain was identical to that observed following culture without nukacin ISK-1 (Fig. 4B and C). On the other hand, the growth levels of E85Q- and H191A-expressing strains decreased drastically, and E85A- and E158Q-expressing strains showed growth levels almost similar to that of the nukEGH-expressing strain (Fig. 4B and C).
FIG. 3.
Immunity assay of recombinant L. lactis using agar-well diffusion. Serially diluted nukacin ISK-1 (40 μM to 0.625 μM) was added to the holes on the agar seeded with recombinant L. lactis strains. After overnight culture at 30°C, the growth inhibition zones formed on the plate were observed. “8048” represents a control strain harboring pNZ8048, “NukF” represents a strain harboring pNZFHisEGH, “E85Q” represents a strain harboring pNZF(E85Q)HisEGH, “E85A” represents a strain harboring pNZF(E85A)HisEGH, “E158Q” represents a strain harboring pNZF(E158Q)HisEGH, “H191A” represents a strain harboring pNZF(H191A)HisEGH, and “EGH” represents a strain harboring pNZEGH.
FIG. 4.
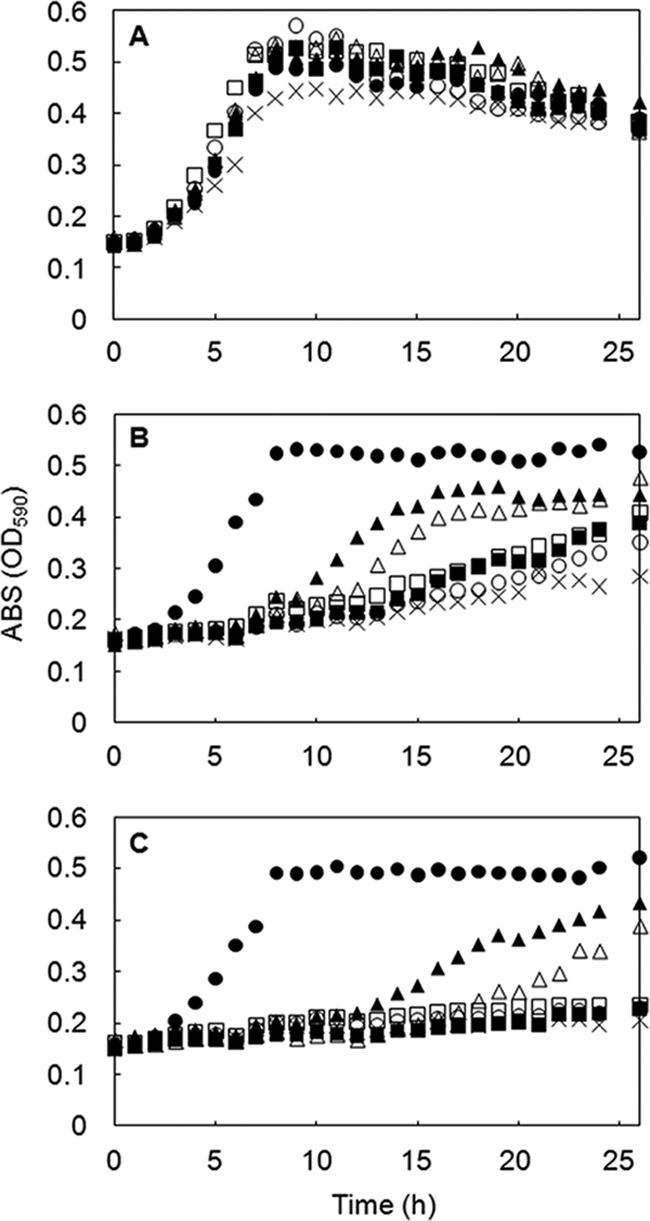
Immunity assay of recombinant L. lactis on microwell plates. Recombinant L. lactis strains were cultured in GM17 broth in the absence (A) or presence (B, C) of nukacin ISK-1 at final concentrations of 5 μM (B) or 10 μM (C). The growth of recombinant L. lactis strains was monitored by measuring OD590. Symbols: •, strain harboring pNZFHisEGH; ▴, strain harboring pNZF(E85Q)HisEGH; ▪, strain harboring pNZF(E85A)HisEGH; ○, strain harboring pNZF(E158Q)HisEGH; ▵, strain harboring pNZF(H191A)HisEGH; □, strain harboring pNZEGH; and ×, strain harboring pNZ8048. The values represent averages of results from 2 independent experiments.
Effects of mutations on the transport activity.
The NBDs of ABC transporters generate driving force for the transport by hydrolyzing ATP. We hypothesized that the decreased immunity levels of NukF mutants observed in this study were due to a decrease in the transport activities caused by mutations of the conserved motifs. Earlier, we showed that nukH-expressing cells bind nukacin ISK-1 and that nukacin ISK-1 bound to NukH is transported by NukFEG in an energy-dependent manner (22). The expression of NukH increases the amount of cell-associated nukacin ISK-1 drastically (2) and enables easy detection of nukacin ISK-1 transported by NukFEG. Therefore, we investigated the effects of mutations on transport activity by using cells coexpressing NukF mutants with NukEGH. We prepared energy-deprived cells of recombinant L. lactis and incubated them with purified nukacin ISK-1. Subsequently, the cell suspensions were spotted on agar plates and seeded with the indicator strain, Lactobacillus sakei subsp. sakei JCM 1157T. We expected that cell-associated nukacin ISK-1 is released from the cell membrane, accompanied with the activation of transporters during cell growth. According to this idea, the transported nukacin ISK-1 can be detected by observing the growth inhibition zone against the indicator strain. We termed this assay the “bioassay-based transport assay.” We observed a clear growth inhibition zone around the area where the nukF-expressing cells had been spotted but not around the areas where E85A-, E158Q- and EGH-expressing cells and control cells (8048) had been spotted (Fig. 5). Also, we could find small growth inhibition zones around the area where E85Q- and H191A-expressing cells had been spotted, even though these zones were not clear (Fig. 5).
FIG. 5.
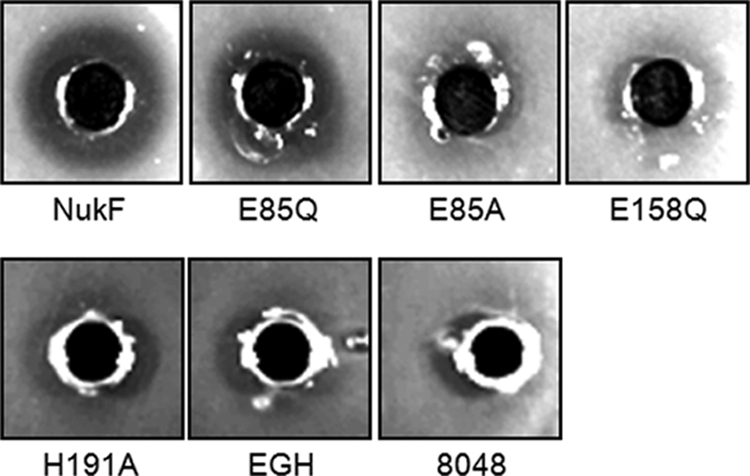
Bioassay-based transport assay. Energy-deprived cells of recombinant L. lactis were preincubated with 2 μM nukacin ISK-1 and then washed and resuspended in buffer. Subsequently, they were added to the holes on the agar plate seeded with L. sakei subsp. sakei JCM 1157T as an indicator strain. After overnight culture at 30°C, the growth inhibition zones were observed. The designations represent the same strains as those indicated in the legend to Fig. 3.
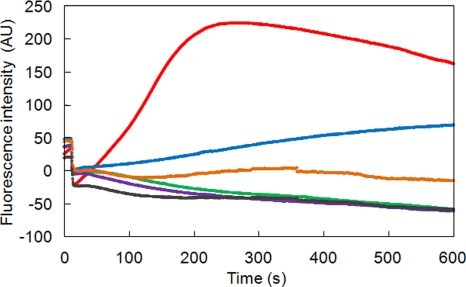
Next, we investigated the transport activities of cells expressing NukF mutants in more detail by using fluorescently labeled nukacin ISK-1 (FITC-nuk). Recently, we demonstrated the real-time monitoring of the transport reaction by using FITC-nuk and L. lactis cells expressing nukFEGH (22). This method enables us to perform the transport assay in real time by observing the increase in fluorescence. Therefore, the transport activities of each NukF mutant were analyzed using this method (Fig. 6). The fluorescence of nukEGH-expressing cells (as a negative control) slowly decreased after the addition of glucose, presumably due to the degradation of FITC. The fluorescence of E85Q-expressing cells increased slowly in comparison to that of nukF-expressing cells. This suggests that the E85Q mutation suppressed the transport activity of NukFEG, leading to a decreased immunity level, as can be observed in Fig. 4. The transport activity of H191A-expressing cells was suppressed more than that of E85Q-expressing cells. On the other hand, E85A- and E158Q-expressing cells, which are low-immunity mutants, did not show any transport activity at all. These phenomena correspond well with the results observed of the bioassay-based transport assay (Fig. 5). Additionally, we found a good correlation between the order of the transport activities and that of immunity levels, as seen in Fig. 4 (i.e., NukF > E85Q > H191A > E85A, E158Q, and EGH). Therefore, we concluded that the decreased immunity levels of cells expressing NukF mutants resulted from suppression of the transport activities.
FIG. 6.
Real-time monitoring of FITC-nuk transport. After washing and resuspending of the energy-deprived cells of recombinant L. lactis treated with 2.5 mM FITC-nuk, the transport reactions were initiated by the addition of glucose to achieve a final concentration of 1%. Fluorescence was monitored for 600 s by using a fluorimeter. The red line represents a strain harboring pNZFHisEGH, the blue line represents a strain harboring pNZF(E85Q)HisEGH, the green line represents a strain harboring pNZF(E85A)HisEGH, the gray line represents a strain harboring pNZF(E158Q)HisEGH, the orange line represents a strain harboring pNZF(H191A)HisEGH, and the purple line represents a strain harboring pNZEGH. The fluorescence level of each cell suspension at the time of glucose addition was defined as 0.
ATPase activities of purified NukF and its mutants.
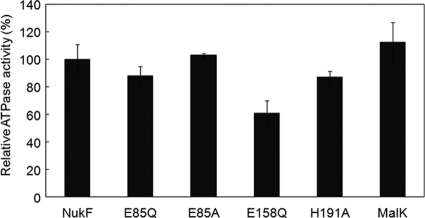
E85Q-expressing cells retained some of the immunity and transport activity; on the other hand, the E85A mutation resulted in a complete loss of these activities. To determine the factor that affected NukF function, we measured the ATPase activities of purified NukF and its mutants. NukF and its mutants were coexpressed with NukEGH and purified from inverted membrane vesicles of L. lactis, because when NukF was expressed independently in E. coli, most of the expressed NukF formed an inclusion body (data not shown). All proteins were purified using Ni-affinity chromatography, and were detected as single bands on the Coomassie brilliant blue stained gel after SDS-PAGE analysis (data not shown). After removal of imidazole from the purified samples, ATPase activities were measured using malachite green methods (Fig. 7). MalK, which is an NBD in the maltose transport system (MalFGK2), was used as a positive control. NukF and MalK showed similar activities in this assay. Several studies indicate that conserved glutamate in the Walker B motif is a catalytic base for ATP hydrolysis (19, 25). Consistent with this rule, the ATPase activity of E158Q decreased drastically (61% of the NukF level). On the other hand, the ATPase activities of E85Q, E85A, and H191A were comparable to that of NukF (88%, 103%, and 87% of the NukF level, respectively). These results are consistent with a previous study on MalK, in which mutations had been introduced in the Q loop and H loop (37). Therefore, we concluded that the functional defects of NukF mutants, with the exception of E158Q, are not directly related to their ATPase activities.
FIG. 7.
ATPase activities of purified NBDs. The reaction mixtures containing 60 μg/ml purified NBDs, 50 mM Tris-HCl (pH 7.4), 5 mM MgSO4, and 5 mM ATP were incubated for 20 min at 30°C. The inorganic phosphate released after ATP hydrolysis was quantified using the malachite green method. The ATPase activity level of purified NukF was defined as 100%. This assay was performed in duplicate, and the standard values are represented with error bars.
DISCUSSION
Mechanisms of immunity against lantibiotics have been studied for more than a decade. Even though the lantibiotic-binding mechanisms of LanI and LanH have been actively studied, the molecular mechanism of lantibiotic transport is still unclear. Several studies have demonstrated that the expression of LanFEG increases the immunity level of host cells and decreases the amount of lantibiotics bound to cells (2, 26, 33, 34). These results suggested that LanFEG transports lantibiotics from the membrane to the extracellular space. In this study, to elucidate the function of the E loop, a novel motif found in LanF, we replaced the E loop with the generally conserved Q loop. It is noteworthy that the E loop is conserved not only in the LanF proteins but also in BcrA, which is the NBD of the bacitracin resistance ABC transporter (Fig. 1). High sequence similarity between LanF proteins and BcrA makes us speculate that LanFEG and the bacitracin resistance ABC transporter share a molecular mechanism of transport.
Mutational studies of NukF clearly demonstrated that the E loop is important for its function. Interestingly, although the E85A mutation in the E loop resulted in severely impaired immunity and transport activity, the E85Q mutation retained a relatively high level of these activities. Additionally, there was good correlation between the results of the bioassay and those of the transport assay (Fig. 4 and 6). Therefore, we presumed that the long lag phase in the growth of E85Q- and H191A-expressing strains that occurred only in the presence of nukacin ISK-1 is due to decreased transport efficiency (Fig. 4). Considering the mode of action of lantibiotics, this speculation seems to be reasonable. It has been shown that several lantibiotics, including nisin (11, 14), LtnAI (α-peptide of lacticin 3147) (38), and mersacidin (3, 13), use lipid II, a cell wall precursor located on the membrane, as a docking molecule. After docking, these lantibiotics form pores on the cytoplasmic membrane and/or inhibit cell wall biosynthesis. Additionally, it was speculated that type A(II) lantibiotics, including nukacin ISK-1 and lacticin 481, also bind to lipid II because their sequences contain the same lipid II-binding motifs found in LtnAI and mersacidin (9, 15). Recently, it was demonstrated that nukacin ISK-1 has bacteriostatic activity causing cell wall width reduction and incomplete septum formation (1). Since most lantibiotics are believed to act on the membrane, rapid transport of lantibiotic molecules from the membrane should directly lead to an increase in the immunity level. Our results provide the first experimental evidence of the correlation between the immunity level and the transport rate of LanFEG.
Measurement of the ATPase activities of purified NBDs revealed that all NukF mutants, except for E158Q, show ATPase activity comparable to that of NukF. Therefore, we concluded that the functional defects of NukF mutants, except for E158Q mutants, are not due to the suppression of ATPase activity. The key factor affecting the function of the NukFEG system needs to be determined. Recently, mutational studies of the Q loop have been carried out while studying DrrA, an NBD of an ABC transporter involved in self-resistance to anticancer antibiotics (doxorubicin and daunorubicin) (30). Nonconservative mutations of the Q loop (Q88S and Q88N) resulted in great reductions in the resistance levels; however, the Q88E mutation retained the resistance to a high degree. An important observation is that the phenotype of the Q88E mutation in DrrA corresponds well to that of the E85Q mutation in NukF. This strongly suggests that the functions of the Q loop and E loop can complement one another, even if they do not complement fully. In other words, the Q loop and E loop might have similar functions.
So far, many reports have suggested that the Q loop acts as a mediator for the conformational change of transmembrane subunits. Molecular dynamics simulation of HisP, a well-studied NBD of bacterial histidine permease, helped in elucidating the conformational transition of the Q loop and the coordination of this transition with the conformational changes of transmembrane subunits and NBDs (16). Similarly, it was suggested that the interaction of the Q loop in DrrA with the N-terminal cytoplasmic tail of DrrB, which is a transmembrane subunit, is involved in transmitting conformational changes between DrrA and DrrB (30). The X-ray structure of the fully assembled maltose transport system showed that the Q loops of MalK are in close proximity to the transmembrane subunit MalFG (24). In the structure of the vitamin B12 transporter, the NBD subunit ButD is bound to the transmembrane subunit BtuC via the Q loop (18). On the basis of the structural similarity of NBDs, Chen et al. proposed that the mechanism involving an interaction between the L-formed cytoplasmic loop of the transmembrane subunit and the cleft formed by the amino acid residues surrounding the Q loop (Q loop cleft) is universal despite sequence variations in the ABC transporters (5). To determine the structure surrounding the E loop of NukF, we performed homology modeling of NukF. The structure of TM0544, which is an NBD of ABC transporters from Thermotoga maritima MSB8, was used as a template for the modeling. As a result, it was predicted that the amino acid residues surrounding the E loop form a cleft in a manner similar to that observed for the Q loop (Fig. 8). Taken together, these results support the hypothesis that the E loop is a specific motif involved in the conformational change of the transmembrane subunits (LanE and LanG) that is accompanied with ATP hydrolysis. In conclusion, the E loop, which is a variant of Q loop, appeared to be a key factor that contributes to the lantibiotic-transport mechanism of LanFEG.
FIG. 8.
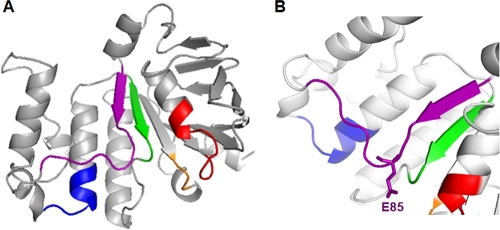
Structural model of NukF. (A) The structure of NukF was built by homology modeling, using the SWISS-MODEL server (http://swissmodel.expasy.org/). The structure of TM0544 (Protein Data Bank [PDB] accession no. 1VPL) was used as a template for the modeling. The figure was prepared using PyMOL (www.pymol.org). The color coding is as follows: red, Walker A; green, Walker B; blue, Signature motif; purple, E loop; and orange, H loop. (B) View focused on the E loop of NukF.
Supplementary Material
Acknowledgments
This work was partially supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (JSPS), from the Japan Science Society, from the Novartis Foundation (Japan) for the Promotion of Science, from the Novozymes Japan Research Fund, and from the Nagase Science and Technology Foundation and by JSPS research fellowships.
Footnotes
Published ahead of print on 9 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Asaduzzaman, S. M., J. Nagao, H. Iida, T. Zendo, J. Nakayama, and K. Sonomoto. 2009. Nukacin ISK-1, an evidence of bacteriostatic lantibiotic. Antimicrob. Agents Chemother. 53:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aso, Y., K. Okuda, J. Nagao, Y. Kanemasa, N. T. B. Phuong, H. Koga, K. Shioya, T. Sashihara, J. Nakayama, and K. Sonomoto. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403-1410. [DOI] [PubMed] [Google Scholar]
- 3.Brötz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H.-G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633-684. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., G. Lu, J. Lin, A. L. Davidson, and F. A. Quiocho. 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell 12:651-661. [DOI] [PubMed] [Google Scholar]
- 6.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draper, L. A., R. P. Ross, C. Hill, and P. D. Cotter. 2008. Lantibiotic immunity. Curr. Protein Pept. Sci. 9:39-49. [DOI] [PubMed] [Google Scholar]
- 9.Dufour, A., T. Hindre, D. Haras, and J. P. Le Pennec. 2007. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol. Rev. 31:134-167. [DOI] [PubMed] [Google Scholar]
- 10.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasper, H. E., N. E. Kramer, J. L. Smith, J. D. Hillman, C. Zachariah, O. P. Kuipers, B. de Kruijff, and E. Breukink. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636-1637. [DOI] [PubMed] [Google Scholar]
- 12.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 13.Hsu, S. T., E. Breukink, G. Bierbaum, H.-G. Sahl, B. de Kruijff, R. Kaptein, N. A. van Nuland, and A. M. Bonvin. 2003. NMR study of mersacidin and lipid II interaction in dodecylphosphocholine micelles. Conformational changes are a key to antimicrobial activity. J. Biol. Chem. 278:13110-13117. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, S. T., E. Breukink, E. Tischenko, M. A. Lutters, B. de Kruijff, R. Kaptein, A. M. Bonvin, and N. A. van Nuland. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11:963-967. [DOI] [PubMed] [Google Scholar]
- 15.Islam, M. R., K. Shioya, J. Nagao, M. Nishie, H. Jikuya, T. Zendo, J. Nakayama, and K. Sonomoto. 2009. Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol. Microbiol. 72:1438-1447. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. M., and A. M. George. 2002. Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. Proc. Natl. Acad. Sci. U. S. A. 99:12639-12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi, N., T. Takenouchi, S. Endo, and E. Munekata. 1992. 1H NMR study on the conformation of bacitracin A in aqueous solution. FEBS Lett. 305:105-109. [DOI] [PubMed] [Google Scholar]
- 18.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 19.Moody, J. E., L. Millen, D. Binns, J. F. Hunt, and P. J. Thomas. 2002. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277:21111-21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagao, J., Y. Aso, K. Shioya, J. Nakayama, and K. Sonomoto. 2007. Lantibiotic engineering: molecular characterization and exploitation of lantibiotic-synthesizing enzymes for peptide engineering. J. Mol. Microbiol. Biotechnol. 13:235-242. [DOI] [PubMed] [Google Scholar]
- 21.Okuda, K., Y. Aso, J. Nagao, K. Shioya, Y. Kanemasa, J. Nakayama, and K. Sonomoto. 2005. Characterization of functional domains of lantibiotic-binding immunity protein, NukH, from Staphylococcus warneri ISK-1. FEMS Microbiol. Lett. 250:19-25. [DOI] [PubMed] [Google Scholar]
- 22.Okuda, K., Y. Aso, J. Nakayama, and K. Sonomoto. 2008. Cooperative transport between NukFEG and NukH in immunity against the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. J. Bacteriol. 190:356-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuda, K., S. Yanagihara, K. Shioya, Y. Harada, J. Nagao, Y. Aso, T. Zendo, J. Nakayama, and K. Sonomoto. 2008. Binding specificity of the lantibiotic-binding immunity protein NukH. Appl. Environ. Microbiol. 74:7613-7619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oldham, M. L., D. Khare, F. A. Quiocho, A. L. Davidson, and J. Chen. 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450:515-521. [DOI] [PubMed] [Google Scholar]
- 25.Orelle, C., O. Dalmas, P. Gros, A. Di Pietro, and J. M. Jault. 2003. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J. Biol. Chem. 278:47002-47008. [DOI] [PubMed] [Google Scholar]
- 26.Otto, M., A. Peschel, and F. Götz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol. Lett. 166:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Peschel, A., and F. Götz. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J. Bacteriol. 178:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Podlesek, Z., A. Comino, B. Herzog-Velikonja, and M. Grabnar. 2000. The role of the bacitracin ABC transporter in bacitracin resistance and collateral detergent sensitivity. FEMS Microbiol. Lett. 188:103-106. [DOI] [PubMed] [Google Scholar]
- 29.Podlesek, Z., A. Comino, B. Herzog-Velikonja, D. Zgur-Bertok, R. Komel, and M. Grabnar. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969-976. [DOI] [PubMed] [Google Scholar]
- 30.Rao, D. K., and P. Kaur. 2008. The Q-loop of DrrA is involved in producing the closed conformation of the nucleotide binding domains and in transduction of conformational changes between DrrA and DrrB. Biochemistry 47:3038-3050. [DOI] [PubMed] [Google Scholar]
- 31.Rees, D. C., E. Johnson, and O. Lewinson. 2009. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10:218-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sashihara, T., H. Kimura, T. Higuchi, A. Adachi, H. Matsusaki, K. Sonomoto, and A. Ishizaki. 2000. A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and identification of the structure. Biosci. Biotechnol. Biochem. 64:2420-2428. [DOI] [PubMed] [Google Scholar]
- 33.Stein, T., S. Heinzmann, S. Dusterhus, S. Borchert, and K. D. Entian. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187:822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein, T., S. Heinzmann, I. Solovieva, and K. D. Entian. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89-94. [DOI] [PubMed] [Google Scholar]
- 35.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 68:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storm, D. R., and J. L. Strominger. 1973. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248:3940-3945. [PubMed] [Google Scholar]
- 37.Walter, C., S. Wilken, and E. Schneider. 1992. Characterization of site-directed mutations in conserved domains of MalK, a bacterial member of the ATP-binding cassette (ABC) family. FEBS Lett. 303:41-44. [DOI] [PubMed] [Google Scholar]
- 38.Wiedemann, I., T. Bottiger, R. R. Bonelli, A. Wiese, S. O. Hagge, T. Gutsmann, U. Seydel, L. Deegan, C. Hill, P. Ross, and H.-G. Sahl. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol. Microbiol. 61:285-296. [DOI] [PubMed] [Google Scholar]
- 39.Wilaipun, P., T. Zendo, K. Okuda, J. Nakayama, and K. Sonomoto. 2008. Identification of the nukacin KQU-131, a new type-A(II) lantibiotic produced by Staphylococcus hominis KQU-131 isolated from Thai fermented fish product (Pla-ra). Biosci. Biotechnol. Biochem. 72:2232-2235. [DOI] [PubMed] [Google Scholar]
- 40.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.