Abstract
BacA is an integral membrane protein, the mutation of which leads to increased resistance to the antimicrobial peptides bleomycin and Bac71-35 and a greater sensitivity to SDS and vancomycin in Rhizobium leguminosarum bv. viciae, R. leguminosarum bv. phaseoli, and Rhizobium etli. The growth of Rhizobium strains on dicarboxylates as a sole carbon source was impaired in bacA mutants but was overcome by elevating the calcium level. While bacA mutants elicited indeterminate nodule formation on peas, which belong to the galegoid tribe of legumes, bacteria lysed after release from infection threads and mature bacteroids were not formed. Microarray analysis revealed almost no change in a bacA mutant of R. leguminosarum bv. viciae in free-living culture. In contrast, 45 genes were more-than 3-fold upregulated in a bacA mutant isolated from pea nodules. Almost half of these genes code for cell membrane components, suggesting that BacA is crucial to alterations that occur in the cell envelope during bacteroid development. In stark contrast, bacA mutants of R. leguminosarum bv. phaseoli and R. etli elicited the formation of normal determinate nodules on their bean host, which belongs to the phaseoloid tribe of legumes. Bacteroids from these nodules were indistinguishable from the wild type in morphology and nitrogen fixation. Thus, while bacA mutants of bacteria that infect galegoid or phaseoloid legumes have similar phenotypes in free-living culture, BacA is essential only for bacteroid development in indeterminate galegoid nodules.
Bacteria of the family Rhizobiaceae are alphaproteobacteria, which form a species-specific symbiotic relationship with leguminous plants. Plants release flavonoids that typically induce the synthesis of lipochitooligosaccharides by rhizobia, which in turn initiate a signaling cascade in the plant, leading to nodule formation (34). Rhizobia become trapped by curling root hairs, which they enter via infection threads that grow and ramify into the root cortex, where newly induced meristematic cells form the nodule (34). Bacteria are released from infection threads and engulfed by a plant-derived symbiosome membrane. In galegoid legumes (a clade in the subfamily Papilionoideae, such as Medicago, Pisum, or Vicia), which form indeterminate nodules that have a persistent meristem, bacteria undergo the endoreduplication of their chromosome, resulting in dramatic increases in size, shape, and DNA content to become terminally differentiated bacteroids (32). However, in phaseoloid legumes (e.g., lotus, bean, and soybean), which form determinate nodules with a transient meristem, bacteria do not undergo endoreduplication and therefore do not enlarge substantially. These bacteroids retain a normal DNA content and can regrow after isolation from nodules (32). The endoreduplication of bacteroids is controlled by the plant, and it is believed that nodule-specific cysteine-rich (NCR) peptides, which are made in indeterminate, but not in determinate, nodules, may be responsible for inducing and maintaining bacteroid development (31, 32). Finally, mature bacteroids receive dicarboxylic acids from the plant, which they use as a carbon, reductant, and energy source for the reduction of N2 to ammonia (38). The ammonia is secreted to the plant, where it is assimilated into amino acids or ureides, depending on the legume, for export to the shoot.
Sinorhizobium meliloti BacA protein was the first bacterial factor identified to be essential for bacteroid development (15). More recently, it also has been shown to be essential for the Mesorhizobium-Astragalus symbiosis (42). S. meliloti elicits the formation of indeterminate nodules on alfalfa, and while S. meliloti bacA null mutants induce nodule formation, bacteria lyse soon after endocytosis but prior to bacteroid differentiation (15, 20). BacA is a cytoplasmic membrane protein that shares 64% identity with SbmA from Escherichia coli (15, 25). SbmA/BacA proteins belong to the ATP binding cassette (ABC) superfamily and share sequence similarity with a family of eukaryotic peroxisomal membrane proteins, including the human adrenoleukodystrophy protein, which is required for the efficient transport of very-long-chain fatty acids (VLCFAs) out of the cytoplasm (9). Consistent with this, S. meliloti BacA is required for the complete modification of lipid A with VLCFAs (9). However, since S. meliloti mutants, which are directly involved in the biosynthesis of VLCFA-modified lipid A, show bacteroid abnormalities but still can form a successful alfalfa symbiosis, the effect of BacA on lipid A VLCFA modification does not fully account for its essential role in bacteroid development (10, 11, 16). Strains mutated in bacA also have an increased resistance to the glycopeptide bleomycin, a low-level resistance to aminoglycoside antibiotics, and an increased sensitivity to ethanol, sodium dodecyl sulfate (SDS), and deoxycholate relative to the sensitivities of the parent strain (12, 18, 25). More recently it has been shown that an S. meliloti bacA null mutant has an increased resistance to a truncated form of a eukaryotic proline-rich peptide, Bac71-16, and was unable to accumulate a fluorescently labeled form of this peptide (28). This finding, combined with the increased resistance of an S. meliloti bacA null mutant to bleomycin, led to the hypothesis that BacA is itself a putative peptide transporter (BacA mediated) or able to alter the activity of such a transporter (BacA influenced) (11, 15, 18, 28).
As the increased resistance of the S. meliloti bacA null mutant to bleomycin and Bac71-16 appears to be independent of the VLCFA modification of lipid A (11, 28), this suggested that either BacA-mediated or BacA-influenced peptide uptake into S. meliloti plays a role in bacteroid development. Since indeterminate galegoid nodules contain hundreds of NCR peptides, whereas determinate phaseoloid nodules lack these host peptides (31), we considered it important to assess the role of BacA in bacteroid development during the formation of both nodule types.
Here, we show that bacA mutants of Rhizobium leguminosarum bv. viciae strains 3841 and A34 failed to develop bacteroids and did not fix nitrogen in indeterminate pea (Pisum sativum) nodules. However, bacA mutants of both R. leguminosarum bv. phaseoli 4292 and Rhizobium etli CE3 formed normal bacteroids and fixed nitrogen at wild-type rates in determinate bean (Phaseolus vulgaris) nodules. This is consistent with BacA being a key component of bacteroid development in indeterminate galegoid nodules that is not required for functional bacteroid formation in determinate phaseoloid nodules.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are detailed in Table 1. It should be noted that strain A34 (renamed from strain 8401 pRL1JI) is a streptomycin derivative of strain 8400. Strain 8400 was derived by curing the sym plasmid from 8002 and conjugating in pRL1JI. Rhizobium strains were grown at 28°C on either tryptone yeast extract (TY) (2), acid minimal salts medium (AMS), global minimal salts (GMS), acid minimal salts agar (AMA), or global minimal salts agar (GMA) (29, 36) with either 10 mM d-glucose, 10 mM d-mannitol, 10 mM l-arabinose, 20 mM succinate, 20 mM malate, 20 mM fumarate, 20 mM myo-inositol, or 30 mM pyruvate as the carbon source and 10 mM ammonium chloride. S. meliloti Rm1021 was grown in either Luria-Bertani medium (LB) (40) supplemented with 2.5 mM calcium chloride and 2.5 mM magnesium sulfate (LB/MC) and resuspended in LB or in GMS medium. The recovery of cultures was performed on either LB agar (1.5%, wt/vol) or on GMA. Antibiotics were used at the following concentrations (in μg ml−1): ampicillin, 100; gentamicin, 5; kanamycin, 20; rifampin, 10; spectinomycin, 50; streptomycin, 500 or 250 (bacA mutant of R. leguminosarum 3841); tetracycline, 10 (E. coli) or 1 (bacA mutant of R. leguminosarum 3841).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic | Source or reference |
|---|---|---|
| R. leguminosarum bv. viciae | ||
| 3841 | Derivative of R. leguminosarum bv. viciae strain 300; Smr | 19 |
| LMB132 | 3841 with ΩTet cassette inserted into unique EcoRI site of bacA; Tcr | This work |
| RU4040 | 3841 bacA::pK19; Kmr | 22 |
| A34 | R. leguminosarum bv. viciae formerly known as 8401/pRL1JI; Smr | 7 |
| RU4043 | A34 bacA::pK19; Kmr | This work |
| R. leguminosarum bv. phaseoli | ||
| 4292 | Derivative of field bean isolate 8002 with sym plasmid pRP2J1; Rifr | 24 |
| RU4109 | 4292 bacA::pK19; Kmr | This work |
| R. etli | ||
| CE3 | Derivative of R. etli CFN42; Smr | 33 |
| LMB128 | CE3 with ΩSpec cassette inserted into unique StuI site of bacA; Spr | This work |
| S. meliloti | ||
| Rm1021 | Derivative of SU47; Smr | 30 |
| SmGF1 | Rm1021, bacA654::Spc (bacA null); Spr | 12 |
| E. coli | ||
| Fusion-Blue | endA1 hsdR17(rk12− mk12+) supE44 thi-1 gyrA96 relA1 lacF′[proA+B+lacIqZΔM15::Tn10(tet)] | Clontech Laboratories |
| DH5α | F−φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | Invitrogen |
| pHP45 | Plasmid containing ΩSpec or ΩTet cassette; Spr or Tcr | 8 |
| pJET1.2/blunt | PCR product cloning vector; Apr | Fermentas |
| pJQ200SK | pACYC derivative, P15A origin of replication; Gmr | 39 |
| pK19mob | pK19mob pUC19 derivative lacZ mob; Kmr | 41 |
| pLMB42 | pJET1.2/blunt containing R. etli CE3 bacA; Apr | This work |
| pLMB43 | pJET1.2/blunt containing R. leguminosarum bv. viciae 3841 bacA; Apr | This work |
| pLMB44 | R. etli CE3 bacA::ΩSp cloned in pJET2.1/blunt; Apr Spr | This work |
| pLMB45 | R. leguminosarum bv. viciae 3841 bacA::ΩTc cloned in pJET2.1/blunt; Apr Tcr | This work |
| pLMB46 | XhoI/XbaI fragment of pLMB44 cloned in pJQ200SK; Gmr Spr | This work |
| pLMB48 | XhoI/XbaI fragment of pLMB45 cloned in pJQ200SK; Gmr Tcr | This work |
| pRK2013 | ColEI replicon with RK2 tra genes, helper plasmid used for mobilizing plasmids; Kmr | 13 |
| pRU2048 | pK19mob containing internal fragment of R. leguminosarum bv. viciae 3841 bacA; Kmr | This work |
| pRU2049 | pK19mob containing internal fragment of R. leguminosarum bv. phaseoli 4292 bacA; Kmr | This work |
| pRK404 | Broad-host-range control plasmid; Tcr | 6 |
| pJG51A | pRK404 with S. meliloti Rm1021 bacA; Tcr | 15 |
Construction of bacA mutants.
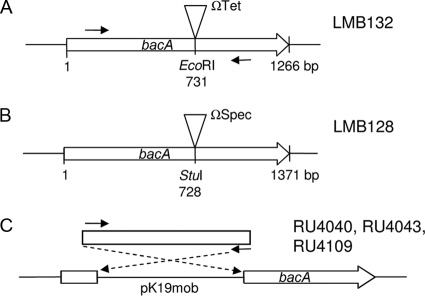
A double-crossover integration mutant in bacA of R. leguminosarum 3841 was made by PCR amplifying bacA (RL3557) from genomic DNA with primers pr0077 and pr0078 (Table 2). The 1.8-kb PCR product was cloned into pJET1.2/blunt, giving plasmid pLMB43. The HP45-ΩTet cassette from pHP45 was cloned into the unique EcoRI site of pLMB43, which is within the bacA gene, to produce pLMB45. The XhoI/XbaI fragment from pLMB45 was cloned into pJQ200SK to create pLMB48. Plasmid pLMB48 was conjugated into R. leguminosarum 3841 and a bacA mutant isolated by selecting for recombination using the sac mutagenesis strategy (23). The insertion of the ΩTet cassette into bacA strain LMB132 (Fig. 1A) was confirmed by PCR mapping using primers pr0075 and pOTfarforward.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Function |
|---|---|---|
| BDbacAfor | TGATTACGCCAAGCTCAGCAGCCGATCGACCTTTCG | bacA PCR primer for internal gene fragment |
| BDbacArev | GCAGGCATGCAAGCTTTGCTGACCTGTCCGAAGGC | bacA PCR primer for internal gene fragment |
| p1060 | TTTACTAGTCGACACTCCCGAGACTCACAGT | Mapping primer for pK19 bacA and bacA |
| p1310 | GCGATGACCTTCTCGGCATCA | RL4439 (mdh) qRT-PCR primer |
| p1311 | CATGGCGTCGAGCGGATTG | RL4439 (mdh) qRT-PCR primer |
| p1346 | AAAGGGCGGCATTGGCAAG | pRL100162 (nifH) qRT-PCR primer |
| p1347 | AAGATCAGGCGCGTAGAATCGG | pRL100162 (nifH) qRT-PCR primer |
| p1348 | CAACGGAAACGATGGTGATCGC | pRL100205 (fixN1) qRT-PCR primer |
| p1349 | GCGGCAAGGCAGAAGCAAAGTA | pRL100205 (fixN1) qRT-PCR primer |
| pK19A | ATCAGATCTTGATCCCCTGC | pK19mob mapping primer |
| pK19B | GCACGAGGGAGCTTCCAGGG | pK19mob mapping primer |
| pOTfarforward | GACCTTTTGAATGACCTTTA | PCR mapping primer specific to sequences on Ω cassette |
| pr0074 | TGCCACCGCAATCCTGCTTG | Forward PCR primer for bacA |
| pr0075 | ACGGCATCAAGGGCTTTCGC | Reverse PCR primer for bacA and mapping PCR primer |
| pr0076 | TAGGATCACGAGTTGCAGAT | Mapping PCR primer for bacA |
| pr0077 | GATCGTCGGGCCTGTTGTCG | Forward PCR primer for bacA |
| pr0078 | GAGGACCGCGCCTATGCCGC | Reverse PCR primer for bacA |
| pr0195 | CTCTGATACCGGCGGCATTCTC | RL1651 (ubiG) qRT-PCR primer |
| pr0196 | TGACATCGGTCGCCTTCATCG | RL1651 (ubiG) qRT-PCR primer |
FIG. 1.
Genetic map of insertions in bacA. (A) Stable tetracycline-resistant interposon insertion in R. leguminosarum 3841 bacA (RL3557) forming LMB132. (B) Stable spectinomycin-resistant interposon insertion in R. etli CE3 bacA (RHE_CH03112) forming LMB128. (C) Single-crossover insertions in bacA of R. leguminosarum strains 3841 (bv. viciae), A34 (bv. viciae), and 4292 (bv. phaseoli), forming RU4040, RU4043, and RU4109, respectively. A triangle is used to indicate the site of interposon insertion. Solid arrows indicate the binding position of PCR primers BDbacAfor and BdbacArev, which were used to amplify an internal fragment of bacA. Fragments were cloned into pK19mob and used to construct integration mutants of R. leguminosarum strains. In strain 3841, bp 139 to 1103 of bacA was cloned in pK19mob. All pK19mob insertions have bacA and the lacZ promoter in the same orientation. The Tc resistance gene is oriented in the same direction as bacA.
To construct a double-crossover integration mutant in bacA of R. etli CE3, primers pr0074 and pr0075 were used to PCR amplify bacA (RHE_CH03112) from genomic DNA. The 3.0-kb PCR product was cloned into the vector pJET1.2/blunt, resulting in pLMB42. The ΩSpec cassette from pHP45 was cloned into the unique StuI site of pLMB42, within the bacA gene, to produce pLMB44. The XhoI/XbaI fragment from pLMB44 was cloned into pJQ200SK, resulting in pLMB46. Plasmid pLMB46 was conjugated into R. etli CE3, and a bacA mutant was isolated by selecting for recombination using the sac mutagenesis strategy as previously described (23). The insertion of the ΩSpec cassette into the bacA gene, giving strain LMB128 (Fig. 1B), was confirmed by PCR mapping using primers pr0076 and pOTfarforward.
In addition, a single-crossover integration mutation in bacA was made in the R. leguminosarum biovars viciae (strains 3841 and A34) and phaseoli (strain 4292). Internal gene fragments were PCR amplified from strains 3841 and 4292 using primers BDbacAfor and BDbacArev (Fig. 1). The 964-bp bacA fragment from each strain was recombined directly into HindIII-digested pK19mob using a BD In-fusion PCR Cloning kit (Clontech) according to the manufacturer's instructions. Plasmid pRU2048 (containing R. leguminosarum bv. viciae 3841 bacA) was conjugated into 3841 and A34, while pRU2049 (containing R. leguminosarum bv. phaseoli 4292 bacA) was conjugated into 4292. Plasmid pRK2013 was used as a helper plasmid, as previously described (35). Insertions into bacA genes of strains 3841, A34, and 4292, confirmed by PCR using p1060- and a pK19mob-specific primer (either pK19A or pK19B), were named RU4040, RU4043, and RU4109, respectively (Table 1, Fig. 1C).
Antimicrobial agent assay.
Cultures were grown in TY overnight (with antibiotics as appropriate) and adjusted to an optical density at 600 nm (OD600) of 0.2, and cells were washed with 0.85% saline as previously published (26). Aliquots (100 μl) of diluted cultures were mixed with 3 ml soft TY agar (0.7%) and poured onto TY agar plates. After 30 min, sterile filter discs (6 mm diameter) were placed onto the top agar, and 5 μl of each test compound was added. Compounds tested in triplicate were bialaphos (1 mg ml−1), bleomycin (10 to 100 mg ml−1), SDS (10%), and vancomycin (10 mg ml−1). After 1 to 3 days of incubation at 28°C, the size of the zone of inhibition was measured from the edge of the filter paper disc. For the measurement of sensitivity to Bac71-16 and Bac71-35, stationary-phase cultures were diluted to an OD600 of ∼0.05 and then grown to mid-exponential phase in defined medium. Cultures then were washed and diluted to an OD600 of ∼0.05 in fresh medium, and then 50-μl aliquots were treated with and without Bac7 peptide for 1 h at 30°C (at 60 rpm) in microtiter plates. Cultures then were serially diluted into unsupplemented medium, 10-μl aliquots were spotted on defined agar, and CFU were determined after incubation at 30°C for 48 h (R. leguminosarum and R. etli) and 96 h (S. meliloti).
To check the MIC for the CRAMP peptide (GLLRKGGEKIGEKLKKIGQKIKNFFQKLVPQPEQ; Biomatik Corporation), growth was recorded in bacterial cultures grown on AMS, 10 mM glucose, and 10 mM ammonium chloride with CRAMP peptide (0.5 to 15 μg ml−1 in 50 mM phosphate buffer, pH 5) and shaking at 28°C for 1 to 3 days.
Plant growth and analysis.
Surface-sterilized dwarf bean (P. vulgaris cultivar Tendergreen) seeds were inoculated with R. etli strains CE3 and LMB128 (CE3 bacA) as well as R. leguminosarum bv. phaseoli strains 4292 and RU4109 (4292 bacA). Surface-sterilized pea (P. sativum cultivar Avola) seeds were inoculated with R. leguminosarum bv. viciae strains 3841, A34, RU4040 (3841 bacA), LMB132 (3841 bacA), and RU4043 (A34 bacA). All seeds were germinated in 1-liter pots of vermiculite watered with sterile nitrogen-free rooting solution, as described previously (35). Three weeks postinoculation, the acetylene reduction of plants was determined in 95% air-5% acetylene for 1 h in 250-ml Schott bottles as described previously (1). Plants were grown for 7 days before inoculation with rhizobia, and after a further 7 days nodules were prepared for microscopy and thin sectioned for electron microscopy as described previously (27).
Microarrays and analysis.
Microarrays were carried out as described previously (14). Total RNA was prepared from laboratory cultures grown on AMS-10 mM d-glucose-10 mM ammonium chloride as described previously (14) and from nodules of pea plants inoculated with R. leguminosarum bv. viciae 3841 and bacA mutant RU4040 at 7 days postinoculation (p.i.) of 7-day-old plants, as described previously (22). Genes upregulated ≥3-fold in bacA nodule bacteria relative to the wild type are shown in Table S1 in the supplemental material, and those downregulated ≥3-fold are shown in Table S2 in the supplemental material. Microarray results were confirmed for RL4439 (mdhA), pRL100162 (nifH), pRL100205 (fixN1), and RL1651 (ubiG) using quantitative reverse transcription-PCR (qRT-PCR) performed in triplicate using a QuantiTect SYBR green PCR kit (Qiagen) on an MJ Mini cycler MiniOpticon Real-Time PCR Detection System (Bio-Rad) as previously described (21) (data not shown). Primers are given in Table 2. Data were analyzed by the relative quantification method (comparative cyclic threshold method [ΔΔCT]) to calculate the fold expression (3, 4).
Microarray data accession numbers.
Results were deposited in MIAMexpress with accession numbers E-MEXP-1965 and E-MEXP-1966 for free-living bacteria and bacteroids, respectively.
RESULTS AND DISCUSSION
Mutants in bacA show altered resistance to antimicrobial compounds in their free-living state.
The principal purpose of this study was to determine whether there is a difference in symbiotic phenotypes of bacA mutants that infect galegoid plants, which form indeterminate nodules, and those of phaseoloid legumes, which form determinate nodules. A series of bacA knockout mutants were made (Fig. 1) to investigate this. Indeterminate nodule formation in peas was studied with three bacA mutants of R. leguminosarum bv. viciae strains 3841 and A34, while for beans, which form determinate nodules, bacA mutants of R. leguminosarum bv. phaseoli 4292 and R. etli CE3 were used.
Since it is well documented that S. meliloti bacA null mutants have altered sensitivity to a number of different antimicrobial compounds (15, 18), a range of such compounds were tested on bacA mutants from strains 3841, A34, 4292, and CE3 (Table 3). Sensitivity to the detergent SDS and the glycopeptide vancomycin increased slightly in each mutant compared to that of their parent strain. The only exception was that the bacA mutant of strain A34 did not show increased sensitivity to SDS (Table 3). However, sensitivity to another glycopeptide antibiotic, bleomycin, was reduced in each of the bacA mutants. Strain 3841 bacA mutants were resistant to bleomycin, even showing bacterial growth to the edge of a disc impregnated with 500 μg bleomycin, 10-fold higher than the amount used for Table 3. However, bacA mutants of the bean-nodulating species (4292 and CE3) showed only a very small increase in resistance to bleomycin. R. leguminosarum bv. viciae A34 also had a small increase in resistance to bleomycin, which is similar to that of R. leguminosarum bv. phaseoli 4292. These strains have the same genetic background except for their symbiotic plasmids (pRL1JI and pRP2JI, respectively).
TABLE 3.
Sensitivity of Rhizobium strains and bacA mutants to antimicrobial compoundsc
| Strain | Genotype | Inhibition by antimicrobial agenta: |
CRAMP MICb(μg) | |||
|---|---|---|---|---|---|---|
| SDS | Van | Bleo | Bial | |||
| R. leguminosarum bv. viciae | ||||||
| A34 | WT | 7 ± 0.6 | ND | 8 ± 0.6 | ND | ND |
| 3841 | WT | 9 ± 1 | 6 ± 0 | 4 ± 0.5 | 13 ± 0 | 1 |
| LMB132 | 3841 bacA::Ωtet | 12 ± 0 | 12 ± 0.5 | Resistant | 14 ± 1 | 0.5 |
| RU4040 | 3841 bacA::pK19 | 11 ± 0.1 | 11 ± 0.5 | Resistant | ND | ND |
| RU4043 | A34 bacA::pK19 | 7 ± 0.6 | ND | 4 ± 0.6 | ND | ND |
| R. leguminosarum bv. phaseoli | ||||||
| 4292 | WT | 8.3 ± 1.1 | 6.3 ± 0.28 | 10 ± 0 | 18 ± 0.57 | ND |
| RU4109 | bacA::pK19 | 13 ± 1 | 14 ± 0.5 | 7 ± 0.5 | 16 ± 0 | ND |
| R. etli | ||||||
| CE3 | WT | 9 ± 0 | 6 ± 0 | 23 + 0.5 | 11 ± 0.51 | 1 |
| LMB128 | bacA::Ωspec | 13 ± 1.5 | 11 ± 0.5 | 21 ± 1 | 10 ± 0 | 0.5 |
The inhibition of bacterial growth in the presence of an antimicrobial agent is measured in mm from the edge of the impregnated disc. SDS, 0.5 mg; vancomycin (Van), 50 μg; bleomycin (Bleo), 50 μg; bialaphos (Bial), 5 μg.
The MIC for CRAMP was determined by growing bacteria in liquid culture in the presence of 0.5 to 15 μg ml−1 CRAMP.
The values shown are the averages from three experiments ± the standard errors of the means. ND, not determined. Plasmid pJG51A (S. meliloti bacA in pRK404) was conjugated into all bacA strains and restored wild-type growth on SDS, vancomycin, and bleomycin, while pRK404 did not.
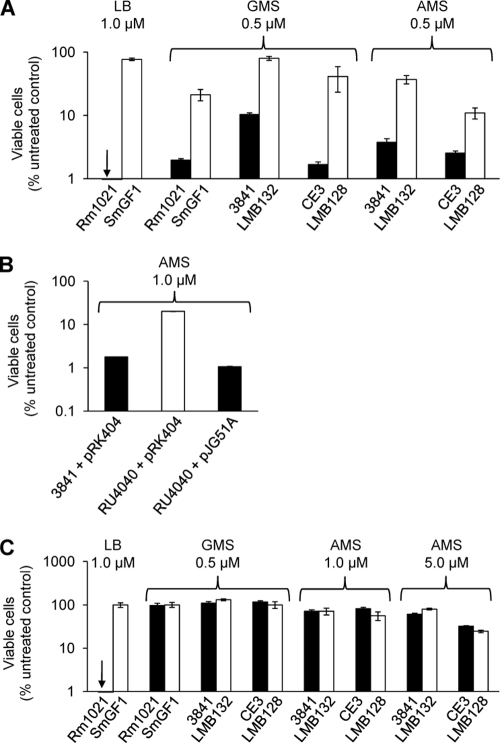
S. meliloti Rm1021 bacA null mutants were shown to be resistant to the toxic effects of 1 μM Bac71-16 (28) and have a greatly increased resistance to 1 μM Bac71-35 relative to that of the parent strain in LB medium (Fig. 2A). R. leguminosarum 3841 and R. etli CE3 bacA mutants also had an increased resistance to Bac71-35 (Fig. 2A). The sensitivity of bacA strain RU4040 to Bac71-35 was restored by complementation with bacA from S. meliloti cloned in pJG51A (Fig. 2B). However, R. leguminosarum 3841 and R. etli CE3 bacA mutants showed no change in their resistance to Bac71-16 in either AMS or GMS medium (Fig. 2C). When the S. meliloti bacA mutant and parent strain were retested on minimal medium, the S. meliloti bacA null also showed no change in resistance to Bac71-16 (Fig. 2C), unlike the situation for cells grown on LB. Thus, the BacA-mediated sensitivity of S. meliloti toward Bac71-16 is dependent upon the nature of the growth medium. This may result from changes in any one of several components of the cell surface, including LPS, exopolysaccharide, and porins, that make them less permeable to Bac71-16. If correct, then the increased size and associated physical properties of Bac71-35 may make it more permeable than Bac71-16 to cells grown in minimal medium.
FIG. 2.
BacA increases the sensitivity of S. meliloti Rm1021, R. leguminosarum 3841, and R. etli CE3 toward Bac71-35. (A and C) Strains of either the wild type (filled bars) or bacA mutants (open bars) were treated with defined concentrations of either Bac71-35 (A) or Bac71-16 (C) for 1 h at 30°C. (B) Wild-type strain with control plasmid (pRK404) or bacA mutant with pJG51A (S. meliloti bacA gene in pRK404) (filled bars) or bacA mutant with control plasmid (open bar) were treated as described for panel A. The CFU were determined after 48 (R. leguminosarum and R. etli), 72 (S. meliloti on LB), or 96 h (S. meliloti on GMS). All datasets shown are representative of trends observed in two independent experiments, and in each case the error bars represent the standard deviations from the means (n = 3) for one experiment.
An R. leguminosarum acpXL mutant that completely lacks the VLCFA modification of lipid A in its free-living state (45) was unaffected in its sensitivity to Bac71-35 compared to that of the parent strain (data not shown). Therefore, consistent with S. meliloti (28), the increased resistance of an R. leguminosarum bacA mutant to Bac71-35 is not due to a reduction in its lipid A VLCFA content.
Since the mutation of BacA reduces sensitivity to peptide antibiotics such as bleomycin and Bac71-35, other peptide antibiotics were tested to see if this is a general property. However, resistance to the tripeptide bialaphos was almost unchanged in mutant strains and their parents (Table 3). In addition, the mouse α-helical antimicrobial peptide CRAMP, which inhibits Z-ring formation and causes cell elongation in Escherichia coli (17), was more inhibitory to bacA mutants of both 3841 and CE3 than to their respective wild-type strains (Table 3). This shows that the reduced sensitivity of bacA mutants to Bac71-35 and bleomycin is not a general property applying to other classes of antimicrobial peptides. Thus, sensitivity to vancomycin and CRAMP is increased rather than decreased in bacA strains, while sensitivity to tripeptide bialaphos is unchanged.
Rhizobium, but not Sinorhizobium, strains mutated in bacA have an elevated calcium requirement for growth on dicarboxylic acids.
Wild-type strains (3841, 4292, and CE3) and bacA mutants (LMB132, RU4040, RU4109, and LMB128) were grown on minimal medium with either d-glucose, d-mannitol, l-arabinose, myo-inositol, pyruvate, or succinate added as the sole carbon source. The growth of each bacA mutant resembled that of parental wild type on every carbon source except succinate, where bacA mutants grew very poorly (data not shown). R. leguminosarum 3841 bacA strains LMB132 and RU4040 also failed to grow on l-malate and fumarate. The inability of bacA mutants to grow on dicarboxylates was overcome by increasing the level of calcium in the medium by 10-fold, to 1.7 mM (data not shown). This is similar to the situation for R. leguminosarum 3841 lpcB mutants, which have a truncated lipopolysaccharide core and require increased calcium levels for growth with organic acids such as succinate (36). Overall, calcium has a profound effect on the structural integrity of the cell wall and outer membrane of rhizobia and is required in higher concentrations for growth on dicarboxylates than on sugars by wild-type 3841 (34). The fact that bacA mutants need more calcium than wild-type Rhizobium strains for growth on succinate is consistent with the ability of BacA to alter components of the cell surface such as VLCFA.
Since the calcium level affected the growth of bacA mutants, a wide range of calcium and magnesium concentrations were tested, but none were found to alter the sensitivity of R. leguminosarum 3841 or bacA mutant LMB132 to Bac71-16 or Bac71-35 (data not shown). The growth of LMB132, RU4040, RU4109, and LMB128 on dicarboxylates was restored by complementation with bacA cloned in pJG51A, demonstrating that the effect is due to the mutation of bacA (data not shown). The S. meliloti bacA mutant SmGF1 was not altered for growth on dicarboxylates, so this effect is specific to the Rhizobium strains tested.
BacA is not essential for bacteroid development in phaseoloid determinate nodules.
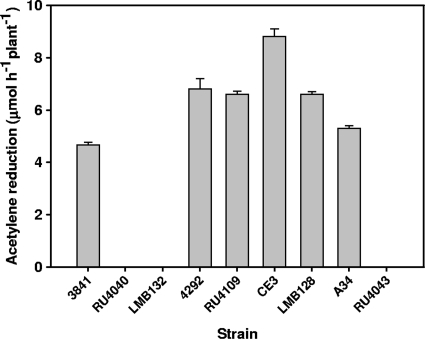
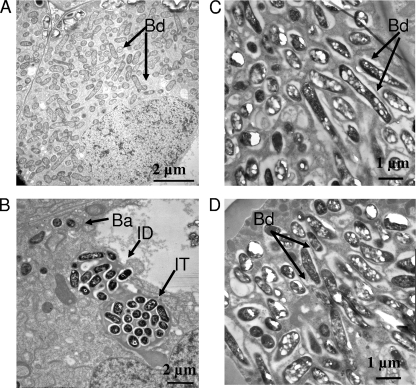
Although bacA mutants of free-living R. leguminosarum biovars viciae and phaseoli, as well as R. etli and S. meliloti, showed broadly similar changes in sensitivity to antimicrobial agents, plant phenotypes of the mutants were dramatically different. Pea plants infected with R. leguminosarum strain 3841 or A34 reduced acetylene, but no acetylene reduction was detectable in the bacA mutants (LMB132, RU4040, and RU4043) (Fig. 3). At 4 weeks, pea plants inoculated with bacA strains were yellow and had white nodules (data not shown). In contrast, plants inoculated with wild-type R. leguminosarum 3841 were green and had red nodules that were full of developed bacteroids (Fig. 4A). Nodules from plants infected with RU4040 (bacA) had no mature bacteroid-filled nodules, with bacteria only detectable in infection threads with few infection droplets and enveloped cells (Fig. 4B). Thus, although peas inoculated with bacA mutants initiate nodule formation and bacteria grow down infection threads, bacteroid formation is blocked at this early stage.
FIG. 3.
Acetylene reduction of wild-type and bacA rhizobia. R. leguminosarum bv. viciae strains 3841 and A34 (wild types), RU4040 (bacA), LMB132 (bacA), and RU4043 (bacA) were inoculated onto pea plants. R. leguminosarum bv. phaseoli 4292 (wild type), R. leguminosarum bv. phaseoli RU4109 (bacA), R. etli CE3 (wild type), and R. etli LMB128 (bacA) were inoculated onto beans. Error bars indicate the standard errors of the means (n = 6).
FIG. 4.
Electron micrographs of nodules harvested 7 days after inoculation with rhizobia. (A) Peas infected with wild-type R. leguminosarum bv. viciae 3841. (B) Peas infected with RU4040 (bacA). (C) Beans infected with wild-type 4292. (D) Beans infected with RU4109 (bacA). Wild-type 3841 shows a mixture of developing and mature bacteroids (A), while for RU4040 the only bacteria we observed were in infection threads (shown), with only a few cells being released into infection droplets (B). Note that there are no mature bacteroids in nodules infected with RU4040, while those inoculated with 3841 are full of them. In contrast, the electron micrographs of wild-type and bacA strains from bean nodules are essentially indistinguishable, with numerous mature bacteroids. Abbreviations: IT, infection thread; ID, infection droplet; Ba, released bacteria; Bd, bacteroid.
In stark contrast, bean plants inoculated with bacA mutants of R. leguminosarum bv. phaseoli 4292 (RU4109) or R. etli CE3 (LMB128) were green and had red nodules that were indistinguishable from those of plants inoculated with the respective wild-type strains (data not shown). In addition, all bean-nodulating bacA mutants reduced acetylene at rates similar to that of wild-type strains (Fig. 3). Electron micrographs of bean nodules infected with wild-type strain 4292 or RU4109 were indistinguishable from each other (Fig. 4C and D, respectively). Thus, while bacteroid development is blocked in bacA mutants of species that form indeterminate nodules on peas, a mutation in bacA has no effect on determinate nodule formation or function in beans. R. leguminosarum bv. viciae A34 and R. leguminosarum bv. phaseoli 4292 have the same genetic background with the exception of their sym plasmids (pRL1JI and pRP2JI). Differences in the symbiotic performance of these strains on their hosts emphasizes the specific requirement for BacA in galegoid but not phaseoloid legumes.
Bacteroids of bacA mutant show differences in gene expression relative to that of wild type.
Gene expression was compared between wild-type 3841 and the bacA mutant RU4040. Initially, cultured free-living cells were analyzed, and there was very little difference in transcription. In RU4040, only the expression of RL3384 (3.7-fold upregulated) was ≥3-fold elevated (P ≤ 0.05), while no gene was ≥2-fold downregulated (P ≤ 0.05). RL3384 encodes a conserved exported protein. In short, the mutation of R. leguminosarum 3841 bacA has almost no effect on gene expression in free-living cultured bacteria.
In a recent study, microarrays were used to analyze the gene expression of R. leguminosarum bv. viciae 3841 in developing pea nodules at 7, 15, and 21 days p.i. (22). Gene expression measured early (7 days p.i.) was due to a mixture of developing bacteroids and cells in infection threads, while by 15 days, gene expression was essentially that of mature bacteroids. Expression of bacA in R. leguminosarum, recovered from nodules 7 days p.i., was 5.3-fold upregulated (P = 0.075) compared to the level of free-living cells but was not differentially expressed at 15 and 21 days. Thus, bacA expression is developmentally regulated.
On examining the effect of BacA on developing pea bacteroids, it was found that at 7 days p.i., 48 genes were ≥3-fold upregulated (P ≤ 0.05) in bacA mutant RU4040 relative to the level for wild-type 3841 (see Table S1 in the supplemental material), and 81 genes were ≥3-fold downregulated (P ≤ 0.05) (see Table S2 in the supplemental material). Of the 48 genes upregulated and the 129 genes up- or downregulated, approximately one-half and one-third, respectively, are concerned with cell membrane components and transport across membranes. Thus, the lack of a functional BacA has an enormous effect on bacterial cell membranes during early stages of symbiosis. In addition to changes in inner membrane components, TonB (RL3694), the periplasmic space-spanning protein that interacts with outer membrane proteins, was 4.8-fold downregulated (P ≤ 0.05) in RU4040. In RU4040, the expression of outer membrane proteins also was affected, with RopA3 (pRL110375) being 10-fold downregulated (P ≤ 0.05) and RopB (RL1589) 5.5-fold upregulated (P ≤ 0.05). There were changes in the expression of enzymes that may affect membrane and peptidoglycan structure; a patatin-like phospholipase (RL2533) was 3-fold downregulated (P ≤ 0.05), peptidoglycan transglycosylase (MtgA, RL4627) was 5-fold downregulated (P ≤ 0.05), and putative penicillin-binding transpeptidase/transglycosylase protein (RL0153) was 3.4-fold upregulated (P ≤ 0.05) in RU4040 (see Tables S1 and S2 in the supplemental material).
In microarray results of RU4040 compared to those of wild-type R. leguminosarum 3841 at 7 days p.i., there was the downregulation of many genes that were elevated in wild-type cells undergoing symbiotic development; e.g., nodO (3.7-fold downregulated), nif genes (3.5 to 12.5-fold downregulated), and fix genes (>14-fold downregulated) (see Table S2 in the supplemental material). Also in this category is the gene encoding 4-aminobutyrate transferase gabT (RL0102), which was highly elevated (35-fold, P = 0.001) in 7-day bacteroids in 3841 (22, 37) but downregulated in RU4040 by 15-fold (P ≤ 0.05). Although already slightly downregulated (1.4- to 2-fold) in wild-type 3841 developing bacteroids compared to the level for free-living cells, the expression of genes encoding the components of ATP synthase were further downregulated in RU4040: RL0924-7, 1.6- to 4.2-fold downregulated, P ≤ 0.05, and RL4405-9, 2.9- to 5-fold downregulated, P ≤ 0.2. Also downregulated were components of the electron transport chain: ferredoxin (fdxB1, pRL100156), 7-fold downregulated (P ≤ 0.05); petA and petB (RL3486-5), 6- to 10-fold downregulated (P ≤ 0.05) (see Table S2 in the supplemental material).
In addition, there was the downregulation of genes that code for proteins involved in lipid metabolism, which may be significant, as bacA mutants have an altered VLCFA modification of lipid A (10, 11). The genes downregulated encode a 3-oxoacyl (acyl carrier protein [ACP]) reductase (RL4430) (2.9-fold downregulated, P = 0.004), ACP (AcpP, RL1559) (2.7-fold downregulated, P = 0.03), lipoyl synthase (RL2249) (3.1-fold downregulated, P = 0.01), lipopolysaccharide biosynthesis O-acetyltransferase (RL0241A) (3.7-fold downregulated, P = 0.03), lipid A biosynthesis UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase (RL2229) (3.4-fold downregulated, P = 0.06), and lipid A oxidase (LxpQ, RL0868) (3-fold downregulated, P = 0.04). The expression of 3-oxoacyl (ACP) reductase, RL4430, was 2.8-fold upregulated in wild-type R. leguminosarum 3841 root nodule bacteria at 7 days p.i. relative to that of cultured bacteria but was not upregulated at 15 and 21 days p.i. (22). Thus, like bacA itself, RL4430 is developmentally regulated and expressed during early bacteroid formation. In the bacA mutant RU4040 inoculated onto peas, the expression of RL4430 was downregulated compared to that of wild-type 3841 at 7 days p.i.
A key protein involved in VLCFA formation in free-living 3841 is ACP AcpXL (RL2817), and the mutation of acpXL abolished the VLCFA modification of lipid A in free-living cells (44). It has been shown that the VLCFA modification of lipid A in an acpXL mutant of R. leguminosarum 3841 was at least partially restored by the pea nodule environment (44), although nodule formation on peas by the acpXL mutant was delayed (43). The expression of acpXL was downregulated 5.1-fold (P = 0.0003) in wild-type R. leguminosarum 3841 bacteria in root nodules at 7 days and also was repressed at 15 and 21 days (22). The ACPs whose expression increased in wild-type bacteroids are NodF (pRL100183) and pRL100144, both of which are approximately 3-fold elevated at 7 days, with levels falling as bacteroids mature. Vedam et al. (44) proposed that pRL100144 substituted for AcpXL in symbiosis, acting as an alternative ACP, and its elevated expression during early bacteroid formation fits with this suggestion. It is important to appreciate that BacA is highly unlikely to be regulating any of these genes directly. Instead, BacA almost certainly alters membrane function, and this is particularly crucial to bacteroid formation. Changes in gene expression in a bacA mutant are probably a consequence of the failure to develop into bacteroids. This also explains why there are so few transcriptional changes in bacA mutants in free-living bacteria; it is only during a large developmental change that transcriptional activity is altered as a secondary consequence of the absence of BacA.
Microarray data from alfalfa nodules infected with an S. meliloti bacA mutant (5) showed the upregulation of genes, many of which were members of the same functional groups as those for strain 3841. Five genes were found to be upregulated in both R. leguminosarum and S. meliloti bacA mutants, but in four cases the proteins encoded were of unknown function. The single commonly upregulated gene of known function is an exported serine protease, DegP (RL1251 and SMc02365).
Conclusions.
This work confirms for several rhizobia that bacA mutants are impaired in membrane function in a number of ways, including VLCFA incorporation into lipid A and peptide uptake. Taken alone this simply reinforces what is already known from extensive studies of S. meliloti (5, 9, 10, 11, 13, 16, 18, 26); however, the importance of this work is in showing that although bacA mutants of rhizobia that nodulate galegoid and phaseoloid legumes are similarly altered in free-living culture phenotypes in the laboratory, the mutation leads to completely different effects on symbiotic development and N2 fixation. BacA is required for bacteroid development in galegoid but not phaseoloid legumes.
It has been shown that bacA mutants of S. meliloti are altered in the accumulation of the Bac7 antimicrobial peptide (28), which is very interesting given the proposed importance of plant NCR peptides in bacteroid development (32). In addition, these plant peptides are present in indeterminate but not determinate nodules, and thus bacA mutants of pea or alfalfa bacteroids might be unable to transport them, leading to subsequent failure to develop into bacteroids. In determinate bean nodules where NCR plant peptides are not present, BacA-dependent changes in the accumulation of peptides may be irrelevant. However, there are two characterized site-directed mutants of the S. meliloti BacA protein (Q193G and R389G) that still are capable of Bac7 peptide uptake but are unable to form successful nodule infections (28). This suggests that the role of BacA in symbiosis is not restricted to alterations in peptide uptake. It also is known that BacA mutants have a ∼50% reduction in the VLCFA decoration of lipid A (9, 11). The mutation of acpXL and lpxXL loci in S. meliloti, which prevents the formation of VLCFA-modified lipid A in free-living cells, affects, but is not essential for, bacteroid development (10, 11, 16). Additionally, it was found that S. meliloti lpxXL mutant bacteroids completely lack the lipid A VLCFA modification, show bacteroid abnormalities, but are symbiotically competent (16). This provides evidence that VLCFA modification is important but not essential for bacteroid development. Thus, neither the effect of BacA on peptide uptake nor that of VLCFA modification alone can explain the symbiotic defect of a bacA mutation.
In free-living culture, the mutation of bacA altered the transcription of only one gene, suggesting that BacA is unlikely to directly regulate other genes. However, given that bacA strains are altered in many properties, this suggests it is the biochemical function of BacA that is important. This might be via its ability to either directly or indirectly alter VLCFA incorporation into lipid A or peptide uptake. However, BacA also may alter the composition or function of the inner or outer membrane in other as-yet undetermined ways. In contrast to free-living cultures, the loss of BacA in R. leguminosarum bacteroids leads to extensive changes in the transcription of genes that encode membrane transport proteins, inner and outer cell membrane components, and enzymes that affect peptidoglycan and membrane phospholipid structure, including lipid A modification (see Tables S1 and S2 in the supplemental material). Consequently, the inability of bacA mutant strains to differentiate into bacteroids within indeterminate nodules may be due to combined effects of the absence of BacA on lipid A modification, response to eukaryotic peptides, dicarboxylic acid utilization, and further effects on the cell envelope as a result of gene expression changes. This is consistent with observations that the bacterial cell membrane and its dynamics (especially transport) are crucially important for differentiation into bacteroids. Changes in the expression of these groups of genes are among the largest seen in pea bacteroid development (22). Given the results for free-living cells, it is highly unlikely that BacA directly affects the expression of so many genes early in bacteroid development; instead, the effect is more likely to be an indirect one causing bacteroids to fail to develop properly. During the development of bacteroids in indeterminate nodules, there are so many changes occurring in the bacterial cell surface that indirect results of a bacA mutation may be particularly profound. The fact that bacteroids forming determinate nodules undergo far fewer fundamental changes in membrane structure and lack endoreduplication (32) may explain why a mutation of bacA is tolerated. Overall, data in this study cannot resolve the function of BacA, but they do highlight a profound difference between bacteroids from different nodule types. Explaining this difference is an important goal for future mechanistic studies.
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (grant numbers P19406 and D000564) and a Medical Research Council New Investigator award (G0501107) to G.P.F.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allaway, D., E. Lodwig, L. A. Crompton, M. Wood, T. R. Parsons, T. Wheeler, and P. S. Poole. 2000. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 36:508-515. [DOI] [PubMed] [Google Scholar]
- 2.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 3.Bustin, S. A. 2000. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25:169-193. [DOI] [PubMed] [Google Scholar]
- 4.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 5.Capela, D., C. Filipe, C. Bobik, J. Batut, and C. Bruand. 2006. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: a transcriptomic dissection. Mol. Plant Microbe Interact. 19:363-372. [DOI] [PubMed] [Google Scholar]
- 6.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 7.Downie, J. A., G. Hombrecher, Q. S. Ma, C. D. Knight, B. Wells, and A. W. B. Johnston. 1983. Cloned nodulation genes of Rhizobium leguminosarum determine host range specificity. Mol. Genet. Genomics 190:359-365. [Google Scholar]
- 8.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, G. P., A. Datta, J. Baumgartner, R. M. Roop, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. USA 101:5012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson, G. P., A. Datta, R. W. Carlson, and G. C. Walker. 2005. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 56:68-80. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson, G. P., A. Jansen, V. L. Marlow, and G. C. Walker. 2006. BacA-mediated bleomycin sensitivity in Sinorhizobium meliloti is independent of the unusual lipid A modification. J. Bacteriol. 188:3143-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson, G. P., R. M. Roop, and G. C. Walker. 2002. Deficiency of a Sinorhizobium meliloti bacA mutant in Alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox, M. A., R. Karunakaran, M. E. Leonard, B. Mouhsine, A. Williams, A. K. East, J. A. Downie, and P. S. Poole. 2008. Characterization of the quaternary amine transporters of Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Lett. 287:212-220. [DOI] [PubMed] [Google Scholar]
- 15.Glazebrook, J., A. Ichige, and G. C. Walker. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7:1485-1497. [DOI] [PubMed] [Google Scholar]
- 16.Haag, A. F., S. Wehmeier, S. Beck, V. L. Marlow, V. Fletcher, E. K. James, and G. P. Ferguson. 2009. The Sinorhizobium meliloti LpxXL and AcpXL proteins play important roles in bacteroid development within alfalfa. J. Bacteriol. 191:4681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handler, A. A., J. E. Lim, and R. Losick. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol. 68:588-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichige, A., and G. C. Walker. 1997. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J. Bacteriol. 179:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston, A. W. B., and J. E. Beringer. 1975. Identification of the Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87:343-350. [DOI] [PubMed] [Google Scholar]
- 20.Jones, K. M., H. Kobayashi, B. W. Davies, M. E. Taga, and G. C. Walker. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karunakaran, R., K. Ebert, S. Harvey, M. E. Leonard, V. Ramachandran, and P. S. Poole. 2006. Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 188:6661-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karunakaran, R., V. K. Ramachandran, J. C. Seaman, A. K. East, B. Moushine, T. H. Mauchline, J. Prell, A. Skeffington, and P. S. Poole. 2009. Transcriptomic analysis of Rhizobium leguminosarum b.v. viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J. Bacteriol. 191:4002-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., A. Bourdes, and P. S. Poole. 2005. De novo alanine synthesis by bacteroids of Mesorhizobium loti is not required for nitrogen transfer in the determinate nodules of Lotus corniculatus. J. Bacteriol. 187:5493-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb, J. W., G. Hombrecher, and A. W. B. Johnston. 1982. Plasmid-determined nodulation and nitrogen-fixation abilities in Rhizobium phaseoli. Mol. Gen. Genet. 186:449-452. [Google Scholar]
- 25.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 26.LeVier, K., and G. C. Walker. 2001. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J. Bacteriol. 183:6444-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodwig, E. M., M. Leonard, S. Marroqui, T. R. Wheeler, K. Findlay, J. A. Downie, and P. S. Poole. 2005. Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol. Plant Microbe Interact. 18:67-74. [DOI] [PubMed] [Google Scholar]
- 28.Marlow, V. L., A. F. Haag, H. Kobayashi, V. Fletcher, M. Scocchi, G. C. Walker, and G. P. Ferguson. 2009. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191:1519-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauchline, T. H., J. E. Fowler, A. K. East, A. L. Sartor, R. Zaheer, A. H. F. Hosie, P. S. Poole, and T. M. Finan. 2006. Mapping the Sinorhizobium meliloti 1021 solute-binding protein-dependent transportome. Proc. Natl. Acad. Sci. USA 103:17933-17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mergaert, P., K. Nikovics, Z. Kelemen, N. Maunoury, D. Vaubert, A. Kondorosi, and E. Kondorosi. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mergaert, P., T. Uchiumi, B. Alunni, G. Evanno, A. Cheron, O. Catrice, A.-E. Mausset, F. Barloy-Hubler, F. Galibert, A. Kondorosi, and E. Kondorosi. 2006. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA 103:5230-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noel, K. D., A. Sanchez, L. Fernandez, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oldroyd, G. E. D., and J. A. Downie. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59:519-546. [DOI] [PubMed] [Google Scholar]
- 35.Poole, P. S., A. Blyth, C. J. Reid, and K. Walters. 1994. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv viciae. Microbiology 140:2787-2795. [Google Scholar]
- 36.Poole, P. S., N. A. Schofield, C. J. Reid, E. M. Drew, and D. L. Walshaw. 1994. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology 140:2797-2809. [DOI] [PubMed] [Google Scholar]
- 37.Prell, J., A. Bourdès, R. Karunakaran, M. L. Gomez, and P. Poole. 2009. Pathway of γ-aminobutyrate (GABA) metabolism in Rhizobium leguminosarum 3841 and its role in symbiosis. J. Bacteriol. 191:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prell, J., and P. Poole. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 14:161-168. [DOI] [PubMed] [Google Scholar]
- 39.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19-selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 42.Tan, X.-J., Y. Cheng, Y.-X. Li, Y.-G. Li, and J.-C. Zhou. 2009. BacA is indispensable for successful Mesorhizobium-Astragalus symbiosis. Appl. Microbiol. Biotechnol. 84:519-526. [DOI] [PubMed] [Google Scholar]
- 43.Vedam, V., J. G. Haynes, E. L. Kannenberg, R. W. Carlson, and D. J. Sherrier. 2004. A Rhizobium leguminosarum lipopolysaccharide lipid A mutant induces nitrogen-fixing nodules with delayed and defective bacteroid formation. Mol. Plant Microbe Interact. 17:283-291. [DOI] [PubMed] [Google Scholar]
- 44.Vedam, V., E. Kannenberg, A. Datta, D. Brown, J. G. Haynes-Gann, D. J. Sherrier, and R. W. Carlson. 2006. The pea nodule environment restores the ability of a Rhizobium leguminosarum lipopolysaccharide acpXL mutant to add 27-hydroxyoctacosanoic acid to its lipid A. J. Bacteriol. 188:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vedam, V., E. L. Kannenberg, J. G. Haynes, D. J. Sherrier, A. Datta, and R. W. Carlson. 2003. A Rhizobium leguminosarum acpXL mutant produces lipopolysaccharide lacking 27-hydroxyoctacosanoic acid. J. Bacteriol. 185:1841-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.