Abstract
Dehalogenases play key roles in the detoxification of halogenated aromatics. Interestingly, only one hydrolytic dehalogenase for halogenated aromatics, 4-chlorobenzoyl-coenzyme A (CoA) dehalogenase, has been reported. Here, we characterize another novel hydrolytic dehalogenase for a halogenated aromatic compound from the 2,4,5,6-tetrachloroisophthalonitrile (chlorothalonil)-degrading strain of Pseudomonas sp. CTN-3, which we have named Chd. Chd catalyzes a hydroxyl substitution at the 4-chlorine atom of chlorothalonil. The metabolite of the Chd dehalogenation, 4-hydroxy-trichloroisophthalonitrile, was identified by reverse-phase high-performance liquid chromatography (HPLC), tandem mass spectrometry (MS/MS), and nuclear magnetic resonance (NMR). Chd dehalogenates chlorothalonil under anaerobic and aerobic conditions and does not require the presence of cofactors such as CoA and ATP. Chd contains a putative conserved domain of the metallo-β-lactamase superfamily and shows the highest identity with several metallohydrolases (24 to 29%). Chd is a monomer (36 kDa), and the isoelectric point (pI) of Chd is estimated to be 4.13. Chd has a dissociation constant (Km) of 0.112 mM and an overall catalytic rate (kcat) of 207 s−1 for chlorothalonil. Chd is completely inhibited by 1,10-phenanthroline, diethyl pyrocarbonate, and N-bromosuccinic acid. Site-directed mutagenesis of Chd revealed that histidines 128 and 157, serine 126, aspartates 45, 130 and 184, and tryptophan 241 were essential for the dehalogenase activity. Chd differs from other reported hydrolytic dehalogenases based on the analysis of amino acid sequences and catalytic mechanisms. This study provides an excellent dehalogenase candidate for mechanistic study of hydrolytic dehalogenation of halogenated aromatic compound.
Halogenated aromatic compounds are widely used in agriculture and industry as solvents, defatting agents, herbicides, and fungicides. A variety of these compounds have been identified as priority organic pollutants by the United Nations and the U.S. Environmental Protection Agency. Therefore, the remediation of these pollutants is desirable. Microorganisms play key roles in the detoxification of halogenated aromatics. The use of microbial enzymes for bioremediation has received increasing attention as an efficient and cost-effective biotechnological approach. Halogen removal from halogenated aromatics reduces both the recalcitrance to biodegradation and the risk of forming toxic intermediates during subsequent metabolic steps. As a result, the key reaction for microbial detoxification of halogenated aromatics is the actual dehalogenation (35).
Investigation of the microbial degradation of different halogenated aromatics has led to the detection and elucidation of various dehalogenases that catalyze the removal of the halogen atom under aerobic and anaerobic conditions (8, 11, 17). Four dehalogenation mechanisms of halogenated aromatics are known, including reductive, thiolytic, oxidative, and hydrolytic mechanisms (41). Reductive dehalogenation plays important roles in the degradation of chlorinated aromatics under anaerobic conditions (36, 44). Several anaerobic bacteria are capable of using chlorinated benzenes (2) or polychlorinated dibenzodioxins (5) as the terminal electron acceptors in their energy metabolism. These bacteria couple reductive dehalogenation to electron transport phosphorylation (15). Several enzymes catalyzing the respiratory reductive dechlorination of halogenated aromatics have also been characterized (1, 4, 7, 20, 28, 38, 40). Under aerobic conditions, some chlorinated aromatics can also be reductively dehalogenated by thiolytic substitution in the presence of glutathione (12, 19, 25, 46, 48). In this dehalogenation system, chlorine atoms are displaced by the nucleophilic attack of the thiolate anion of glutathione. The nucleophilic attack of the thiolate anion is catalyzed by glutathione S-transferases (43). Besides the two well-characterized mechanisms for aryl halide reductive dehalogenation, two other mechanisms have been reported, including reduced NADPH-dependent reductive dechlorination of 2,4-dichlorobenzoyl-coenzyme A (CoA) to 4-chlorobenzoyl-CoA in Corynebacterium sepedonicum KZ-4 and coryneform bacterium strain NTB-1 (31), as well as a CoA-mediated reductive dehalogenation of 3-chlorobenzoate in Rhodopseudomonas palustris RCB100 using 3-chlorobenzoate as the carbon source rather than as a terminal electron acceptor (13). Oxidative dehalogenation of halogenated aromatics is catalyzed by monooxygenase (29), dioxygenases (34, 37, 39, 45), and peroxidase (30).
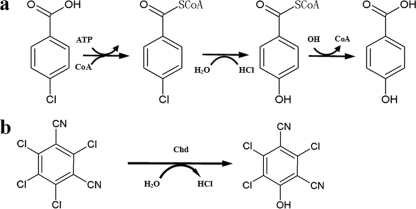
Even though several hydrolytic dehalogenases involved in dehalogenation of halogenated aliphatic hydrocarbons and halogenated carboxylic acids have been characterized, only one kind of hydrolytic dehalogenase for halogenated aromatics has been reported. The only hydrolytic dehalogenase identified to date is 4-chlorobenzoyl-CoA dehalogenase in the 4-chlorobenzoate degradation system (32, 33). For the hydrolytic substitution of the chlorine atom of 4-chlorobenzoate with a hydroxyl group, activation by the CoA thioester formation is required. Initially, 4-chlorobenzoate-CoA ligase adenylates the carboxyl group in a reaction requiring ATP, followed by the replacement of AMP with CoA and the formation of a thioester. This intermediate is sufficiently energized to facilitate the replacement of the hydroxyl group with a 4-chlorine atom; this is catalyzed by 4-chlorobenzoyl-CoA dehalogenase. Finally, 4-hydroxybenzoyl-CoA thioesterase removes the CoA (Fig. 1a). Three separate enzymes are involved in this system, and cofactors including CoA and ATP are needed (32, 33).
FIG. 1.
Dechlorination mechanism of 4-chlorobenzoate in Pseudomonas sp. CBS3 and Acinetobacter sp. strain 4-CB1 (a) and the first-step dechlorination mechanism of 2,4,5,6-tetrachloroisophthalonitrile (chlorothalonil) in Pseudomonas sp. CTN-3 (b).
Chlorothalonil (2,4,5,6-tetrachloroisophthalonitrile), a broad-spectrum chlorinated aromatic fungicide, is the second most widely used agricultural fungicide in the United States, with 5 million kilograms applied annually (9). Chlorothalonil is highly toxic to fish, birds, and aquatic invertebrates (6) and is commonly detected in ecosystems (18). The bacterial strain Pseudomonas sp. CTN-3, capable of efficiently transforming chlorothalonil, was isolated in our laboratory from long-term chlorothalonil-contaminated soil in the Jiangsu Province in China. In Ochrobactrum anthropi SH35B, a glutathione-dependent glutathione S-transferase was reported to be able to catalyze the nucleophilic substitution of chlorine atoms of chlorothalonil (19). The glutathione S-transferase in our CTN-3 bacterial strain showed 84% identity with that of O. anthropi SH35B. However, the glutathione S-transferase from CTN-3 was not functionally expressed. Here, we report for the first time the characterization of a novel chlorothalonil hydrolytic dehalogenase (Chd) that contains a conserved domain of the metallo-β-lactamase superfamily. Chd is the second hydrolytic dehalogenase for chlorinated aromatic compounds to be identified. The hydrolytic dehalogenation of chlorothalonil catalyzed by Chd is independent of CoA and ATP. Based on the analysis of amino acid sequences and catalytic mechanisms, Chd is unique from the other reported hydrolytic dehalogenases.
MATERIALS AND METHODS
Chemicals.
Chlorothalonil (99.3% purity), diethyl pyrocarbonate (DEPC), and 1,10-phenanthroline were purchased from Sigma-Aldrich (St. Louis, MO). N-Bromosuccinic acid (NBS) and hydroxylamine were purchased from Hanhong Chemical Co., Ltd. (Shanghai, China). All other reagents used in this study were of analytical reagent grade.
Cloning of the chlorothalonil dehalogenase gene.
The cloning of the chlorothalonil dehalogenase gene was carried out using the shotgun method. Genomic DNA of Pseudomonas sp. CTN-3 was prepared by a high-salt method (24) and subjected to partial digestion with Sau3AI. Fractions containing approximately 2- to 4-kb DNA fragments were pooled, ligated into the BamHI site of the plasmid pUC118, and transformed into competent Escherichia coli DH5α. The library was plated onto Luria-Bertani (LB) agar plates containing 100 mg liter−1 of ampicillin and 0.4 mM chlorothalonil. The plates were incubated at 37°C for approximately 10 h and then stored at 16°C for 48 h. Colonies producing a clear transparent halo due to chlorothalonil transformation were screened and further tested for their chlorothalonil-transforming abilities by gas chromatography. The positive clones were sequenced by Shanghai Invitrogen Biotechnology Co., Ltd. Nucleotide and deduced amino acid sequence analysis, open reading frame (ORF) determination, multiple alignment, and molecular mass determination were performed using Omiga software (version 2.0). BlastP was used for a deduced amino acid identity search (www.ncbi.nlm.nih.gov/Blast). Solvent accessibility and the secondary structure of the protein were predicted by the modeling server JPred (10). Multiple protein sequence alignments were constructed using Clustal X. Phylogenetic analyses of the protein sequences were performed using the software MEGA 3.1. Distances (distance options according to the Kimura two-parameter mode) were calculated, and clustering was performed with the neighbor-joining method (21).
Gene expression and purification of the recombinant enzyme.
The positive ORF without its translation stop codon was amplified by PCR and inserted into the NdeI and XhoI sites of the expression vector pET29a(+) to generate the recombinant plasmid pET-chd. The enzyme was overexpressed in E. coli BL21(DE3) using the His-Bind protein fusion and purification system. Harvested cells were washed and disrupted by using a French press (Thermo Spectronic). Cell debris and insoluble proteins were removed by centrifugation (12,000 ×g for 10 min at 4°C). The supernatant was loaded onto a His-Bind resin (Novagen). After elution of nontarget proteins with 25 mM imidazole in 20 mM Tris-HCl (pH 6.5) and 50 mM NaCl, the target fusion protein was eluted with 100 mM imidazole in 20 mM Tris-HCl (pH 6.5) and 50 mM NaCl. The enzyme was dialyzed against phosphate-buffered saline (PBS; 50 mM; pH 7.0) for 24 h and concentrated using an Amicon ultrafiltration tube. Protein concentrations were quantified by the Bradford method using bovine serum albumin as the standard.
Chlorothalonil determination by gas chromatography.
The solution mixture was extracted with an equal volume of dichloromethane. The organic layer was dehydrated with anhydrous Na2SO4 and dried under reduced pressure using a centrifugal evaporator at room temperature. The residual organic material was redissolved in n-hexane. Chlorothalonil was detected by gas chromatography (Shimadzu GC-14B) coupled with an electron capture detector and an SPB-5 column (30 m by 250 μm by 0.25 μm). Nitrogen was used as the carrier gas (1 ml min−1), and the temperatures of the injector/interface, column, and detector were set at 190°C, 250°C, and 250°C, respectively.
Metabolite identification.
Five hundred milliliters of PBS (50 mM; pH 7.0) containing 300 mg of chlorothalonil was incubated with 3 ml of the purified enzyme (1 mg ml−1) at 37°C for 36 h. For detection of chloride production, the enzymatic reaction was terminated by adding 0.1 ml of 30% HNO3. The mixture was centrifuged, and the supernatant was mixed with mercuric thiocyanate and ferric ammonium sulfate and then monitored spectrophotometrically at 460 nm (16). Abiotic dehalogenation was tested in the same buffer without adding the enzyme. For identification of the metabolite produced during chlorothalonil transformation by the purified enzyme, the enzymatic solution was acidified to a pH of 3 with HCl and extracted with an equal volume of ethyl acetate. The organic layer was dehydrated with anhydrous Na2SO4 and dried as described above. The residual organic material was redissolved in acetonitrile and separated by thin-layer chromatography (TLC) on a silica gel G using a hexane-isopropanol-methanol (5:3:2, vol/vol/vol) solvent system. The metabolite was visualized under UV light at 254 nm. The spots corresponding to the metabolite were scratched from the plates, extracted in acetonitrile, and dried as described above. The metabolite was first identified by reverse-phase high-performance liquid chromatography (HPLC; Shimadzu LC-20AD, Waters 2487 Dual λ absorbance detector) with a Nova-Pak C18 cartridge (3.9 by 150 mm). The metabolite was further confirmed by tandem mass spectrometry (MS/MS; Finnigan TSQ Quantum Ultra AM). Nuclear magnetic resonance (NMR) analysis was carried out to determine the position of the dehalogenated chlorine atom. The 1H and 13C NMR spectra were recorded on a JOEL JNM ECA-600 spectrometer operating at 600.17 MHz and 150.91 MHz with CDCl3 or dimethyl sulfoxide inside, using the standard pulse sequence at room temperature. Assignment of the signals was carried out by considering their relative heights and by comparison with the calculated chemical shift values using substituent shift parameters.
Dehalogenation assay under anaerobic conditions.
Anaerobic dehalogenation experiments were performed in 25-ml Erlenmeyer flasks that were closed gas-tight by headspace caps. Trace elements of oxygen in the solution were removed by the addition of a reduced form of glutathione to a concentration of 10 mM. Resazurin (0.002 mM) was used as an indicator of anoxic conditions in the assay. The dehalogenation activity was monitored by gas chromatography.
Determination of the molecular mass and pI.
The molecular mass of the denatured enzyme was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was stained with Coomassie brilliant blue G250 (Amresco). A zymograph assay of the enzyme was conducted as previously described, except that chlorothalonil was used as the substrate (14). To determine the molecular mass of the native enzyme, the enzyme was analyzed by using an Ultraflex II matrix-assisted laser desorption-ionization-time-of-flight mass spectrometer (MALDI-TOF MS; Bremen, Germany) equipped with a solid-state smart beam under the control of the FlexControl 2.4 software (Bruker Daltonics GmbH). The pI of the enzyme was estimated by PAGE with 6.25% Ampholine (pH 3.5 to 10) in a gel rod (0.5 by 1.0 cm) using a kit for isoelecric focusing calibration (Pharmacia LKB) according to the supplier's recommendations.
Enzyme assay.
All enzyme assays were performed in PBS (50 mM; pH 7.0), and no more than 10% of the substrate was dehalogenated during the assay. Because Chd had the highest catalytic activity at 50°C, all dehalogenase activities were assayed at 50°C unless otherwise stated. For dehalogenase activity assays, 50 μl of the enzyme was mixed with 0.2 mM chlorothalonil in 1 ml of PBS and the reaction mixture was incubated at 50°C for 10 min. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of the metabolite 4-hydroxy-2,5,6-trichloroisophthalontrile (4-TPN-OH) per min from chlorothalonil. 4-TPN-OH was obtained from the transformation of chlorothalonil by the purified enzyme, and the purity (>99.99%) was detected by reverse-phase HPLC. For kinetic studies, chlorothalonil was appropriately diluted into at least five different concentrations around the dissociation constant (Km) value. Initial reaction velocities measured at various concentrations of substrate were fitted to the Lineweaver-Burk transformation of the Michaelis-Menten equation. Kinetic analyses by curve fitting were performed with SigmaPlot software.
Biochemical characterization.
The pH range of the enzyme was determined by incubating the enzyme with 0.2 mM chlorothalonil for 30 s at pHs from 4 to 10. For the pH stability determination, the enzyme was incubated in PBS (50 mM) at pHs from 4 to 10 at 50°C for 10 min. For determination of the thermostability, the enzyme was preincubated in a water bath at different temperatures for 1 h, and then the remaining activity was assayed. The effects of potential inhibitors on the enzyme were determined by the addition of various chemical agents to the reaction mixture preincubated at 50°C for 10 min. Reaction of dehalogenase with hydroxylamine and NBS was carried out as previously described (47).
Site-directed mutagenesis.
Mutagenesis of the enzyme was performed using rapid PCR site-directed mutagenesis. The entire plasmid containing the chd gene was amplified by using Primestart DNA polymerase and phosphorylated primers (synthesized in GenScript) that introduced the desired changes. The amplified linear PCR product containing the desired mutation was circularized with T4 DNA ligase and T4 polynucleotide kinase. The resulting plasmid was transformed into competent E. coli cells. Twenty-six mutants (C25A, H38Q, D45A, H63Q, D67A, S81T, S126H, H128Q, D130A, T150I, H157Q, D184A, E203V, D215A, W219F, W227F, W241F, D244A, D264A, H271Q, W282F, H283Q, D323A, W324F, H326Q, and D337A) were constructed using this technique. The DNA sequences of these resulting mutants were verified by double sequencing.
Nucleotide sequence accession numbers.
The nucleotide sequences of the chlorothalonil hydrolytic dehalogenase genes were deposited in the GenBank database under accession numbers GQ292539 and GQ485642.
RESULTS
Cloning of the chlorothalonil dehalogenase gene and purification of the recombinant enzyme.
Two positive clones, designated as 1-10 and 2-1, were screened from a library containing approximately 15,000 transformants. Clone 2-1 was found to show only about 50% of the transformation capability of clone 1-10. The inserted fragments in the two positive clones had a 1,243-bp overlap in sequence that nearly covered a 1,026-bp ORF. In the inserted fragment of clone 2-1, 19 bp at the 3′ end of the 1,026-bp ORF were not included; this may explain the lower transformation ability of clone 2-1. The 1,026-bp ORF was expressed in E. coli DH5α, and the capacity to degrade chlorothalonil was confirmed. The cloned gene encodes a 342-amino-acid protein with a calculated molecular mass of 36,823 Da. A putative ribosomal binding site (AGGTAA) was located 6 bp upstream of the ATG start codon. The G+C content was 58.6%. No potential signal sequence was found. The enzyme encoded by this gene was overexpressed in E. coli BL21(DE3) and purified from the crude extract using Ni-nitrilotriacetic acid affinity chromatography.
Identification of the metabolite produced during chlorothalonil transformation.
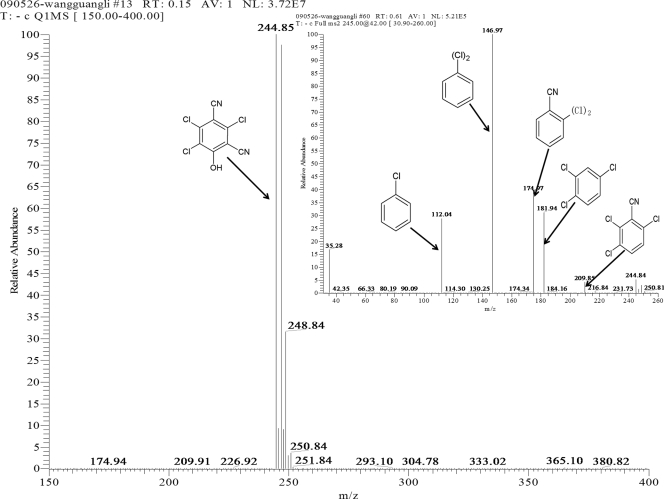
Chloride production was detected spectrophotometrically during the transformation of chlorothalonil by the purified enzyme, indicating that a dehalogenation reaction was involved. Only one metabolite was detected by TLC and reverse-phase HPLC after the complete transformation of chlorothalonil. MS/MS identification of the metabolite showed prominent protonated molecular ions at m/z = 244.85 [M-H]− in the standard MS, enabling the assignment of the molecular ion (M−) at m/z = 246. The characteristic fragment ion peaks of the metabolite in second-order MS were at m/z = 209.85, 181.94, 146.97, 112.04, and 35.28. Therefore, the metabolite was identified as hydroxy-trichloroisophthalonitrile (Fig. 2).
FIG. 2.
Standard MS profile and second-order mass spectrum (inset) of the metabolite produced during chlorothalonil transformation by Chd.
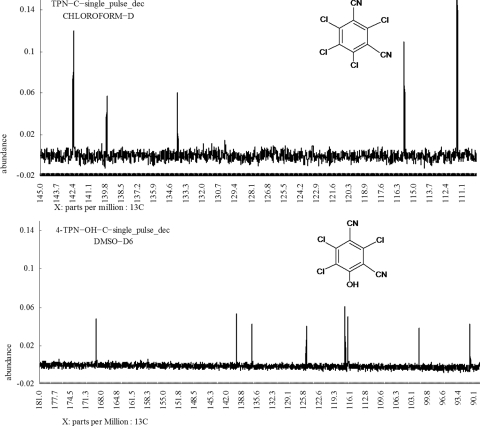
NMR analysis was performed to determine the position of the hydroxyl group in hydroxy-trichloroisophthalonitrile. The 13C NMR spectrum of chlorothalonil revealed five signals at 111.54, 115.79, 133.97, 139.66, and 142.34 ppm. The identification of these five signals was in agreement with the estimation result of chlorothalonil (Fig. 3, top panel). The 13C NMR spectrum of hydroxy-trichloroisophthalonitrile revealed eight signals at 90.94, 101.64, 116.45, 117.04, 125.21, 136.60, 139.74, and 169.10 ppm. These signals indicate that a 4- or 6-chlorine atom was replaced by a hydroxyl group, because only five signals would be detected if a 2- or 5-chlorine atom was substituted (Fig. 3, lower panel). Due to the symmetry of the 4- and 6-chlorine atom in chlorothalonil, the metabolite was identified as 4-TPN-OH.
FIG. 3.
13C NMR spectra of chlorothalonil (top panel) and 4-hydroxy-trichloroisophthalonitrile (lower panel).
Hydroxyl group substitution of the chlorine atom in chlorothalonil has been reported for soil microorganisms (26). The 4-chlorine atom was presumed to be the most susceptible chlorine atom to be replaced in chlorothalonil (3). However, there has been no convincing evidence to confirm the position of the chlorine atom that is replaced. Here, for the first time, we provide authentic evidence to prove that the 4-chlorine atom was dehalogenated.
The substrate specificity of the enzyme was tested using various chloroaromatics, including chlorobenzene, p-dichlorobenzene, 4-chloronitrobenzene, pentachloronitrobenzene, 4-chlorobenzoate, 4-chlorophenylacetic acid, m-chloroaniline, p-chloroaniline, and pentachlorophenol. None of the other chloroaromatics tested was found to be dehalogenated by the enzyme. Interestingly, only one chlorine atom of chlorothalonil could be dehalogenated, even though the 4- and 6-chlorine atoms are symmetrical. The reactivity of 4-TPN-OH at a particular site is commonly affected by the electron-withdrawing properties of nitrile and other chlorine atoms at the ortho or para positions; therefore, the bond dissociation energy (BDE) of the 6-position carbon-chlorine will be affected by the 4-hydroxyl group. However, whether the dehalogentaion of only one chlorine atom from chlorothalonil is due to an effect on BDE or the enzyme's stereochemical selection of substrate needs further investigation.
Dehalogenation under anaerobic conditions.
The purified enzyme transformed chlorothalonil to 4-TPN-OH, indicating that the dehalogenation reaction was either oxidative or hydrolytic. To identify whether an oxygenase or a hydrolytic dehalogenase was responsible for the dehalogenation, dehalogenation by the enzyme in the absence of oxygen was carried out to further confirm the mechanism of the dehalogenation reaction. Nearly no differences between the dehalogenation activities of the enzyme in the presence or absence of oxygen were found, demonstrating that the hydroxyl group of 4-TPN-OH was derived from water and not from molecular oxygen. Therefore, the conversion of chlorothalonil to 4-TPN-OH was a hydrolytic process, as opposed to an oxidative one (Fig. 1b). According to the above data, the characterized enzyme was designated as chlorothalonil hydrolytic dehalogenase (EC 3.8.1.12).
Characteristics of Chd.
The sequence of Chd was compared with other known enzymes available in the NCBI database. Chd contains a putative conserved domain of the metallo-β-lactamase superfamily and shows the highest identity with several metallohydrolases, including a cyclase from Streptomyces coelicolor (29% identity), a Zn-dependent hydrolase from Stackebrandtia nassauensis (27% identity), and a β-lactamase-like protein from “Candidatus Koribacter versatilis” (24% identity). The secondary structure of Chd, predicted by the modeling server JPred, shows that Chd has a characteristic fold of the metallo-β-lactamases. The characteristic metallo-β-lactamase fold consists of an αβ/βα sandwich consisting of a core of β-sheets surrounded by α-helices (Fig. 4). Furthermore, His-X-His-X-Asp-His, the most characteristic signature of the metallo-β-lactamase superfamily, was also found. However, the first histidine was replaced by serine (Fig. 4, shaded).
FIG. 4.
Sequence of Chd aligned with other metallohydrolases. Chd was aligned with COG0491(GloB), a Zn-dependent hydrolase, and smart00849 (lactamase B) of the metallo-β-lactamase superfamily. The secondary structure was predicted using the modeling server JPred and is shown in the alignment. Helices are indicated with an H, and strands (extended) are indicated by an E. Conserved characteristic signatures G-X-S-X-G and H-X-H-X-DH are shaded. Residues essential for Chd catalysis are marked with asterisks. Residues with some effect or without significant effect on Chd catalysis are marked with a triangle or a dot, respectively. The potential Zn(II) ligands of COG0491 and smart00849 are underlined.
The molecular mass of the denatured recombinant Chd on SDS-PAGE was determined to be approximately 36 kDa. The determined molecular mass is in good agreement with the molecular mass deduced from the amino acid sequence (36,823 Da). Zymographic analysis of Chd on agar plates containing 0.4 mM chlorothalonil showed a single hydrolytic band corresponding to the same position as that on SDS-PAGE. The molecular mass of native Chd was estimated using MALDI-TOF MS. Only one mass peak was found, corresponding to a molecular mass of 33,886 Da. These data indicate that Chd is a monomer. The pI value of Chd was estimated to be 4.13. For chlorothalonil, the overall catalytic rate (kcat) of Chd was 207 s−1 with a dissociation constant (Km) of 0.112 mM. The catalytic efficiency value (kcat/Km) under optimal conditions was 1.85 μM−1 s−1. These data indicate that chlorothalonil is a good substrate for Chd.
The optimal pH of Chd was observed to be approximately 7.0, and the enzyme was fairly stable at pHs between 6.0 and 9.0. The enzyme retained more than 95% of its original activity after preincubation in that pH range for 30 min. Chd was fairly stable up to 40°C and retained more than 95% of its activity at 50°C for 10 min. Chd retained 18% of its residual activity at 50°C for 1 h and completely lost activity at 60°C for 1 h. Only 3 to 5% of the enzymatic activity of Chd was lost when stored at 4°C for 8 weeks, and no significant loss of activity was found when it was stored at −20°C over a 3-month period in PBS (50 mM; pH 7.0) supplemented with 10% (vol/vol) glycerol. The surfactant SDS (10 mM) had a strong inhibitory effect (more than 90% inhibition) on Chd activity, while Tween 80 and Triton X-100 (10 mM) showed 50 to 60% inhibition of Chd activity. Chd catalytic activity was completely inhibited by the Zn2+-chelating metalloprotease inhibitor 1,10-phenanthroline (1 mM), and the catalytic activity of Chd was recovered by the subsequent supplementation with Zn2+. However, subsequent supplementation with Cu2+, Ba2+, Mg2+, or Ni2+ could not recover the catalytic activity of Chd.
Identification of active residues essential for Chd catalysis.
Deletion of the first 15 amino acids (MPPGCSGLCSFVGLT) in the amino terminus of Chd did not significantly affect the catalytic activity of Chd. However, deletion of the first 50 amino acids in the amino terminus resulted in a complete loss of Chd catalytic activity. These data indicate that residues from positions 16 to 50 are essential for Chd function or configuration. Group-directed chemical modification reagents were used to probe for active residues in Chd essential for substrate binding and/or catalysis. Treatment of the Chd with 0.5 mM DEPC resulted in the complete loss of catalytic activity, which was fully regained by subsequent treatment with hydroxylamine (0.1 M), indicating the involvement of His residues in the active sites of the enzyme, because other nucleophilic residues, Cys and Lys, would not be reversed by treatment with hydroxylamine (47). Chemical modification of Chd with 1 mM NBS resulted in the complete loss of catalytic activity, and this was prevented by the saturation of the active site with the product ligand 4-TPN-OH (1 mM). These data provide evidence favoring an essential role of a tryptophan in the active site of Chd (47). Site-directed mutagenesis was used to identify the specific residues essential for catalysis. Replacement of Cys25 with alanine did not significantly affect the catalytic activity of Chd, indicating that the nucleophilic residue cysteine is not involved in catalysis. There are only two cysteine residues in Chd, and the other residue is within the first 15 amino acids of the amino terminus. Next, we replaced the histidine residues at positions 38, 63, 128, 157, 271, 283, and 326 with glutamine. Significant loss of catalytic activity was observed only when histidines 63, 128, and 157 were mutated. The H63Q mutant was still capable of transforming chlorothalonil, but its Km was increased to 0.154 mM. However, the H128Q and H157Q mutants completely lost their catalytic activity. All five tryptophan residues were replaced with phenylalanine residues, respectively. Only the W241F mutant completely lost its catalytic activity, and other tryptophan mutants were not found to have a significant loss of catalytic activity. Further, we mutated some aspartate residues to alanines. The D45A, D130A, and D184A mutants were found to completely lose their catalytic activities. The Km for the D337A mutant Chd increased to 0.176 mM, and it showed about 50% transformation activity compared to the wild-type Chd. This Km change in the D337A mutant might explain the lower transformation ability of clone 2-1. All other mutants were not found to have a significant loss of catalytic activity. His127 and Asp129 in Chd seem to be involved in metal coordination, because most of the amino acids believed to serve as Zn(II) ligands for the metallo-β-lactamase superfamily are conserved within the consensus His-X-His-X-Asp-His signature. Mutation of the conserved His271, a potential metal binding ligand in the metallo-β-lactamase superfamily (Fig. 4, underlined), to glutamine did not significantly affect the catalytic activity of Chd.
DISCUSSION
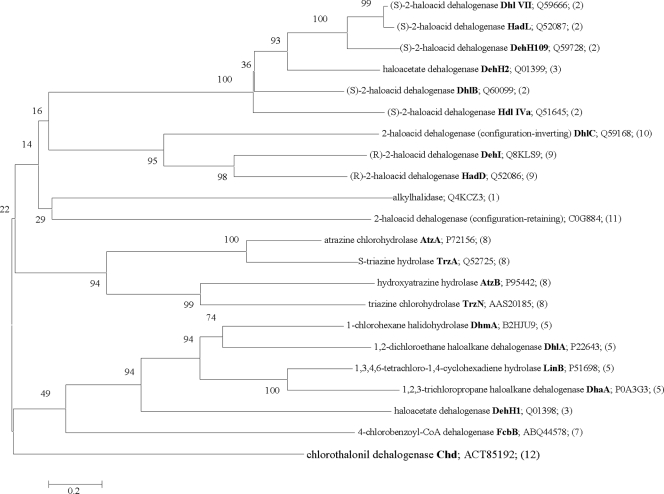
Hydrolytic dehalogenation is commonly observed as the first step in the degradation of synthetic halogenated compounds. Hydrolytic dehalogenases catalyze the replacement of halogen with a hydroxyl group from water as the nucleophile, resulting in the formation of a primary alcohol, a halide ion, and a proton. To date, several hydrolytic dehalogenases have been reported, including alkylhalidase (EC 3.8.1.1), (S)-2-haloacid dehalogenase (EC 3.8.1.2), haloacetate dehalogenase (EC 3.8.1.3), haloalkane dehalogenase (EC 3.8.1.5), 4-chlorobenzoyl-CoA dehalogenase (EC 3.8.1.7), atrazine chlorohydrolase (EC 3.8.1.8), (R)-2-haloacid dehalogenase (EC 3.8.1.9), 2-haloacid dehalogenase (configuration inverting; EC 3.8.1.10), and 2-haloacid dehalogenase (configuration retaining; EC 3.8.1.11). All these halidohydrolase-type dehalogenases seem to prefer short-chain halogenated aliphatic hydrocarbons and halogenated carboxylic acids. Until now, only one hydrolytic dehalogenase for halogenated aromatics has been reported: the 4-chlorobenzoate dehalogenase complex. The hydrolytic dehalogenation activity of the 4-chlorobenzoate dehalogenase complex is comprised of three separate enzymes, 4-chlorobenzoate-CoA ligase, 4-chlorobenzoyl-CoA dehalogenase, and 4-hydroxybenzoyl-CoA thioesterase, and needs CoA and ATP. The chlorothalonil dehalogenase reported here is the second hydrolytic dehalogenase for halogenated aromatic compounds to be identified. The notable difference between Chd and the 4-chlorobenzoate dehalogenase complex is that no other cofactors are required for hydrolytic dehalogenation by Chd. Chd is a novel dehalogenase that shares very low amino acid sequence identity with other reported dehalogenases. Phylogenetic analysis based on the amino acid sequences revealed that Chd forms a distinct subclade with other hydrolytic dehalogenases (Fig. 5).
FIG. 5.
Phylogenetic tree of the hydrolytic dehalogenases constructed according to the neighbor-joining method. Statistically significant bootstrap values are shown at the nodes and are expressed as a percentage from 500 replications. Sequence accession numbers of the hydrolytic dehalogenases follow the enzyme symbol (in bold). The number in parentheses indicates the EC classification of the hydrolytic dehalogenases (i.e., classification numbers are EC 3.8.1.*, with the number in parentheses in place of the asterisk).
Besides the absence of sequence relatedness, the mechanism of catalysis by Chd is different from that of other dehalogenases. Studies of the crystal structures of DhlA from Xanthobacter autotrophicus GJ10 (42), DhaA from Rhodococcus sp. (27), and LinB from Sphinomonas paucimobilis UT26 (23) show that these haloalkane dehalogenases belong to the α/β-hydrolase superfamily. The catalytic triad (Asp-His-Asp/Glu) of haloalkane dehalogenases is located within an internal, predominantly hydrophobic cavity, occurring at the same topological positions as in other members of the α/β-hydrolase superfamily and containing the conserved feature sequence Gly-X-Ser-X-Gly at the nucleophile elbow. The central serine of the feature sequence is the terminal residue of the catalytic triad (Ser/Asp/Cys-His-Asp/Glu), whose function consists of orienting and activating the Serine-Oγ to act as the nucleophile. In Chd, the conserved catalytic residues (Asp-His-Asp/Glu) of the haloalkane dehalogenases were not found in the sequence alignment. Interestingly, the Gly-X-Ser-X-Gly feature signature of the α/β-hydrolase superfamily was found in Chd (Fig. 4, shaded), even though poor conservation at the nucleophile elbow of haloalkane dehalogenases was reported. Site-directed mutagenesis yielded nearly no reduction in the activity of Chd upon the replacement of the serine within the Gly-X-Ser-X-Gly motif with threonine. However, complete loss of activity was seen as a result of the corresponding exchange within the His-X-His-X-Asp-His motif. Thus, the catalytic function of Chd appears to be located within the metallo-β-lactamase motif, instead of the α/β-hydrolase motif. In the most typical signature of the metallo-β-lactamase superfamily, His-X-His-X-Asp-His, the first histidine and the aspartic acid are reported to be invariant. However, in Chd, the first histidine of the signature is replaced with a serine. Interestingly, the reverse replacement of the serine with histidine in the signature resulted in the complete loss of the catalytic activity of Chd.
The catalytic mechanism of Chd may also differ from that of 4-chlorobenzoyl-CoA dehalogenase, which contains a crotonase-like superfamily domain. In 4-chlorobenzoyl-CoA dehalogenase, Asp145 functions as the active site nucleophile, Trp137 serves as a hydrogen bond donor to the Asp145 C=O, and His90 serves to deprotonate the bound H2O molecule (22, 47). Even though 4-chlorobenzoyl-CoA dehalogenase also catalyzes the hydrolytic dehalogenation of the 4-chlorine atom of halogenated aromatics and Asp45, His63, His157, Asp183, Trp241, and Asp337 in Chd proved to be important residues for catalysis, no conserved catalytic residues were found between 4-chlorobenzoyl-CoA dehalogenase and Chd. The catalytic mechanism by which Chd dehalogenates chlorothalonil needs further investigation, and the specific roles of these essential catalytic residues in Chd remain to be elucidated by crystal structure studies. Collectively, we have identified a good dehalogenase candidate for study of the hydrolytic dehalogenation mechanism for halogenated aromatic compounds.
Acknowledgments
This work was supported by grants from National Programs for High Technology Research and Development of China (2007AA10Z405), the Chinese National Natural Science Foundation (30600016), and the Key Technology R&D Program of Jiangsu Province (BE2008669).
We gratefully acknowledge Haijun Yang of the Analysis Center, Department of Chemistry, Tsinghua University, for excellent assistance in NMR spectra analyses.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Adrian, L., J. Rahnenfuhrer, J. Gobom, and T. Holscher. 2007. Identification of a chlorobenzene reductive dehalogenase in Dehalococcoides sp. strain CBDB1. Appl. Environ. Microbiol. 73:7717-7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrian, L., S. Ulrich, W. Joerg, and G. Helmut. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Binkley, R. W., G. L. Kirstner, V. C. Opaskar, and P. Olynyk. 1977. Photochemical reaction of 2,4,5,6-tetrachloroisophthalonitrile. Chemosphere 6:163-166. [Google Scholar]
- 4.Boyer, A., R. Page-BeLanger, M. Saucie, R. Villemur, F. Lepine, P. Juteau, and R. Beaudet. 2003. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6-trichlorophenol from Desulfitobacterium frappieri PCP-1. Biochem. J. 373:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunge, M., L. Adrian, A. Kraus, M. Opel, W. G. Lorenz, J. R. Andreesen, H. Gorisch, and U. Lechner. 2003. Reductive dehalogenation of chlorinated dioxins by an anaerobic bacterium. Nature 421:357-360. [DOI] [PubMed] [Google Scholar]
- 6.Caux, P. Y., R. A. Kent, G. T. Fan, and G. L. Stephenson. 1996. Environmental fate and effects of chlorothalonil: a Canadian perspective. Crit. Rev. Environ. Sci. Technol. 26:45-93. [Google Scholar]
- 7.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4- hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 8.Copley, S. D. 1998. Microbial dehalogenases: enzymes recruited to convert xenobiotic substrates. Curr. Opin. Chem. Biol. 2:613-617. [DOI] [PubMed] [Google Scholar]
- 9.Cox, C. 1997. Chlorothalonil. J. Pest. Reform. 17:14-20. [Google Scholar]
- 10.Cuff, J. A., M. E. Clamp, A. S. Siddigni, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 11.de Jong, R. M., and B. W. Dijkstra. 2003. Structure and mechanism of bacterial dehalogenases: different ways to cleave a carbon-halogen bond. Curr. Opin. Struct. Biol. 13:722-730. [DOI] [PubMed] [Google Scholar]
- 12.Dotterer, S. K., and R. L. Harris. 1988. MNDO study of nucleophilic aromatic substitution. J. Org. Chem. 53:777-779. [Google Scholar]
- 13.Egland, P. G., J. Gibson, and C. S. Harwood. 2001. Reductive, coenzyme A-mediated pathway for 3-chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris. Appl. Environ. Microbiol. 67:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, G., Z. Cui, T. Huang, and S. Li. 2004. Expression, purification, and characterization of a novel methyl parathion hydrolase. Protein Express. Purif. 36:170-176. [DOI] [PubMed] [Google Scholar]
- 15.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 16.Iwasaki, I., S. Utsumi, and T. Ozawa. 1952. New colorimetric determination of chloride using mercuric thiocyanate and ferric ion. Bull. Chem. Soc. Jpn. 25:226. [Google Scholar]
- 17.Janssen, D. B., E. O. Jantien, and J. P. Gerrit. 2001. Microbial dehalogenation. Curr. Opin. Biotechnol. 12:254-258. [DOI] [PubMed] [Google Scholar]
- 18.Kazos, E. A., C. G. Nanos, C. D. Stalikas, and C. N. Konidari. 2008. Simultaneous determination of chlorothalonil and its metabolite 4-hydroxychlorothalonil in greenhouse air: dissipation process of chlorothalonil. Chemosphere 72:1413-1419. [DOI] [PubMed] [Google Scholar]
- 19.Kim, Y. M., K. Park, G. J. Joo, E. M. Jeong, J. E. Kim, and I. K. Rhee. 2004. Glutathione-dependent biotransformation of the fungicide chlorothalonil. J. Agric. Food Chem. 52:4192-4196. [DOI] [PubMed] [Google Scholar]
- 20.Krasotkina, J., T. Walters, K. A. Maruya, and S. W. Ragsdale. 2001. Characterization of the B12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276:40991-40997. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 22.Lau, E. Y., and T. C. Bruice. 2001. The active site dynamics of 4-chlorobenzoyl-CoA dehalogenase. Proc. Natl. Acad. Sci. U. S. A. 98:9527-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marek, J., J. Vevodova, I. Kuta-Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 24.Miller, S. A., D. D. Dykes, and H. F. Polesky. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyachi, K., S. K. Suh, T. Nagata, and M. Takagi. 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motonaga, K., K. Takagi, and S. Matumoto. 1996. Biodegradation of chlorothalonil in soil after suppression of degradation. Biol. Fertil. Soils 23:340-345.
- 27.Newman, J., T. S. Peat, R. Richard, L. Kan, P. E. Swanson, J. A. Affholter, I. H. Holmes, J. F. Schindler, C. J. Unkefer, and T. C. Terwilliger. 1999. Haloalkane dehalogenase: structure of a Rhodococcus enzyme. Biochemistry 38:16105-16114. [DOI] [PubMed] [Google Scholar]
- 28.Ni, S., J. K. Fredrickson, and L. Xun. 1995. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy, G. V. B., and M. H. Gold. 2000. Degradation of pentachlorophenol by Phanerochaete chrysosporium: intermediates and reactions involved. Microbiology 146:405-413. [DOI] [PubMed] [Google Scholar]
- 31.Romanov, V., and R. P. Hausinger. 1996. NADPH-dependent reductive ortho dehalogenation of 2,4-dichlorobenzoic acid in Corynebacterium sepedonicum KZ-4 and coryneform bacterium strain NTB-1 via 2,4-dichlorobenzoyl coenzyme A. J. Bacteriol. 178:2656-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz, A., K. Gartemann, J. Fiedler, E. Grundt, and R. Eichenlaub. 1992. Cloning and sequence analysis of genes for dehalogenation of 4-chlorobenzoate from Arthrobacter sp. strain SU. Appl. Environ. Microbiol. 58:4068-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholten, J. D., K. H. Chang, P. C. Babbit, H. Charest, M. Sylvestre, and D. Dunaway-Mariano. 1991. Novel enzymic hydrolytic dehalogenation of a chlorinated aromatic. Science 253:182-185. [DOI] [PubMed] [Google Scholar]
- 34.Schweizer, D., A. Markus, M. Seez, H. H. Ruf, and F. Lingens. 1987. Purification and some properties of component B of the 4-chlorophenylacetate 3,4-dioxygenase from Pseudomonas species strain CBS3. J. Biol. Chem. 262:9340-9346. [PubMed] [Google Scholar]
- 35.Slater, J. H., A. T. Bull, and D. J. Hardman. 1995. Microbial dehalogenation. Biodegradation 6:181-189. [DOI] [PubMed] [Google Scholar]
- 36.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 37.Stefan, B., R. M. Jeremy, N. T. Kenneth, and H. P. Dietmar. 1998. Identification of chlorobenzene dioxygenase sequence elements involved in dechlorination of 1,2,4,5-tetrachlorobenzene. J. Bacteriol. 180:520-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thibodeau, J., A. Gauthier, M. Duguay, R. Villemur, F. Lepine, P. Juteau, and R. Beaudet. 2004. Purification, cloning, and sequencing of a 3,5-dichlorophenol reductive dehalogenase from Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 70:4532-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van de Pas, B. A., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 41.van Pee, K., and S. Unversucht. 2003. Biological dehalogenation and halogenation reactions. Chemosphere 52:299-312. [DOI] [PubMed] [Google Scholar]
- 42.Verschueren, K. H. G., F. Seljee, H. J. Rozeboom, K. H. Kalk, and B. W. Dijkstra. 1993. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature 363:693-698. [DOI] [PubMed] [Google Scholar]
- 43.Wilce, M. C., and M. W. Parker. 1994. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta 1205:1-18. [DOI] [PubMed] [Google Scholar]
- 44.Wohlfarth, G., and G. Diekert. 1997. Anaerobic dehalogenases. Curr. Opin. Biotechnol. 8:290-295. [DOI] [PubMed] [Google Scholar]
- 45.Xun, L., J. Bohuslavek, and M. Cai. 1999. Characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) of Sphingomonas chlorophenolica ATCC 39723. Biochem. Biophys. Res. Commun. 266:322-325. [DOI] [PubMed] [Google Scholar]
- 46.Xun, L., T. Edward, and S. O. Cindy. 1992. Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J. Bacteriol. 174:8003-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, G., R. Q. Liu, K. L. Taylor, H. Xiang, J. Price, and D. Dunaway-Mariano. 1996. Identification of active site residues essential to 4-chlorobenzoyl-coenzyme A dehalogenase catalysis by chemical modification and site directed mutagenesis. Biochemistry 35:10879-10885. [DOI] [PubMed] [Google Scholar]
- 48.Zheng, Y. J., and R. L. Ornstein. 1997. Mechanism of nucleophilic aromatic substitution of 1-chloro-2,4-dinitrobenzene by glutathione in the gas phase and in solution. Implications for the mode of action of glutathione S-transferases. J. Am. Chem. Soc. 119:648-655. [Google Scholar]