Abstract
More than 200 direct CodY target genes in Staphylococcus aureus were identified by genome-wide analysis of in vitro DNA binding. This analysis, which was confirmed for some genes by DNase I footprinting assays, revealed that CodY is a direct regulator of numerous transcription units associated with amino acid biosynthesis, transport of macromolecules, and virulence. The virulence genes regulated by CodY fell into three groups. One group was dependent on the Agr system for its expression; these genes were indirectly regulated by CodY through its repression of the agr locus. A second group was regulated directly by CodY. The third group, which includes genes for alpha-toxin and capsule synthesis, was regulated by CodY in two ways, i.e., by direct repression and by repression of the agr locus. Since S. aureus CodY was activated in vitro by the branched chain amino acids and GTP, CodY appears to link changes in intracellular metabolite pools with the induction of numerous adaptive responses, including virulence.
Bacterial survival depends upon the ability to sense and respond to environmental stresses, such as changes in temperature, pH, osmolarity, cell population density, and nutrient availability. Staphylococcus aureus has a well-characterized ability to survive when faced with suboptimal conditions, highlighted by the ability of S. aureus to persist in mammalian hosts both as a commensal and as a pathogen. Many regulators of S. aureus virulence gene expression have been characterized (6). With the exception of the stress-dependent activation of σB and the link between CcpA (catabolite control protein A) and select virulence factor expression (46), however, the specific mechanisms of virulence regulation in response to changes in nutrient availability are largely unknown. The best-characterized regulator of S. aureus virulence in response to environmental changes is the Agr (accessory gene regulator) system. This system, encoded at the agr locus, includes a quorum-sensing mechanism that activates a two-component system that controls synthesis of a regulatory RNA, RNAIII (for a review, see reference 33).
CodY, a highly conserved regulatory protein of stationary-phase adaptation in low-G+C Gram-positive bacteria, is emerging as a regulator of virulence in S. aureus (28, 38, 47) as well as in other Gram-positive pathogens (4, 13, 19, 20, 28-30). First discovered in two nonpathogenic species, Bacillus subtilis and Lactococcus lactis, CodY senses nutrient availability by direct interaction with metabolite effectors. CodY homologs define a unique, winged helix-turn-helix-containing family of transcription factors. For B. subtilis CodY, as well as for CodY proteins from Clostridium difficile, Listeria monocytogenes, and Bacillus cereus, the effectors are GTP and the branched chain amino acids (BCAAs; isoleucine, leucine, and valine) (4, 13, 20, 31, 39, 43). GTP and the BCAAs increase synergistically the affinity of CodY for its DNA target sites (17, 50). CodY proteins from L. lactis and Streptococcus pneumoniae, however, respond only to BCAAs (19, 37). A region of B. subtilis CodY that is important for DNA binding (22) and a second region of the protein that is important for BCAA interaction (23, 50) are highly conserved in CodY proteins from many species, including S. aureus (23).
We recently showed that codY null mutations in two clinical isolates of S. aureus resulted in overexpression of the agr locus, as well as the genes that encode alpha-toxin (hla) and the proteins that synthesize polysaccharide intercellular adhesin (PIA) (icaADBC), a major component of staphylococcal biofilm matrices (28). We have now used global transcriptional analyses and genome-wide in vitro DNA binding analysis to determine the extent to which CodY-dependent regulation of metabolic genes and virulence factors is mediated by the Agr system and whether the specific genes affected by a codY mutation are direct or indirect targets of CodY. Transcriptional analysis revealed that S. aureus CodY contributes to the regulation of many virulence genes in strain UAMS-1, as well as genes involved in amino acid metabolism, carbon flow, nitrogen assimilation, and transport systems. While this work was in progress, Pohl et al. (38) reported that a codY mutation in S. aureus strain Newman causes differential regulation of many genes involved in central metabolism and virulence and that the overexpression of some of these genes is likely due to the overexpression of the agr locus.
We extended this analysis by determining that dozens of genes whose expression is affected by a codY mutation are direct targets of CodY binding. Additionally, DNase I footprinting revealed that purified S. aureus CodY binds to several of these promoters at predicted CodY boxes and that the binding is dependent on the presence of the BCAAs and GTP.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. aureus strains were grown on tryptic soy agar (TSA) or in tryptic soy broth (TSB; Becton Dickinson Co.) with shaking (250 rpm) at 37°C, maintaining a flask-to-medium volume ratio of 10:1. Antibiotics were used as needed at the following concentrations: erythromycin, 10 μg/ml; kanamycin, 50 μg/ml; neomycin, 50 μg/ml; tetracycline, 3 μg/ml (all obtained from Sigma-Aldrich, St. Louis, MO).
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant genotype and/or characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1 TOPO | Cloning vector | Invitrogen |
| pCM22 | pCR2.1 TOPO containing ilvD promoter from SA564 | This study |
| pCD2 | pCR2.1 TOPO containing ilvD promoter from NCTC 8325 | This study |
| pCM24 | pCR2.1 TOPO containing capA promoter from SA564 | This study |
| pBAD30 | Expression vector with araBAD promoter | 16 |
| pKM1 | pBAD30 with S. aureus codY with five additional histidine codons at the C terminus | McCarty and Sonenshein, unpublished |
| S. aureus strains | ||
| UAMS-1 | Clinical isolate | 14 |
| MS1 | UAMS-1 ΔcodY::ermC | 28 |
| SA564 | Clinical isolate | 45 |
| CDM7 | SA564 ΔcodY::ermC | 28 |
| RN6911 | RN690 Δagr::tetM | 49 |
| CM18 | UAMS-1 Δagr::tetM | This study |
| CM19 | UAMS-1 ΔcodY::ermC Δagr::tetM | This study |
| UAMS929 | UAMS-1 sarA::kan | 5 |
| CM20 | UAMS-1 sarA::kan | This study |
| CM21 | UAMS-1 Δagr::tetM sarA::kan | This study |
| CM22 | UAMS-1 ΔcodY::ermC sarA::kan | This study |
| CM23 | UAMS-1 ΔcodY::ermC Δagr::tetM sarA::kan | This study |
Construction of agr and sarA mutant strains.
The Δagr::tetM mutation from strain RN6911 (49) was introduced into strains UAMS-1 and MS1 (ΔcodY::ermC) (28) by phage φ11-mediated transduction, as described by others (32). Transductants resistant to tetracycline were characterized by PCR analysis to confirm the replacement of the agr locus by the tetM gene. The resulting strains were CM18 (Δagr::tetM) and CM19 (ΔcodY::ermC Δagr::tetM). The sarA::kan mutation from UAMS929 (5) was introduced into strains UAMS-1, MS1, CM18, and CM19 by phage φ11-mediated transduction, generating strains CM20 (sarA::kan), CM21 (Δagr::tetM sarA::kan), CM22 (ΔcodY::ermC sarA::kan), and CM23 (ΔcodY::ermC Δagr::tetM sarA::kan). PCR analysis confirmed the insertion of kan within the sarA gene. All the codY mutant strains were confirmed to be unable to produce CodY protein by immunoblotting with an antibody against B. subtilis CodY (39).
Microarray analysis.
S. aureus colonies from TSA plates were used to inoculate a 3-ml culture in TSB. This flask and four additional 3-ml cultures with successive 100-fold dilutions of the initial inoculum were incubated overnight. The next day, flasks were selected that contained cultures with an optical density at 600 nm (OD600) of 3 to 7 and used to inoculate fresh TSB to a starting OD600 of 0.05. After the bacteria reached an OD600 of between 0.5 and 2, they were diluted again in fresh TSB to an OD600 of 0.05 to ensure that the experimental cultures were adapted to the exponential growth phase. RNA was extracted from exponential-phase samples (OD600, 0.6 to 0.8) and post-exponential-phase samples (OD600, 8 to 11), as previously described (28).
cDNA synthesis, biotin labeling, hybridization, and microarray analysis using Affymetrix S. aureus GeneChips were used as described previously (2). Samples from UAMS-1, MS1, CM18, and CM19 were prepared in duplicate. Data from duplicate experiments were normalized, averaged, and analyzed with GeneSpring GX software (Agilent). A gene was defined as differentially expressed if it exhibited at least a 2-fold change in transcript level in different strains or under different conditions and if the change was statistically significant by Student's t test (P < 0.05).
RT-PCR analysis.
Semiquantitative reverse transcription-PCR (RT-PCR) analysis was done as previously described (28). The following primers were used for cDNA synthesis of each respective gene (see also Table S1 in the supplemental material): OCM74 for oppB, OCM72 for sspA, OCM78 for tst-1, OCM76 for capA, OCM82 for sak, OCM80 for spa, OCM53 for ilvD, and OCD15 for 16S rRNA. The following additional primers were included for PCR amplification of the cDNA: oppB (OCM73), sspA (OCM71), tst-1 (OCM77), capA (OCM75), sak (OCM81), spa (OCM79), ilvD (OCM54), and 16S rRNA (OCD14 or OCD15).
CodY overexpression and purification.
The S. aureus CodY protein with five additional histidine residues appended to the C terminus was purified from Escherichia coli DH5α containing the pKM1 plasmid. To construct pKM1 (K. McKarty and A. L. Sonenshein, unpublished data), the codY gene was amplified from S. aureus genomic DNA (gDNA) by PCR with primers OKM1 and OKM2 (see Table S1) such that a SacI site, followed by the codY ribosome binding site, preceded the codY gene. The reverse primer incorporated the five additional histidine residues followed by a stop codon and an SphI site at the 3′ end of the gene. The resulting PCR product was cloned in pBAD30, placing the S. aureus codY gene under the control of the araBAD promoter (16). E. coli(pKM1) was grown to mid-exponential phase in Luria broth containing ampicillin (50 μg/ml) and then induced with 0.2% l-arabinose and incubated for 4 h. Cells were harvested by centrifugation, and the His6-tagged CodY protein was purified as previously described (44) using Talon Co2+ metal affinity resin (Novagen). The concentration of CodY was determined in a Bio-Rad assay using bovine serum albumin as the standard.
DNase I footprinting analysis.
Oligonucleotides OCM86 (for ilvD), OCM83 (for oppB), and OCM107 (for katA) were 5′ end labeled with radioactive [γ-32P]ATP (Perkin-Elmer) using T4 polynucleotide kinase (New England BioLabs, Inc.) according to the manufacturer's recommendations. The 32P-labeled primer was then purified with a QIAquick nucleotide removal kit (Qiagen). To generate the 32P-end-labeled promoter fragment for the footprinting reactions, the katA or oppB promoter was PCR amplified from UAMS-1 gDNA with 32P-labeled OCM107 and OCM108 or 32P-labeled OCM83 and OCM84, respectively. The ilvD promoter region was PCR amplified from pCM22 using 32P-labeled OCM86 and P-ilvD1. The ilvD region in pCM22 was generated by PCR amplification of SA564 gDNA using primers OCM86 and P-ilvD1 and introduced into the pCR2.1 TOPO vector according to the manufacturer's recommendations (Invitrogen). Following amplification of the 32P-end-labeled DNA fragment, 40,000 cpm of the end-labeled PCR product was used for each footprinting reaction. CodY was mixed at various concentrations (25 to 800 nM) with the 32P-end-labeled promoter DNA in binding buffer (20 mM Tris [pH 8.0], 50 mM KCl, 2 mM MgCl2, 5% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, 0.05% Nonidet P-40, 25 μg/ml calf thymus DNA). When indicated, 10 mM (each) isoleucine, leucine, and valine or 2 mM GTP or both were added. Binding reaction mixtures were incubated at room temperature for 20 min and digested for 30 s with 2 μl of RQ1 DNase I (Promega) containing 0.25 to 0.5 units of enzyme. DNase I digestion and sample purification were as described by Handke et al. (17). A sequence ladder was created using pCM22 or pCM24 DNA and the primers OCM86 or OCM88, respectively, using the Sequenase kit (USB) and [α-35S]dATP (Perkin-Elmer). The plasmid pCM24 was created by ligating the capA promoter (generated by PCR amplification of SA564 gDNA using primers OCM66 and OCM67) to the pCR2.1 TOPO vector according to the manufacturer's recommendations (Invitrogen). The pCM24 sequence (generated with OCM88) was run with the oppB and katA footprints.
DNase I digestion products and sequencing reaction products were separated on a denaturing 8 M urea-6% polyacrylamide gel by electrophoresis at 1,200 V for 2.5 h. The dried gel was exposed to a phosphor screen (Kodak) and analyzed with a Storm 820 scanner and software (Amersham Biosciences).
Primer extension analysis to map the transcriptional start site of ilvD.
Twenty micrograms of RNA isolated from exponentially growing cells of S. aureus strain NCTC 8325 and purified as described previously (28) was incubated with 5′-end-labeled oligonucleotide P-ilvD2 and SuperScript II reverse transcriptase (SSII; Invitrogen) according to the manufacturer's recommendations. The reverse transcription products were collected by precipitation with ethanol, redissolved, and separated on a denaturing 8 M urea-6% polyacrylamide gel as above. A sequencing ladder was prepared as described above using pCD2 as the template and P-ilvD2 as primer. The ilvD region in pCD2 was generated by PCR amplification of NCTC 8325 genomic DNA using primers P-ilvDseq and P-ilvD2 and cloning of the PCR product in the pCR2.1 TOPO vector.
Affinity purification of CodY-DNA complexes and analysis with the Illumina Genome Analyzer II.
Since the genome of UAMS-1 has not yet been sequenced, we used genomic DNA (two independent preparations) from the sequenced S. aureus strain NCTC 8325. Cells were scraped from a TSA plate grown overnight at 37°C, washed with 0.1 M NaCl, and lysed by incubation for 30 min at 37°C in 0.5 ml of Tris-EDTA containing 100 μg lysostaphin. After extraction with phenol-chloroform-isoamyl alcohol and ethanol precipitation, each DNA pellet was resuspended in 500 μl sterile deionized water and sheared by sonication (Branson 250 Sonifier with a microtip) to a fragment size of 300 to 1,000 bp by four 30-s sonication cycles on ice; each sonication cycle was separated by a 30-s incubation on ice. After addition of 10 μg of RNase A, the samples were incubated at 37°C for 20 min. The DNA was repurified by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation, resuspended in 100 μl 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and cleaned by using the Qiagen PCR purification kit, according to the manufacturer's recommendations. To create separately bar-coded DNA populations, four different bar-coded adapters were created. To do so, equal amounts of complementary oligonucleotides (e.g., BC1a and BC1b for bar code 1) were mixed and allowed to anneal by heating to 95°C and slow cooling to <40°C. (The b series of oligonucleotides were phosphorylated at the 5′ ends after synthesis.) Samples (5 μg) of blunt-ended gDNA were prepared by treatment with the Quick Blunting kit (New England BioLabs), according to the manufacturer's recommendations, followed by incubation at 75°C for 30 min. A dA residue was added to the 3′ end of each fragment by incubation with 1 μl of DNA polymerase I Klenow fragment (lacking exonuclease activity; New England BioLabs) and 0.5 μl dATP (100 mM) for 45 min at 37°C, followed by a 30-min incubation at 75°C. The sheared, blunt-ended DNA was then ligated to the bar-coded adapters (0.125 nmol per reaction mixture) using the Quick Ligation kit. Each bar-coded sample was run on a 2% agarose gel, and DNA bands corresponding to the range of 400 to 500 bp were recovered from the gel using the Qiagen QIAquick gel extraction kit. Each purified, bar-coded DNA sample was then split into seven aliquots, each of which was subjected to 15 cycles of PCR amplification using primers Olj139 and Olj140 (see Table S1 in the supplemental material). The seven reaction mixtures with the same bar code were pooled, extracted with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol. Each DNA pellet was dissolved in 60 μl of sterile, deionized water and quantitated by measuring absorbance at 260 nm.
Fifty micrograms of bar-coded DNA, in a total volume of 250 μl, was then used in a binding reaction mixture with 200 nM purified His6-tagged S. aureus CodY or His-tagged B. subtilis aconitase (42) in a buffer containing 20 mM Tris-Cl (pH 8.0), 50 mM sodium glutamate, 10 mM MgCl2, 0.05% Nonidet P-40, 5% glycerol, and 25 μg/ml E. coli tRNA, supplemented with 10 mM (each) isoleucine, leucine, and valine (ILV) and 2 mM GTP. After gentle mixing at room temperature for 25 min, the sample was mixed with Talon Co2+ metal affinity resin (0.1-ml packed bed volume) that had been preequilibrated with the supplemented buffer. The resin was washed six times with 0.25 ml of supplemented buffer, resuspended in 100 μl of 10 mM Tris (pH 8.0), boiled for 5 min, and then incubated with proteinase K (100 μg/ml) for 2 h at 65°C, followed by boiling for 5 min. After centrifugation for 1 min at 3,000 × g, 100 μl of the supernatant fluid was purified by using a PCR purification kit (Qiagen) and then subjected to 10 cycles of PCR amplification with primers Olj139 and Olj140, repurified, quantified, and subjected to analysis with the Illumina Genome Analyzer II at the Tufts University Core Facility. Each affinity purification experiment was done in duplicate. Forty cycles of sequencing were performed for each fragment end. Sequence alignment and assembly were performed using MAQ (24) against the S. aureus NCTC 8325 complete genome sequence (NCBI accession number CP000253.1). Only valid pairs that contained no more than three mismatches with respect to the aligned sequence and had a paired insert size of less than 600 bases were included in the analysis. Using BioPerl scripts, a coverage value (corresponding to the number of sequenced overlapping DNA fragments at a given position) was generated for every position of the genome for each set, creating a coverage map for each sample. The CodY/aconitase (Acn) enrichment ratio at every position was then calculated. To eliminate bias due to differences in the number of mapped reads between samples, mapped reads were randomly discarded from the larger set until each had the same number. Regions of enrichment in the CodY sample were defined as sequences for which the CodY samples showed an average enrichment ratio greater than 5-fold over a length of more than 100 consecutive positions. The coverage maps were compared, and the R statistical computing environment was used to calculate the Pearson correlation coefficient for the CodY and Acn samples. The CodY samples had a correlation of 0.98, the Acn samples had a correlation of 0.97, and the CodY-Acn correlation values ranged from 0.24 to 0.27. The enrichment values (CodY/Acn) had a correlation of 0.84.
The nucleotide sequences from the enriched regions comparing bar codes 1 and 2 were used as a training set for finding motifs by using MEME 4.1.0 (1). We allowed MEME to find motifs that occurred zero times or once per fragment on either strand of DNA and that had a P value of ≤0.003. A script was created that searched for canonical CodY boxes using the consensus sequence AATTTTCWGAAAATT (12, 15), within the identified regions, allowing up to three mismatches.
Identification of a capA promoter binding protein.
Total cellular extract was isolated from lysostaphin-treated cells of an overnight culture of strain Becker as previously described (27). The protein extract was fractionated by ammonium sulfate precipitation. Precipitated protein fractions collected with 45% ammonium sulfate were dissolved and dialyzed overnight in Tris-EDTA (pH 7.5). The dialysate was applied to a G150 gel filtration column (45 cm by 1.7 cm) that had been equilibrated with Tris-HCl (pH 8.0) and 0.9% NaCl. Protein fractions (∼1 ml) were collected in the same buffer and quantified by bicinchoninic acid assay (Pierce). Each fraction was tested for binding to a [γ-32P]ATP-labeled 155-bp DNA fragment containing the capA promoter region amplified by PCR from strain Becker using primer pairs cp8gs3 and cp8gs6 (see Table S1 in the supplemental material). Fractions that caused a band shift were pooled and used in a large-scale gel shift assay. The shifted protein-DNA complex was extracted from the polyacrylamide gel, boiled for 5 min in SDS sample buffer, and resolved by SDS-PAGE. The gel was stained with SYPRO Ruby (Bio-Rad), and the prominent band corresponding to an apparent molecular mass of 29 kDa was excised and identified by mass spectrophotometric analysis.
Gel shift assay.
C-terminal His6-tagged CodY was purified on a nickel column (Novagen) and used in the gel shift assay. A 139-bp fragment containing the capA promoter was PCR amplified from strain Newman gDNA using primer pairs cp8gs6 and cp8gs15 and labeled using a digoxigenin labeling kit (Roche). The gel shift assay was carried out by mixing the probe (∼5 pM) with increasing amounts of CodY protein in a total reaction volume of 20 μl as previously described (35). For competition experiments, an unlabeled DNA fragment was used as a specific competitor, and a 161-bp DNA fragment containing the promoter of the 16S rRNA gene, amplified using primers P16S1 and P16S2, was used as a nonspecific competitor.
Microarray data accession number.
All of the microarray data used for this paper have been deposited in the GEO database (http://ncbi.nlm.nih.gov/geo) under accession number GSE20973.
RESULTS
The CodY regulon of strain UAMS-1.
To assess the breadth of the CodY regulon in the S. aureus osteomyelitis isolate UAMS-1, RNA extracted from exponential- and post-exponential-phase cultures in TSB medium of UAMS-1 and its codY mutant, MS1, was subjected to microarray analysis. The concentration of CodY in exponential- and post-exponential-phase cultures is the same (28), but CodY was expected to be most active under conditions of nutrient excess. In fact, when exponential-phase samples were compared, 179 genes were more than 2-fold overexpressed (Table 2) and 24 genes were more than 2-fold underexpressed (Table 3) in the codY mutant, relative to its parent. For comparison, 106 genes were 2-fold overexpressed and 18 genes were 2-fold underexpressed in a codY mutant of strain Newman grown in a defined medium containing the BCAAs (38). Thus, S. aureus CodY, like B. subtilis CodY (31), acts predominantly as a negative regulator of transcription. In post-exponential phase, only 30 of the 109 genes that were differentially regulated in the codY mutant of UAMS-1 were overexpressed.
TABLE 2.
Select genes that are overexpressed in the codY mutant of UAMS-1
| Gene group and annotationa | Gene | Genetic organizationb | Assignmentc | Expression ratio |
||||
|---|---|---|---|---|---|---|---|---|
| Expd |
Pxpe |
|||||||
| codY | agr codY | codY | agr | agr codY | ||||
| Amino acid biosynthesis | ||||||||
| Aspartate family | ||||||||
| SA1360 | thrA | thrA | Aspartate kinase | 54.2 | 37.8 | |||
| SA1362 | hom | SA1361, hom, thrCB, SA1365 | Homoserine dehydrogenase | 45.3 | 36.2 | |||
| SA1363 | thrC | Threonine synthase | 51.4 | 33.5 | ||||
| SA1364 | thrB | Homoserine kinase | 54.6 | 41.4 | ||||
| SA1365 | Hydrolase, haloacid dehalogenase-like family | 5.8 | 4.5 | |||||
| SA1428 | lysC | lysC, asd, dapABD, SA1433, dal, lysA | Aspartokinase, alpha- and beta- subunits | 21.9 | 12.9 | |||
| SA1429 | asd | Aspartate-semialdehyde dehydrogenase | 27 | 21.3 | ||||
| SA1430 | dapA | Dihydrodipicolinate synthase | 28.8 | 20.3 | ||||
| SA1431 | dapB | Dihydrodipicolinate reductase | 28.5 | 19.4 | ||||
| SA1432 | dapD | 2,3,4,5-Tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | 24.9 | 17.1 | ||||
| SA1433 | Peptidase, M20/M25/M40 family | 16.8 | 11.7 | |||||
| SA1434 | Da | Alanine racemase family protein | 12.7 | 10.2 | ||||
| SA1435 | lysA | Diaminopimelate decarboxylase | 7 | 4.3 | ||||
| SA0012 | SA0012 | Homoserine O-acetyltransferase | 6.8 | 5.3 | ||||
| BCAAs | ||||||||
| SA1477 | ilvA1 | ald1, ilvA1, SA1476, SA1475 | Threonine dehydratase, catabolic | 4.4 | ||||
| SA2042 | ilvD | ilvDBHC, leuABCD, ilvA | Dihydroxy-acid dehydratase | 202 | 144.6 | |||
| SA2043 | ilvB | Acetolactate synthase, large subunit, biosynthetic type | 233.2 | 155 | ||||
| SA2044 | ilvN | Acetolactate synthase, small subunit, truncation | 489.9 | 353.2 | ||||
| SA2045 | ilvC | Ketol-acid reductoisomerase | 236.3 | 189.7 | ||||
| SA2046 | leuA | 2-Isopropylmalate synthase | 190.4 | 143.1 | ||||
| SA2047 | leuB | 3-Isopropylmalate dehydrogenase | 145.6 | 123.1 | ||||
| SA2048 | leuC | 3-Isopropylmalate dehydratase, large subunit | 117.6 | 99.1 | ||||
| SA2049 | leuD | 3-Isopropylmalate dehydratase, small subunit | 157.3 | 153.2 | ||||
| SA2050 | ilvA | Threonine dehydratase | 46.8 | 48.8 | ||||
| SA0600 | ilvE | ilvE | BCAA aminotransferase | 6.2 | 4 | |||
| Methionine | ||||||||
| SA0431 | SA0431, SA0430, SA0429, metE, SA0427 | trans-Sulfuration enzyme family protein | 86.2 | 53.7 | ||||
| SA0430 | trans-Sulfuration enzyme family protein | 89.4 | 60.1 | |||||
| SA0429 | 5-Methyltetrahydrofolate-homocysteine methyltransferase, putative | 88.6 | 57 | 2.8 | ||||
| SA0428 | metE | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 48 | 29.4 | 2.4 | 2.7 | ||
| SA0427 | Conserved hypothetical protein | 21.1 | 12.9 | |||||
| SA0870 | SA0870 | LysE/YggA family protein | 7.5 | 6 | ||||
| SA2575 | SA2575, 2574 | Aspartate aminotransferase, class I | 10.5 | 6.8 | ||||
| SA2574 | d-Isomer-specific 2-hydroxyacid dehydrogenase family protein | 7.9 | 4.3 | |||||
| Histidine | ||||||||
| SA2704 | hisZ | hisZGD, SA2701, BHAFI | Conserved hypothetical protein | 17.3 | 11 | |||
| SA2703 | hisG | ATP phosphoribosyltransferase | 7.5 | 5.3 | ||||
| SA2702 | hisD | Histidinol dehydrogenase | 13.8 | 7.8 | ||||
| SA2700 | hisB | Imidazole glycerol phosphate dehydratase | 6.4 | |||||
| SA2699 | hisH | Imidazole glycerol phosphate synthase, glutamine amidotransferase subunit | 4.1 | 2.9 | ||||
| SA2698 | hisA | Phosphoribosylformimino-5-aminoimidazole carboxamide ribotide isomerase | 11.8 | 7 | ||||
| SA2697 | hisF | HisF protein (cyclase) | 8.4 | 5.8 | ||||
| SA2696 | hisI | Phosphoribosyl-ATP pyrophosphatase/phosphoribosyl-AMP cyclohydrolase | 6.9 | 4.8 | ||||
| SA0784 | hisC | hisC | Histidinol-phosphate aminotransferase | 7.3 | 6 | |||
| Serine | ||||||||
| SA1772 | SA1772, serA | Aminotransferase, class V | 67.7 | 39.7 | ||||
| SA1773 | serA | d-3-Phosphoglycerate dehydrogenase | 72.1 | 44.3 | ||||
| SA1774 | SA1776-SA1774 | Hydrolase, haloacid dehalogenase-like family | 9.7 | 8.4 | ||||
| Aromatic amino acids | ||||||||
| SA1403 | trpE | trpEGDCFBA | Anthranilate synthase component I | 2.9 | ||||
| SA1406 | trpC | Indole-3-glycerol phosphate synthase | 34.6 | 27.5 | ||||
| SA1407 | trpF | N-(5-Phosphoribosyl)anthranilate isomerase | 21.8 | 16.9 | ||||
| SA1408 | trpB | Tryptophan synthase, beta subunit | 28.2 | 19.9 | 3 | |||
| SA1409 | trpA | Tryptophan synthase, alpha subunit | 8.3 | 5.6 | ||||
| SA1505 | aroB | aroCBA, SA1503-SA1500 | 3-Dehydroquinate synthase | 3.2 | 2.9 | |||
| SA1504 | aroA | 3-Phosphoshikimate 1-carboxyvinyltransferase | 2.8 | 2.7 | ||||
| SA1787 | SA1787 | Chorismate mutase/phospho-2-dehydro-3-deoxyheptonate aldolase | 5.8 | 4.4 | ||||
| Glutamate | ||||||||
| SA0514 | gltB | gltBD | Glutamate synthase, large subunit | 41.7 | 38 | |||
| SA0515 | gltD | Glutamate synthase, small subunit | 28 | 22.5 | ||||
| Tyrosine | ||||||||
| SA1401 | tyrA | tyrA | Prephenate dehydrogenase | 9.6 | 6.8 | |||
| Purine | ||||||||
| SA1074 | purK | purEKCSQLFMNHD | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | 2.7 | 3.3 | |||
| SA1075 | purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 4.9 | 5 | ||||
| SA1076 | purS | Phosphoribosylformylglycinamidine synthase | 4.5 | 5.9 | ||||
| SA1077 | purQ | Phosphoribosylformylglycinamidine synthase I | 6.7 | 7.3 | ||||
| SA1078 | purL | Phosphoribosylformylglycinamidine synthase II | 4.7 | 5.5 | ||||
| SA1079 | purF | Amidophosphoribosyltransferase | 5.5 | 6.7 | ||||
| SA1080 | purM | Phosphoribosylformylglycinamidine cyclo-ligase | 5.2 | 6.8 | ||||
| SA1081 | purN | Phosphoribosylglycinamide formyltransferase | 5.1 | 6.2 | ||||
| SA0925 | purH | Phosphoribosylamine-glycine ligase | 5.3 | 5.6 | ||||
| SA1083 | purD | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | 5.3 | 5 | ||||
| Transport | ||||||||
| Amino acid and peptide transport | ||||||||
| SA0185 | SA0185, SA0186, SA0187, ggt | Peptide ABC transporter, permease protein | 28.7 | 25.5 | ||||
| SA0186 | Peptide ABC transporter, permease protein | 29.6 | 26.8 | 12.3 | 13.8 | |||
| SA0187 | Hypothetical protein | 29.4 | 23.2 | |||||
| SA0188 | ggt | Gamma-glutamyltranspeptidase | 15.7 | 12.1 | 9.2 | 8 | ||
| SA0991 | oppB | oppBCDF, SA0995 | Oligopeptide ABC transporter, permease protein | 35.9 | 25.9 | |||
| SA0992 | oppC | Oligopeptide ABC transporter, permease protein | 44.3 | 35.2 | ||||
| SA0993 | oppD | Oligopeptide ABC transporter, ATP binding protein | 42.6 | 32.1 | ||||
| SA0994 | oppC | Oligopeptide ABC transporter, ATP binding protein | 5.4 | 5.3 | ||||
| SA0995 | Oligopeptide ABC transporter, oligopeptide binding protein | 52.9 | 48.3 | |||||
| SA2476 | SA2476-SA2471 | Peptide ABC transporter, peptide-binding protein | 3.5 | 2.5 | ||||
| SA0302 | brnQ2 | brnQ2-0303 | BCAA transport system II carrier protein | 8.9 | 5.8 | |||
| SA0171 | brnQ1 | brnQ1 | BCAA transport system II carrier protein | 10.9 | 8.5 | |||
| SA2619 | SA2619 | Amino acid permease | 61.6 | 48.1 | ||||
| SA1963 | SA1963 | Proline permease | 4.8 | 3.1 | ||||
| SA0010 | SA0010, SA0011 | AzlC family protein | 7.2 | 4.8 | ||||
| SA0011 | Conserved hypothetical protein | 13.9 | 11.7 | |||||
| PTS | ||||||||
| SA0228 | SA0228-SA0232 | Transcriptional antiterminator | 2.7 | 1.7 | ||||
| SA0230 | PTS system, sorbitol-specific IIB component | 2.9 | 1.4 | |||||
| Other transporters | ||||||||
| SA0882 | SA0882-SA0884 | ABC transporter, ATP binding protein | 6.8 | 4.8 | ||||
| SA0883 | ABC transporter, permease protein | 7.7 | 5.7 | |||||
| SA0884 | ABC transporter, substrate binding protein | 9.5 | 8 | |||||
| SA0504 | SA0504-SA0506 | ABC transporter, ATP binding protein | 42.8 | 41.1 | ||||
| SA0505 | ABC transporter, permease protein | 49.8 | 48.4 | |||||
| SA0506 | ABC transporter, substrate binding protein | 46.4 | 48.1 | |||||
| SA1018 | SA1018-1019 | Sodium:alanine symporter family protein | 41.9 | 33.3 | ||||
| SA1019 | Conserved hypothetical protein | 8.3 | 7.4 | |||||
| SA1038 | SA1038-SA1040 | Membrane protein | 26 | 20.9 | ||||
| SA1039 | Conserved hypothetical protein | 10.2 | 9.8 | |||||
| SA1040 | ABC transporter, ATP binding protein | 15.9 | 12.4 | |||||
| SA2144 | SA2144-SA2143 | ABC transporter, ATP binding protein | 6.9 | 6.5 | ||||
| SA2314 | SA2314-SA2312 | Sodium/bile acid symporter family protein | 12.7 | 10.8 | ||||
| SA2312 | Conserved hypothetical protein | 4.6 | 3.6 | |||||
| SA2525 | SA2525, SA2526 | ABC transporter, ATP-binding protein | 8 | 7.7 | ||||
| SA1144 | isdF | isdCDEF, srtB, isdG | Iron compound ABC transporter | 2.4 | 2.4 | |||
| SA2483 | SA2483 | Transporter, putative | 9.9 | 5.9 | ||||
| SA0126 | SA0127-SA0125 | Phosphonate ABC transporter, permease protein | 3.0 | 1.9 | ||||
| SA0128 | SA0128 | Phosphonate ABC transporter, phosphonate binding protein | 3.3 | 2.4 | ||||
| SA0267 | SA0267-SA0264 | Hypothetical protein | 6.1 | 4.6 | ||||
| SA0264 | ABC transporter, ATP binding protein | 3.6 | 2.7 | |||||
| SA0303 | SA0303 | Hypothetical protein, similar to sodium-coupled permease | 4.3 | 3.1 | ||||
| SA1963 | Proline permease | 4.8 | 3.1 | |||||
| Krebs cycle and carbon overflow | ||||||||
| SA1123 | pyc | pyc | Pyruvate carboxylase | 5.9 | 4.3 | |||
| SA1449 | sucA | sucAB | 2-Oxoglutarate dehydrogenase, E1 component | 4.6 | 3.4 | |||
| SA1448 | sucB | 2-Oxoglutarate dehydrogenase, E2 component | 3.5 | 2.6 | ||||
| SA0222 | ldh1 | ldh1 | l-Lactate dehydrogenase | 22.9 | 11.7 | |||
| Environmental stress response | ||||||||
| SA0118 | sodA | sodA | Superoxide dismutase | 5.8 | 4.2 | |||
| SA2641 | gpxA2 | gpxA2 | Glutathione peroxidase | 6.7 | 5.4 | |||
| Regulators | ||||||||
| SA2585 | SA2585 | Regulatory protein, putative | 133.2 | 88.4 | ||||
| SA0767 | SA07607, saeRS | 4.8 | 3.2 | |||||
| SA0765 | saeS | Sensor histidine kinase SaeS | 2.4 | |||||
| Virulence factors | ||||||||
| SA2026 | agrA | agrBDCA | Accessory gene regulator protein A | 6.4 | ||||
| SA2022 | hld | hld/RNAIII | Delta-hemolysin | 26 | ||||
| SA0136 | cap5A | capABCDEFGHIJKLMNOP | Capsular polysaccharide biosynthesis | 5.5 | 0.11 | |||
| SA0138 | cap5C | Capsular polysaccharide biosynthesis | 5.2 | 0.21 | ||||
| SA0139 | cap5D | Capsular polysaccharide biosynthesis | 5.7 | 0.16 | ||||
| SA0141 | cap5F | Capsular polysaccharide biosynthesis | 5 | 0.18 | ||||
| SA0142 | cap5G | UDP-N-acetylglucosamine 2-epimerase | 4.1 | 0.23 | ||||
| SA0148 | cap5 M | Galactosyltransferase | 3.9 | 0.27 | ||||
| SA0149 | cap5N | Capsular polysaccharide biosynthesis | 3.7 | |||||
| SA0151 | cap5P | UDP-N-acetylglucosamine 2-epimerase | 2.6 | |||||
| SAR2036 | chp | chp | Chemotaxis-inhibiting protein, MRSA252 | 21.7 | 16.9 | |||
| SA1168 | efb | efb | Fibrinogen binding protein | 8.2 | 8.5 | |||
| SA2694 | geh | geh, SACOL2693 | Glycerol ester hydrolase | 10.3 | 5 | 0.07 | 0.09 | |
| SA1173 | hla | hla | Alpha-hemolysin precursor | 5.5 | 0.26 | |||
| SA2003 | hlb | hlb | Phospholipase C | 3.2 | ||||
| SA2689 | icaA | icaADBC | Intercellular adhesion protein A | 75.6 | 56.8 | |||
| SA2691 | icaB | Intercellular adhesion protein B | 75.4 | 52.4 | ||||
| SA2462 | icaC | Intercellular adhesion protein C (N315) | 15 | 11.7 | ||||
| SA2690 | icaD | Intercellular adhesion protein D | 109.2 | 77.3 | ||||
| SA1170 | katA | katA | Catalase (N315) | 4.3 | 3.1 | |||
| SA0248 | lrgB | lrgAB | lrgB protein | 2.9 | ||||
| SA0860 | nuc | nuc | Thermonuclease precursor | 19.3 | 8.2 | 4.9 | 2.4 | |
| SA1812 | rot | rot | Repressor of toxins | 2.3 | 2.6 | |||
| SA1758 | sak | sak | Staphylokinase precursor (N315) | 3 | ||||
| SA0901 | sspA | sspABC | Serine protease, V8 | 14.6 | ||||
| SA1056 | sspB1 | sspAB1C | Cysteine protease precursor | 10.2 | ||||
| SA1055 | sspC | SspC protein | 7.7 | |||||
| SA1970 | sspB2 | sspB2, SA1971 | Cysteine protease precursor | 5.6 | 3.6 | 5.4 | ||
| SA1971 | Hypothetical protein | 4.8 | 3.4 | 6.7 | 2.6 | |||
| SA1819 | tst-1 | tst-1 | Toxic shock syndrome toxin 1 (N315) | 5.7 | 0.14 | |||
| SA1186 | SA1186-SA1188 | Antibacterial protein (phenol-soluble modulin) | 3.9 | 0.01 | 0.01 | |||
| SA0480 | SA0480 | Hypothetical protein | 40.7 | 36.8 | ||||
| SA1164 | SA1164 | Fibrinogen binding-related protein | 4.7 | 6 | ||||
| SA2418 | SA2418 | IgG binding protein SBI | 3.9 | 2.6 | ||||
| SA0270 | SA0270 | Staphyloxanthin biosynthesis protein, putative | 5.6 | |||||
| SA0390 | SA0390 | Lipase precursor (Mu50, MW2, MSSA476, MRSA252, N315 COL) | 7.3 | 3.3 | ||||
| SA0119 | sasD | sasD | Cell wall surface anchor family protein | 4.4 | 3.3 | |||
Boldface annotations indicate that a region of enrichment was found within the highlighted gene or within the intergenic region upstream of the highlighted gene.
Putative transcriptional unit.
Functional assignment based on the S. aureus COL strain annotation, unless otherwise indicated.
Expression ratio during exponential growth. The mutant strain is indicated in the table. The expression ratio of the mutant strain (codY, agr, or agr codY) relative to the wild type is shown.
Expression ratio during post-exponential growth. The mutant strain is indicated in the table. The expression ratio of the mutant strain (codY, agr, or agr codY) relative to the wild type is shown.
TABLE 3.
Select genes that are underexpressed in the codY mutant of UAMS-1
| Gene group and annotationa | Gene | Genetic organizationb | Assignmentc | Expression ratio |
||||
|---|---|---|---|---|---|---|---|---|
| Expd |
Pxpe |
|||||||
| codY | agr codY | codY | agr | agr codY | ||||
| Sugar utilization and degradation | ||||||||
| SA1042 | SA1042-SA1043 | Hypothetical protein | 3.3 | 2.2 | ||||
| SA1043 | Glycosyltransferase, group 1 family protein | 3.5 | 2.5 | |||||
| Environmental stress response | ||||||||
| SA2057 | rsbU | rsbUVW, rpoF | σB regulator protein | 2.5 | 2.7 | |||
| SA2731 | SA2731 | Cold shock protein, CSD family | 3.1 | 3.3 | ||||
| Regulators | ||||||||
| SA0096 | sarS | sarS | Staphylococcal accessory regulator S | 3 | ||||
| SA0120 | SA0120 | Transcriptional regulator, GntR family | 2.2 | 3.5 | ||||
| SA0245 | lytS | lytSR | Sensor histidine kinase | 2.7 | 4.1 | |||
| SA0307 | pfoR | pfoR | Perfringolysin O regulator protein | 2.4 | ||||
| SA1060 | SA1060 | Transcriptional regulator, MarR family | 2.8 | 2.3 | ||||
| SA2308 | SA2308 | Phosphosugar binding transcriptional regulator, RpiR family | 2.3 | 2.8 | 5.2 | |||
| SA2349 | SA2349 | Transcriptional regulator, TetR family | 3.2 | 2.4 | 3.6 | |||
| Virulence factors | ||||||||
| SA2294 | SA2294 | Conserved hypothetical protein | 2.4 | 2.6 | ||||
| SA2295 | SA2295 | Staphyloxanthin biosynthesis protein | 3.3 | 4.4 | ||||
| SA2584 | isaA | isaA | Immunodominant antigen A | 3.2 | ||||
| SA0901 | SA0901-SA0906 | Pathogenicity island protein | 3.7 | 3 | ||||
| SA0902 | Pathogenicity island protein | 2.2 | ||||||
| SA0903 | Pathogenicity island protein | 2 | ||||||
| SA1758 | sak | sak | Staphylokinase precursor (N315) | 2.2 | ||||
| SAR2040 | SAR2041-SAR2040 | Autolysin (MRSA252) | 4.1 | |||||
| SAV1954 | SAV1955-SAV1950 | φ PVL ORF 18-19-like protein (Mu50) | 4.4 | |||||
| SA2511 | fnbA | fnbA | Fibronectin binding protein A | 2.4 | ||||
| SA0023 | orfX | orfX | OrfX (N315) | 2.8 | 2.5 | |||
Boldface indicates that a region of enrichment was found within the highlighted gene or within the intergenic region upstream of the highlighted gene.
Putative transcriptional unit.
Functional assignment based on the S. aureus COL strain, unless otherwise indicated.
Expression ratio during exponential growth. The mutant strain is indicated in the table. The expression ratio of the wild type relative to the mutant strain (codY, agr, or agr codY) is shown.
Expression ratio during post-exponential growth. The mutant strain is indicated in the table. The fold expression of the wild type relative to the mutant strain (codY, agr, or agr codY) is shown.
The CodY regulon of S. aureus has considerable overlap with the CodY regulons of B. subtilis (31), L. lactis (15), L. monocytogenes (4), S. pneumoniae (19), and C. difficile (S. S. Dineen, S. McBride, and A. L. Sonenshein, unpublished data), especially with respect to metabolic genes. For instance, in S. aureus, as in most of the other species, the genes encoding the enzymes of the BCAA biosynthetic operon (ilvDBNC leuABCD ilvA) and the biosynthetic pathways for certain other amino acids (cysteine, glutamate, histidine, serine, tryptophan, tyrosine, and the aspartate family amino acids [aspartate, lysine, threonine and methionine]) were overexpressed in the codY mutants of both UAMS-1 (Table 2) and Newman (38). Similarly, many genes encoding amino acid or peptide transporters, various permeases, and several peptidases were differentially regulated in the codY mutant of UAMS-1 compared to the parental strain (Table 2).
The members of the CodY regulon that are unique to S. aureus are primarily genes whose products have been linked to pathogenesis (Tables 2 and 3) (38). Our previous work identified the hla (alpha-toxin), ica (PIA synthesis), and agr loci as targets of negative regulation by CodY (28). A total of 50 genes associated with virulence were differentially expressed in the codY mutant of UAMS-1 during either the exponential or post-exponential growth phase. During exponential phase, 28 characterized or putative virulence genes, organized in 20 apparent transcription units, were overexpressed in the codY mutant of UAMS-1 (Table 2). These genes included agrA, hld, capACDFG, chp, efb, geh, hla, hlb, icaADBC, katA, geh, nuc, rot, sspB2, SA1971, and sodA, as well as other genes located within a pathogenicity island or that share similarity with characterized adhesins or phenol-soluble modulins. The genes capA- to -N, geh, katA, and hld, and a phenol-soluble modulin were also differentially expressed in the codY mutant of strain Newman (38). Semiquantitative RT-PCR analysis confirmed the transcription profiles of selected genes for UAMS-1 and its codY mutant and extended the findings to the clinical isolate SA564 and its codY mutant (see Fig. S1 in the supplemental material).
The role of agr in the CodY regulon of UAMS-1.
The effect of a codY mutation on the agr locus (40-fold) is much more severe in strains UAMS-1 and SA564 (Table 2 and reference 28) than in strain Newman (2- to 3-fold) (38). Since the basal level of agr expression during exponential growth seems to be higher in Newman than in UAMS-1 or SA564, the derepressing effect of a codY mutation may be less pronounced in Newman. It is not known what causes the strain-to-strain variability in the activity of the agr locus during exponential growth.
As the agr locus is a known regulator of virulence genes (6, 9, 14, 34), we sought to determine the extent to which the overexpression of virulence genes in the codY mutant was caused by increased transcription of the agr locus by comparing transcript levels in the isogenic strains UAMS-1, MS1 (ΔcodY::ermC), CM18 (Δagr::tetM), and CM19 (ΔcodY::ermC Δagr::tetM). (We made the assumption that an agr mutation has no effect on CodY function, since the level of the codY transcript was not affected by an agr mutation in strain UAMS-1 [data not shown].)
An agr mutation by itself resulted in no differences in the abundance of specific transcripts in exponential-phase cells, as previously reported (8). Moreover, comparison of transcript levels in the codY mutant and the agr codY double mutant showed that the agr locus has little impact on the expression of metabolic genes that are CodY regulated (Tables 2 and 3). Similarly, a group of virulence factors that includes katA, chp, efb, geh, icaADBC, nuc, rot, sspB2, SA0480 (a pathogenicity island gene encoding a hypothetical protein), and SA1164 (a gene encoding a fibrinogen binding-related protein) was altered in expression to essentially the same extent in both the codY mutant and the codY agr double mutant strains (relative to the wild-type parent) (Table 2). In strain Newman, the virulence-related genes katA and sodM and some genes of the cap locus were affected by a codY mutation in an agr mutant strain (38). We conclude that CodY regulates transcription of the majority of its regulon in an agr-independent manner.
However, for other members of the CodY regulon, including the virulence-associated genes capACDFG, hla, hlb, SA1186 (phenol-soluble modulin), and SA0270 (putative staphyloxanthin biosynthesis protein), the effect of a codY mutation was suppressed by an agr mutation (Table 2). That is, in the agr single mutant and in the agr codY double mutant, the transcript levels for these genes fell below (or were at) the limit of detection. These results suggest that CodY-dependent regulation of these genes is mediated primarily through the effect of CodY on the agr locus, but we cannot rule out the possibility that in the absence of positive regulation by the Agr system the level of transcription of these genes is so low that any independent effect of a codY mutation would be undetectable.
Role of sarA in codY-mediated overexpression of the agr locus and the ica locus.
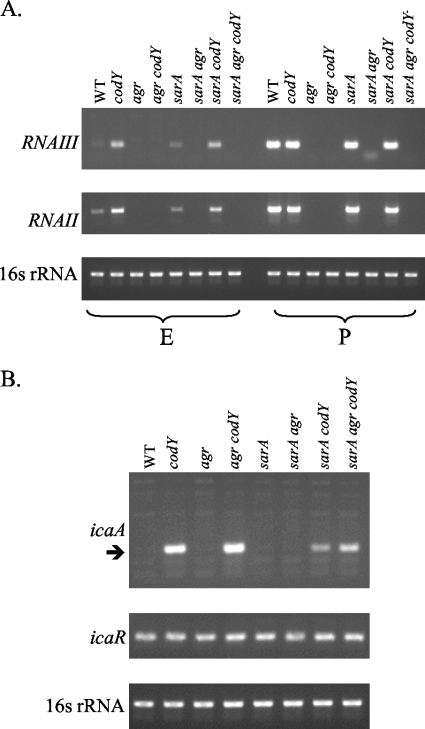
Since SarA is a known activator of both the agr (10, 18) and ica loci (21, 48), we asked if CodY was acting through sarA to cause overexpression of some virulence genes. Semiquantitative RT-PCR analysis showed that RNAII and RNAIII were overexpressed in a codY mutant independently of the status of the sarA gene (Fig. 1A), but sarA may be needed for maximal expression of these genes. Similar analysis (Fig. 1B) showed that icaA was overexpressed in codY, agr codY, and agr sarA codY mutants, confirming and extending the microarray results and demonstrating that repression of the icaA promoter by CodY is independent of both SarA and RNAIII. Additionally, Fig. 1B shows that the abundance of icaR mRNA was unaffected by any of the mutations tested, a result consistent with our previous finding that the effect of a codY mutation on expression of the ica locus is independent of icaR transcript levels (28). During post-exponential growth, the icaA transcript levels in all strains were at similarly low, undetectable levels (data not shown).
FIG. 1.
Effects of sarA and agr mutations on CodY-regulated genes. (A) Transcripts corresponding to RNAIII, RNAII, and 16S rRNA were analyzed by RT-PCR from samples extracted during exponential (E) and post-exponential (P) phases of the wild-type (WT) and mutant strains of UAMS-1, as indicated. (B) Transcripts corresponding to icaA, icaR, and 16S rRNA were analyzed by RT-PCR from samples extracted from exponential-phase cells of the WT and mutant strains of UAMS-1, as indicated.
Genome-wide identification of CodY binding sites in S. aureus.
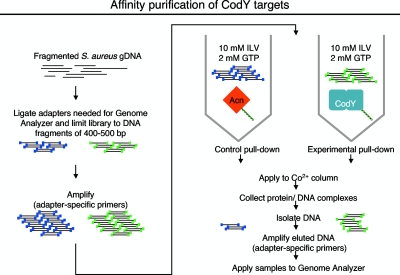
To determine the direct targets of CodY, we designed an affinity purification (pull-down) experiment using purified His6-tagged S. aureus CodY to identify all regions of the S. aureus genome that interact with CodY in vitro. gDNA from the S. aureus strain NCTC 8325 was used in the pull-down experiment because the genome of this strain has been fully sequenced, a prerequisite for effective resequencing using the Illumina Genome Analyzer II (GA II). The protocol for this experiment is outlined in Fig. 2. Binding reactions were carried out with 200 nM His6-tagged S. aureus CodY, a fragmented, adapter-ligated gDNA library, and a buffer containing the effectors of CodY activity (10 mM ILV and 2 mM GTP). A control pull-down experiment with 200 nM purified His-tagged B. subtilis aconitase (Acn) was carried out in parallel. The DNA-protein complexes were isolated from solution using a Co2+ resin, amplified, and subjected to high-throughput paired-end sequencing. That is, the sequences of the first and last ∼40 bp of each DNA fragment were determined directly. Then, for each fragment, the sequence between the fragment ends was filled in based on the published sequence of the NCTC 8325 genome. In this way, a coverage map of the entire genome in each purification experiment was created. The coverage map represents the number of times a specific base pair on the chromosome is represented in the sequencing of each DNA sample. An enrichment value was then generated for every position on the chromosome by comparing the coverage map of the CodY pull-down assay to the coverage map of the Acn pull-down assay. Enriched regions were arbitrarily defined as regions longer than 100 bp that showed 5-fold or higher enrichment throughout their length in the CodY pull-down assay relative to the Acn pull-down assay. From this collection of regions, a subset in which the position (bp) of maximum enrichment in the CodY sample was 20-fold or higher was selected for further analysis. Table S4 in the supplemental material lists all 231 positions of high enrichment that were identified, the maximum enrichment value found in that region, the local gene structure near the position of enrichment in NCTC 8325, and the corresponding annotation and gene function. Fifty-eight percent (134/231) of the positions of maximal enrichment mapped to intergenic regions. In the NCTC 8325 genome, as well as many other S. aureus genomes, only 15% of the nucleotides map to intergenic regions (http://cmr.jcvi.org/tigr-scripts/CMR/GenomePage.cgi?org=ntsa05). Thus, CodY binding sites show a preference for intergenic regions compared to coding sequences.
FIG. 2.
Protocol for genome-wide identification of CodY binding sites. Briefly, gDNA was sheared by sonication. Adapters were ligated to the sheared DNA and then gel purified to obtain a DNA library containing fragments of 400 to 500 bp in length. The DNA library was then subjected to PCR amplification using adapter-specific primers to generate enough DNA for the pull-down experiment. A 200 nM concentration of His-tagged CodY or 200 nM His-tagged Acn was incubated with 50 μg sheared, adapter-ligated DNA and the effector molecules (2 mM GTP and 10 mM ILV). Protein-DNA complexes were purified with a Co2+ resin. Following elution of the protein from the resin, the DNA was isolated, PCR amplified again, and subjected to analysis with a Illumina Genome Analyzer II.
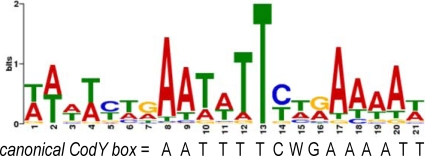
Using the MEME suite (http://meme.sdsc.edu/meme4_1_1/cgi-bin/meme.cgi), we found a 21-bp conserved motif (referred to here as the de novo motif) (Fig. 3) in 223 of the 231 regions of high enrichment. A 15-bp conserved sequence (the canonical CodY box, AATTTTCWGAAAATT) was previously described based on analysis of L. lactis and B. subtilis CodY-regulated genes (3, 12, 15). The canonical CodY box is contained almost completely within the de novo motif (Fig. 3). The enriched regions were also searched for the previously described CodY binding box, allowing up to three mismatches. The locations of the first nucleotide of each 21-bp de novo (i.e., newly identified) motif and of each 15-bp CodY box, relative to the position of maximal enrichment, are shown in Table 4 and in the supplemental information. Ninety-seven CodY boxes were found within the enriched regions and usually overlapped with the newly identified motifs.
FIG. 3.
S. aureus CodY binding motif. A search for common motifs in the 231 regions enriched in the CodY pull-down experiment was conducted using the MEME suite and the parameters described in Materials and Methods. The motif shown was found in 223 of the 231 enriched regions, with a P value ≤0.003. The canonical CodY box previously identified (12, 15) is shown below the S. aureus de novo binding motif for reference.
TABLE 4.
Regions of the S. aureus genome selected by binding to CodY in vitro
| Function of group and canonical box(es)a | De novo motif b | Enrichment factorc | Genome positiond | Putative transcriptional unite | Gene descriptionf | Expression ratiog | |||
|---|---|---|---|---|---|---|---|---|---|
| Amino acid metabolism | |||||||||
| −40 | 3 | 48, −, TTTTCTGATTTTTTAGAAAGG | 27 | 1263288 | thrA(←)IG(→)SA1361, hom, thrCB, SA1365 | Aspartate kinase, homoserine dehydrogenase | 54.2, 45.3 | ||
| −6 | 3 | 4, +, TTTTCAGGTTATTCCAAAAAA | 31 | 1336491 | IGlysC, IGasd, dapABD, SA1433, dal, lysA | Aspartokinase | 21.9 | ||
| 3 | 2 | 4, +, TTGTTAAAATATTCTAAAAAT | 22 | 1338048 | IGlysC, IGasd, dapABD, SA1433, dal, lysA | Aspartate-semialdehyde dehydrogenase | 27 | ||
| 28 | 3 | −29, −, ATATCAGAATTTTCTGATTAC | 22 | 15763 | IGSA0012 | Homoserine O-acetyltransferase | 6.8 | ||
| 52 | 2 | −13 | 2 | −53, −, TAGTTAGAATTGTCTAAAAAT | 89 | 2111452 | IGilvDBHC, leuABCD, ilvA | Dihydroxy-acid dehydratase | 202 |
| −3, +, ATTTCAGAATTTTCGAAACAC | 20 | 2115627 | IGilvDBHC, leuABCD, ilvA | Ketol-acid reductoisomerase | 236 | ||||
| 21 | 2119424 | IGilvDBHC, leuABCD, ilvA | 3-Isopropylmalate dehydratase | 118 | |||||
| 38 | 1 | −31, +, AATTTTGAATTTTTAGAAAAT | 20 | 541652 | IGilvE | BCAA aminotransferase | 6.2 | ||
| −61 | 2 | 60, −, AAATCAAAATTTTCTGAATAA | 49 | 356777 | IGSA0431, SA0430, SA0429, metE, SA0427 | trans-Sulfuration enzyme family protein | 86.2 | ||
| 14 | 2 | −7, +, ATGTCTAAATTTTCTAATAAT | 26 | 355320 | IGSA0431, SA0430, SA0429, metE, SA0427 | trans-Sulfuration enzyme family protein | |||
| −51 | 3 | 58, +, AACATAGAATATTCTGAAAAA | 72 | 806009 | IGSA0870 | LysE/YggA family protein | 7.5 | ||
| −8 | 3 | 16, −, TAATCTGAAAATTCCTAATAT | 32 | 2650538 | IGSA2575, SA2574 | Aspartate aminotransferase, class I | 10.5 | ||
| −8 | 2 | 7, −, TACACCGAATTTTCTGAAAAA | 51 | 2789232 | IGhisZGD, SA2701, BHAFI | Conserved hypothetical protein | 17.3 | ||
| −2 | 3 | 7 | 3 | 1, −, TTAACTGAAATTTCAAAAAAC | 26 | 718943 | IGhisC | Histidinol phosphate aminotransferase | 7.3 |
| −10 | 0 | 9, −, TTTAGCAAATTTTCTGAAAAT | 25 | 1736733 | IGSA1772, serA | Aminotransferase | 67.7 | ||
| 24 | 1738502 | IGSA1772, serA | d-3-Phosphoglycerate dehydrogenase | 72.1 | |||||
| −7 | 3 | 6, −, ATATCTGAATTTTCAGAAATG | 31 | 1740826 | SA1776-SA1774 | Hydrolase haloacid dehalogenase-like family | 9.7 | ||
| −3 | 3 | 6 | 3 | 2, −, TTAACCGAATATTCGAAATAT | 29 | 1310343 | IGtrpEGDCFBA | Anthranilate synthase component I | 2.9 |
| 17, +, AAGTGTGAAGATTCAGAATAA | 28 | 1313511 | IGtrpEGDCFBA | Indole-3-glycerol phosphate synthase | 34.6 | ||||
| −22 | 3 | 29, +, ATGTCAGAATTATCTTAATAT | 78 | 1441733 | IGaroCBA,SA1503-SA1500 | 3-Dehydroquinate synthase | 3.2 | ||
| −113 | 3 | 157, +, GTTAAAAAATATTCTAACAAT | 21 | 1759183 | IGSA1787 | Chorismate mutase/phospho-2-dehydro-3-deoxyheptonate aldolase | 5.8 | ||
| 4, +, ATATCAGAATATTCTAAAAAA | 25 | 433302 | IGgltAB | Glutamate synthase, large subunit | 41.7 | ||||
| 0 | 2 | 8, +, TATTCTGAAAATTTCGAAATA | 34 | 1308768 | IGtyrA | Prephenate dehydrogenase | 9.6 | ||
| 5, +, TATTCTGAAATATGTGAATAA | 42 | 982377 | purEKCSQLFMNHD | Phosphoribosylformylglycinamidine synthase I | 6.7 | ||||
| −6 | 3 | 5, −, AATTCTTAATTATCAGAATAT | 59 | 2691012 | IGSA2620 (SA2397 in N315) | Similar to pyridoxal phosphate-dependent aminotransferase | 50.1 | ||
| Transporters | |||||||||
| 240, +, TTATCGGATAGTTCTGGAAAT | 26 | 2395061 | IGSA2322 | Peptidase | 9.9 | ||||
| 14 | 3 | −7, +, AATTCTGAAATTTCAGAAATT | 28 | 182228 | IGSA0185, SA0186, SA0187, ggt | Peptide ABC transporter, permease protein | 28.7 | ||
| −99 | 2 | 97, +, ATTTCAGAAAATTTTGAAGGG | 54 | 894043 | IGoppBCDF, SA0995 | Oligopeptide ABC transporter, permease protein | 35.9 | ||
| −28 | 2 | −97, −, TTGTCTAAAAATTCTGAAAAA | 35 | 2687792 | IGSA2619 | Amino acid permease | 61.6 | ||
| 7 | 3 | 0, +, TTTTCCTACTTTTCTAAACAT | 31 | 1991788 | IGSA1963 | Proline permease | 4.8 | ||
| 3, +, AAAATTGAAAATTCTCAAAAA | 22 | 14546 | IGSA0010, SA0011 | AzlC family protein | 7.2 | ||||
| 0 | 3 | 8, +, TTAACTGAATATTTCGGAAAA | 45 | 233878 | IGSA028-SA0232 | Transcriptional antiterminator, BglG family | 2.7 | ||
| 72, +, TAATTTAAAAAATCTGAAAAA | 29 | 298820 | IGbrnQ2, SACOL0203 | BCAA transport system II carrier protein | 8.9 | ||||
| −10 | 2 | 9, +, TTTTTCGAATTTTCCGAAATC | 30 | 814085 | IGSA0882-SA0884 | ABC transporter, ATP binding protein | 6.8 | ||
| −10 | 3 | 9, −, TATTTTAAATTTTCTAACAAA | 27 | 812857 | |||||
| 1 | 2 | −2, −, TTTGCCGAATTTTCTAAAAAA | 34 | 424606 | IGSA0504-SA0506 | ABC transporter, ATP binding protein | 42.8 | ||
| −1 | 1 | 0, −, TAATCTTAATTTTCAGAAAAG | 43 | 2385954 | IGSA2314-SA2312 | Sodium/bile acid symporter family protein | 12.7 | ||
| 5, +, AAATCTTAAGATTCAGATTAT | 35 | 2598042 | IGSA2525, SA2626 | ABC transporter, ATP binding protein | 8 | ||||
| Carbon flow | |||||||||
| 10, +, TTATCTAATTAGTTAGAAAAT | 39 | 1028771 | IGpyc | Pyruvate carboxylase | 5.9 | ||||
| −6, +, GTTTGCGAATATTCTGATAAG | 35 | 1358559 | sucAB | 2-Oxoglutarate dehydrogenase, E1 component | 4.6 | ||||
| Virulence | |||||||||
| 19, +, AATTTCGAATATTTAAATTTT | 39 | 99390 | IGsodA | Superoxide dismutase | 5.8 | ||||
| 25, +, ATCACCAAAATTTCTGATAGT | 21 | 2094935 | agrBDCA | Accessory gene regulator A | 6.4 | ||||
| 5, −, AATTGCTAATTTTCTAATTTT | 26 | 121388 | capABCDEFGHIJKLMNOP | Capsular polysaccharide biosynthesis | 5.7 | ||||
| 40, +, TTTAAGGATTTATCTGAATGT | 28 | 130063 | capABCDEFGHIJKLMNOP | Capsular polysaccharide biosynthesis | 3.9§ | ||||
| −55 | 3 | 54, −, TACTCAAAATTTTCTTATAAT | 29 | 1078012 | IGhla | Alpha-hemolysin precursor | 5.5 | ||
| 6 | 3 | 9, −, AAAACTTAAAATTTCGAATAA | 32 | 2029853 | IGhlb | Phospholipase C | 3.2 | ||
| −19 | 2 | 26, +, AAAAGCGAATTTTCTGAATAA | 44 | 2776864 | icaADBC | Intercellular adhesion protein B | 75.4 | ||
| −51, +, AAATTTGAAATTTTAGTTAAA | 32 | 1070247 | IGSA1164 | Fibrinogen binding-related protein | 4.7 | ||||
| 8, +, TACGCTGAATGTTCTGAATTA | 27 | 2487229 | IGSA2418 | IgG binding protein SBI | 3.9 | ||||
| Miscellaneous | |||||||||
| −6, +, TTTTTAGAATGTTCTGATAAA | 44 | 2713447 | IGgpxA2 | Glutathione peroxidase | 6.7 | ||||
| 2 | 3 | 5, +, AATTCTGAATTTTTAGATAAA | 35 | 2663219 | IGSA2585 | Regulatory protein, putative | 133.2 | ||
| −25 | 2 | 23, +, TTTACCGAAAATTGAAGAAAT | 20 | 51514 | SA0076 | Hypothetical protein | 4.2 | ||
| −22 | 3 | 20, +, ATATCTGAATATTTTGTTAAA | 30 | 92377 | IGSA0111 | Acetoin reductase | 12.8 | ||
| 26 | 3 | −27, −, ATCAATGAATATTCTGACTAT | 21 | 939605 | IGSA1033 | Hypothetical protein | 14.3 | ||
| −7 | 1 | 14, +, TACACTGACTTTTCCGAAAAT | 31 | 1724742 | IGSA1759 | Universal stress protein family | 8.4 | ||
| 14, −, TAAAATGAAAAATCAAAAAAT | 27 | 2678850 | IGSA2608 | Hypothetical protein | 4.5 | ||||
| −1, −, AAATTAGATTATTTAAAATTA | 48 | 946756 | IGSA1042 | Hypothetical protein | 0.3 | ||||
The first number shown indicates the position of the first nucleotide in the canonical CodY box (≤3 mismatches) relative to the position of maximum enrichment and is followed by a boldface number that indicates the number of mismatches relative to the consensus sequence AATTTTCWGAAAATT. The negative or positive value associated with the number indicates whether the first nucleotide of the box was found upstream (−) or downstream (+) of the position of maximum enrichment. When multiple boxes were found, they appear in consecutive columns.
The number shown indicates the position of the first nucleotide in the de novo motif closest to the position of maximum enrichment. A negative or positive value for this first nucleotide in the motif indicates whether the motif was found upstream (−) or downstream (+) of the position of maximum enrichment, respectively. The plus (+) or minus (−) value following the position of the motif indicates on which strand the motif is found, relative to the genome. The sequence corresponding to the motif is shown.
The enrichment factor is for the position of maximum enrichment in the CodY pull-down sample (relative to the Acn pull-down sample) and corresponds to the identified motif(s) or the canonical box(es) indicated in the row.
The position in the NCTC 8325 genome of maximum enrichment in the CodY pull-down assay.
The putative transcriptional unit shown is based on the COL genome; the underlined regions correspond to the locations of the positions of maximum enrichment. The boldface text indicates the gene closest to the position of maximum enrichment whose overexpression ratio is displayed in the right-most column. IG, intergenic region.
The annotated function of the gene shown in boldface nearest the position of maximum enrichment is shown. When multiple gene names are shown in boldface, the function occurs in the order in which the genes occur within the transcriptional unit.
Overexpression during exponential growth in the codY mutant (relative to wild type) of the gene nearest to the position of maximum enrichment as identified in the CodY pull-down assay. When multiple expression ratios are shown, they refer to multiple genes and are listed in the order in which the underlined genes appear in the transcriptional unit. §, the overexpression value for the codY mutant during post-exponential growth (relative to wild type) of the gene nearest the position of maximum enrichment.
We also asked whether the enriched positions were near genes whose expression is affected by a codY mutation. Of the 231 enriched positions, 63 fell within or upstream of genes that were overexpressed in the codY mutant of UAMS-1, and 9 were associated with genes that were underexpressed in the codY mutant. The 59 enriched positions that corresponded to genes regulated by CodY during the exponential growth phase mapped to a total of 49 transcriptional units that are strong candidates for direct regulation by CodY (Table 4). Ninety-six percent (56/59) of the enriched regions that are associated with genes regulated by CodY during exponential growth have a newly identified motif. Sixty-two percent of these regions of enrichment have at least one CodY box, which is always in close proximity to, if not overlapping the location of, a newly identified motif. Interestingly, the de novo motif is not symmetrical and is therefore specific to one strand, whereas the CodY box is palindromic.
Based on this analysis, many genes that are strong candidates for direct repression by CodY are involved in amino acid biosynthesis or macromolecular transport (Table 4). In addition, nine transcription units associated with virulence are likely to be directly regulated by CodY. The promoter regions of hlb, hla, SA1164 (encoding a fibrinogen binding-related protein), and SA2419 (encoding IgG-binding protein SBI) were bound by CodY in vitro and repressed by codY in vivo. In the case of hla, which encodes alpha-toxin, the site of maximum enrichment in the CodY pull-down assay lies about 700 bp upstream of the transcriptional start site; this site corresponds to the location of a CodY box (with three mismatches) and a de novo motif. The hlb gene, encoding beta-toxin, has a CodY box (with three mismatches) located 319 bp upstream of the translational start site at a position that corresponds to a determined site of maximum enrichment (Table 4). Interestingly, the hla gene is subject to regulation by the Agr system; the impact of a codY mutation was lost in an agr mutant. The fact that hla appears to be a direct target of CodY binding suggests that hla is subject to two forms of regulation, positive regulation mediated by RNAIII and negative regulation by CodY.
One position of maximum enrichment corresponds to a gene that was underexpressed in a codY mutant during exponential growth. This gene, SA1042, encodes a hypothetical protein of unknown function with homologs present only in other S. aureus strains. SA1043, which encodes a glycosyltransferase group I family protein, is immediately downstream of SA1042 and is probably within the same transcription unit. Both SA1042 and SA1043 were underexpressed in the codY mutant during exponential growth. This operon may be the only example in S. aureus of direct positive regulation by CodY during the exponential growth phase. Surprisingly, the region of enrichment associated with SA1042 does not contain a canonical CodY box (with fewer than five mismatches), but it does contain a newly identified motif.
Nine positions of enrichment are associated with genes underexpressed in the codY mutant during post-exponential growth and include positions near the following genes: SA1042, purQ, SA0788, SA2309, SA1883, SA1664, SA0088, SA0662, SA1042, and SA1883. Three enriched regions are associated with genes overexpressed in the codY mutant during post-exponential growth and include positions near capK, SA2491 (a conserved hypothetical protein), and sspB1 (a cysteine protease precursor). The fact that the expression of some genes that appear to be direct targets of CodY is only affected by a codY mutation in post-exponential-phase cells, i.e., in cells in which CodY is expected to be inactive even in the wild-type strain, has two implications. First, such genes are probably not affected by a codY mutation during exponential growth because of overriding regulation by other factors. Second, CodY must retain some activity in wild-type post-exponential-phase cells.
Additionally, a considerable number of enriched regions were identified within or near genes whose transcript abundance is not affected by a codY mutation in UAMS-1. This list includes sasG, several surface proteins (including isaB, clfB, and ebh), and numerous other genes (for a complete list, see Table S4 in the supplemental material). The failure to observe in vivo CodY-mediated regulation near in vitro CodY targets can be due to several factors, including the activities of other regulators at a given locus and the genetic variation in S. aureus isolates. For instance, Pohl et al. recently reported that a codY mutation in strain Newman causes underexpression of the sasG gene during exponential growth (38). Moreover, the CodY pull-down experiment identified a position of maximum enrichment within the sasG gene of NCTC 8325. Because the UAMS-1 genome lacks the region (RD5) that contains the sasG gene (7), the sasG gene did not show up among those affected by a codY mutation in that isolate.
CodY-dependent regulation of the agr locus.
Although a codY null mutation clearly causes overexpression of the agr locus in strains UAMS-1, SA564 (28), and Newman (38), gel mobility shift analysis showed only a low-affinity interaction of CodY with the P2 and P3 promoter regions of the agr locus, which drive expression of all agr genes (data not shown), and the CodY pull-down experiment did not show any significant enrichment of the P2-P3 region. The CodY pull-down assay, however, did show binding of CodY to the agrC gene (Table 3). When the agr locus was first described, a weak promoter (termed P1) was detected in this region through a gene fusion with a promoterless lacZ gene (36). Thus, CodY may directly regulate the P1 promoter of the agr locus or block elongation of transcripts from the major P2 promoter upstream of agrB. Future work will be directed toward understanding how the agr locus is regulated by CodY.
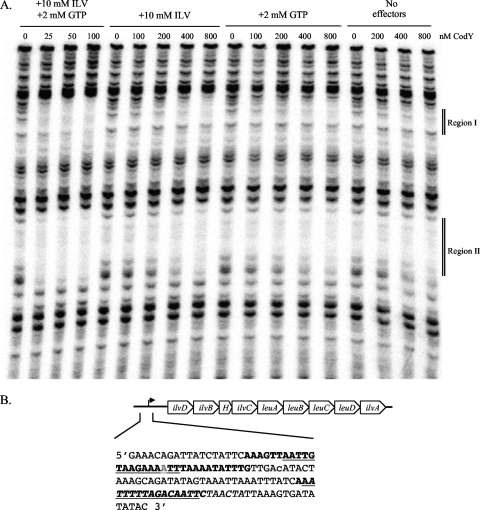
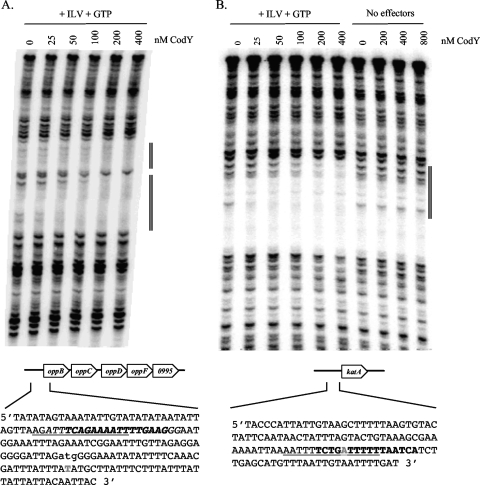
CodY binding sites in the ilvD promoter region.
Because the ilvD promoter has two CodY boxes (11, 38), was overexpressed in codY mutants of strains Newman (38), UAMS-1, and SA564, and was enriched in the pull-down assay, it is very likely to be a direct CodY target. To confirm such an interaction, we used DNase I footprinting analysis to locate the CodY binding sites(s) within the ilvD promoter region. As shown in Fig. 4, we found two high-affinity binding sites that correlated well with the positions of the CodY boxes. Moreover, the binding sites defined by footprinting analysis overlapped with the region enriched in the pull-down experiment (Fig. 4B). Primer extension analysis was used to map the 5′ end of the ilvD mRNA (Fig. 4B and data not shown). The apparent transcriptional start site of the ilvD promoter fell between the two sites of CodY binding. Given that the ilv operon is strongly overexpressed in a codY mutant, the sequence-specific binding of CodY leads us to conclude that ilvD is directly repressed by CodY.
FIG. 4.
Analysis of the ilvD promoter region. (A) DNase I footprinting analysis of CodY binding to the ilvD promoter in the presence and absence of 10 mM ILV (isoleucine, leucine, and valine) or 2 mM GTP or both. The concentration of CodY used in each reaction mixture is listed above each lane. (B) Annotation of the ilvD promoter sequence. Nucleotides in bold type represent DNA that was protected by CodY in the footprinting analysis. Predicted CodY binding sites are indicated (the canonical CodY box, AATTTTCNGAAAATT, is underlined, and the de novo motif is indicated in italics). The nucleotide in gray represents the position of maximal enrichment (89-fold) in the ilvD promoter, as determined in the CodY pull-down experiment. The apparent transcriptional start site mapped by primer extension lies 336 nucleotides upstream of the ilvD start codon and is indicated as a lowercase letter.
S. aureus CodY is activated synergistically by the BCAAs and GTP.
CodY proteins from B. subtilis (17, 43), C. difficile (13), and L. monocytogenes (4) are activated as DNA binding proteins by the BCAAs and GTP. The CodY proteins of L. lactis and S. pneumoniae, however, are only activated by the BCAAs (19, 37). To test the response of S. aureus CodY to these metabolites, we measured binding to the ilvD promoter in the absence of effectors or in the presence of 2 mM GTP or 10 mM ILV or both. When GTP and BCAAs were both present, CodY bound to the ilvD promoter with high affinity (50% protection was seen at 25 to 50 nM CodY). When either the BCAAs or GTP was absent, the affinity was greatly reduced but still higher than that of CodY alone (compare protection levels seen at 800 nM CodY with and without effectors).
CodY binding sites in the oppB and katA promoters.
Footprinting analysis was also used to locate the sites of CodY interaction with the promoter regions of oppB and katA. CodY bound to the oppB promoter at a position that overlaps with both a CodY box and the sequence enriched in the pull-down experiment (Fig. 5A). Similarly, CodY interacted with the katA promoter in a region that includes a CodY box (with three mismatches) and overlaps with the region enriched in the pull-down experiment (Fig. 5B). Binding of CodY was enhanced in the presence of the BCAAs and GTP (Fig. 5B). The footprinting experiment provides strong evidence that the katA promoter region is a direct target of CodY even though the maximum enrichment in the pull-down experiment was only 9-fold. Moreover, expression of the katA gene was derepressed by a codY mutation in both agr wild-type and mutant backgrounds.
FIG. 5.
Analysis of the katA and oppB promoter regions. (A) DNase I footprinting analysis of CodY binding to the oppB promoter in the presence of 10 mM ILV and 2 mM GTP. (B) Footprinting analysis of CodY binding to the katA promoter in the presence or absence of 10 mM ILV and 2 mM GTP. The concentration of CodY used in each reaction mixture is listed above each lane. The corresponding transcriptional unit and promoter sequence are shown below each footprint. In each case, predicted CodY binding sites are indicated (the canonical CodY box, AATTTTCNGAAAATT, is underlined, and the de novo motif is indicated in italics), the sequence corresponding to the region of protection by CodY in the footprinting analysis is shown in bold, and the position of maximum enrichment (54-fold for oppB and 9-fold for katA) defined by the CodY pull-down experiment is indicated by the gray nucleotide.
The cap locus.
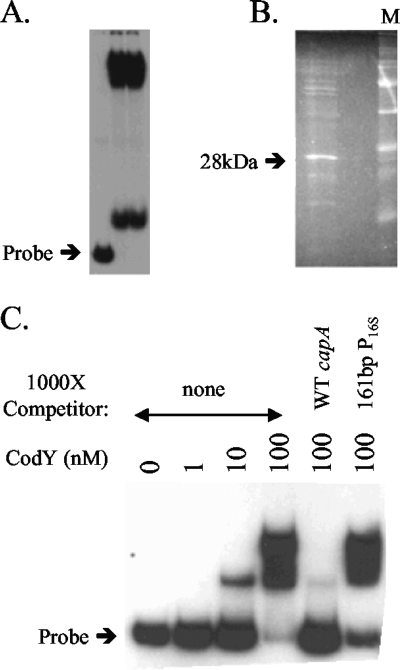
Pohl et al. (38) reported that the cap genes of strain Newman, which are responsible for capsule biosynthesis (25, 35), are regulated by CodY in both agr-positive and agr-negative strains, indicating that in strain Newman CodY regulates the cap locus through an agr-independent mechanism. The capA promoter has a CodY box that overlaps with the −10 region of the promoter (35, 38), reinforcing the likelihood that CodY directly regulates this operon.
Strong evidence that CodY directly represses the capA promoter of strain Newman came from experiments designed to identify in an unbiased way proteins that bind to the promoter. Protein extracts were partially purified by differential ammonium sulfate precipitation and then fractionated by gel filtration chromatography. Each fraction of the elution was tested for binding to the capA promoter by using a gel mobility shift assay. Fractions that caused a change in mobility of a capA promoter probe were pooled and used in a large-scale gel shift experiment. Two large bands corresponding to the shifted probe (Fig. 6A) were excised and subjected to SDS-PAGE analysis (Fig. 6B). A major protein band of about 29 kDa was detected after staining with SYPRO Ruby and transferred to a polyvinylidene difluoride membrane for N-terminal sequencing. The resulting sequence identified CodY as the major component of this sample. Gel shift analysis with purified CodY-His6 showed that CodY bound with high affinity to a 139-bp fragment containing the capA promoter. A complete shift occurred between 10 and 100 nM CodY (Fig. 6C). The shifted bands were competed away with a 1,000-fold excess of specific, cold competitor (the same 139-bp fragment from the capA promoter region), but not with a similar excess of nonspecific cold competitor (the promoter of the 16S rRNA gene) (Fig. 6C).
FIG. 6.
CodY binding to the capA promoter. (A) Gel shift analysis of the capA promoter from strain Newman. (B) The protein-DNA complexes associated with both shifted bands were extracted and subjected to SDS-PAGE analysis. (C) A fragment of the capA promoter region from strain Newman was tested for binding to purified S. aureus CodY by gel shift analysis. CodY binding reactions were also carried out in the presence of 100 nM CodY and nonspecific (a 161-bp fragment of the 16S rRNA promoter region) or specific (the wild-type [WT] capA promoter) cold competitors.
Despite the clear interaction of CodY with the capA promoter of strain Newman, the cap operon of strain UAMS-1 was overexpressed in exponential-phase cells of the codY mutant (Table 1), but not in strains lacking the agr locus (CM18 or CM19). Thus, the overexpression of the agr locus in the UAMS-1 codY mutant is at least partially responsible for the effect of a codY mutation on cap operon expression, in keeping with the fact that agr is known to activate cap transcription (26). Our data, however, did not allow us determine whether in UAMS-1 CodY has any effect on cap operon expression that is independent of agr, because the level of expression in the agr mutant was below detection. Interestingly, the capA promoter region of strain NCTC 8325 was not enriched in the CodY pull-down experiment, but two other regions within the cap operon, including sites within capD and capK, were enriched in the pull-down experiment (Table 4). Perhaps in NCTC 8325 and UAMS-1 CodY regulates one of the previously identified, internal promoters of the capA-P operon (41), or it might regulate this locus by blocking elongation of transcription from the capA promoter.
DISCUSSION
The present study, in conjunction with previous findings reported by Pohl et al. (38), shows that CodY directly or indirectly regulates more than a hundred genes in S. aureus. Many of these genes encode enzymes of amino acid biosynthesis, amino acid and peptide transport, and other nutrient transporters and show considerable overlap with members of the CodY regulons of other bacteria. By integrating in vivo expression data and in vitro DNA binding results, we have identified a number of genes for which CodY most likely acts as a direct repressor of transcription under conditions of nutrient excess.
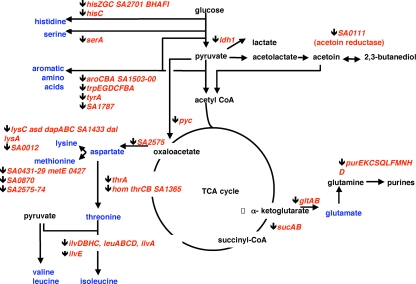
CodY also regulates central metabolic pathways that determine the synthesis and distribution of pyruvate and 2-oxoglutarate, including the conversion of 2-oxoglutarate to glutamate, the step that couples carbon and nitrogen metabolism. The central metabolic pathways that appear to be directly regulated by CodY are depicted in Fig. 7. S. aureus encodes three lactate dehydrogenases, ldh1, ldh2, and ldh3 (40), that can, in principle, interconvert pyruvate and lactate. The conversion of pyruvate to lactate regenerates NAD+ from NADH; the reverse reaction allows lactate to be utilized as a carbon source. Interestingly, strains lacking ldh1 are attenuated in virulence, and strains lacking both ldh1 and ldh2 are nearly avirulent (40), linking pathogenesis to central metabolism. Since ldh1 is a direct target of repression by CodY, the product of ldh1 is likely to serve primarily as a lactate utilization enzyme.
FIG. 7.
Genes of central metabolism that are direct targets of S. aureus CodY. The genes and operons that are direct targets of CodY are shown in italics and in association with the pathways they encode. Each gene or operon is preceded by an arrow that indicates whether the gene is repressed (downward pointing) or activated (upward pointing) by CodY. TCA, tricarboxylic acid; CoA, coenzyme A.
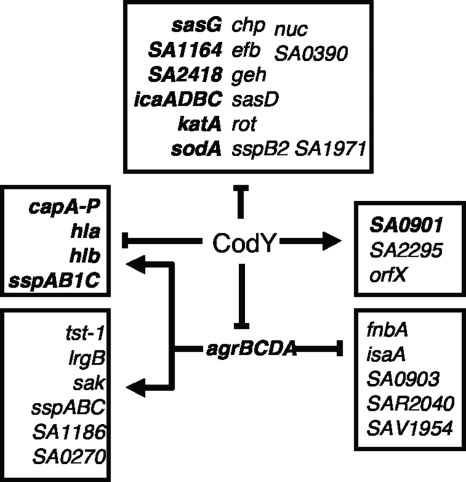
Of the many S. aureus virulence factor genes whose expression is affected by a codY mutation in strains UAMS-1, SA564, and Newman (28, 38), about half are overexpressed in a codY mutant strain mostly, if not solely, because they are positively regulated by the Agr system and CodY is a repressor of the agr locus. A second large group of virulence genes and operons that is regulated by CodY independently of agr includes icaADBC, ldh1, isdF, sodA, SA2585, chp, geh, katA, nuc, rot, sspB2, SA1971, SA0480, SA1164, SA2418, rsbU, lytS, SA2294, SA2295, and SA0901.
The distinction between genes repressed by CodY through its effects on the agr locus and genes repressed directly by CodY is not always strict (Fig. 8). That is, some genes appear to be regulated by CodY both directly and indirectly. For instance, in both UAMS-1 and Newman, a codY mutation causes overexpression of cap operon genes during the exponential growth phase, and this overexpression is at least partially attenuated in an agr codY double mutant (this work and reference 38), consistent with the observation that the capA promoter is activated by RNAIII (26). However, during the exponential phase in strain Newman and in post-exponential phase in UAMS-1, the cap genes are significantly derepressed in the agr codY double mutant compared to an agr single mutant. Taken together, the data from UAMS-1 and Newman suggest that codY represses some genes of the cap locus through both agr-independent and agr-dependent mechanisms. Moreover, our results demonstrate that CodY binds to the capA promoter region of strain Newman and to sites within the cap operon in strain NCTC 8325.
FIG. 8.
Direct and indirect effects of CodY on S. aureus virulence gene expression. Genes shown in boldface are direct targets of CodY-mediated regulation, based on in vitro CodY binding. Connecting lines indicate whether CodY (or an agrBCDA product) regulates these genes negatively (T-junction) or positively (arrowhead). Genes regulated by CodY indirectly through its effect on the agr locus or on other, unknown regulatory genes are shown in normal typeface.
Two other virulence genes that appear to be direct targets of CodY, sodA and katA, encode superoxide dismutase and catalase, respectively, which neutralize superoxides and hydrogen peroxide generated by reactive oxygen bursts produced by the host immune system. Evidence for direct repression by CodY is especially strong for the katA promoter, since footprinting analysis showed that CodY binds with high affinity to the katA promoter close to the position of maximum enrichment in the pull-down experiment. Positions of high enrichment were also found in or upstream of the virulence genes hla, hlb, sspB1, SA1164, SA2418, SA2585, and sasG. The hla (alpha-toxin) gene is interesting because its mRNA is dependent on RNAIII for translation and the gene is repressed by CodY at the level of transcription. SA2585, which encodes a putative member of the PfoR superfamily and has homologs in many Staphylococcus species, was overexpressed in codY mutants of both UAMS-1 (Table 2) and Newman, in which it was annotated as a hypothetical protein (38); this derepression is independent of the agr locus. It would be very interesting to determine the role of this transcriptional regulator in S. aureus and whether it contributes to the expression of genes of the CodY regulon.
Although CodY strongly represses the agrBDCA locus during exponential growth, we still do not know the exact mechanism of CodY regulation at this locus. In vitro binding assays show that CodY does not interact with the P2/P3 promoter region but instead binds within the agrC gene. One interpretation of these data presupposes that promoter P1, which lies near the end of agrC but has not been precisely mapped, is necessary for synthesis of a basal level of AgrA (the response regulator), without which the quorum-sensing system cannot respond to its peptide signal. Repression of P1 by CodY prevents any signal transduction when cells are in a nutrient-replete environment. Alternatively, CodY may achieve the same effect by acting as a transcription roadblock for mRNA initiated at P2. The two models are not mutually exclusive. Future work is aimed at addressing the role of the P1 promoter in CodY regulation of the agr locus. The presence of apparent CodY binding sites within other operons, such as the cap and ica operons, may also reflect roadblock mechanisms of regulation.
Many virulence determinants in S. aureus and other pathogens can be viewed as an extension of the stationary-phase response characteristic of many bacteria, including both pathogens and nonpathogens. That is, many bacterial pathogens induce the production of virulence factors under conditions of nutrient limitation and environmental stress, in conjunction with other stationary-phase-associated adaptive responses. In many cases, the benefit to the bacterium may be its more effective competition for limited nutrients. In S. aureus, as in other Gram-positive species, the CodY protein, through its monitoring of intracellular metabolite pools, appears to link the expression of many virulence genes to the control of central metabolic pathways and to the nutritional status and metabolic potential of the cell.
Supplementary Material
Acknowledgments
We thank Katie McCarty for construction of pKM1, Mark Smeltzer for providing strain UAMS929 and antibody to SarA, David Lazinski for helpful advice in designing the protocol for the affinity purification experiment, Kieran Pechter for providing purified B. subtilis aconitase, and Boris Belitsky for important suggestions and detailed criticism of the manuscript.
This work was supported by research grants (GM042219 to A.L.S. and AI037027 to C.Y.L.) from the U.S. National Institutes of Health and by funds from the University of Nebraska Medical Center awarded to P.M.D.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bailey, T., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. AAAI Press 2:28-36. [PubMed] [Google Scholar]
- 2.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4664-46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., and A. L. Sonenshein. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, H., D. Pearce, S. Glenn, C. Taylor, M. Kuhn, A. Sonenshein, P. Andrew, and I. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453-1467. [DOI] [PubMed] [Google Scholar]
- 5.Blevins, J., K. Beenken, M. Elasri, B. Hurlburt, and M. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 7.Cassat, J., P. Dunman, F. McAleese, E. Murphy, S. Projan, and M. Smeltzer. 2005. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J. Bacteriol. 187:576-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassat, J., P. Dunman, E. Murphy, S. Projan, K. Beenken, K. Palm, S. Yang, K. Rice, K. Bayles, and M. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clincial isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075-3090. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, A., A. Bayer, G. Zhang, H. Gresham, and Y. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Cheung, A., M. Bayer, and J. Heinricks. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Hengst, C. D., S. A. F. T. van Hijum, J. M. W. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon, identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332-34342. [DOI] [PubMed] [Google Scholar]
- 13.Dineen, S., A. Villapakkam, J. Nordman, and A. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206-219. [DOI] [PubMed] [Google Scholar]
- 14.Gillaspy, A., S. Hickmon, R. Skinner, J. Thomas, C. Nelson, and M. Smeltzer. 1995. Role of the accessory gene regulaory (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guedon, E., B. Sperandio, N. Pons, S. Dusko Ehrlich, and P. Renault. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models of CodY regulation in Firmicutes. Microbiology 151:3895-3909. [DOI] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Handke, L. D., R. P. Shivers, and A. L. Sonenshein. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinrichs, J., M. Bayer, and A. Cheung. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendriksen, W., H. Bootsma, S. Estevao, T. Hoogenboezem, A. de Jong, R. de Groot, O. Kuipers, and W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsueh, Y., E. Somers, and A. Lee Wong. 2008. Characterization of the codY gene and its influence on biofilm formation in Bacillus cereus. Arch. Microbiol. 189:557-568. [DOI] [PubMed] [Google Scholar]
- 21.Jefferson, K., D. Pier, D. Goldman, and G. Pier. 2004. The teichoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph, P., M. Ratnayake-Lecamwasam, and A. L. Sonenshein. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J. Biol. Chem. 281:11366-11373. [DOI] [PubMed] [Google Scholar]
- 24.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luong, T., S. Sau, M. Gomez, J. Lee, and C. Lee. 2002. Regulation of Staphylococcus aureus capsular polysaccharide expression by agr and sarA. Infect. Immun. 70:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmood, R., and S. Khan. 1990. Role of upstream sequence in the expression of the staphylococcal enterotoxin B gene. J. Biol. Chem. 265:4652-4666. [PubMed] [Google Scholar]
- 28.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malke, H., and J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56:707-714. [DOI] [PubMed] [Google Scholar]
- 30.Malke, H., K. Steiner, W. McShan, and J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 31.Molle, V., Y. Nakaura, R. Shivers, H. Yamaguchi, R. Losik, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick, R. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 33.Novick, R., and E. Geisinger. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541-564. [DOI] [PubMed] [Google Scholar]
- 34.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang, S., S. Sau, and C. Lee. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J. Bacteriol. 181:2492-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, H., R. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613-621. [DOI] [PubMed] [Google Scholar]
- 38.Pohl, K., P. Francois, L. Stenz, F. Schlink, T. Geiger, S. Herbert, C. Goerke, J. Schrenzel, and C. Wolz. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratnayake-Lecamwasam, M., P. Serror, K. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson, A., S. Libby, and F. Fang. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672-1676. [DOI] [PubMed] [Google Scholar]
- 41.Sau, S., J. Sun, and C. Lee. 1997. Molecular characterization and transciprtional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serio, A., K. Pechter, and A. Sonenshein. 2006. Bacillus subtilis aconitase is required for efficient late-stage sporulation gene expression. J. Bacteriol. 188:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 44.Shivers, R. P., and A. L. Sonenshein. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 56:1549-1559. [DOI] [PubMed] [Google Scholar]
- 45.Somerville, G., M. Chaussee, C. Morgan, R. Fitzgerlad, D. Dorward, L. Reitzer, and J. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somerville, G., and R. Proctor. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tu Quoc, P., P. Genevaux, M. Pajunen, H. Savilahti, C. Georgopoulos, J. Schrenzel, and W. Kelley. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 74:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle, J., A. Toledo-Arana, C. Berasain, J. Ghigo, B. Amorena, J. Penades, and I. Lasa. 2003. SarA and not sigma B is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 49.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villapakkam, A., L. D. Handke, B. R. Belitsky, V. M. Levdikov, A. J. Wilkinson, and A. L. Sonenshein. 2009. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J. Bacteriol. 191:6865-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.