Abstract
Glucose metabolism in Legionella pneumophila was studied by focusing on the Entner-Doudoroff (ED) pathway with a combined genetic and biochemical approach. The bacterium utilized exogenous glucose for synthesis of acid-insoluble cell components but manifested no discernible increase in the growth rate. Assays with permeabilized cell preparations revealed the activities of three enzymes involved in the pathway, i.e., glucokinase, phosphogluconate dehydratase, and 2-dehydro-3-deoxy-phosphogluconate aldolase, presumed to be encoded by the glk, edd, and eda genes, respectively. Gene-disrupted mutants for the three genes and the ywtG gene encoding a putative sugar transporter were devoid of the ability to metabolize exogenous glucose, indicating that the pathway is almost exclusively responsible for glucose metabolism and that the ywtG gene product is the glucose transporter. It was also established that these four genes formed part of an operon in which the gene order was edd-glk-eda-ywtG, as predicted by genomic information. Intriguingly, while the mutants exhibited no appreciable change in growth characteristics in vitro, they were defective in multiplication within eukaryotic cells, strongly indicating that the ED pathway must be functional for the intracellular growth of the bacterium to occur. Curiously, while the deficient glucose metabolism of the ywtG mutant was successfully complemented by the ywtG+ gene supplied in trans via plasmid, its defect in intracellular growth was not. However, the latter defect was also manifested in wild-type cells when a plasmid carrying the mutant ywtG gene was introduced. This phenomenon, resembling so-called dominant negativity, awaits further investigation.
The Gram-negative bacterium Legionella pneumophila is a facultative intracellular parasite and a clinically important pathogen of human beings. In the natural habitat, it replicates within free-living amoebae in the environment (6). However, once inhaled into humans as contaminated aerosol, it survives and replicates in alveolar macrophages and causes severe pneumonia, Legionnaires' disease (13).
It is generally believed that L. pneumophila utilizes amino acids as carbon and energy sources while it neither ferments nor oxidizes carbohydrates (7). Thus, the formulation of chemically defined media (8, 18, 22) soon led to the discovery that a certain combination of amino acids is sufficient to support the growth of L. pneumophila (21). Almost at the same time, it was also noted that glutamate serves as the principal energy source (23) whereas glucose has no effect on the growth of the bacterium (18, 22). However, despite this widely held notion, there are a few reports that present evidence that glucose is actually metabolized by L. pneumophila, mainly through the Entner-Doudoroff (ED) and/or pentose phosphate pathway (21, 23). In accordance with these observations, the analysis of the genome structure of the bacterium identified the complete set of genes related to the ED pathway (NC_002942), but the physiological significance of this phenomenon remained unclear. Recently, it has been shown that the expression of the L. pneumophila genes involved in the ED pathway is upregulated within amoebic cells (2). Furthermore, Salmonella enterica serovar Typhimurium, another facultative intracellular pathogen, is known to use gluconate and related carbohydrates by the ED pathway when growing within macrophages (5).
In the present work, we tried to clarify the physiological roles of the ED pathway (Fig. 1A) more fully by genetic means, i.e., the construction and characterization of L. pneumophila mutants in which genes essential to the ED pathway are inactive due to insertion mutations. Our first aim was to estimate the degree to which the pathway contributes to glucose metabolism in the bacterium. The results described here clearly showed that the dependence of glucose metabolism on the pathway was nearly complete, thus excluding the possibility of significant contributions by the glycolytic and pentose phosphate pathways. Our second aim was to know whether an active ED pathway was required for intracellular growth of L. pneumophila, as suggested by data obtained with amebic cells (2). In this respect, our results seem to indicate that the functional ED pathway is actually a requirement.
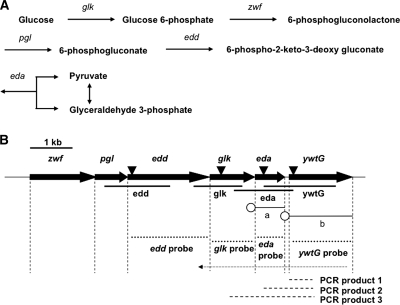
FIG. 1.
(A) The Entner-Doudoroff pathway. The genes zwf, pgl, edd, glk, and eda encode glucose-6-phosphate dehydrogenase, 6-phosphogluconolactonase, phosphogluconate dehydratase, glucokinase, and 2-dehydro-3-deoxy-phosphogluconate aldolase, respectively. (B) Schematic diagram of the DNA fragments used. The large arrows represent genes relevant to this study; vertical dashed lines, ends of coding regions; inverted triangles, sites for insertion of the Kmr cassette; thick lines, DNA fragments for preparation of gene-disrupted mutants; thin lines, DNA inserts in plasmids pMMB207eda (a) and pMMB207ywtG (b) used in complementation; open circles, DNA segments providing a promoter and a ribosome-binding site; dotted lines, probes for Northern hybridization; thin dotted arrow, cDNA synthesized by RT-PCR with primer RT-0; dashed lines, PCR products made on the RT-PCR product as a template with primers RT1 through RT4. For primers and other details, see Materials and Methods and Table S1 in the supplemental material.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Strain AM511, a streptomycin-resistant, restriction-negative derivative of strain Philadelphia 1, was used as the wild type throughout this study. BYE broth contained, per liter, 10 g of yeast extract (Becton Dickinson, Tokyo, Japan), 0.4 g of l-cysteine-HCl, 0.25 g of soluble ferric pyrophosphate, 10 g of ACES [N-(2-acetamido)-2-aminoethanesulfonic acid], and KOH to adjust the pH to 6.9. BCYE agar was BYE broth supplemented with 0.2% activated charcoal and solidified with 1.5% (wt/vol) agar. LB broth and LB agar for Escherichia coli strains contained, per liter, 10 g of Polypeptone (Daigo Eiyo, Osaka, Japan), 5 g of yeast extract, 5 g of NaCl, and NaOH to adjust the pH to 7.2, with or without 1.5% agar. All incubation was done at 37°C. Liquid cultures were grown with gentle shaking, and the growth was monitored by turbidometry at 600 nm. As required, antibiotics were used at the following concentrations: kanamycin (Km), 30 μg/ml; chloramphenicol (Cm), 5 μg/ml (for L. pneumophila) or 25 μg/ml (for E. coli); and ampicillin (Ap), 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype and/or relevant informationa | Source or reference |
|---|---|---|
| L. pneumophila | ||
| AM511 | Strr Res− Mod+ derivative of strain Philadelphia 1 | 11 |
| HRD1 | AM511 edd::Kmr | This study |
| HRD2 | AM511 glk::Kmr | This study |
| HRD3 | AM511 eda::Kmr | This study |
| HRD4 | AM511 ywtG::Kmr | This study |
| HRD300 | AM511(pMMB207) | This study |
| HRD301 | AM511(pMMB207Km) | This study |
| HRD302 | AM511(pMMB207ywtG) | This study |
| HRD303 | AM511(pMMB207ywtG::Kmr) | This study |
| HRD310 | HRD1(pMMB207ywtG) | This study |
| HRD320 | HRD2(pMMB207ywtG) | This study |
| HRD330 | HRD3(pMMB207ywtG) | This study |
| HRD340 | HRD4(pMMB207ywtG) | This study |
| HRD341 | HRD4(pMMB207) | This study |
| HRD331 | HRD3(pMMB207eda) | This study |
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 9 |
| Plasmids | ||
| pGEM-T Easy | Apr | Promega |
| pLAW344 | oriT (RK2) oriR (ColE1) sacB Cmr Apr | 25 |
| pMMB207 | RSF1010 derivative; lncQ lacIq Cmrtacp oriT | 15 |
| pMMB207eda | pMMB207 Ωeda+ with promoter | This study |
| pMMB207ywtG | pMMB207 ΩywtG+ with promoter | This study |
| pMMB207Km | pMMB207 ΩKmr | This study |
| pMMB207ywtG::Kmr | pMMB207 ΩywtG::Kmr with promoter | This study |
| pUT-mini-Tn5 Km | Kmr | 4 |
| pUC19 | Apr | Stratagene |
| pUC19 ywtG | pUC19 ΩywtG+ with promoter | This study |
| pUC19 ywtG::Kmr | pUC19 ΩywtG::Kmr with promoter | This study |
| pHRD1 | pGEM-T Easy Ωedd+ | This study |
| pHRD1Km | pHRD1 Ωedd::Kmr | This study |
| pHRD1LAW | pLAW344 Ωedd::Kmr | This study |
| pHRD2 | pGEM-T Easy Ωglk+ | This study |
| pHRD2Km | pHRD2 Ωglk::Kmr | This study |
| pHRD2LAW | pLAW344 Ωglk::Kmr | This study |
| pHRD3 | pGEM-T Easy Ωeda+ | This study |
| pHRD3B | pGEM-T Easy Ωeda with BamHI site | This study |
| pHRD3Km | pHRD3B Ω eda::Kmr | This study |
| pHRD3LAW | pLAW344 Ωeda::Kmr | This study |
| pHRD4 | pGEM-T Easy ΩywtG+ | This study |
| pHRD4B | pGEM-T Easy ΩywtG with BamHI site | This study |
| pHRD4Km | pHRD4B ΩywtG::Kmr | This study |
| pHRD4LAW | pLAW344 ΩywtG::Kmr | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance.
DNA manipulations.
PCR was performed with Ex Taq or LA Taq DNA polymerase (Takara Bio, Shiga, Japan) or KOD FX DNA polymerase (Toyobo, Osaka, Japan) in a T1 Thermocycler (Biometra, Goettingen, Germany). Blunting of DNA ends was achieved with a DNA Blunting Kit (Takara Bio). Restriction endonucleases were purchased from Toyobo. All enzyme reactions were carried out as recommended by the suppliers. Purification of PCR products and preparation of plasmid DNA were carried out by DNA with a Gel Band Purification Kit (GE Healthcare, Buckinghamshire, United Kingdom) and a Wizard Plus Kit (Promega, Madison, WI), respectively.
Oligonucleotide primers.
The primers used are listed in Table S1 in the supplemental material. They were designed according to the genomic sequence of strain Philadelphia 1 (NC_002942).
Construction of gene-disrupted mutants.
The construction of gene-disrupted mutants was carried out by inserting a Kmr cassette, Mini-Tn5 Km, excised from pUT-mini-Tn5 Km (4), into each of the target genes, i.e., edd, glk, eda, and ywtG, in the following steps. (i) Appropriate DNA segments were amplified with the primers 1F/1R, 2F/2R, 3F/3R, and 4F/4R (see Table S1 in the supplemental material) to give the products edd, glk, eda, and ywtG, respectively (Fig. 1B); (ii) TA cloning of the PCR products with pGEM-T Easy gave pHRD1, pHRD2, pHRD3, and pHRD4, respectively; (iii) for pHRD1 and pHRD2, insertion of the SmaI-excised Kmr cassette into their blunted BanIII sites gave pHRD1Km and pHRD2Km; in pHRD3 and pHRD4, BamHI sites were introduced using the primers 5F/5R or 6F/6R and a Quick Change Site-Directed Mutagenesis Kit (Stratagene, CA) to give pHRD3B and pHRD4B, which in turn gave rise to pHRD3Km and pHRD4Km by accepting the BamHI-excised Kmr cassette at their BamHI sites; (iv) ligation of NotI-digested pHRD1Km, pHRD2Km, pHRD3Km, and pHRD4Km to the NotI site of the allelic-exchange vector pLAW344 (25) gave pHRD1LAW, pHRD2LAW, pHRD3LAW, and pHRD4LAW, respectively; (v) each of these plasmids was introduced into cells of strain AM511 by electroporation, and transformants were selected for on BCYE agar plates supplemented with Km (3); (vi) finally, cells of the Kmr transformants were grown and subjected to selection for possession of the Kmr cassette and loss of the integrated pLAW344 on BCYE agar containing Km and 2% sucrose. The strains thus obtained, HRD1, HRD2, HRD3, and HRD4, were examined by PCR and Southern hybridization for their authenticity.
Construction of other plasmids.
To make plasmids carrying the eda+ or ywtG+ gene to be used for complementation, a functional set of a promoter and a ribosome-binding site had to be placed ahead of their coding sequences because of their locations in the terminal portion of an operon (Fig. 1B). Hence, we separately amplified a region containing both the promoter of the operon and the ribosome-binding site for the zwf gene with primers 7F and 7R-1 (for eda) or 7R-2 (for ywtG) and the coding region of each gene with primers 8F and 8R (for eda) or 9F and 9R (for ywtG). By using an overlap consisting of the ribosome-binding site and the proximal portion of either gene, we then fused the PCR products by the recombinant-PCR technique. The final products were ligated to pMMB207 to make pMMB207eda and pMMB207ywtG using the BamHI and PstI recognition sequences derived from the primers. pMMB207Km was constructed by excision of the cassette from pUT-mini-Tn5 Km with BamHI digestion and its ligation to the BamHI site of pMMB207. pMMB207ywtG::Kmr was made as follows. The ywtG segment of pMMB207ywtG was recloned with pUC19 at its BamHI and PstI sites to generate pUC19ywtG. The Kmr cassette excised from pUT-mini-Tn5 Km with HindIII digestion was blunted and ligated to the MscI site of pUC19ywtG to generate pUC19ywtG::Kmr. The ywtG::Kmr segment from pUC19ywtG::Kmr was then recloned with pMMB207 at its BamHI and HindIII sites.
Isolation of RNA and Northern hybridization.
Total RNA was isolated from test strains by the standard procedure (1). The isolated RNA was treated with DNase I (Takara Bio) and stored at −80°C until it was used. For Northern blotting, 20 μg of the RNA was size fractionated by electrophoresis through a 1% agarose gel containing 2.2 M formaldehyde. The RNA was transferred overnight to a nylon membrane, and hybridization was conducted overnight with the edd, glk, eda, and ywtG probes (Fig. 1B) generated by PCR with primers N1F/N1R through N4F/N4R labeled with digoxigenin-dUTP by using a PCR DIG Probe Synthesis Kit (Roche, Tokyo, Japan). The labeled probes were visualized using a DIG DNA Labeling and Detection Kit (Roche).
RT-PCR.
Reverse transcription (RT)-PCR was performed using the Superscript III First-Strand Synthesis System (Invitrogen, Tokyo, Japan) with primer RT-0 according to the manufacturer's instructions, with 2 μg of total RNA in a final volume of 20 μl. The cDNA produced was subjected to amplification with primers RT-1 through RT-4 to give PCR products 1, 2, and 3 (Fig. 1B). The DNA size markers used were 100-bp and 1-kb DNA ladders (Toyobo).
Spectrophotometric measurements.
The Spectronic Genesys 5 spectrophotometer (Milton Roy, Rochester, NY) was used for all photometric measurements, including turbidometry. The light path was 1 cm in length.
Determination of the glucose concentration.
The glucose concentration was determined using the Wako Glu 2 kit (L type; Wako, Osaka, Japan).
Assay of glucose uptake.
Glucose uptake was assayed by growing cells in the presence of 14C-labeled glucose and following the incorporation of radioactivity into the acid-insoluble fraction. Cells of L. pneumophila were grown in BYE broth supplemented with 0.2% d-[U-14C]glucose (specific activity, 3.3 kBq/μmol) at 37°C with gentle shaking. Samples (0.3 ml each) were taken at intervals and mixed into 10 ml of chilled 5% (wt/vol) trichloroacetic acid. After standing for 30 min on ice, they were filtered through nitrocellulose filters (0.45-μm pore size; Millipore, Tokyo, Japan), which were rinsed three times with chilled 5% trichloroacetic acid. The radioactivity held on the filters was determined in a liquid scintillation counter (LSC 5100; Aloka, Tokyo, Japan) with the Clear-sol scintillation cocktail (Nacalai Tesque; Kyoto, Japan).
Preparation of permeabilized cells for enzyme assays.
Bacterial cells were collected from 50 ml of log-phase cultures (turbidity, ∼0.6) and washed twice with the same buffer used in the assay by centrifugation at 9,400 × g for 5 min. The cells were resuspended in the buffer and incubated for 1 h at 37°C with gentle shaking to exhaust the substrate remaining in the cells. After centrifugation at 9,400 × g for 5 min, the cells were resuspended in 0.2 ml of the buffer. The cells were then permeabilized by treatment with toluene essentially as previously described (10).
Enzyme assays.
Glucokinase activity was assessed by measuring ATP-dependent phosphorylation of d-[U-14C]glucose essentially as described previously (20, 24). The complete reaction mixture consisted of 30 μl of the permeabilized cell suspension and 150 μl of a solution containing 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 20 mM ATP, and 0.3 mM d-[U-14C]glucose (specific activity, 132 kBq/μmol). After incubation at 37°C for 30 min with shaking, a 50-μl aliquot of each reaction mixture was mixed with 0.1 ml of isopropyl alcohol to stop the reaction and 0.5 ml of a 10% glucose solution. This mixture was applied to a column containing 0.2 ml of DEAE cellulose (DE52; Whatman, Tokyo, Japan). After the column was washed with 1 ml of water three times, the phosphorylated sugar was eluted with 1.5 ml of 1 M HCl directly into a scintillation vial. The radioactivity was measured as described above.
The combined activities of phosphogluconate dehydratase and 2-dehydro-3-deoxy-phosphogluconate aldolase were determined by two-step procedures essentially as described previously (19). The reaction mixture consisted of 50 μl of the permeabilized cell suspension and 200 μl of a solution containing 50 mM Tris-HCl, pH 7.2, 10 mM MgCl2, 5 mM 6-phosphogluconate, and 1 mM EDTA. After being incubated at 37°C for 30 min, the mixtures were heated in a boiling water bath for 2 min to inactivate all enzymes, including intrinsic NADH oxidase. After being chilled on ice for 2 min, the mixtures were centrifuged at 16,000 × g for 5 min and the supernatants were saved for quantitation of the pyruvate formed, which was carried out as follows. The reaction mixture consisted of 0.2 ml of the supernatant and 1.8 ml of a solution containing 50 mM Tris-HCl, pH 7.2, 10 mM MgCl2, 1 mM EDTA, 0.1 mM NADH, and 0.25 units/ml l-lactate dehydrogenase from bovine heart (Sigma, St. Louis, MO). Incubation was done at 37°C with shaking until decrease in absorbance at 340 nm ceased. The amount of pyruvate formed was calculated using the molar extinction coefficient of NADH.
Intracellular growth of L pneumophila.
Intracellular growth of L. pneumophila was studied by the standard assay (12, 16, 17) using A549 human epithelial cells, peritoneal macrophages from A/J mice, and Acanthamoeba culbertsoni. Cells of L. pneumophila strains grown in BYE broth to a cell density of approximately 2 × 1010 CFU/ml were pelleted by centrifugation at 9,400 × g for 5 min and resuspended in 500 times the original volume of RPMI 1640 tissue culture medium containing 10% fetal bovine serum, which was unable to support the growth of the bacterial cells. A549 cells were maintained in the tissue culture medium under 5% CO2 in air. Peritoneal macrophages of A/J mice were prepared by lavaging the peritoneal cavities of the mice 4 days after an intraperitoneal injection of 2.5 ml of 4% (wt/vol) thioglycolate medium (Becton Dickinson, Tokyo, Japan). The bacteria (0.5 ml of a 4 × 107-CFU/ml suspension) were added to monolayers of A549 cells or peritoneal macrophages (∼2 × 105 cells in 0.5 ml of the tissue culture medium per well) in 24-well dishes to give a multiplicity of infection (MOI) of about 100. The infected cells were incubated at 37°C under 5% CO2 in air for 1.5 h and washed three times with phosphate-buffered saline (PBS) to remove unadsorbed bacteria. To measure the total number of bacteria associated with the host cells, 1 ml each of sterile distilled water was added to control wells, the cells were scraped from the bottoms of the wells with a rubber policeman to release all bacteria from the host cells, and dilutions of the lysates were plated on BCYE agar plates. To each of the remaining wells, 0.5 ml of fresh tissue culture medium was added, and the dishes were incubated at 37°C under 5% CO2 in air. At 24-h intervals, the total number of bacterial cells in each well was determined as described above. Infection of A. culbertsoni was carried out in a similar manner, but bacteria were suspended in peptone-yeast extract-glucose (PYG) buffer instead of RPMI 1640, and AC buffer (14) containing 0.05% Triton X-100 was added to release intracellular bacteria.
To assess the degree of adhesion or internalization of bacteria, A549 cells were infected with bacteria in RPMI 1640 at an MOI of about 1,000, incubated for 1.5 h, and washed three times with PBS to remove unadsorbed bacteria. Then, the total number of cell-associated bacteria was determined as described above to estimate the efficiency of adhesion. For internalization, the infected A549 cells were washed and incubated in RPMI 1640 containing 100 μg of gentamicin per ml for 1 h to kill extracellular bacteria. Then, the cells were washed three times with PBS to remove the antibiotic, and the total number of viable bacteria was determined as described above.
RESULTS
Evidence for utilization of exogenous glucose in the wild-type strain.
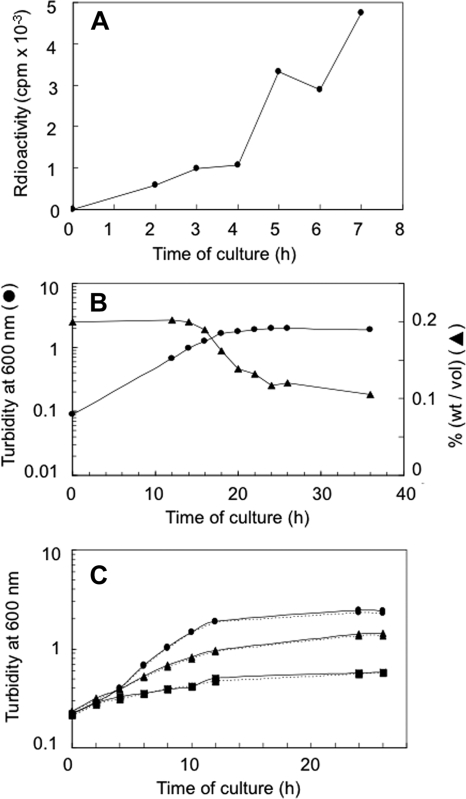
Since 14C-labeled glucose has been shown to be metabolized in L. pneumophila (21, 23), we first tried to confirm the previous findings by growing cells of strain AM511 in the presence of uniformly labeled [14C]glucose and monitoring the incorporation of radioactivity into the acid-insoluble fraction of the cells. The uptake of radioactivity was clearly detectable, indicating that glucose actually served as a precursor of cellular macromolecules (Fig. 2A). In accordance with this finding, when cells of the same strain were grown in BYE broth supplemented with glucose at 0.2%, the glucose concentration in the medium started decreasing after the culture entered the late exponential phase (Fig. 2B). Perhaps the rate of glucose uptake by each cell was so low that the decrease in the glucose concentration remained undetectable until the culture attained a high enough cell density in the late exponential phase. On the other hand, the addition of glucose to the culture brought about no detectable increase either in the growth rate or in the final cell density, even when diluted BYE medium was used (Fig. 2C). Conceivably, this absence of a growth-promoting effect of glucose may reflect its minute role as a nutrient, particularly an energy source, under the overwhelming contribution by amino acids.
FIG. 2.
Glucose utilization by strain AM511. The experiment was performed at least three times, and typical results are shown. (A) Incorporation of 14C into the acid-insoluble fraction from uniformly labeled glucose. (B) Consumption of exogenous glucose. •, turbidity of the culture; ▴, glucose concentration in the culture medium. (C) Bacterial growth with (solid lines) or without (dotted lines) glucose (0.2%) added to undiluted BYE medium (•); 30% BYE medium in ACES buffer, pH 6.9 (▴); or 10% BYE medium in ACES buffer (▪).
Demonstration of enzyme activity of the ED pathway in the wild-type strain.
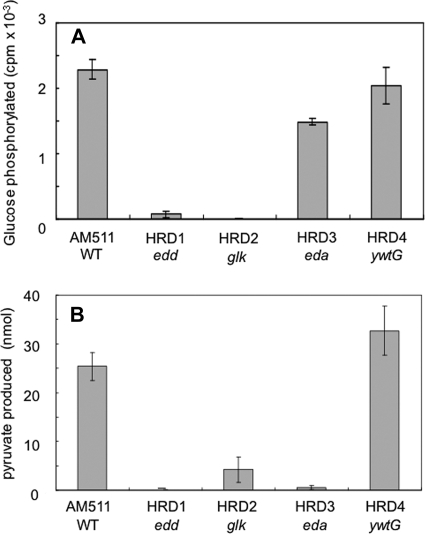
Tracer studies had previously implicated the ED pathway in glucose utilization by L. pneumophila (21, 23), and the recent analysis of its genomic structure had revealed the presence of an operon-like cluster of genes, i.e., zwf-edd-glk-eda-ywtG, apparently containing all the genes that are required for the operation of the pathway (Fig. 1B). However, the activity of the gene products had not yet been demonstrated in vitro. Using permeabilized cells of strain AM511 as the enzyme source, we assessed the activities of the signature enzymes of the pathway, i.e., phosphogluconate dehydratase (Edd protein) and 2-dehydro-3-deoxy-phosphogluconate aldolase (Eda protein), as well as of glucokinase (Glk protein).
As for the first two enzymes, the formation of pyruvate from 6-phosphogluconate, reflecting the combined actions of both enzymes, was measured. The results clearly demonstrated that the conversion actually took place. The amount of pyruvate produced was significant (25.4 ± 2.9 nmol) under the conditions used, while it was undetectable in the absence of 6-phosphogluconate (data not shown), a finding consistent with the notion that glucose is metabolized through the ED pathway. On the other hand, the assay of glucokinase showed that about 4% of the glucose in each reaction mixture (2,285 ± 152 cpm) became phosphorylated in the presence of ATP under the conditions used, but not in its absence (data not shown), indicating that the enzyme is likely to be involved in phosphorylation.
Construction of mutants deficient in ED pathway proteins.
To clarify the physiological roles played by the ED pathway, we constructed gene-disrupted mutants of strain AM511 by inserting the Kmr cassette into each of the edd, glk, eda, and ywtG genes (Fig. 1B and Table 1). The authenticity of the edd, glk, and eda mutants obtained was confirmed by assaying the enzyme activity as follows. The activity of glucokinase was defective, not only in the glk mutant HRD2, but also in the edd mutant HRD1 (Fig. 3A). On the other hand, the conversion of 6-phosphogluconate to pyruvate, representing the combined activities of Edd and Eda, was diminished, not only in the edd mutant HRD1 and the eda mutant HRD3, but also in the glk mutant HDR2 (Fig. 3B). We interpret these results as indicating that these three genes actually constitute part of an operon and that a polar effect due to a strong transcription terminator in the Kmr cassette abolished the expression of the downstream genes.
FIG. 3.
Enzyme activities in different strains. The experiment was performed at least three times, and the results are shown as means and standard deviations. (A) Glucokinase. (B) Combination of phosphogluconate dehydratase and 2-dehydro-3-deoxy-phosphogluconate aldolase. WT, wild type.
Further evidence for an operon.
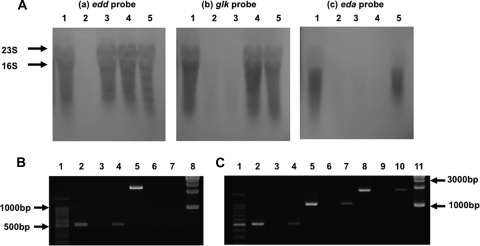
To substantiate the above-mentioned interpretation, we determined the presence or absence of the transcript for each gene by Northern hybridization against RNA extracted from the mutants (Fig. 4A). The results demonstrated that the edd, glk, and eda probes each showed a characteristic pattern of hybridization with respect to the test RNAs. Thus, the strains whose RNAs gave negative hybridization were the edd mutant for the edd probe; the edd and glk mutants for the glk probe; and the edd, glk, and eda mutants for the eda probe. These results clearly indicated the existence of a single transcriptional unit comprising at least the edd, glk, and eda genes arranged in that order, which accounts for the apparent polar effect mentioned above.
FIG. 4.
Evidence for an operon. (A) Northern hybridization with edd (a), glk (b), and eda (c) probes (cf. Fig. 1B). Lane 1, AM511 (wild type); lane 2, HRD1 (edd mutant); lane 3, HRD2 (glk mutant); lane 4, HRD3 (eda mutant); lane 5, HRD4 (ywtG mutant). 23S and 16S indicate the positions of rRNA markers. (B) Demonstration of the ywtG gene transcript. Lane 1, 100-bp DNA ladder; lane 2, PCR product 1 (cf. Fig. 1B) made with primers RT-1 and RT-2 and genomic DNA from the wild type as a template (positive control); lane 3, the same as lane 2 but with total RNA from the wild type as a template (negative control); lane 4, the same as lane 2 but with cDNA for wild-type RNA (synthesized using primer RT-0) as a template; lanes 5, 6, and 7, the same as lanes 2, 3, and 4, respectively, but with the ywtG mutant HRD4 instead of the wild type; lane 8, 1-kb DNA ladder. (C) Transcription of the glk-eda-ywtG region in the wild type. Lanes 1 through 4, the same as the corresponding lanes in panel B, serving as references; lanes 5, 6, and 7, the same as lanes 2, 3, and 4, respectively, but for demonstrating PCR product 2 (cf. Fig. 1B) made with primers RT-1 and RT-3; lane 8, 9, and 10, the same as lanes 2, 3, and 4, respectively, but for demonstrating PCR product 3 (cf. Fig. 1B) made with primers RT-1 and RT-4; lane 11, 1-kb DNA ladder.
In the case of the ywtG gene, which is situated at the distal end of the putative operon, the hybridization signal was too weak to be discernible, even in the parental strain. Hence, we subjected the extracted RNA to cDNA synthesis, followed by PCR amplification of the cDNA. The cDNA synthesis was carried out using primer RT-0, which hybridizes with the ywtG gene at its subterminal location downstream of the cassette insertion site. When the cDNA thus formed was amplified with the ywtG-specific primers RT-1 and RT-2, PCR product 1 (cf. Fig. 1B) was detectable with cDNA from the wild type (Fig. 4B, lane 4), but not with that from the ywtG mutant, HRD4 (Fig. 4B, lane 7). Furthermore, when primer RT-1 was used in combination with the eda-specific primer RT-3 or the glk-specific primer RT-4, PCR product 2 or 3 (cf. Figure 1B) was observed (Fig. 4C, lanes 7 and 10). These results clearly identified the ywtG gene as the terminal component of the operon.
Physiological effects of the mutations.
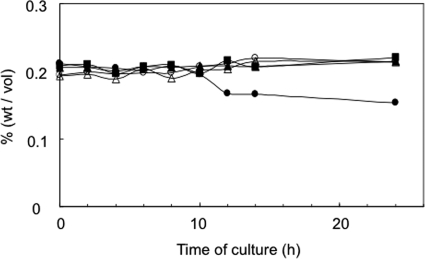
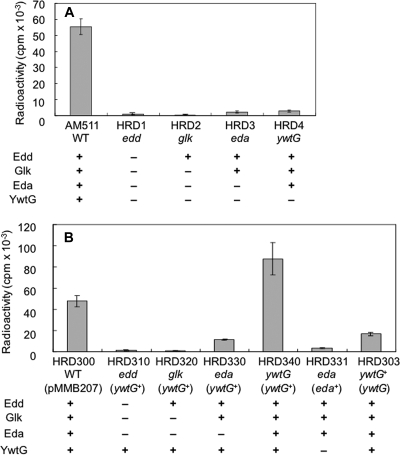
As expected, all of the mutants were indistinguishable from the wild type with respect to growth in BYE medium (data not shown). In contrast, it was noted that the consumption of glucose in the culture medium was diminished in all of the mutants (Fig. 5). In addition, when the mutants were fed with 14C-labeled glucose, the incorporation of radioactivity into the acid-insoluble fraction of the cells was practically negligible (Fig. 6A). Overall, these results seemed to indicate that the ED pathway is responsible for the utilization of glucose.
FIG. 5.
Consumption of exogenous glucose by gene-disrupted mutants. •, AM511; ▴, HRD1 (edd); ▪, HRD2 (glk); ○, HRD3 (eda); ▵, HRD4 (ywtG). Representative results of at least three independent experiments are shown.
FIG. 6.
Incorporation of radioactivity from 14C-labeled glucose into the acid-insoluble cellular fractions of L. pneumophila strains. Cells of gene-disrupted mutants (A) and various partial heterozygotes (B) were incubated for 24 h under the conditions described in Materials and Methods. The matrix below each graph shows the presence (+) or absence (−) of the relevant enzymes in each strain. In panel B, pMMB207 represents the vector used to construct the heterozygotes. The means and standard deviations of at least three independent determinations are shown.
Especially worth mentioning is the observation that the ywtG mutant was defective in metabolizing exogenous glucose. Since this defect was manifested in the presence of the complete set of the ED pathway enzymes, it strongly indicated that the ywtG gene product, previously annotated as a xylose proton symporter (NC_002942), was responsible for glucose uptake by the bacterium. Because the ywtG gene is situated at the distal end of the operon, there is no possibility of a polar effect causing this phenotype.
The above interpretation was supported by complementation experiments. When each mutant was supplied with the ywtG+ gene in trans through the introduction of plasmid pMMB207ywtG, the glucose uptake capacity was restored in the ywtG mutant but not in the others (Fig. 6B). This observation not only confirmed the identity of the YwtG protein as the glucose transporter, but also lent further support to the exclusive role of the ED pathway in glucose metabolism in the bacterium. Moreover, when the eda mutant was supplied with the eda+ gene in trans through the introduction of plasmid pMMB207eda, the enzyme activity of the ED pathway was recovered (data not shown), but the glucose uptake capacity was not restored (Fig. 6B). This finding was consistent with the notion that the ywtG gene is the terminal component of the operon.
Effect on growth in eukaryotic cells.
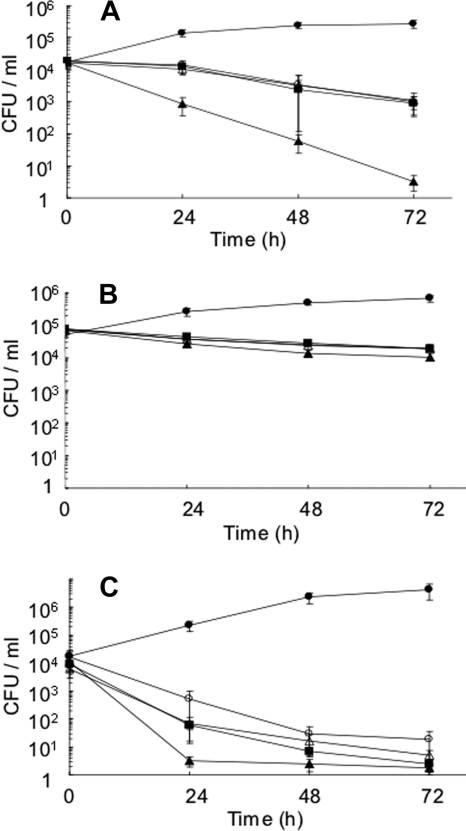
According to a recent report (2), the expression of the genes related to the ED pathway is upregulated when L. pneumophila cells are growing within amoebae. Therefore, we looked at the behavior of our mutants within A549 human epithelial cells, peritoneal macrophages from A/J mice, and A. culbertsoni. The results showed that all of the mutants tested were defective in growth compared with the wild type in the three types of eukaryotic cells (Fig. 7A, B, and C). In contrast, all of the mutants were indistinguishable from the wild type with respect to adhesion and internalization (data not shown). These observations strongly suggested that the functional ED pathway was required for active intracellular growth of L. pneumophila.
FIG. 7.
Intracellular growth of gene-disrupted mutants. The host cells used were A549 human epithelial cells (A), A/J mouse peritoneal macrophages (B), and A. culbertsoni (C). •, AM511; ▴, HRD1 (edd); ▪, HRD2 (glk); ○, HRD3 (eda); ▵, HRD4 (ywtG). The means and standard deviations of at least three independent determinations are shown.
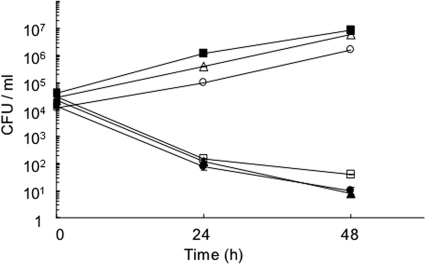
In an attempt to further substantiate the apparent correlation between glucose-utilizing ability and the capacity for intracellular growth, we looked at the behavior of strain HRD340, in which the deficiency in glucose utilization due to the ywtG mutation had been reversed by the ywtG+ gene supplied in trans (see above). Unexpectedly, however, this strain was incapable of growing in A. culbertsoni (Fig. 8); essentially the same results were obtained in A549 human epithelial cells and peritoneal macrophages from A/J mice (data not shown). Interestingly, strain HRD303, which has the opposite arrangement of the ywtG alleles, i.e., ywtG+ on the chromosome and mutant ywtG on the plasmid, was similarly defective in intracellular growth (Fig. 8), whereas it was capable of metabolizing glucose in vitro, albeit less efficiently than HRD340 (Fig. 6B). In other words, the complementation of the ywtG mutation by the wild-type allele was totally negative for growth capacity within eukaryotic cells but did occur with respect to the ability to metabolize glucose to different degrees, depending on the positions of both alleles.
FIG. 8.
Intracellular growth of ywtG+-ywtG::Kmr partial heterozygotes in A. culbertsoni. •, HRD340 (ywtG on chromosome-ywtG+ on vector); ▴, HRD303 (ywtG+-ywtG); ▪, HRD300 (ywtG+-blank vector); ○, HRD301 (ywtG+-Kmr cassette on vector); ▵, HRD302 (ywtG+-ywtG+); □, HRD341 (ywtG-blank vector). The experiments were repeated at least three times, and typical results are shown.
DISCUSSION
In L. pneumophila, carbohydrate catabolism has remained an ill-explored area. Growth stimulation by glucose has never been demonstrated with this bacterium, despite the evidence that glucose is actually metabolized (21, 23). After confirming that strain AM511 exhibits the ability to metabolize glucose, we intended to solve unsettled problems with respect to glucose utilization by the bacterium. In doing so through a genetic approach, we focused our attention on the ED pathway for two reasons. First, the pathway has been implicated in glucose metabolism based on the metabolic fate of 14C in differentially labeled glucose (21, 23); second, the intracellular growth of the bacterium reportedly causes the upregulation of the genes apparently involved in the pathway, i.e., zwf, pgl, edd, glk, eda, and ywtG (2).
First of all, the results of enzyme assay, Northern hybridization, and RT-PCR with the gene-disrupted mutants provided corroborative evidence that the genes edd, glk, eda, and ywtG actually belong to an operon. In addition, the genomic sequence strongly suggested that the genes zwf and pgl, situated upstream of the above-mentioned four genes, are also part of the operon. This notion is undoubtedly compatible with the observed upregulation of all the genes mentioned above (2). On the other hand, these observations raise the question of what serves as the inducing stimulus. We are presently unable to give a definite answer to this question, but we consider that neither glucose itself nor its metabolites, e.g., 6-phosphogluconate, is likely to be the inducer, because addition of glucose to the culture medium increased neither the activity of the glucokinase nor the combined activities of Edd and Eda (our unpublished results).
Our observations clearly indicate that the membrane transport and phosphorylation of glucose are carried out by the YwtG protein, a member of the major facilitator superfamily, and the Glk protein, a glucokinase, respectively. Thus, the disruption of the ywtG gene resulted in a total loss of glucose uptake, which was restored by complementation with the wild-type gene supplied in trans. In the case of Glk, the gene disruptant was shown to exhibit practically no activity of glucose phosphorylation in vitro.
Our results also indicate that glucose is metabolized at least predominantly via the ED pathway. The critical finding to support this notion is that the diminished ability of the eda mutant to metabolize glucose was recovered only slightly, even when the coexisting deficiency in YwtG due to polar effect was compensated for by complementation with the ywtG+ gene. In other words, the decreased utilization of glucose by the ywtG+-complemented eda mutant was actually due to the loss of activity of the Eda protein, which is specific to, and essential for, the ED pathway. Obviously, this conclusion substantiates previous reports that suggested the importance of the ED pathway (21, 23). Additionally, the small amount of glucose utilization observed in the ywtG+-complemented eda mutant may be accounted for by the glycolytic pathway and the accompanying nonoxidative branch of the pentose phosphate cycle leading to the formation of ribose 5-phosphate. It is pertinent to mention that the presence of the enzymes involved in these pathways was predicted by the genome annotation (NC_002942).
The gene-disrupted mutants were indistinguishable from the wild type with respect to the growth rate in culture, which is consistent with the observation that exogenously added glucose did not influence the growth of wild-type cells. In sharp contrast, however, all mutants examined were defective in multiplication within eukaryotic cells. In most strains, parallelism was shown to exist between the ability to metabolize glucose and the ability to multiply within eukaryotic cells, but this relationship apparently broke down in the ywtG-ywtG+ partial heterozygotes HRD303 and HRD340. Although this phenomenon involving the mutant and wild-type genes suggests the possibility that the disrupted ywtG gene has a dominant-negative effect, we have so far been unable to reproduce this state with various fragments of the ywtG gene (our unpublished results). Although the functional ED pathway is highly likely to be a requirement for successful intracellular growth of the bacterium, this phenomenon must be accounted for before a complete understanding of the connection is attained.
Apart from the above problem, the most crucial point that remains to be understood is how the operation of the ED pathway supports the intracellular growth of L. pneumophila. The simplest possibility would be that the intraphagosomal or intravacuolar environment may pose a nutritional difficulty for L. pneumophila cells, e.g., some sort of amino acid deficiency, which makes glucose an indispensable substrate for growth. If this is the case, carefully designed in vitro experiments may be sufficient to answer the question.
Supplementary Material
Acknowledgments
We thank Hiroaki Taniai and Qin Tian for their useful suggestions, as well as Hideko Kajiwara for her technical assistance. E.H. is particularly indebted to Masaki Fujita and Yo-Ichi Nakanishi for permission to join the present work. We thank L. Saza for her helpful comments on the English of an earlier version of the manuscript.
This work was supported by grant-in-aid 21390131 for scientific research from the Ministry of Education, Sports, Culture, Science, and Technology, Japan.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, NY.
- 2.Brüggemann, H., A. Hagman, M. Jules, O. Sismeiro, M. A. Dillies, C. Gouyette, F. Kunst, M. Steinert, K. Heuner, J. Y. Coppee, and C. Buchrieser. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 8:1228-1240. [DOI] [PubMed] [Google Scholar]
- 3.Chen, D. Q., S. S. Huang, and Y. J. Lu. 2006. Efficient transformation of Legionella pneumophila by high-voltage electroporation. Microbiol. Res. 161:246-251. [DOI] [PubMed] [Google Scholar]
- 4.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 6.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrity, G. M., J. A. Bell, and T. Lilburn. 2005. Order VI. Legionellales ord. nov., p. 210-236. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 8.George, J. R., L. Pine, M. W. Reeves, and W. K. Harrell. 1980. Amino acid requirements of Legionella pneumophila. J. Clin. Microbiol. 11:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg, H. L., and R. E. Reeves. 1972. Correlation between hexose transport and phosphotransferase activity in Escherichia coli. Biochem. J. 126:1241-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marra, A., and H. A. Shuman. 1989. Isolation of a Legionella pneumophila restriction mutant with increased ability to act as a recipient in heterospecific matings. J. Bacteriol. 171:2238-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruta, K., H. Miyamoto, T. Hamada, M. Ogawa, H. Taniguchi, and S. Yoshida. 1998. Entry and intracellular growth of Legionella dumoffii in alveolar epithelial cells. Am. J. Respir. Crit. Care Med. 157:1967-1974. [DOI] [PubMed] [Google Scholar]
- 13.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 14.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa, M., S. Yoshida, and Y. Mizuguchi. 1994. 2-Deoxy-D-glucose inhibits intracellular multiplication and promotes intracellular killing of Legionella pneumophila in A/J mouse macrophages. Infect. Immun. 62:266-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnishi, H., Y. Mizunoe, A. Takade, Y. Tanaka, H. Miyamoto, M. Harada, and S. Yoshida. 2004. Legionella dumoffii DjlA, a member of the DnaJ family, is required for intracellular growth. Infect. Immun. 72:3592-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pine, L., J. R. George, M. W. Reeves, and W. K. Harrell. 1979. Development of a chemically defined liquid medium for growth of Legionella pneumophila. J. Clin. Microbiol. 9:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponce, E., M. Garcia, and M. E. Munoz. 2005. Participation of the Entner-Doudoroff pathway in Escherichia coli strains with an inactive phosphotransferase system (PTS− Glc+) in gluconate and glucose batch cultures. Can. J. Microbiol. 51:975-982. [DOI] [PubMed] [Google Scholar]
- 20.Romano, A. H., J. D. Trifone, and M. Brustolon. 1979. Distribution of the phosphoenolpyruvate: glucose phosphotransferase system in fermentative bacteria. J. Bacteriol. 139:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesh, M. J., S. A. Morse, and R. D. Miller. 1983. Intermediary metabolism in Legionella pneumophila: utilization of amino acids and other compounds as energy sources. J. Bacteriol. 154:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiss, E., M. G. Peacock, and J. C. Williams. 1980. Glucose and glutamate metabolism of Legionella pneumophila. Curr. Microbiol. 4:1-6. [Google Scholar]
- 24.Whitesell, R. R., H. Ardehali, R. L. Printz, J. M. Beechem, S. M. Knobel, D. W. Piston, D. K. Granner, W. Van Der Meer, L. M. Perriott, and J. M. May. 2003. Control of glucose phosphorylation in L6 myotubes by compartmentalization, hexokinase, and glucose transport. Biochem. J. 370:47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.