Abstract
Agrobacterium tumefaciens VirB proteins assemble a type IV secretion apparatus and a T-pilus for secretion of DNA and proteins into plant cells. The pilin-like protein VirB3, a membrane protein of unknown topology, is required for the assembly of the T-pilus and for T-DNA secretion. Using PhoA and green fluorescent protein (GFP) as periplasmic and cytoplasmic reporters, respectively, we demonstrate that VirB3 contains two membrane-spanning domains and that both the N and C termini of the protein reside in the cytoplasm. Fusion proteins with GFP at the N or C terminus of VirB3 were fluorescent and, like VirB3, localized to a cell pole. Biochemical fractionation studies demonstrated that VirB3 proteins encoded by three Ti plasmids, the octopine Ti plasmid pTiA6NC, the supervirulent plasmid pTiBo542, and the nopaline Ti plasmid pTiC58, are inner membrane proteins and that VirB4 has no effect on membrane localization of pTiA6NC-encoded VirB3 (pTiA6NC VirB3). The pTiA6NC and pTiBo542 VirB2 pilins, like VirB3, localized to the inner membrane. The pTiC58 VirB4 protein was earlier found to be essential for stabilization of VirB3. Stabilization of pTiA6NC VirB3 requires not only VirB4 but also two additional VirB proteins, VirB7 and VirB8. A binary interaction between VirB3 and VirB4/VirB7/VirB8 is not sufficient for VirB3 stabilization. We hypothesize that bacteria use selective proteolysis as a mechanism to prevent assembly of unproductive precursor complexes under conditions that do not favor assembly of large macromolecular structures.
Bacteria use type IV secretion (T4S) to deliver macromolecules to prokaryotes and eukaryotes (12). Animal and human pathogens deliver proteins to their eukaryotic hosts to affect cellular processes causing disease. The plant-pathogenic bacterium Agrobacterium tumefaciens delivers both proteins and DNA to plants and other eukaryotes. DNA delivered by Agrobacterium directs constitutive synthesis of phytohormones in a transformed plant cell, promoting cancerous growth (56). The Ptl toxin of Bordetella pertussis modifies G proteins by ADP-ribosylation, affecting intracellular cell signaling, and CagA of Helicobacter pylori disrupts epithelial cell polarity by inhibiting PAR1 kinase activity (37, 44, 47). T4S is ancestrally related to bacterial conjugation, a mechanism used by bacteria for interbacterial plasmid transfer, enabling them to acquire novel genes for antibiotic resistance, degradation of organic molecules, toxin production, and other virulence traits (29).
The VirD4/VirB family of proteins, found conserved in many alphaproteobacteria, mediates T4S (12). The Ti plasmid-encoded Agrobacterium T4S system requires VirD4 and 11 VirB proteins, VirB1 to VirB11, for efficient DNA transfer (7, 54). The membrane and membrane-associated VirB proteins assemble a macromolecular structure at the cell membrane to promote substrate transfer (12). The octopine Ti plasmid pTiA6NC-encoded VirB6 to VirB11 proteins assemble the T4S apparatus at a cell pole (34, 35, 39). The VirD4 coupling protein targets the VirE2 substrate protein to the cell pole (4). A recent study found that the nopaline Ti plasmid pTiC58 T4S system (T4SS) and its substrates form a helical array around the cell circumference (1). Structural studies using Escherichia coli conjugative plasmid pKM101-encoded VirB homologues showed that TraN (VirB7), TraO (VirB9), and TraF (VirB10) form the core complex and that TraF forms a channel at the outer membrane (11, 23). The Agrobacterium VirB proteins assemble a T-pilus, an appendage composed primarily of VirB2, with VirB5 and VirB7 as its minor constituents (38, 40, 41, 48, 50, 55). VirB3, a homolog of the pilin-like TraL protein encoded in E. coli plasmids, is postulated to function in T-pilus assembly (52). Three ATP-utilizing proteins, VirB4, VirB11, and VirD4, supply energy for substrate translocation (3, 9, 34).
The membrane topology of all the VirB proteins, except for VirB3, was determined by analyses of random phoA insertion mutants, targeted phoA fusions, and targeted bla fusions (6, 14, 15, 21, 22, 31, 35, 53). phoA and bla, which encode alkaline phosphatase and β-lactamase, respectively, serve as excellent markers for periplasmic proteins, as they are enzymatically active only when targeted to the cell periplasm (8, 30). Green fluorescent protein (GFP) is an ideal cytoplasmic marker because it fluoresces only when located in the cytoplasm (19, 20). When GFP is targeted to the periplasm through fusion with a membrane-spanning domain (MSD), it fails to fold properly and does not fluoresce.
The prevailing view, based on in silico analysis, is that VirB3 is a bitopic membrane protein with a periplasmic C terminus. No phoA-positive insertions in virB3, however, were identified in two random mutagenesis studies of the virB operon (6, 15). The small size of VirB3, a polypeptide of 108 amino acids (aa), could be a contributing factor to the negative findings. Yet several PhoA-positive insertions in two smaller VirB proteins, VirB2 (74-aa mature peptide) and VirB7 (41-aa mature peptide), were successfully obtained in both studies. Therefore, the negative findings may also be indicative of the presence of a small periplasmic domain in VirB3. Biochemical studies showed that the nopaline Ti plasmid pTiC58-encoded VirB3 protein (pTiC58 VirB3) associates with the bacterial outer membrane, while VirB2 associates with both the inner and outer membranes (52). The pTiC58 VirB4 protein is required for localization of VirB3 to the outer membrane (33). VirB4 is also required for VirB3 stability (33, 55). A low level of VirB3 accumulated in a nonpolar pTiC58 virB6 deletion mutant; however, addition of virB6 in trans did not restore the level of the protein, even though it restored tumorigenicity (27). VirB3 participates in the formation of protein complexes with the T-pilus proteins VirB2 and VirB5 (55).
Homologues of VirB3 are found in many alphaproteobacteria with a T4SS. While most VirB3 homologues are small proteins, several recently identified homologues are fusions of VirB3 and the immediate downstream protein VirB4 (5, 10, 24). These fusion homologs, which include Actinobacillus MagB03 (GenBank accession no. AAG24434), Campylobacter CmgB3/4 (EAQ71805), Yersinia pseudotuberculosis TriC (CAF25448), Citrobacter koseri PilX3-4 (ABV12046), and Klebsiella pneumoniae PilX3-4 (BAF49490), have VirB3 at the N terminus and VirB4 at the C terminus. Agrobacterium VirB4 is an integral membrane protein with a cytoplasmic N terminus (14). Its homologues are expected to have a similar topology. The prevailing view that pTi VirB3 has a periplasmic C terminus is inconsistent with the cytoplasmic location of the N terminus of VirB4 in the VirB3-VirB4 fusion protein homologues.
In this study, we report the membrane topology of Agrobacterium VirB3 and demonstrate that the C terminus of the protein resides in the cytoplasm. We also demonstrate that VirB3 is an inner membrane protein, not an outer membrane protein as previously reported (52). The octopine Ti plasmid pTiA6NC VirB4 protein does not affect membrane localization of VirB3 but does stabilize VirB3. VirB4, however, is not sufficient for pTiA6NC VirB3 stabilization. Two additional proteins, VirB7 and VirB8, are required for the stabilization of pTiA6NC VirB3.
MATERIALS AND METHODS
Strains, cell growth, and virulence assays.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains of E. coli were grown at 37°C in Luria broth, and Agrobacterium strains were grown in AB minimal medium at 28°C. Antibiotics were used at the following concentrations: for E. coli, carbenicillin at 60 μg/ml, tetracycline at 10 μg/ml, and gentamicin at 20 μg/ml; and for Agrobacterium, carbenicillin at 60 μg/ml and tetracycline at 7.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αF′ | F′ endA1 hsdR17 recA1 gyrA Δ(lacZYA-argF)U169 φ80dlacZΔM15 | Laboratory stock |
| CJ236 | FΔ(HindIII)::cat ung-1 dut-1 | Laboratory stock |
| Agrobacterium tumefaciens | ||
| C58 | Harbors nopaline Ti plasmid pTiC58 | Laboratory stock |
| A136 | C58 cured of pTiC58 | Laboratory stock |
| A348 | A136 with octopine Ti plasmid pTiA6NC | Laboratory stock |
| A281 | A136 with succinamopine Ti plasmid pTiBo542 | Laboratory stock |
| AD802 | phoA derivative of A136 | 15 |
| A348ΔB | A348 with a deletion of pTiA6NC virB operon (PC1001 to PC1011) | 2 |
| A348ΔB1 to -B11 | A348 strains with nonpolar deletion of each of the 11 pTiA6NC virB genes | 7 |
| Plasmids | ||
| pUC118 | ColE1 cloning vector | Laboratory stock |
| pBSIIKS+ | ColE1 cloning vector | Laboratory stock |
| pBSKSgn | Gentamicin-resistant derivative of pBSIIKS+ | F. White |
| pAD1448 | phoA fusion vector; phoA codon 11 to end of gene in pUC118 | 15 |
| pGEX2T | ColE1 vector for GST fusion construction | GE Healthcare |
| pTJS75 | Wide-host-range IncP cloning vector | Laboratory stock |
| pAD1708 | pTJS75 derivative with the constitutive vir gene regulator virG(N54D) mutant | This study |
| pAD1412 | pTJS75Δ1 derivative with the virG(N54D) mutant | 15 |
| pAD1756 | virDp in pBSKSgn | This study |
| pAD1757 | virB3 as a SalI-XhoI fragment in pAD1756 | This study |
| pAD1758 | pAD1412 fused to pAD1757 | This study |
| pB3n-phoA | pAD1448 with a virB3-phoA fusion at the nth codon of virB3 | This study |
| pWB3n-phoA | pAD1412 fused to pB3n-phoA | This study |
| pGFP2 | virDp-gfp mut3 in pUC118 | This study |
| pGFP5 | pGFP2 with XbaI site filled in | This study |
| pGfp-virB3 | virB3 as an XbaI fragment in pGFP2 | This study |
| pVirB3-gfp | virB3 as an XhoI fragment in pGFP5 | This study |
| pAD1765 | pAD1708 fused to pGfp-virB3 | This study |
| pAD1784 | pAD1708 fused to pVirB3-gfp | This study |
| pGST-virB2 | virB2 codon 48 to end of gene in pGEX2T | This study |
| pAD1287 | pTiA6NC virB operon in pBSKSII+ | 2 |
| pTJ12 | virB1 to virB5 in pBSIIKS+ | This study |
| pAD1882 | virB1 to virB4 in pBSIIKS+ | This study |
| pAD1887 | pAD1412 fused to pAD1882 | This study |
| pAD1885 | virB1, virB2, and virB3-virB4 fusion in pBSIIKS+ | This study |
| pAD1888 | pAD1412 fused to pAD1885 | This study |
| pAD1889 | virBp-virB4 in pBSIIKS+ | This study |
| pAD1890 | pAD1412 fused to pAD1889 | This study |
| pAD1891 | virDp-virB2 in pGFP5 | This study |
| pAD1892 | pAD1412 fused to pAD1891 | This study |
| pAD1416 | virDp in pUC118 | This study |
| pAD1910 | virDp-virB7 in pAD1416 | This study |
| pAD1916 | pAD1412 fused to pAD1910 | This study |
| pAD1911 | virDp-virB8 in pAD1416 | This study |
| pAD1917 | pAD1412 fused to pAD1911 | This study |
| pAD1915 | virDp-virB7 and virB8 in pAD1416 | This study |
| pAD1918 | pAD1412 fused to pAD1915 | This study |
| pGEX2T | GST fusion vector | GE Healthcare |
| pGST-B2 | virB2 codon 48 to end of gene in pGEX2T | This study |
To induce vir gene expression, Agrobacterium grown overnight in AB minimal medium was diluted 15- to 20-fold with AB induction medium (AB morpholineethanesulfonic acid, pH 5.8, 50 μM acetosyringone, and appropriate antibiotics) and grown at 22°C for 16 to 20 h. Virulence was monitored by tumor formation assays on Kalanchoë daigremontiana leaves as described previously (25, 36).
VirB3 fusion plasmids.
Plasmid pAD1757 contains virB3 under the control of the virD promoter (virDp). It was constructed by cloning the virB3 coding segment (positions −10 to +64; −n and +n indicate the numbers of nucleotides preceding the first base of the start codon and following the last base of the stop codon, respectively), obtained by PCR amplification, as a SalI-XhoI fragment into the SalI site of plasmid pAD1756, a gentamicin-resistant pBSIIKS+ derivative containing virDp. To improve the ribosome binding site sequence of virB3, a C→A substitution (in bold) was incorporated into the forward amplification primer 5′-GGGGGTCGACGAGGTAGTTGATGAATGATCG-3′ (the SalI site and the virB3 start codon are underlined).
To construct virB3-phoA fusions, the desired region of the virB3 coding segment (up to codon 40, 49, 77, 85, or 108) along with virDp of pAD1757 was amplified by PCR and cloned as a BamHI-PstI fragment into the phoA fusion vector pAD1448 (15). The resulting pB3n-phoA plasmids were fused to plasmid pAD1412 to construct pWB340-phoA, pWB349-phoA, pWB377-phoA, pWB385-phoA, and pWB3108-phoA, which are wide-host-range derivatives proficient in replication in Agrobacterium.
To study gfp fusions in Agrobacterium, we constructed the plasmid vector pGFP2, which contains a virDp-gfp mut3 gene (13). The virDp-gfp gene was amplified by PCR from plasmid pAD1730, a plasmid with a virDp-gfp-virD4 gene fusion, and cloned as a BglII-BamHI fragment into the plasmid vector pUC118. The gfp coding region in pGFP2 is flanked by unique XhoI and XbaI sites that permit construction of gene fusions with gfp at either the 3′ end (C-terminal fusion) or the 5′ end (N-terminal fusion). Plasmid pGFP5 is a derivative of pGFP2 in which the XbaI site was filled in to create a translational stop codon for the gfp open reading frame (ORF). To construct a gfp-virB3 fusion, the virB3 coding region (codon 2 to the end) was amplified by PCR and cloned as an XbaI fragment into pGFP2. The virB3-gfp fusion was constructed by cloning virB3 (codons 2 to 108) without the translation stop codon, obtained by PCR amplification, as an XhoI fragment into pGFP5. Plasmids pGfp-VirB3 and pVirB3-Gfp were fused to plasmid pAD1708 at a unique EcoRI site to construct the wide-host-range derivatives pAD1765 and pAD1784, respectively. Plasmid pAD1708 is a derivative of plasmid pTJS75 containing the constitutive vir gene regulatory mutant virG(N54D) (cited in reference 15).
virB expression plasmids.
Plasmid pTJ12 is a colE1 plasmid with pTiA6NC virB1 to virB5 under the control of the native virB promoter. It was constructed by deleting a 5.3-kb SacI fragment carrying the distal virB genes from plasmid pAD1287 (2). Plasmid pAD1882 contains virB1 to virB4 and was constructed by deleting the virB5 ORF (codons 10 to 213) by site-specific mutagenesis with oligonucleotide dB5 (GAGGGCACCAAAGATGACTCTAGACGTTGCAATAAGTTGCGTCG; complementary strand, with new XbaI site at the junction) on the single-stranded (ss) pTJ12 DNA template. Plasmid pAD1885 contains virB1, virB2, and a virB3-virB4 fusion gene (B34). It was constructed by site-specific mutagenesis of ss pTJ12 DNA with two mutagenic deoxyoligonucleotide primers: dB5 and B34 (TTCCACTCGCGCCGAGCTCCACCATTCCTCTCCCT; complementary strand, with SacI site at junction). The oligonucleotide B34 introduces a change in the virB3-virB4 junction region sequence from GTGTAATGCTC to GTGGAGCTC, placing the last virB3 codon, GTG, in frame with the second virB4 codon, CTC, with a glutamic acid codon (GAG) spacer. Plasmid pAD1889 contains virB4 under the control of the virB promoter. It was constructed by site-specific mutagenesis of ss pTJ12 DNA with oligonucleotides dB5 and dB1-3. Mutagenesis with dB1-3 (TCCTCTCCCTCGCGGTCTAGAAAGCGACCCCGATC) introduces a deletion of virB2 and nearly all of both virB1 and virB3 (fuses virB1 codon 9 to virB3 codon 102), placing virB4 next to its native promoter.
Plasmid pAD1891 contains a virDp-virB2 gene. It was constructed by cloning virB2 (positions −50 to +26) downstream from virDp in pGFP5. The virB2 DNA replaced the gfp ORF and was cloned as an XhoI-BamHI fragment. Plasmids pAD1910, pAD1911, and pAD1915 contain virDp-virB7, virDp-virB8, and virDp-virB7-virB8 genes, respectively, and were constructed by cloning DNA fragments encompassing virB7 (positions −30 to +22), virB8 (positions −21 to +28), and both genes (position −30 of virB7 to position +28 of virB8), obtained by PCR amplification, into plasmid pAD1416. Plasmids pAD1757, -1882, -1885, -1889, -1891, -1910, -1911, and -1915 were fused to pAD1412 to construct the wide-host-range derivatives pAD1758, -1887, -1888, -1890, -1892, -1916, -1917, and -1918, respectively. The plasmids were introduced into an appropriate Agrobacterium strain by electroporation (42). Plasmid pGST-B2 expresses a glutathione S-transferase (GST)-VirB2 fusion protein and was constructed by cloning a segment of virB2 (codon 48 to the end) as a BamHI-EcoRI fragment into the vector pGEX2T. All gene fusions and mutants were confirmed by DNA sequence analysis.
Alkaline phosphatase assay.
To measure alkaline phosphatase activity, a 0.5-ml bacterial culture grown in AB induction medium was used. Cells were permeabilized with 1 drop of chloroform and 1 drop of 0.05% SDS. Enzyme activity was determined spectrophotometrically, with p-nitrophenyl phosphate as a substrate, as described previously (15, 43). Data shown are averages for four experiments.
Cell fractionation and membrane purification.
Total membranes were isolated according to the method of de Maagd and Lugtenberg (18), with minor modifications. The bacterial pellet from 500 ml of culture was resuspended in buffer A (25 mM phosphate buffer, pH 7.2, 20% sucrose). After the addition of lysozyme (0.2 mg/ml [final concentration]), the cell mixture was incubated for 30 min at room temperature. DNase I, RNase I, and MgCl2 were then added to final concentrations of 0.2 mg/ml, 0.2 mg/ml, and 10 mM, respectively. Cells were placed on ice for 5 min and lysed by 2 to 3 passages through a French pressure cell at 16,000 to 18,000 lb/in2. The lysate was sonicated for 2 min, and phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM. After 10 min on ice, the lysate was centrifuged twice for 15 min each at 4,000 × g to remove unlysed cells and cell debris. KCl was added to the cell-free supernatant (50 mM final concentration), and the supernatant was centrifuged at 170,000 × g for 1 h. The pellet (membrane fraction) was rinsed with buffer A and resuspended in 0.6 ml buffer A. The high-speed supernatant (cytosolic-plus-periplasmic fraction) was saved for later use.
Inner and outer membranes were purified by density gradient centrifugation according to the method of Hancock and Nikaido (26). To improve the yield of outer membrane and to achieve better separation of the two membranes, EDTA-free buffers were used in all studies (26). The efficiency of separation was determined by assaying the enzyme activity of the inner membrane marker NADH oxidase (45). Samples were layered on top of a step gradient consisting of 1 ml 70%, 3 ml 64%, 3 ml 58%, 3 ml 52%, and 1.5 ml 40% sucrose in 25 mM phosphate buffer, pH 7.2, and centrifuged for 16 to 18 h at 151,000 × g in an SW41 rotor. Three distinct bands, for the inner membrane, an intermediate fraction, and the outer membrane, were evident after centrifugation. The bands were recovered, and membranes were pelleted by centrifugation at 170,000 × g for 1.5 h. Pellets were resuspended in 300 μl buffer A and stored at −70°C.
Protein analysis.
A GST-VirB2 fusion protein containing the C-terminal 74 residues of VirB2 was used as an antigen to raise polyclonal antibodies in rabbits. Antibodies were purified by affinity chromatography on an Affigel-GST-VirB2 fusion column (28). Antibodies against the other VirB proteins have been described previously (34). Antibodies against the nopaline VirB proteins were a generous gift of Christian Baron, University of Montreal, Canada.
VirB proteins were analyzed by Western blot assays following electrophoresis on SDS-Tricine-12.5% polyacrylamide gels (49). After gel electrophoresis, proteins were transferred to a nitrocellulose membrane. The membrane was first incubated with purified VirB2 (1:1,000 dilution), VirB3 (1:3,000), VirB4 (1:1,000), VirB7 (1:1,500), VirB8 (1:1,000), VirB9 (1:2,000), VirB10 (1:3,000), VirB11 (1:3,000), VirB3nop (1:1,000), VirB7nop (1:20,000), VirB11nop (1:10,000), GFP (1:6,000; eBioscience Corp.), or PhoA (1:20,000; 5′-3′, Inc.) antibody, followed by secondary antibodies conjugated to IR dye 680 (Li-Cor Biosciences). Proteins were detected by a Li-Cor Odyssey infrared imaging system, and scans were quantified by measuring the pixel volume of a band, using Odyssey software. To compare VirB3 levels between two strains, we quantified both the VirB3 and VirB10 bands. The ratio of VirB3 to VirB10 was used to calculate the fold difference in the level of VirB3. For quantification of the membrane fractions, the total of the inner and outer membrane fractions was arbitrarily set at 100%.
Epifluorescence microscopy.
GFP fusions were analyzed by epifluorescence microscopy as described earlier (36). A 5-microliter sample of induced bacteria was placed on a polylysine-coated slide, and a coverslip was placed on top. After 10 to 15 min at room temperature to allow cell attachment, samples were viewed with a Leica DM LB2 fluorescence microscope equipped with a Hamamatsu Orca-ER digital charge-coupled device camera. Images were recorded with Openlab software (Improvision Inc.) and processed with Adobe Photoshop CS2 (Adobe Corp.) for presentation.
RESULTS
Topology of VirB3.
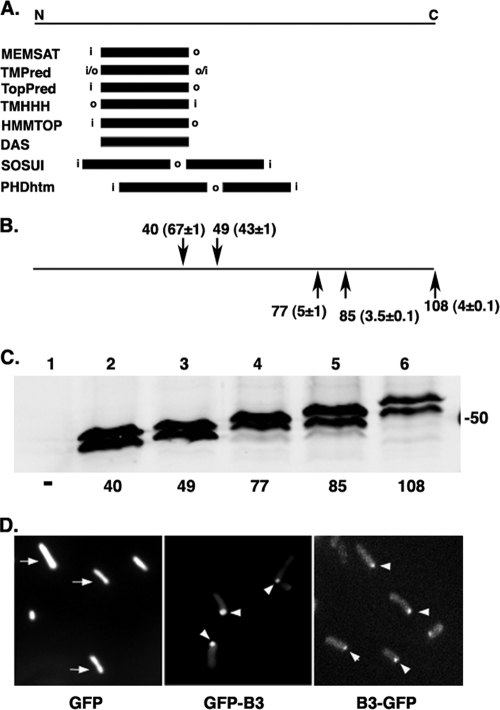
VirB3 is a small protein of 108 amino acid residues with a large hydrophobic region (residues 10 to 76). To investigate whether the hydrophobic region functions in membrane spanning, the primary sequence of VirB3 was analyzed in silico, using several membrane topology prediction programs available free on the Internet. Six of the eight programs predicted that VirB3 contains a single MSD around residues 18 to 43 (Fig. 1A). Two programs predicted that VirB3 contains two MSDs (at aa 13 to 35 and 40 to 62 and aa 23 to 46 and 51 to 68), with a small, four-residue periplasmic loop. Almost all programs predicted that the N terminus resides in the cytoplasm. The C terminus of the protein will therefore lie in the periplasm in the single-MSD model and in the cytoplasm in the two-MSD model.
FIG. 1.
Topology of VirB3. (A) Membrane-spanning domains predicted by the commonly used protein topology programs available free on the Internet. The position of an MSD on a linear map of the protein is shown as a dark box. i, inside (cytoplasmic location); o, outside (periplasmic); N, N terminus; C, C terminus. (B) Locations of targeted PhoA fusions and their alkaline phosphatase activities. The number (mean alkaline phosphatase activity ± standard deviation [SD]) identifies the VirB3 residue at the fusion junction. The PhoA-positive and PhoA-negative fusions are shown above and below the line, respectively. (C) Stability of VirB3-PhoA fusions. The stability of VirB3 fusion proteins was monitored by Western blot assays using anti-PhoA antibodies. Total proteins were separated by electrophoresis in SDS-12.5% polyacrylamide (PA) gels, transferred to nitrocellulose filters, and probed with anti-PhoA antibodies. Lanes 1 to 6, Agrobacterium AD802 expressing no PhoA fusion, B340-PhoA, B349-PhoA, B377-PhoA, B385-PhoA, and B3108-PhoA, respectively. The upper band in lanes 2 to 6 is likely the unprocessed fusion protein. The number on the right (in kDa) indicates the position of a marker protein. (D) Analysis of GFP fusions. Phenotypes of GFP-VirB3 and VirB3-GFP were determined by epifluorescence microscopy (36). Bacteria expressing GFP exhibited fluorescence throughout the cell (arrows), and those expressing GFP-VirB3 (GFP-B3) or VirB3-GFP (B3-GFP) had fluorescence predominantly at a cell pole (arrowheads).
To experimentally determine protein topology, we analyzed fusions of VirB3 with the periplasmic marker protein PhoA. We constructed five fusions where the mature E. coli PhoA protein was fused to amino acid 40, 49, 77, 85, or 108 of VirB3. All five fusions would possess alkaline phosphatase activity if VirB3 has a single MSD and a cytoplasmic N terminus, the preferred in silico topology. In the alternate, two-MSD model, only two fusions, those at aa 40 and 49, would be expected to exhibit alkaline phosphatase activity. Analysis of the fusion proteins by enzyme assays and protein analysis supported the two-MSD model (Fig. 1B and C). Two of the fusions, those at aa 40 and 49, had alkaline phosphatase activity, while the remaining three (C-terminal fusions) exhibited little or no enzyme activity. Western blot assays showed that comparable levels of the fusion proteins accumulated in all strains. These results indicate that VirB3 contains one periplasmic domain and that the C terminus of the protein resides in the cytoplasm. The absence of a signal sequence and the PhoA+ phenotype of VirB340-PhoA and VirB349-PhoA strongly suggest that the N terminus of the protein is cytoplasmic. Therefore, the first hydrophobic segment (within residues 13 to 40) must function in membrane spanning. To have both N and C termini in the cytoplasm, the protein needs to cross the membrane twice, suggesting that VirB3 has two MSDs.
A PhoA-negative phenotype is a negative result and can be suggestive only of a cytoplasmic location of a protein domain. To verify the cytoplasmic assignment of both the N and C termini, we analyzed fusions of VirB3 with the cytoplasmic marker GFP (19, 20). We constructed two gfp fusions, namely, gfp-virB3, where gfp was fused to the second codon of virB3 (N-terminal fusion), and virB3-gfp, in which gfp was fused to the last codon of virB3 (C-terminal fusion). Bacteria expressing GFP-VirB3 or VirB3-GFP exhibited green fluorescence, indicating that both the N and C termini of the protein reside in the cell cytoplasm (Fig. 1D). Both fusions, like wild-type VirB3, localized to a cell pole.
A VirB3-VirB4 fusion is functional in DNA transfer.
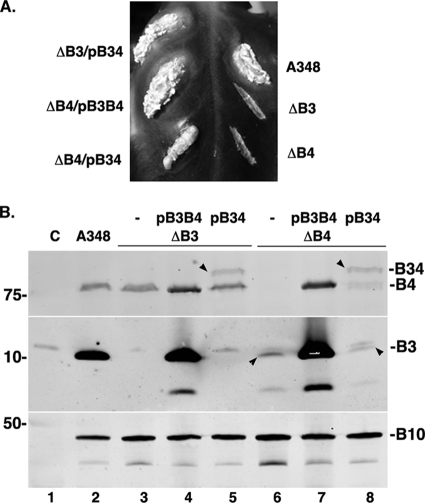
A small subset of VirB3 homologues are large proteins composed of both VirB3 and VirB4. We studied whether a fusion of the Agrobacterium VirB3 and VirB4 proteins would retain biological activities of both components. Using site-specific mutagenesis, the virB3-virB4 intergenic region was modified to construct a virB3-virB4 gene fusion in which the virB3 stop codon was replaced with a glutamic acid codon (to create a scorable SacI restriction site) and was followed by the second codon of virB4. The activity of the VirB3-VirB4 fusion protein was monitored by determining its ability to complement a nonpolar deletion in virB3 or virB4 in tumor formation assays on K. daigremontiana leaves (Fig. 2A). The fusion protein complemented both mutations, albeit with different efficiencies. While it fully complemented the virB3 deletion mutant Agrobacterium A348ΔB3, the virulence of A348ΔB4 was only partially restored. In the latter case, the appearance of tumors was delayed and tumor sizes were small.
FIG. 2.
A VirB3-VirB4 fusion protein is functional in DNA transfer to plants. (A) Complementation of Agrobacterium A348ΔB3 (ΔB3) and A348ΔB4 (ΔB4) was monitored by tumor formation assays. Plasmid pB34 (pAD1888) expresses a virB3-virB4 fusion, and plasmid pB3B4 (pAD1887) expresses wild-type virB3 and virB4. Both plasmids express virB1 and virB2 as well. (B) Expression of VirB3-VirB4 fusion and other VirB proteins was monitored by Western blot assays. Proteins were separated by electrophoresis in SDS-10% PA gels (VirB4 and VirB3-VirB4 fusion) or SDS-12.5% PA-Tricine gels (VirB3 and VirB10). Arrowheads identify the VirB3-VirB4 fusion bands (lanes 5 and 8) and the weak VirB3 bands in two of the A348ΔB4 derivatives (lanes 6 and 8). Note that no VirB3-specific band is detectable for A348ΔB3/pB34, which expresses the VirB3-VirB4 fusion protein (lane 5). A protein migrating slightly slower than VirB3 cross-reacted nonspecifically with the anti-VirB3 antibodies. VirB3 expression was high in wild-type A348 and in bacteria expressing virB3 from a plasmid (lanes 2, 4, and 7). The lower bands in both the VirB3 and VirB10 blots are degradation products. Lane C, induced A348ΔB.
Expression of the virB3-virB4 fusion gene was monitored by Western blot assays. The fusion protein accumulated at a level lower than that of wild-type VirB4 (Fig. 2B, compare lanes 5 and 8 with lane 2). The low level of VirB3-VirB4 in the cell may be a reason for the low efficiency of virB4 complementation. Analysis with anti-VirB3 antibodies showed that bacteria expressing the VirB3-VirB4 fusion did not accumulate wild-type VirB3 (lane 5). These results indicate that the VirB3-VirB4 fusion is responsible for the complementation of the virB3 deletion mutant. VirB4 has a stabilizing effect on VirB3 (discussed later). The VirB3-VirB4 fusion lacks this VirB4 function, as no increase in VirB3 accumulation was observed in Agrobacterium A348ΔB4 expressing the fusion protein (lane 8). The VirB3-VirB4 fusion protein therefore does not interact with the free VirB3 present in A348ΔB4. One possible reason for the loss of interaction is that intramolecular interaction between the VirB3 and VirB4 domains of the VirB3-VirB4 fusion makes the VirB4 domain unavailable for intermolecular interaction with free VirB3.
VirB3 is an inner membrane protein.
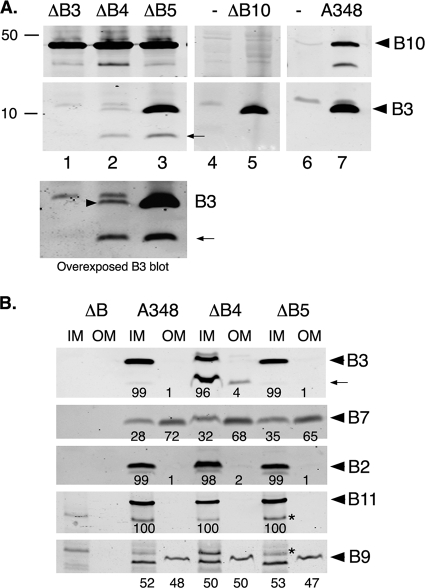
The topology of VirB3 presented in this report suggests that VirB3 is an inner membrane protein. In a previous study, the nopaline Ti plasmid pTiC58 VirB3 protein was found associated primarily with the outer membrane (52). Localization to the outer membrane was dependent on VirB4 (33). To address this apparent anomaly, we determined the subcellular locations of VirB3 proteins from two Ti plasmids, the octopine Ti plasmid pTiA6NC and the leucinopine Ti plasmid pTiBo542, by biochemical methods. The two proteins differed only at three positions near the N-terminal end, and the anti-VirB3 antibodies raised against the C-terminal 15 residues of the pTiA6NC protein recognized both VirB3 proteins (Fig. 3A).
FIG. 3.
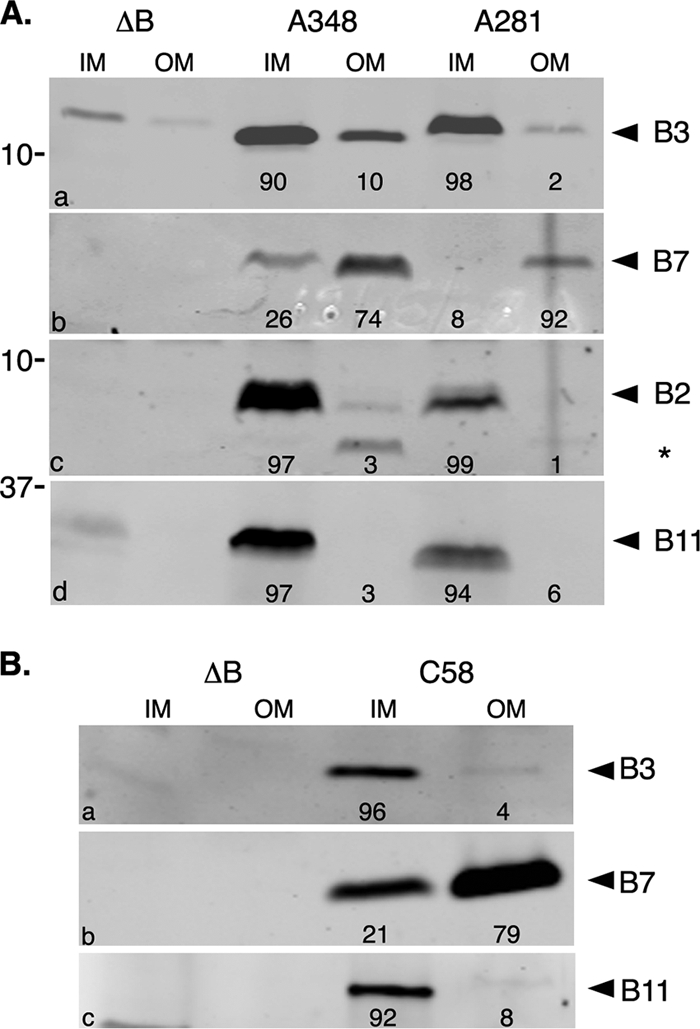
Membrane localization of VirB3. Subcellular locations of VirB3 expressed from the Ti plasmids pTiA6NC (A), pTiBo542 (A), and pTiC58 (B) were determined by Western blot assays. Inner (IM) and outer (OM) membranes were separated by sucrose density gradient centrifugation, and the presence of VirB proteins was monitored by Western blot assays. Proteins were separated in SDS-12.5% PA-Tricine gels and transferred to nitrocellulose filters. (a) The filter was probed with anti-VirB3 antibodies. For control experiments, the filter in panel a was probed with anti-VirB11 antibodies (d), and a duplicate filter was probed sequentially with anti-VirB7 (b) and anti-VirB2 (c) antibodies. The residual VirB7-specific band in panel c is marked with an asterisk. Bands were quantified by measuring pixel volume, using Li-Cor Odyssey software. The number below each lane indicates the percentage of the protein present in that fraction. ΔB, A348ΔB; A348, octopine strain with pTiA6NC; A281, supervirulent strain with pTiBo542; C58, nopaline strain with pTiC58.
Inner and outer membranes were purified by sucrose density gradient centrifugation as described by Hancock and Nikaido (26). The efficiency of separation was determined by assaying the enzyme activity of the inner membrane marker NADH oxidase. About 95% of the NADH oxidase fractionated with the inner membrane fraction. Protein profile analysis by SDS-PAGE showed that the outer membrane fraction contained the unique Omp proteins (data not shown). Localization of several Vir proteins by Western blot assays provided additional support for relatively low-level or no cross-contamination between the two membrane fractions. The inner membrane proteins VirB8, VirB11, and VirD4 fractionated exclusively with the inner membrane fraction, and the VirB7 lipoprotein localized primarily to the outer membrane (Fig. 3A and data not shown). Analysis using anti-VirB3 antibodies showed that almost all of the VirB3 (≥90%) of both the octopine and leucinopine Ti plasmids localized to the inner membrane. VirB2, like VirB3, localized exclusively to the inner membrane. The pTiC58 VirB2 protein was found associated with both membranes (52).
These results do not agree with a previous report that the nopaline Ti plasmid pTiC58 VirB3 protein is an outer membrane protein (52). To address this difference, we determined the subcellular location of pTiC58 VirB3 (Fig. 3B). Like its counterparts expressed from the other two Ti plasmids, the nopaline Ti plasmid VirB3 protein localized exclusively to the inner membrane. In control experiments, the majority of pTiC58 VirB7 protein localized exclusively to the outer membrane, and the pTiC58 VirB11 protein localized to the inner membrane, as expected.
VirB4 does not affect membrane localization of pTiA6NC VirB3.
Both accumulation and membrane localization of pTiC58 VirB3 are affected by VirB4 (33). To study the effect of virB4 on pTiA6NC virB3, we analyzed Agrobacterium A348ΔB4, the octopine strain with a nonpolar deletion in virB4 (7). In agreement with previous observations, this strain accumulated a very low level of VirB3 (Fig. 4A). It also accumulated a significant amount of a degradation product (marked with an arrow in Fig. 4A and B). In contrast, strains with a nonpolar deletion in either virB5 or virB10, like the wild-type parent, accumulated a high level of VirB3. Some accumulation of the VirB3 degradation product was observed in strain A348ΔB5; however, this represented a small percentage (<4%) of the total cellular VirB3. Comparable levels of VirB10 and other VirB proteins accumulated in the virB3, virB4, and virB5 deletion strains, indicating that the mutations did not have a global effect on VirB protein levels (Fig. 4) (reference 7 and data not shown).
FIG. 4.
Effect of VirB4 on stability and membrane localization of VirB3. The effect of VirB4 on VirB3 was monitored by Western blot assay. (A) VirB4 is required for VirB3 stability. Total proteins from Agrobacterium strains were separated by electrophoresis on SDS-PA-Tricine gels and then transferred to a nitrocellulose filter. The filter was first probed with anti-VirB3 antibodies and then with anti-VirB10 antibodies. Agrobacterium A348ΔB4 expresses a low level of VirB3 (arrowhead in lane 2 of overexposed panel) and accumulates a significant amount of a degradation product (arrow). (B) VirB3 localized to the inner membrane in both the presence and absence of VirB4. Subcellular localization of VirB proteins was monitored as described in the legend to Fig. 3. Similar amounts of protein were used for all analyses, except for the VirB3 blot. To detect the small amount of VirB3 present in A348ΔB4, an eightfold excess of both membrane fractions was loaded on the gel. The number below each lane indicates the percentage of the protein present in that particular fraction. The VirB9-specific band in the OM fraction migrated slightly slower than that in the IM fraction, probably because of the high carbohydrate content of the OM. The band marked with an asterisk in the VirB9 and VirB11 blots is a nonspecific band found in all blots probed with antibodies raised in rabbits. −, no Ti plasmid; A348, wild-type strain; ΔB, A348ΔB; ΔB3, A348ΔB3; ΔB4, A348ΔB4; ΔB5, A348ΔB5; ΔB10, A348ΔB10.
Analysis of purified inner and outer membrane fractions by Western blot assays using anti-VirB3 antibodies showed that VirB4 had no effect on membrane localization of pTiA6NC VirB3. VirB3 in Agrobacterium A348ΔB4 localized primarily to the inner membrane (Fig. 4B). A similar inner membrane localization of VirB3 was observed in the VirB4-containing deletion mutant Agrobacterium A348ΔB5 and in three wild-type strains (Fig. 3 and 4B).
In both Agrobacterium A348ΔB4 and A348ΔB5, the majority of the VirB7 lipoprotein was found associated with the outer membrane, while VirB9 fractionated almost equally with both the inner and outer membranes. The inner membrane proteins VirB11 and VirB2 localized exclusively to that membrane. The absence of VirB4 had no significant effect on either the accumulation or membrane localization of these proteins. Comparable levels of VirB2, VirB7, VirB9, and VirB11 were found in the two strains.
Three VirB proteins, VirB4, VirB7, and VirB8, are required for stabilization of pTiA6NC VirB3.
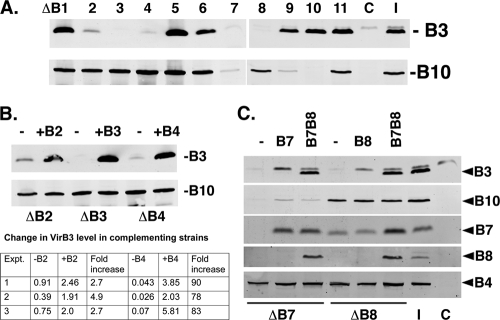
VirB4 is required for stabilization of VirB3 (Fig. 4A) (33, 55). We investigated whether other VirB proteins are required for VirB3 stability. Eleven Agrobacterium mutants with a nonpolar deletion in each of the pTiA6NC virB genes (7) were analyzed by Western blot assays (Fig. 5A). Three mutants, Agrobacterium A348ΔB4, A348ΔB7, and A348ΔB8, accumulated little or no VirB3. One mutant, A348ΔB2, accumulated a low level of VirB3. To ensure that the differences in levels of VirB3 in the mutants did not result from variation in vir gene induction, we determined the cellular level of a distal virB gene product, VirB10. All mutants except for Agrobacterium A348ΔB7 and A348ΔB9 accumulated significant amounts of VirB10. Only one of the two exceptions, A348ΔB7, was deficient in VirB3 accumulation. The mutation in Agrobacterium A348ΔB7 is pleiotropic in nature, and the accumulation of all downstream proteins was greatly reduced in this mutant (Fig. 5C) (7, 22). A nonpolar deletion in virB9, virB10, or virB11, however, had no effect on VirB3 accumulation. Therefore, the very low level of VirB3 in Agrobacterium A348ΔB7 must be due to the absence of VirB7, VirB8, or both proteins.
FIG. 5.
VirB4, VirB7, and VirB8 are required for stability of VirB3. (A) VirB3 accumulation in virB mutants. The level of VirB3 in Agrobacterium strains A348ΔB1 to -B11, with a nonpolar deletion of each of the 11 virB genes, was monitored by Western blot assays. The level of VirB10 was monitored as a control for virB expression. (B) VirB3 level in A348ΔB4 is restored by virB4 provided in trans. The levels of VirB3 and VirB10 in A348ΔB2 to -B4 and their derivatives harboring an expression plasmid containing the missing gene were monitored. Increases in the VirB3 level upon complementation of the A348ΔB2 and A348ΔB4 strains were determined by quantifying the VirB3- and VirB10 (both induction and loading controls)-specific bands. (C) Both VirB7 and VirB8 are required for VirB3 accumulation in A348ΔB7 and A348ΔB8. Levels of VirB3 and other relevant VirB proteins in Agrobacterium A348ΔB7, A348ΔB8, and their derivatives harboring a plasmid containing virB7 (lane B7), virB8 (lane B8), or both virB7 and virB8 (lanes B7B8) were monitored. C, induced A348ΔB; I, induced A348.
The low level of VirB3 in Agrobacterium A348ΔB2 suggests a role of VirB2 in VirB3 stabilization. However, the efficiency of virB3 translation initiation may have inadvertently been altered during the construction of Agrobacterium A348ΔB2. Translation of virB3 mRNA from the polycistronic virB mRNA is under the control of virB2, not virB3, sequences in this mutant, because to introduce a complete deletion of virB2 the virB2 start codon was fused to the second codon of virB3. Since ribosome binding site sequences and their locations relative to the initiator ATG codon are important determinants of the efficiency of translation initiation, an alteration in these sequences is likely to affect protein levels in a cell. In addition, virB2-virB3 shares overlapping stop and start codons, suggesting that translational coupling may play a role in virB3 translation (17). To investigate the effect of VirB2 on VirB3 stability, we determined whether virB2 supplied in trans would restore VirB3 to a near-wild-type level in the A348ΔB2 host. If inefficient translation was the reason for the low VirB3 level, no major change in protein level would be expected. In strain A348ΔB2/pvirB2, only a small (∼3- to 5-fold) increase in the VirB3 level was observed, suggesting that VirB2 probably has no significant effect on VirB3 stability (Fig. 5B). For comparison, the addition of virB4 to A348ΔB4 led to an ∼75-fold increase in VirB3 level.
In addition to Agrobacterium A348ΔB4, two other mutants, A348ΔB7 and A348ΔB8, accumulated very low levels of VirB3. Agrobacterium A348ΔB7 did not accumulate an appreciable level of VirB8 and is thus deficient in both VirB7 and VirB8 (Fig. 5C) (7). The introduction of a virB7 overexpression plasmid into A348ΔB7 restored VirB7 protein but did not restore accumulation of VirB3 or VirB8. Coexpression of virB7 and virB8 led to accumulation of VirB3 along with VirB7 and VirB8. Therefore, both VirB7 and VirB8 are required for VirB3 stability, and VirB7 is incapable of stabilizing VirB3 in the absence of VirB8. Analysis of Agrobacterium A348ΔB8 harboring the virB7- and virB8-overexpressing plasmid supported this conclusion. The VirB3 level was restored in Agrobacterium A348ΔB8 harboring the virB7 and virB8 overexpression plasmid. Agrobacterium A348ΔB8 with a plasmid containing only the virB8 ORF did not stabilize VirB3, but this strain failed to accumulate VirB8 (Fig. 5C, lane 5). This result was not unexpected, because during characterization of the A348ΔB8 mutant, it was noted (7) that virB8 expression requires virB7 and that full complementation of the mutant requires coexpression of virB7 and virB8.
DISCUSSION
Agrobacterium VirB3 contains a large hydrophobic region. Our topology analysis with targeted PhoA and GFP fusions indicates that this region traverses the cytoplasmic membrane twice, placing the VirB3 C terminus in the cell cytoplasm. Functional analysis of a chimeric VirB3-VirB4 fusion protein provided additional support for the deduced topology. To retain both VirB3 and VirB4 functions in the VirB3-VirB4 fusion protein, the C-terminal end of VirB3 and the N-terminal end of VirB4 must be targeted to their proper subcellular locations, because mislocalization of either domain is likely to be detrimental to protein function. The N terminus of VirB4 is cytoplasmic (14). Only a cytoplasmic location of the VirB3 C terminus will target the VirB4 N-terminal region in a fusion protein to the same cellular compartment, its proper subcellular location. The VirB3-VirB4 fusion retained both VirB3 and VirB4 functions, as it fully complemented the nonpolar virB3 deletion mutant A348ΔB3 and was moderately competent in complementing the nonpolar virB4 deletion mutant A348ΔB4 (Fig. 2).
The biochemical fractionation studies presented here show that VirB3 is an inner membrane protein (Fig. 3). The VirB2 pilin protein of the pTiA6NC and pTiBo542 Ti plasmids also localized to the inner membrane, suggesting that the T-pilin monomers accumulate at this membrane (Fig. 3). The monomers need to be extracted from the inner membrane and transported through the T4S apparatus for T-pilus assembly. VirB3 may play a role in this process. A similar inner membrane localization of unassembled pilin of the F plasmid as well as the VirB3 homolog TraL has been observed (29, 46).
Three VirB proteins, VirB4, VirB7, and VirB8, are required for VirB3 stability (Fig. 4 and 5). Both VirB4 and VirB8 must be present in a cell for VirB3 stabilization, since a strain lacking either protein, e.g., A348ΔB4 (lacks VirB4) or A348ΔB8, A348ΔB7/pB7, or A348ΔB8/pB7 (all lack VirB8), failed to accumulate VirB3. Whether there is a similar requirement for VirB7 could not be addressed because the virB7 deletion mutant A348ΔB7 lacks VirB8 as well, and virB8 could not be expressed in trans without virB7 (Fig. 5) (7, 22). The role of VirB7 may simply be to ensure virB8 expression, or like VirB4 and VirB8, VirB7 may have a direct effect on VirB3 stability. The three proteins are unable to substitute for one another, as overexpression of virB4 in A348ΔB7 and A348ΔB8 or of virB7 and virB8 in A348ΔB4 had no stabilizing effect on VirB3 (data not shown). Binary interaction of VirB3 and VirB4/VirB7/VirB8 is therefore not sufficient for VirB3 stabilization, and all four proteins must be present in the bacterium for accumulation of wild-type levels of VirB3.
The requirement of three proteins for the stabilization of VirB3 suggests that VirB3 needs to interact with proteins already in a complex for its stability. The VirB proteins assemble two structures: the T4S channel and the T-pilus. Two VirB3-stabilizing proteins, VirB7 and VirB8, are components of the T4S channel (11, 22, 23, 34). Other components of the secretion channel, VirB6, VirB9, and VirB10, however, are not required for VirB3 stabilization, indicating that channel assembly is not necessary for VirB3 stability (Fig. 5). The stabilizing effect of VirB7, VirB8, and VirB4 on VirB3 is therefore likely to be required for T-pilus biogenesis.
Structural studies with the E. coli pKM101 proteins showed that TraE (VirB8) does not stably associate with the TraN-TraO-TraF (VirB7-VirB9-VirB10) T4S core complex (11, 23). TraE either is not a structural component of this T4S apparatus or is loosely associated with the core complex. Studies with the VirB proteins suggest that the latter is probably the case in Agrobacterium. The association of pTiA6 VirB8 with the T-strand DNA before it makes contact with the proteins of the core complex suggests a role for VirB8 early in the DNA translocation process (9). VirB8 also targets the core T4S apparatus proteins, VirB7, VirB9, and VirB10, to the cell pole during T4S apparatus assembly and is involved in interactions with several VirB proteins, including VirB9 and VirB10 (9, 16, 34, 38). Recently, the nopaline Ti plasmid T4SS proteins were found to localize to the cell circumference, not to a cell pole (1). The reason for the difference is not apparent. Several pTiA6 VirB proteins did localize to multiple sites on the cell membrane, but only in strains that lacked one or more VirB proteins (34). In wild-type bacteria and bacteria expressing a defined subset of the VirB proteins, the pTiA6 proteins localized to a cell pole. Future studies will determine whether the subcellular location of the T4SS differs in the two Ti plasmids.
In addition to its role in the formation of the T4SS core complex, VirB7 plays a role in pilus assembly, since small amounts of VirB7 are found associated with the T-pilus (48). While VirB3 is not a component of the T-pilus, it probably is required for pilus biogenesis, as it lies within the VirB2-VirB3-VirB4-VirB5 pilus cluster, interacts with the pilin components VirB2 and VirB5, and participates in the formation of a VirB2-VirB3-VirB5 protein complex (52, 55). Stabilization of VirB3 is likely to precede the formation of the VirB2-VirB3-VirB5 complex, because constituents of this complex have no significant effect on VirB3 stability. The requirement of VirB8 for VirB3 stabilization suggests that in addition to its role in T4S apparatus assembly, VirB8 has an additional role in T-pilus biogenesis. Formation of a protein complex involving VirB3, VirB4, VirB7, and VirB8 may be the first step in T-pilus assembly. This is followed by the recruitment of the pilin components VirB2 and VirB5 for delivery to the pilus assembly site. VirB3 thus can serve as a coupling protein to deliver VirB2 and VirB5 monomers to the T4S machinery for pilus assembly.
The present study suggests that Agrobacterium may use selective proteolysis as a mechanism to regulate the assembly of complex structures. We postulate that VirB3 serves as a sensor for the pilus assembly pathway. Insufficient VirB3, resulting from an unavailability of sufficient VirB4, VirB7, or VirB8, will shut off pilus assembly. Assembly of the T-pilus requires the T4S channel (40). VirB8, a central component of the channel, probably serves as a monitor for channel assembly. The absence of VirB8, indicative of the lack of a secretion channel, prevents assembly of abortive prepilus complexes by not stabilizing VirB3.
Acknowledgments
We are grateful to Christian Baron for generously providing antibodies against the nopaline VirB proteins and to Peter Christie for the Agrobacterium virB deletion mutants.
This work was supported by a grant from the University of Minnesota Agricultural Experiment Station.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Aguilar, J., J. Zupan, T. A. Cameron, and P. C. Zambryski. 2010. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U. S. A. 107:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. U. S. A. 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmakuri, K., Z. Ding, and P. J. Christie. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 6.Beijersbergen, A., S. Smith, and P. J. Hooykaas. 1994. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32:212-218. [DOI] [PubMed] [Google Scholar]
- 7.Berger, B., and P. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broome-Smith, J., and B. Spratt. 1986. A vector for the construction of translational fusions to TEM beta-lactamase and the analysis of protein export signals and membrane protein topology. Gene 49:341-349. [DOI] [PubMed] [Google Scholar]
- 9.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandran, V., R. Fronzes, S. Duquerroy, N. Cronin, J. Navaza, and G. Waksman. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormack, B., R. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 14.Dang, T., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, A., and Y. H. Xie. 1998. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol. Microbiol. 27:405-414. [DOI] [PubMed] [Google Scholar]
- 16.Das, A., and Y. H. Xie. 2000. Agrobacterium tumefaciens T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, A., and C. Yanofsky. 1984. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 12:4757-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Maagd, R. A., and B. Lugtenberg. 1986. Fractionation of Rhizobium leguminosarum cells into outer membrane, cytoplasmic membrane, periplasmic, and cytoplasmic components. J. Bacteriol. 186:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drew, D., D. Sjöstrand, J. Nilsson, T. Urbig, C. N. Chin, J. W. deGier, and G. von Heijne. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl. Acad. Sci. U. S. A. 99:2690-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feilmeier, B., G. Iseminger, D. Schroeder, H. Webber, and G. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez, D., T. A. Dang, G. Spudich, X.-R. Zhou, B. Berger, and P. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez, D., G. Spudich, X.-R. Zhou, and P. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fronzes, R., E. Schäfer, L. Wang, H. Saibil, E. Orlova, and G. Waksman. 2009. Structure of a type IV secretion system core complex. Science 323:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galli, D. M., J. Chen, K. F. Novak, and D. J. Leblanc. 2001. Nucleotide sequence and analysis of conjugative plasmid pVT745. J. Bacteriol. 183:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garfinkel, D., and E. Nester. 1980. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J. Bacteriol. 144:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock, R. E. W., and H. Nikaido. 1978. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hapfelmeier, S., N. Domke, P. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlaw, E., and D. Lane. 1988. Antibodies—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Hazes, B., and L. Frost. 2008. Towards a systems biology approach to study type II/IV secretion systems. Biochim. Biophys. Acta 1778:1839-1850. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman, C. S., and A. Wright. 1985. Fusions of secreted proteins to alkaline phosphatase: an approach for studying protein secretion. Proc. Natl. Acad. Sci. U. S. A. 82:5107-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubowski, S. J., V. Krishnamoorthy, E. Cascales, and P. J. Christie. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J. Mol. Biol. 341:961-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Jones, A. L., K. Shirasu, and C. Kado. 1994. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J. Bacteriol. 176:5255-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Judd, P. K., R. B. Kumar, and A. Das. 2005. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc. Natl. Acad. Sci. U. S. A. 102:11498-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judd, P. K., R. B. Kumar, and A. Das. 2005. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol. Microbiol. 55:115-124. [DOI] [PubMed] [Google Scholar]
- 36.Judd, P. K., D. Mahli, and A. Das. 2005. Molecular characterization of the Agrobacterium tumefaciens DNA transfer protein VirB6. Microbiology 151:3483-3492. [DOI] [PubMed] [Google Scholar]
- 37.Katada, T., M. Tamura, and M. Ui. 1983. The A protomer of islet-activating protein, pertussis toxin, as an active peptide catalyzing ADP-ribosylation of a membrane protein. Arch. Biochem. Biophys. 224:290-298. [DOI] [PubMed] [Google Scholar]
- 38.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar, R. B., and A. Das. 2002. Functional domains and polar location of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 40.Lai, E.-M., O. Chesnokova, L. Banta, and C. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mersereau, M., G. Pazour, and A. Das. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149-151. [DOI] [PubMed] [Google Scholar]
- 43.Miller, J. 1992. A short course in bacterial genetics: laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Moss, J., S. J. Stanley, D. L. Burns, J. Hsia, D. Yost, G. Myers, and E. Hewlett. 1983. Activation by thiol of the latent NAD glycohydrolase and ADP-ribosyltransferase activities of Bordetella pertussis toxin (islet-activating protein). J. Biol. Chem. 258:11879-11882. [PubMed] [Google Scholar]
- 45.Osborn, M. J., J. E. Gander, and E. Parisi. 1972. Mechanism of assembly of the outer membrane of Salmonella typhimurium: site of synthesis of lipopolysaccharide. J. Biol. Chem. 247:3973-3986. [PubMed] [Google Scholar]
- 46.Paiva, W. D., T. Grossman, and P. M. Silverman. 1992. Characterization of F-pilin as an inner membrane component of Escherichia coli K12. J. Biol. Chem. 267:26191-26197. [PubMed] [Google Scholar]
- 47.Saadat, I., H. Higashi, C. Obuse, M. Umeda, N. Murata-Kamiya, Y. Saito, H. Lu, N. Ohnishi, T. Azuma, A. Suzuki, S. Ohno, and M. Hatakeyama. 2007. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447:330-334. [DOI] [PubMed] [Google Scholar]
- 48.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schagger, H. 2006. Tricine-SDS-PAGE. Nat. Protoc. 1:18-22. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Shirasu, K., and C. Kado. 1993. Membrane localization of the Ti plasmid VirB proteins involved in biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111:287-294. [DOI] [PubMed] [Google Scholar]
- 53.Thorstenson, Y. R., and P. C. Zambryski. 1994. The essential virulence protein VirB8 localizes to the inner membrane of Agrobacterium tumefaciens. J. Bacteriol. 176:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, Q., A. Carle, C. Gao, D. Sivanesan, K. A. Aly, C. Hoppner, L. Krall, N. Domke, and C. Baron. 2005. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J. Biol. Chem. 280:26349-26359. [DOI] [PubMed] [Google Scholar]
- 56.Zupan, J., T. Muth, O. Draper, and P. Zambryski. 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23:11-28. [DOI] [PubMed] [Google Scholar]