Abstract
The purple nonsulfur photosynthetic bacterium Rhodobacter capsulatus has been extensively studied for its metabolic versatility as well as for production of a gene transfer agent called RcGTA. Production of RcGTA is highest in the stationary phase of growth and requires the response regulator protein CtrA. The CtrA protein in Caulobacter crescentus has been thoroughly studied for its role as an essential, master regulator of the cell cycle. Although the CtrA protein in R. capsulatus shares a high degree of sequence similarity with the C. crescentus protein, it is nonessential and clearly plays a different role in this bacterium. We have used transcriptomic and proteomic analyses of wild-type and ctrA mutant cultures to identify the genes dysregulated by the loss of CtrA in R. capsulatus. We have also characterized gene expression differences between the logarithmic and stationary phases of growth. Loss of CtrA has pleiotropic effects, with dysregulation of expression of ∼6% of genes in the R. capsulatus genome. This includes all flagellar motility genes and a number of other putative regulatory proteins but does not appear to include any genes involved in the cell cycle. Quantitative proteomic data supported 88% of the CtrA transcriptome results. Phylogenetic analysis of CtrA sequences supports the hypothesis of an ancestral ctrA gene within the alphaproteobacteria, with subsequent diversification of function in the major alphaproteobacterial lineages.
The purple nonsulfur bacterium Rhodobacter capsulatus is a model organism for various aspects of bacterial physiology, such as bioenergetics and N2 fixation, and also engages in an unusual mechanism of genetic exchange, carried out by a bacteriophage-like element called the R. capsulatus gene transfer agent (RcGTA) (34, 56). The production of RcGTA is maximal in the stationary phase of growth of R. capsulatus cultures (49) and is regulated by at least 2 distinct signaling systems, one through quorum sensing of a long chain acyl-homoserine lactone (43) and the other involving the response regulator protein CtrA (30).
The CtrA protein was first characterized for Caulobacter crescentus (41), where it is essential for viability and acts as a master regulator of the cell cycle (reviewed in reference 45), controlling at least 25% (144 of 553) of the genes involved in cell cycle progression (31). Despite sharing remarkable sequence identity (71%) with the CtrA protein from C. crescentus, the R. capsulatus protein has a very different role because it is not essential and does not appear to be involved in cell cycle processes. One function of CtrA in common to the two species is the regulation of expression of genes that encode the flagellum (29, 41). The ctrA genes of Sinorhizobium meliloti (3), Brucella abortus (6), and Ruegeria sp. strain TM1040 (36) have also been studied. Similarly to C. crescentus and R. capsulatus CtrA, Ruegeria CtrA controls motility (36). A search of the GenBank database reveals that convincing homologs which share >50% identity with the C. crescentus protein are present in all alphaproteobacterial complete genome sequences, with the exception of Pelagibacter ubique (17).
A portion of the R. capsulatus genome sequence was described previously (21, 53), and the complete annotated sequence is now available (GenBank accession no. CP001312 and CP001313; http://rhodo.img.cas.cz/). The availability of this sequence has allowed us to identify the genes dysregulated by loss of CtrA through comparisons of transcriptomic and proteomic data from wild-type and ctrA mutant cell cultures. Because RcGTA production is CtrA dependent and changes over a culture growth phase, measurements of the transcriptome and proteome of cultures in the logarithmic and stationary phases of the population growth cycle were compared. Therefore, we have also analyzed growth phase differences in gene expression in R. capsulatus. Our transcriptome results show that CtrA is an important regulator in R. capsulatus because it is required for proper expression of more than 225 genes, including those predicted to control motility, gene exchange, pilus, and gas vesicle formation as well as those for expression of many putative signal transduction proteins and transcriptional regulators. Of the genes dysregulated by the loss of CtrA, proteins for 58 were observed and 51 of these quantitatively validated the transcriptome data (88%). Our analyses also demonstrate that CtrA is not responsible for growth phase-dependent gene expression in R. capsulatus and that CtrA does not control R. capsulatus genes involved in cell cycle events, such as DNA replication or cell division. Despite almost universal conservation of CtrA in the alphaproteobacteria, a bioinformatic analysis of representative organisms suggests that CtrA has different functions that fall along phylogenetic lines in the different major taxonomic orders.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Cultures of R. capsulatus were grown anaerobically in YPS medium (54) at 37°C with illumination from standard 60-W incandescent light bulbs. YPS is a complex medium and was chosen because of its use in previous studies on CtrA and RcGTA (29, 30, 43). The strains used were the genome-sequenced SB1003 strain (57) and a ctrA mutant, SBRM1. SBRM1 was constructed from SB1003 by GTA transduction of a disrupted version of ctrA (29) into the chromosome. Growth was monitored over time by turbidity, and samples were collected in the logarithmic growth phase and early in the stationary phase of growth (see Fig. S1 in the supplemental material).
Iron supplementation experiments were conducted by preparing YPS medium with the addition of the salt and trace element components of RCV medium [EDTA, MgSO4, CaCl2, FeSO4, thiamine HCl, MnSO4, H3BO3, Cu(NO3)2, ZnSO4, and NaMoO4] (5) and also with the same components except for FeSO4. All of these were added together to prevent the iron from precipitating, which happens if FeSO4 is added on its own. This addition increased the Fe2+ concentration from ∼7 μM in YPS (7) to ∼50 μM.
RNA isolation and microarray analyses.
RNA was extracted using an RNeasy kit (Qiagen, Inc., Mississauga, Canada) according to the manufacturer's recommendations. RNA was isolated from both strains in both growth phases from three growth experiments on different days, yielding 12 total samples. Whole-genome expression arrays (100-3660 format) were constructed by Affymetrix, Inc. (Santa Clara, CA), through a custom design. The arrays were designed based on a preliminary version of the complete genome sequence and contain oligonucleotide probes for 3,635 open reading frames (ORFs) and 1,452 intergenic regions greater than 90 bp, with each ORF represented by 11 probe pairs. RNA samples and arrays were processed at the Michael Smith Genome Science Center (Vancouver, Canada), with cDNA synthesis, labeling, and target hybridization performed as described in the Affymetrix Expression Analysis Technical Manual for prokaryotic samples.
Raw data from scanned arrays were robust multiarray (RMA) normalized (24). By use of GeneSpring version 7.2 (Agilent Technologies, Santa Clara, CA), the signal intensities were further normalized (50th percentile per chip) so that the median normalized signal intensity equals 1. Subsequent data visualization and analyses were performed using GeneSpring version 7.2. Data filtering was set at a raw signal intensity of >50 to remove low-signal-intensity features. Differential expression thresholds were set at a 2-fold limit, and gene lists were created by selecting those genes that differed ≥2-fold for normalized signal intensity in one sample relative to the level for the other within an individual replicate of RNA samples, with subsequent identification of the genes in common to 3 of 3 replicate experiments.
The relationship between the preliminary genome annotation used to design the microarrays and the final genome sequence (GenBank accession no. CP001312 and CP001313; http://rhodo.img.cas.cz/) is provided in Table S1 in the supplemental material.
Protein extraction and digestion.
Global, soluble, and insoluble protein fractions were extracted from cell lysates using established protocols (11). Proteins from the global and soluble preparations were denatured and reduced by adding urea, thiourea, and dithiothreitol (DTT) to give final concentrations of 7 M, 2 M, and 5 mM, respectively. Samples were incubated at 60°C for 30 min and then diluted 10-fold with 100 mM NH4HCO3, pH 8.4. CaCl2 was added to the diluted sample to give a final concentration of 1 mM, and the sample was digested at 37°C using sequencing grade trypsin (Roche, Indianapolis, IN) at a ratio of 1 unit per 50 μg protein. Digested samples were then desalted using a C18 solid phase extraction (SPE) column (Supelco, St. Louis, MO).
For the insoluble protein extract, the cell lysate was ultracentrifuged at 350,000 × g for 10 min at 4°C. The resulting supernatant, containing soluble proteins, was digested as described above. The insoluble pellet was washed in 100 mM NH4HCO3, pH 7.8, and suspended in 7 M urea, 2 M thiourea, 1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonatein}, 50 mM NH4HCO3, pH 7.8. DTT was then added to give a final concentration of 9.7 mM. The sample was incubated and digested as described above, with the exception that 50 mM NH4HCO3, pH 7.8, was used for the 10-fold dilution. Following digestion, removal of salts and detergent was performed using a strong cation exchange (SCX) column (Supelco, St. Louis, MO).
Tandem LC-MS analysis and reference peptide database generation.
Peptides from each protein digest were fractionated (30 fractions each digest) using SCX and reverse-phase high-performance liquid chromatography (HPLC) according to established protocols (1). Peptides from each fraction (10 μg) were analyzed on a quad-column HPLC system coupled to an linear trap quadropole (LTQ) mass spectrometer (ThermoFisher Scientific, San Jose, CA). Reverse-phase separation of peptide digests occurred by way of columns (fused silica capillary tubing 60 cm by 360 μm [outside diameter] and 75 μm [inside diameter]; Pacific Northwest National Laboratory, Richland, WA) packed with 3 μm Jupiter C18 stationary phase (Phenomenex, Torrence, CA). The HPLC system was equilibrated with 100% mobile phase A (0.2% acetic acid and 0.05% trifluoroacetic acid [TFA] in water) at 10,000 lb/in2. Mobile phase B (0.1% TFA in 90% acetonitrile-10% water) displaced mobile phase A 50 min after peptide injection, generating an exponential gradient. Split flow controlled the gradient speed operating under constant pressure (10,000 lb/in2). Separated peptides were ionized (positive) using an electrospray ionization (ESI) interface (Pacific Northwest National Laboratory) with chemically etched electrospray emitters (150 mm [outside diameter] and 20 mm [inside diameter]) (26). The capillary temperature and ESI voltage were 200°C and 2.2 kV, respectively.
Selected parent ion peaks were fractionated using collision-induced dissociation, and tandem mass spectra were matched to theoretical spectra for peptide sequence assignment using the sequence search algorithm X!Tandem (13). A modified parameter file allowed for partial tryptic peptides and static modifications to pass the first round of X!Tandem matching. Theoretical spectra were generated from the translated R. capsulatus SB1003 genome sequence. Peptides with residue lengths of 6 amino acids (aa) or greater, having an E value of ≤−2(log10), were placed in a reference peptide database along with their calculated theoretical masses and normalized elution times (27) for matching to liquid chromatography-mass spectrometry (LC-MS) measurements.
High-mass-accuracy LC-MS and label free quantification.
A modified 9.4 T FTICR mass spectrometer (Bruker Daltonics, Billerica, MA) was used to obtain relative quantitative proteome information. The HPLC conditions were the same those as reported above. Mass spectra from quadruplicate measurements (16 total analyses) were processed with the in-house-developed software program Decon2LS (35), based on the THRASH algorithm. The resulting mono-isotopic masses were clustered into LC-MS features on the basis of neutral mass, charge state, abundance, isotopic fit (i.e., quality of fit between recorded and simulated isotopic patterns), and spectrum number (relating to LC retention time). The assembled set of LC-MS features was then searched against the reference peptide database by use of VIPER (37). A tolerance window representing a mass measurement accuracy of <5 ppm and a normalized elution time error of <1% was applied to ensure reliable peptide identification.
MS peak intensities were used as a measure of the relative peptide abundances. The mean abundance of the LC-MS features was used, and the relative abundances of constituent peptides were averaged to derive the relative abundance of the parent protein (48). Peptide abundances were normalized to a common baseline by use of central tendency normalization (10) and “rolled up” to a protein abundance estimate using the Z-score rollup algorithm available in DAnTE (40). Software tools used for this proteomics analysis are publicly available at http://ncrr.pnl.gov/.
Identification of potential CtrA binding sites.
Consensus CtrA full sites (TTAA-N7-TTAAC) and half-sites (TTAACCAT) were identified using the sequence search function in Artemis (42).
Microarray data accession number.
Transcriptomic data from this work have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession no. GSE18149); proteomic data are available at http://omics.pnl.gov/.
RESULTS
Nucleotide sequence of the Rhodobacter capsulatus genome.
The genome of R. capsulatus strain SB1003 consists of a circular chromosome containing 3,738,958 bp and a circular plasmid of 132,962 bp. The genome has a relatively high GC content (66.6%). The full genome annotation contains 3,531 ORFs identified in the chromosome and 154 ORFs in the plasmid. Functions are assigned to 3,100 ORFs (84.1%). The annotation is accessible at http://rhodo.img.cas.cz/ (GenBank accession no. CP001312 and CP001313).
Growth phase expression analyses.
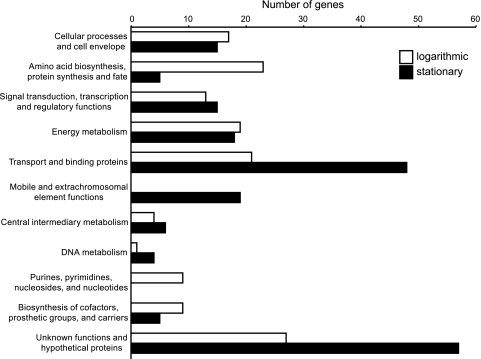
Transcriptomic and proteomic data were gathered during the logarithmic (log) and stationary phases of the growth of cultures for comparative analyses. The transcript comparisons identified 334 genes in the genome (9%) that show significant (≥2-fold) increase or decrease in expression between the log and stationary phases of growth. Of these, quantitative peptide data were obtained for 152 proteins (46%) in the proteome. Transcript levels were ≥2-fold higher (range 2 to 20-fold) in the log phase of growth than in the stationary phase for 143 genes (see Table S2 in the supplemental material). These genes are predicted to be involved in various processes, with most in metabolic categories (Fig. 1). Transcripts of genes encoding ribosomal proteins were as much as 5-fold more abundant in the log-phase cultures. A number of flagellar and chemotaxis genes are also more highly expressed in the log phase of growth than in the stationary phase. Of the 143 genes more highly expressed in the log phase, observed peptides identified 78 of the corresponding proteins (55%). The relative abundance patterns of the proteins agree with the transcript data for 50 of the 78 (64%) genes (see Table S2 in the supplemental material).
FIG. 1.
Growth phase differences in gene expression in R. capsulatus. The numbers of genes that display ≥2-fold-higher transcript levels in the logarithmic and stationary phases that fall within each specified gene categories are shown. The categories are based on the TIGR roles indicated in the genome annotation; for simplicity, we combined the following: cellular processes and cell envelope; amino acid biosynthesis, protein synthesis, and protein fate; signal transduction, transcription, and regulatory functions; and unknown functions and hypothetical proteins. The transcript and proteome data for all genes are in Table S2 (logarithmic phase) and Table S3 (stationary phase) in the supplemental material.
In the stationary phase, 192 genes showed ≥2-fold-higher transcript levels than in the log phase (Fig. 1; see also Table S3 in the supplemental material). The proteome analysis detected 74 of the proteins encoded by the 192 genes more highly expressed in stationary phase (39%), and 66 (89%) quantitatively agreed with transcript levels (see Table S3 in the supplemental material). There is a noticeable shift in the gene category representation numbers in stationary phase away from genes involved in biosynthetic functions, which is accompanied by obvious increases in other categories (Fig. 1). These data reveal that an adaptation of R. capsulatus to stationary phase in this medium involves increasing the expression of a large number (48) of transport system genes, some of which were transcribed as much as 60-fold higher than in the log phase (see Table S3 in the supplemental material). The transport category in stationary phase includes approximately 20 genes predicted to be involved in iron uptake, suggesting that the cells become iron limited under these conditions. We conducted growth experiments to test if iron limitation is the cause of the stationary phase under these growth conditions. Increasing the iron concentration in the medium from ∼7 μM to ∼50 μM resulted in no change in growth rate or increased yield in the cultures (see Fig. S2 in the supplemental material). Therefore, although the cells do increase expression of iron acquisition genes in the stationary phase, iron limitation is not the cause of the stationary phase under these conditions. There are also large increases in the mobile element category (from 0 to 19), mostly consisting of the RcGTA genes, and in the unknown-function and hypothetical-protein categories (from 28 to 58) (Fig. 1).
We identified 8 genes that are predicted to encode sigma factors (Table 1). The transcript levels are increased in the stationary phase for all of these, and rcc00458 and rcc02637 have 2.6- and 3.7-fold-higher transcript levels, respectively. The protein encoded by rcc00458 is the ortholog (the protein sequences are 72% identical) of the Rhodobacter sphaeroides RpoHII stress response sigma factor (18). A bioinformatics analysis indicates that rcc02637 may encode a protein belonging to the FecI-like group of extracytoplasmic function (ECF) sigma factors (ECF05-ECF10), which may indicate a function in iron acquisition (51). Two other ECF sigma factors show higher transcript levels in the stationary phase. Gene rcc02291 encodes an EcfG-like stress response sigma factor (ECF15), as determined by a bioinformatics analysis and its genomic context (51), and gene rcc02811 encodes RpoH, which has been described as a heat shock sigma factor (16). The gene rcc02724 is annotated as a sigma factor and displays sequence similarity to other genes annotated as sigma factors, but a BLAST analysis at the NCBI Conserved Domain Database does not identify any sigma factor domains in the sequence. The gene rcc00568, encoding RpoN, which regulates nitrogen fixation genes (2, 25), has extremely low transcript levels under these growth conditions (<0.6 for all) (Table 1), as expected (14), because the cells are not fixing nitrogen in this medium. Two genes, rcc03323 and rcc03324, are predicted to encode an antisigma and anti-antisigma pair and are 3.3- and 2.1-fold higher in the log phase, respectively; these two genes are also affected by the loss of CtrA (see below).
TABLE 1.
Sigma factor expression in R. capsulatus
| Gene | Product and functiona | Transcript levels (log phase, stationary phase) |
Transcript fold changeb |
|||
|---|---|---|---|---|---|---|
| SB1003 | ctrA | SB1003 vs ctrA (log phase) | SB1003 vs ctrA (stationary phase) | Stationary phase vs log phase (SB1003) | ||
| rcc00458 | RpoHII, stress response ECF sigma factor (18) | 4.60, 12.02 | 3.81, 14.43 | 1.2 | −1.2 | 2.6 |
| rcc00568 | RpoN, nitrogen fixation sigma factor (2, 14, 25) | 0.25, 0.33 | 0.31, 0.55 | −1.2 | −1.7 | 1.3 |
| rcc00699 | σ24 ECF sigma factor | 2.61, 4.25 | 2.64, 4.74 | 0 | −1.1 | 1.6 |
| rcc02291 | EcfG-like sigma factor (ECF15), stress response? (51) | 0.76, 1.29 | 0.82, 1.97 | −1.1 | −1.5 | 1.7 |
| rcc02637 | σ24 ECF sigma factor | 6.12, 22.76 | 8.45, 27.74 | −1.4 | −1.2 | 3.7 |
| rcc02724 | ECF sigma factor ? | 0.35, 0.60 | 0.48, 0.60 | −1.4 | 0 | 1.7 |
| rcc02811 | RpoH, heat shock ECF sigma factor (16) | 8.75, 15.67 | 10.70, 20.88 | −1.2 | −1.3 | 1.8 |
| rcc03054 | RpoD, major vegetative sigma factor (39) | 7.80, 10.80 | 9.35, 12.05 | −1.2 | −1.1 | 1.4 |
The potential functions are discussed further in the text.
Positive values indicate higher transcript levels in the wild type, and negative values indicate higher levels in the ctrA mutant.
For a different perspective on the growth phase gene expression patterns, we also identified the 100 most highly transcribed genes, excluding rRNA genes, in both of the growth phases (see Table S4 in the supplemental material). These represent the upper ∼3% of genes in terms of magnitude of hybridization signal. There is considerable overlap between the growth phases, with 57 genes shared between phases, and many of these genes are related to photosynthetic growth and electron transfer. The most abundant transcript segment observed in these experiments was pucB, encoding the light-harvesting 2 beta (LH2 β) protein, followed by pufB, encoding the LH1 β protein. A striking feature of the 86 genes that are not shared between the phases (i.e., with 43 from each phase that are not in the top 100 for both phases) is that many of them are highly expressed in both growth phases, even though they were not in the top 100 in both growth phases. Of the 43 genes unique to the log-phase top 100, fewer than half (19 genes; 44%) show ≥2-fold difference in expression between the growth phases, while for the stationary phase, only slightly more than half (23 genes; 53%) show ≥2-fold differences. Therefore, under the conditions investigated here, much of the stationary-phase adaptation occurs through genes that are not the most highly expressed. The ctrA gene was among the top 100 genes in both phases, further supporting its role as an important regulator in R. capsulatus, as described below.
CtrA is a positive regulator of gene expression.
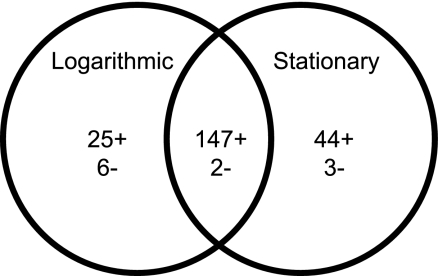
The global regulatory activity of CtrA in R. capsulatus was determined using paired transcriptome and proteome measurements. Comparisons were made between wild-type and ctrA mutant strains and between the log and stationary phases of culture growth. These data demonstrate that regardless of the phase of growth, CtrA is almost exclusively a positive regulator of gene expression. In the log phase, 172 genes had ≥2-fold-lower transcript levels in the ctrA mutant, and 191 genes had ≥2-fold-lower transcript levels in the mutant in stationary phase (Fig. 2). In these two groups of genes, 147 are shared between the growth phases, and therefore, the total number of genes positively regulated by CtrA under the conditions investigated here is 216 (∼6% of the genome). In contrast, only 11 genes show transcript amounts ≥2-fold greater in the ctrA mutant (Fig. 2), accounting for <5% of regulated genes.
FIG. 2.
Effects of loss of CtrA as defined by whole-genome transcript analyses. The Venn diagram shows the number of gene transcripts that were ≥2-fold downregulated for transcript levels in the ctrA mutant (+) and the number of gene transcripts that were ≥2-fold upregulated for transcript levels in the ctrA mutant (−). The numbers of genes found in the logarithmic and stationary growth phases and the amount of overlap between the two lists are indicated. The included genes were identified in 3/3 independent replicate experiments.
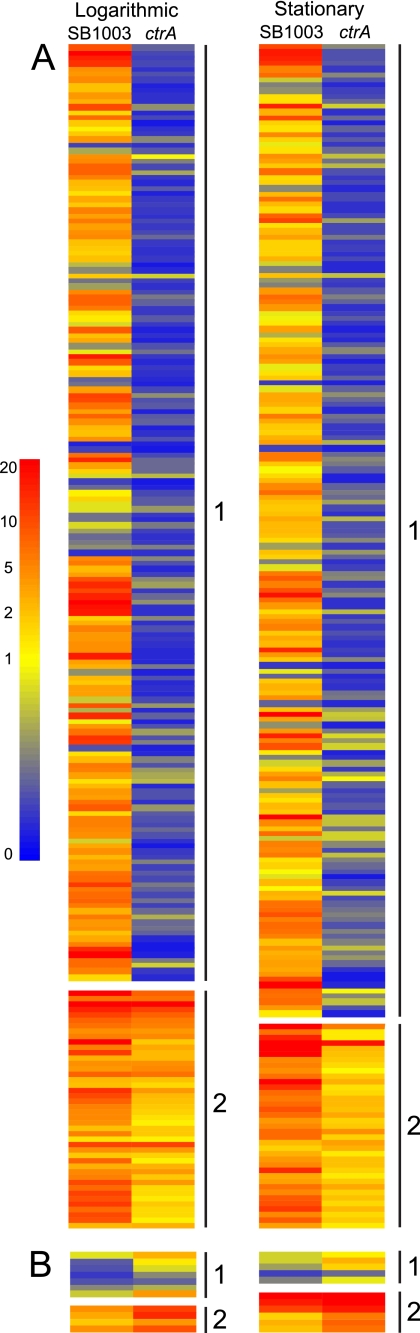
We present the relative gene expression data for the 216 genes negatively affected (Fig. 3A) and 11 genes positively affected (Fig. 3B) by the loss of CtrA as a heat map (the complete numerical transcript and proteome data for these genes are available in Table S5 in the supplemental material). The genes are organized into two groups in each growth phase on the basis of their transcript levels in the ctrA mutant (Fig. 3). We chose to divide the genes on the basis of an intensity value of 1 because an intensity of >1 indicates that the gene is above the median observed for all genes in the genome. For the genes that are negatively affected by the loss of CtrA, group 1 contains genes that have a signal intensity of <1 in the mutant, while those within group 2 have an intensity of >1 in the mutant and therefore appear to be transcribed even in the absence of CtrA (Fig. 3A). Almost all of these genes fall within the same cluster in both phases (see Table S5 in the supplemental material). For the genes that are positively affected by the loss of CtrA, group 1 contains genes that have a normalized signal intensity of <1 in the wild type, while those within group 2 have an intensity of >1 in the wild type and therefore appear to be transcribed even in the presence of CtrA (Fig. 3B). Unlike the trend observed for the negatively affected genes, only 5 of these 11 positively affected genes are in the same cluster in the different growth phases (see Table S5 in the supplemental material). Overall, the range of transcript levels observed in the 227 genes in the absence of CtrA likely indicates that some of these genes are subject to additional mechanisms of regulation, and some may be only indirectly affected by the loss of CtrA.
FIG. 3.
Effects of loss of CtrA on gene expression in R. capsulatus. The quantitative transcript heat map shows the 216 genes negatively affected ≥2-fold by loss of CtrA (A) and the 11 genes positively affected ≥2-fold by loss of CtrA (B). The genes are organized in 2 groups for each category and growth phase on the basis of their transcript levels in the ctrA mutant. For genes negatively affected by loss of CtrA (A), group 1 contains genes that have a signal intensity of <1 in the mutant, while those within group 2 have an intensity of >1 in the mutant. For genes that are positively affected by the loss of CtrA (B), group 1 contains genes that have a signal intensity of <1 in the wild type, while those within group 2 have an intensity of >1 in the wild type. Genes are presented in their order of occurrence in the genome (see Table S1 in the supplemental material) within each group. The scale is colored to indicate relative amounts of the transcripts as indicated in the culture samples indicated above the columns. The numerical transcript and proteomic data for all 227 genes are in Table S5 in the supplemental material.
Transcript levels of the ctrA gene were high in both growth phases for the wild-type cells, and there was an increase in the level of ctrA transcripts in the stationary phase (see Table S4 in the supplemental material), in agreement with previous observations (30). However, a comparison of the proteomes shows a slightly larger amount of CtrA in the log phase than in the stationary phase. Additionally, because of the difficulty of detecting phosphorylated peptides, we were unable to detect the region of CtrA predicted to contain the conserved site of phosphorylation (Asp-51), and so we cannot comment on the relative levels of active protein in the two growth phases. Nevertheless, the decrease in transcript levels for 172 genes in the logarithmic phase caused by the loss of ctrA demonstrates that the CtrA signaling system does not require a stationary-phase signal for activation, in contrast to a previous suggestion (30).
Regulation of flagellar and chemotaxis genes.
Previous work demonstrated that CtrA is required for expression of some flagellar genes in R. capsulatus (29). The data presented in this paper allow an investigation of the function of CtrA in the expression of all flagellar and chemotaxis genes. Genes predicted to be involved in flagellar and chemotaxis functions are a large proportion of the genes negatively affected by loss of CtrA, with 73 of the 216 genes (∼34%) assigned to either flagellar or chemotaxis functions on the basis of homology or residing in apparent operons with flagellar or chemotaxis (che) genes. CtrA is essential for maximal expression of essentially all putative flagellum-dependent motility genes (see Fig. S3 in the supplemental material). The majority of the motility genes (65/73) fall within group 1 (Fig. 3A) and therefore have extremely low (signal intensity < 1) transcript levels in the ctrA mutant. These motility genes include apparent duplications of at least some of the che genes in different locations (rcc01759 to rcc01767 and rcc01352 to rcc01358) and 13 putative methyl-accepting chemotaxis proteins (see Table S6 in the supplemental material); we can identify only one potential che gene, which contains several conserved Che protein domains (rcc00782), that is not CtrA dependent for maximal transcript levels under the conditions investigated here. For the 73 motility genes, 28 proteins (38%) were identified and represent 2.3% of all peptides detected. The quantitative protein data match the pattern found in the transcriptome for 25 of the 28 genes and match the transcriptomic data in one of the two growth phases for the other 3 genes (see Fig. S3 and Table S6 in the supplemental material).
Expression of the gene transfer agent gene cluster.
CtrA was discovered in R. capsulatus because it is required for production of the gene transfer agent RcGTA (30). Transcript levels of the RcGTA genes increase in the stationary phase, and these genes/ORFs constitute most of the genes in the mobile element category (Fig. 1). The proteome analysis detected the 3 RcGTA proteins encoded by transcript segments of the RcGTA gene cluster present at the highest levels, and there were large increases in the amounts of these 3 proteins in the stationary phase of wild-type cultures (see Fig. S4 in the supplemental material). Our results support a model where CtrA is required for transcription of the RcGTA gene cluster (30), and quorum sensing causes an increase in expression of the cluster in stationary phase (43).
Other genes affected by loss of CtrA.
There are 138 genes dysregulated in the ctrA mutant that are not involved in motility or part of the RcGTA gene cluster. Of these genes, 27 (20%) yielded peptides that were identified, and there was complete agreement between the transcript and peptide data for 23 (85%) of the proteins (see Table S5 in the supplemental material).
We identified 22 putative (nonchemotaxis) signal transduction and transcription regulatory proteins that are affected by the loss of CtrA (Table 2). These include proteins containing conserved signaling domains, such as PAS, EAL and GGDEF, phosphorelay histidine kinase, and response regulator receiver, and transcription regulatory domains, such as the helix-turn-helix motif. Although there were no large effects on the transcript levels of the 8 sigma factor genes (Table 1) in the ctrA mutant, there were effects on the expression of 2 sigma factor regulatory genes, rcc03323 and rcc03324 (Table 2).
TABLE 2.
Signal transduction and transcriptional regulator proteins affected by the loss of CtrA
| Gene | Size of protein (aa) | Predicted function (signaling and regulatory domain[s]a) | Transcript fold changeb (log phase, stationary phase) |
|---|---|---|---|
| rcc00180 | 109 | Phosphorelay (HPT) | 26.3, 25.3 |
| rcc00181 | 409 | Response regulator (REC, PP2C) | 17.4, 10.8c |
| rcc00346 | 514 | c-di-GMP signaling (GGDEF, EAL) | 7.6, 4.7c |
| rcc00537 | 247 | Response regulator (REC) | 28.4, 23.7 |
| rcc00620 | 610 | Response regulator and c-di-GMP signaling (REC, GGDEF, EAL) | 14.0, 6.3 |
| rcc00621 | 619 | Sensor kinase (CHASE4, SK) | 5.7, 5.0 |
| rcc00645 | 1,245 | c-di-GMP signaling (GGDEF, EAL, PAS) | 7.7, 25.9c |
| rcc01849 | 253 | Unknown (PAS) | 3.3, 1.7 |
| rcc02075 | 416 | Unknown (PAS) | 5.8, 3.6 |
| rcc02539 | 627 | c-di-GMP signaling (GGDEF, EAL) | 8.1, 4.1 |
| rcc02629 | 353 | c-di-GMP signaling (GGDEF) | 8.1, 2.8 |
| rcc02675 | 170 | Transcriptional regulator (HTH) | 19.3, 5.9 |
| rcc02856 | 412 | Unknown (PAS) | 13.9, 2.7 |
| rcc02857 | 1,158 | c-di-GMP signaling (GGDEF, EAL, PAS) | 12.5, 5.6 |
| rcc03176 | 411 | Unknown (PAS) | 17.1, 10.0c |
| rcc03177 | 284 | c-di-GMP signaling (EAL) | 19.5, 17.6 |
| rcc03301 | 1,284 | c-di-GMP signaling (GGDEF, EAL, PAS) | 4.5, 3.0 |
| rcc03323 | 115 | Anti-antisigma factor (STAS) | 32.3, 8.7 |
| rcc03324 | 163 | Antisigma regulatory factor (HATPase) | 22.0, 9.4 |
| rcc03452 | 743 | Sensor kinase (SK, REC) | 9.8, 3.0 |
| rcp00117 | 280 | c-di-GMP signaling (EAL) | 24.9, 12.1c |
| rcp00137 | 412 | Unknown (PAS) | 9.0, 9.4 |
Conserved domains were identified by BLAST analyses at the NCBI Conserved Domain Database. HPT, histidine phosphotransfer; REC, response-regulator receiver; PP2C, serine/threonine phosphatase; GGDEF, diguanylate cyclase; EAL, c-di-GMP phosphodiesterase; CHASE4, extracellular sensory domain; SK, sensor kinase; PAS, Per-Arnt-Sim; HTH, helix-turn-helix; STAS, antisigma factor antagonist; HATPase, histidine kinase-like ATPase.
Transcript fold change is shown as level for wild type/level for mutant.
Protein detected in proteome and matching transcript trend; data are available in Table S5 in the supplemental material.
An unusual differential expression pattern was observed for a subset of these 138 genes: log-phase transcript levels were similar in the wild-type and ctrA mutant strains, but transcript levels were ≥2-fold lower in the mutant only in the stationary phase. The seven genes having this expression profile (rcc00499 to -00501 and rcc00504 to -00507) fall within group 2 (Fig. 3A) and are part of an 11-gene cluster that contains homologs of the genes that direct synthesis of the CtrA-dependent pili in C. crescentus (46). There are recognizable homologs of the CpaB, CpaC, CpaE, and CpaF proteins, but the R. capsulatus genes in the locations corresponding to C. crescentus pilA, cpaA, and cpaD (rcc00499, -00500, and -00503, respectively) do not have recognizable similarity to the C. crescentus sequences. Five other genes in this cluster contain conserved domains found in pilus assembly proteins (rcc00506 to -00510). In the wild-type strain, gene rcc00499 has the third highest transcript level observed (see Table S4 in the supplemental material); the gene organization context and this high level of expression support assignment of the putative function of the encoded protein as a pilin. Such Flp (fimbrial low-molecular-weight protein) pilus gene clusters are widely distributed in bacteria (52), and the pattern of partial gene cluster conservation described above has been documented as widespread in other members of the order Rhodobacterales (47).
Also of note is a cluster of 26 genes in the genome (rcc01051 through rcc01076), in which 22 have >2-fold-lower transcript levels in the ctrA mutant (see Table S5 in the supplemental material). This cluster contains 10 genes predicted to encode gas vesicle proteins, most of which are in an apparent 12-gene operon (the largest gap between any of these 12 genes is 3 bases). There is another apparent 5-gene operon (rcc01063 to rcc01067) in this cluster, wherein rcc01066 encodes a photoactive yellow protein (PYP) xanthopsin photoreceptor (28). This gene organization has been noted previously, and it was hypothesized that R. capsulatus uses PYP to repress gas vesicle formation under high light intensity (28). Our finding that these genes are coregulated in that they are dependent on CtrA for proper expression further strengthens the hypothesis that there is a connection between PYP and gas vesicle formation. However, the putative gas vesicle genes were transcribed in our high-light-intensity growth conditions, although it is possible they could be more highly expressed under lower-light-intensity conditions.
CtrA binding sites.
CtrA binding sites have been best characterized for C. crescentus and are characterized by the nucleotide consensus sequences TTAA-N7-TTAAC (full site) (41) and TTAACCAT (half-site) (31). However, it was recently shown that CtrA also binds to some TTAA sequences (50). It is thought that CtrA recognizes the same sites in R. capsulatus because of an identical amino acid sequence in the putative helix-turn-helix DNA sequence recognition domain (30). It is also known (6, 9) or thought (3, 20) that the CtrAs of other species bind the same sequences. There are 12 sites in the R. capsulatus genome that perfectly match the full CtrA site (Table 3), and 8 of these are found 5′ of genes that were affected by the loss of CtrA. The four genes not affected ≥2-fold include the cckA gene, but the quantitative protein data suggest that loss of ctrA does lead to smaller amounts of CckA. Also, some genes affected by loss of CtrA, such as rcc00006 and rcc00499, have nearly perfect consensus full sites (TTAA-N7-TTAAG and TTAA-N7-TTTAC, respectively). We found 15 occurrences of the CtrA half-site, and 9 of these occur 5′ of genes that we identified as dysregulated by loss of CtrA (Table 3), but 3 of these 8 genes also have the full sites.
TABLE 3.
Potential CtrA binding sites in the R. capsulatus genome
| Gene | Description | Location of predicted sitea |
Transcript fold changeb (log phase, stationary phase) | Quantitative proteome datac (log phase, stationary phase) | |
|---|---|---|---|---|---|
| TTAA-N7-TTAAC | TTAACCAT | ||||
| rcc00042 | PAS/PAC sensor domain protein | −47 | 12.6, 14.9 | ND | |
| rcc00222 | DNA repair protein RadC | −30 | 7.6, 21.8 | ND | |
| rcc00280 | Hemolysin-type calcium-binding repeat family protein | −100 | 1.2, −1.1 | ND | |
| rcc00393 | UvrABC system protein C | −372 | −1.3, −1.7 | ND | |
| rcc00434 | Hypothetical protein | −57 | −54 | 1.4, 3.9 | ND |
| rcc00459 | Glutathione S-transferase domain proteind | −49 | −1.4, −1.3 | 0.97, −0.43 | |
| rcc00463 | Protein of unknown function UPF0102 | −315 | 4.4, 13.7 | ND | |
| rcc00498 | Transglycosylase, Slt family | −249 | −1.25, −1.1 | ND | |
| rcc00516 | Tyrosine phenol-lyase | −570 | −1.7, −1.4 | −1.43, −1.47 | |
| rcc00645 | Diguanylate cyclase/phosphodiesterase with PAS/PAC sensor | −68 | 7.7, 25.9 | −1.14, 0.74 | |
| rcc00844 | Conserved hypothetical protein | −82 | −90 | 21.3, 28.7 | ND |
| rcc00845 | Hypothetical protein | −273 | −118 | 5.2, 17.8 | |
| rcc00872 | Alcohol dehydrogenase, zinc-binding domain protein | −193 | −1.3, −1.1 | −0.05, −0.50 | |
| rcc01237 | Glycosyl transferase, family 2 | −166 | 2.4, 5.1 | ND | |
| rcc01324 | Type I restriction-modification system, R subunit | −475 | −1.4. −1.1 | ND | |
| rcc01384 | UvrABC system protein B | −65 | −1.4, 0 | −2.42, 1.25 | |
| rcc01462 | Ribonucleoside-diphosphate reductase NrdJ | −491 | −1.3, 1.6 | −2.06, 1.23 | |
| rcc01749 | Histidine kinase CckA | −96 | 1.4, −1.1 | 1.33, 1.12 | |
| rcc01849 | Protein of unknown function DUF1457 | −40 | 3.3, 1.7 | ND | |
| rcc01932 | Glycosyl transferase, family 4 | −183 | −1.7, −1.4 | ND | |
| rcc01940 | Hemolysin-type calcium-binding repeat family protein | −129 | −5.0, −2.5 | ND | |
| rcc03000 | Conserved hypothetical protein | −241 | 2.0. 1.7 | ND | |
| rcc03209 | Protein containing DUF484 | −46 | 2.6, 2.1 | 1.98, 3.09 | |
| rcc03214 | Protein of unknown function DUF1127 | −131 | −1.3, −2.5 | ND | |
Position of the last (3′) base of the predicted binding site relative to the first base of the predicted start codon for the gene.
Positive values indicate higher transcript levels in the wild type, and negative values indicate higher levels in the ctrA mutant.
Absolute difference in Z-score data for each protein, with positive values representing larger amounts in the wild type and negative values representing larger amounts in the ctrA mutant. ND, not detected.
Gene rcc00460 is predicted to be cotranscribed with rcc00459 and is affected by loss of CtrA.
Together, these observations may indicate that full CtrA binding sites, and variations thereof, are more relevant than the half-site as CtrA-dependent regulatory sequences in R. capsulatus, at least under the conditions investigated here. The CtrA-dependent putative regulatory and signaling proteins (Table 2) may provide a link between CtrA and the regulation of many of the genes affected by loss of CtrA that lack recognizable CtrA binding sites.
DISCUSSION
CtrA regulates diverse processes in R. capsulatus.
We have identified 227 genes as being dysregulated by the loss of CtrA in R. capsulatus, and our findings demonstrate that CtrA is almost exclusively a positive regulator of gene expression. Our transcriptomic and proteomic approach has identified putative pilus and gas vesicle formation genes and proteins as requiring CtrA for maximal expression. Additionally, we have validated previously described functions for CtrA in RcGTA production and flagellum biosynthesis (29, 30). Therefore, it appears that CtrA is involved in the control of multiple types of extracellular structures (RcGTA, flagella, and pili) and in the control of movement through distinct mechanisms (flagella and gas vesicles). Gas vesicles have not been reported to occur in R. capsulatus, and the proteins encoded by these genes were not detected in the proteomic work, but this may be due to the difficulty in solubilizing gas vesicle protein membranes (15). Pilus-like structures have been visualized on R. capsulatus cells, including on a ctrA mutant (44). Transcripts of the putative pilus genes identified here were significantly reduced in the ctrA mutant in the stationary phase, but not in the log phase, and so it is possible that these genes are involved in production of the previously observed structures. Additionally, we identified a number of potential signal transduction and transcription regulator proteins that are affected by the loss of CtrA. We expect that alterations in the expression of these genes contribute to the pleiotropic effects that result from loss of CtrA. The relative amounts of peptides were measured for ∼25% of the corresponding proteins, and the quantitative proteomic data validated the CtrA dependence for 88% of these. Therefore, we have observed a high degree of congruence between the RNA- and protein-derived gene expression data.
The transcript profiles indicate that almost all (including putative) genes involved in flagellum-dependent motility in R. capsulatus are CtrA dependent. We found that 35 of the 73 motility genes were downregulated in the ctrA mutant >20-fold relative to the wild type in the log phase of growth, showing a very strong dependence on CtrA. However, the patterns of regulation are not consistent among all of these genes. Some genes display significantly higher transcript levels in the wild-type strain, but their transcripts are still present in the ctrA mutant. These genes are not absolutely dependent on CtrA for expression, although CtrA is clearly required for maximal expression. Other genes that are affected by the loss of CtrA also show this same pattern and continue to be transcribed in the ctrA mutant. These may represent genes that are indirectly affected by loss of CtrA and not direct targets of CtrA regulation.
We identified putative CtrA binding sites 5′ of 24 genes/operons in the R. capsulatus genome, and 14 of these were identified as dysregulated in the transcriptomic analyses. The correlations between these putative CtrA sites and the changes in transcripts detected suggest that both the full and the half-sites are important for regulation of transcription by CtrA. There are other partially conserved sites where CtrA may bind, as found for C. crescentus (31, 41, 50), but the genuine DNA-binding properties of R. capsulatus CtrA remain to be determined. Regardless, it is likely that there are downstream regulatory proteins needed for the control of many of the 227 identified genes, and we found 22 genes that encode putative signal transduction and/or transcription regulatory proteins affected by the loss of CtrA (not counting chemotaxis signal transduction genes). Other than general predictions about the functions of some of these (e.g., sensor kinase or c-di-GMP signaling), we cannot speculate on the specific roles of any of these proteins, because we are unable to identify orthologs of these proteins that have a known, specific function in another species.
Growth phase changes in gene expression.
The data in this paper show that the CtrA signaling system regulates RcGTA and other gene expression in the both log and stationary phases of growth, as opposed to performing solely stationary-phase-dependent regulation of gene expression. Some (∼15%) of the genes affected by loss of CtrA show a ≥2-fold increase in transcript levels in stationary phase relative to those in log phase; this may represent the integration of other regulatory processes, such as quorum sensing, which is known to positively regulate RcGTA gene expression in the stationary phase (43). In addition to increasing the expression of the RcGTA genes, R. capsulatus responded to the stationary phase by shifting transcription away from biosynthesis functions and toward transport functions. In particular, our data indicate that cultures become iron limited in the stationary phase of these growth conditions. It is known that iron is required for maximal photosynthesis pigment production in R. capsulatus (12) and photosynthesis gene expression in the related bacterium Rhodobacter sphaeroides (23). However, iron limitation is not the cause of the stationary phase under these growth conditions, because increasing the iron concentration in the growth medium had no observable effect. There was also a large increase in expression of genes in the unknown-function and hypothetical-protein categories, reflecting a poor understanding of the stationary phase of growth in the alphaproteobacteria in general.
Some of the stationary-phase adaptations appear to be mediated through changes in sigma factor expression, although there is not a homolog of the stationary-phase sigma factor RpoS (22, 33), as found in Escherichia coli. It also appears that some regulation of gene expression by CtrA is through sigma factors, but this is mediated through regulation of sigma-regulatory proteins rather than direct effects on sigma factor gene expression.
CtrA functions vary across the alphaproteobacteria.
The bacterium Ruegeria sp. TM1040, in the marine Roseobacter group, is the closest relative of R. capsulatus for which CtrA and CckA have been studied (36). Similar to R. capsulatus, CtrA is not essential for viability and controls flagellum-dependent motility. Disruption of ctrA causes elongation of Ruegeria cells; however, we have found no effects of ctrA disruption on cell size or shape in R. capsulatus. The role of CtrA has been thoroughly characterized for C. crescentus (31, 32), where 144 genes are affected by loss of CtrA (32). Control of genes involved in flagellum synthesis, chemotaxis, and pilus synthesis by CtrA is conserved between C. crescentus and R. capsulatus. However, a major difference between the two organisms is that R. capsulatus CtrA is not involved in controlling genes for cell division, DNA methylation, DNA replication and repair, ribosomes, RNA polymerase, or NADH dehydrogenase. Although CtrA does affect expression of signal transduction proteins in both species, the genes are not orthologous. Also, there is no indication that R. capsulatus CtrA is involved in regulating the initiation of DNA replication because there are no putative CtrA binding sites near the origin of replication.
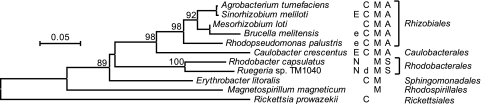
A phylogenetic tree of CtrA sequences from species representing the major taxonomic orders within the alphaproteobacteria for which complete genome sequences were available is shown in Fig. 4, and mapped onto the tree are the known or predicted functions of CtrA in these species. Predicted functions are based on the presence of CtrA binding sites located 5′ of genes involved in the various processes. The relationships of the ctrA sequences match the relationships for these organisms, as determined by other means (19, 55), suggesting a vertical descent of the ctrA gene from the last common ancestor of the alphaproteobacteria. On the basis of the pattern of CtrA functions across these species, we speculate that CtrA was involved in the regulation of the cell cycle and possibly motility in the last common ancestor of these groups. It seems that in the Rhodobacterales (and possibly the Rhodospirillales), a role for CtrA in the cell cycle has been lost and that ctrA is therefore no longer essential. A role for CtrA in motility has been retained in all groups other than the Rickettsiales. There do not appear to be flagellar genes in the Rickettsiales, and so these organisms appear to have lost these genes as a result of genome size reduction in adaptation to an obligately parasitic lifestyle (4, 38).
FIG. 4.
Correlation of CtrA function with phylogeny in the alphaproteobacteria. The (putative) functions of CtrA in the species are indicated (E, essential; e, putatively essential based on the inability to create a mutant strain [Brucella {6} and Rhodopseudomonas {A. S. Lang and J. T. Beatty, unpublished}]; N, nonessential; C, cell cycle; d, possible role in cell division; M, motility). The mode of cell division is indicated where known (A, asymmetrical; S, symmetrical). The indicated CtrA functions are based on previous publications where available (3, 6, 9, 21, 28, 32-34, 38, 42, 43) or the presence of CtrA binding sites upstream of genes involved in the various processes. Note that not all of these species are known to be motile by means of flagella but that they have predicted CtrA binding sites next to flagellar genes. Phylogenetic analyses were conducted with MEGA4 (56), with evolutionary relationships inferred by neighbor joining (46) and bootstrap values based on 10,000 replicates shown next to well-supported branch points (17). Evolutionary distances were computed using the JTT matrix (26), and the scale bar indicates the number of substitutions per site. The GenBank accession numbers for the sequences used are as follows: for Agrobacterium tumefaciens, NP_355385.1; for S. meliloti, NP_386824.1; for Mesorhizobium loti, NP_104064.1; for Brucella melitensis, NP_539340.1; for Rhodopseudomonas palustris, NP_946978.1; for C. crescentus, NP_421829.1; for R. capsulatus, AAF13177.1; for Ruegeria sp. TM1040, YP_613394.1; for Erythrobacter litoralis, YP_459735.1; for Magnetospirillum magneticum, YP_419992.1; and for Rickettsia prowazekii, NP_220465.1.
In C. crescentus, the activity of CtrA is controlled through transcriptional, phosphorylation, and proteolytic regulation (8). There is a complex system controlling the activation and turnover of CtrA, involving the histidine kinase proteins DivJ, DivL, PleC, and CckA, the histidine phosphotransferase ChpT, the response regulators DivK and CpdR, the protease ClpXP, and a protein of unknown function, RcdA. However, of these proteins, we can find convincing homologs of only CckA and ClpXP in the R. capsulatus genome. The Caulobacter model of a control system centered on CtrA has become a paradigm, but our results show that this system is not conserved in all alphaproteobacteria, in agreement with Hallez et al. (20). We suggest that there are (at least) two types of CtrA-centered systems, one (as in Caulobacter and the Rhizobiales) in which the CtrA protein is essential and that, along with CckA, Div, and Ple proteins, forms a regulatory network needed for cell viability and one (as in Rhodobacter) in which the CtrA protein is not essential for cell viability, and control of the cell cycle proceeds through a different regulatory network. These two different systems may relate to fundamental differences in cell division mechanisms between asymmetrically dividing cells of organisms such as Caulobacter and cells of organisms, such as Rhodobacter, that divide by symmetric binary fission (20).
Supplementary Material
Acknowledgments
We thank M. Rise and S. Christian for assistance with microarray data analysis.
Research in A.S.L.'s laboratory is supported by grants from Natural Sciences and Engineering Research Council (NSERC), the Canada Foundation for Innovation, and the Industrial Research and Innovation Fund from the Government of Newfoundland and Labrador. R.G.M. is supported in part by a fellowship from Memorial University. Research in J.T.B.'s laboratory is supported by grants from NSERC, the University of British Columbia, and the Canadian Institutes for Health Research, and J.T.B. is a recipient of a Killam Research Fellowship. A portion of the research described in this paper was funded by the Genomes to Life program sponsored by the U.S. Department of Energy Office of Biological and Environmental Research and performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated by Battelle for the DOE under contract DE-ACO5-76RLO 1830. H.S. and V.P. were supported by Czech grants 1M68378050 and AVOZ50520514.
Footnotes
Published ahead of print on 2 April 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adkins, J. N., H. M. Mottaz, A. D. Norbeck, J. K. Gustin, J. Rue, T. R. Clauss, S. O. Purvine, K. D. Rodland, F. Heffron, and R. D. Smith. 2006. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol. Cell. Proteomics 5:1450-1461. [DOI] [PubMed] [Google Scholar]
- 2.Alias, A., F. J. Cejudo, J. Chabert, J. C. Willison, and P. M. Vignais. 1989. Nucleotide sequence of wild-type and mutant nifR4 (ntrA) genes of Rhodobacter capsulatus: identification of an essential glycine residue. Nucleic Acids Res. 17:5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, M. J., D. Y. Hung, A. Reisenauer, L. Shapiro, and S. R. Long. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol. 183:3204-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batut, J., S. G. E. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the α-proteobacteria. Nat. Rev. Microbiol. 2:933-945. [DOI] [PubMed] [Google Scholar]
- 5.Beatty, J. T., and H. Gest. 1981. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch. Microbiol. 129:335-340. [Google Scholar]
- 6.Bellefontaine, A. F., C. E. Pierreux, P. Mertens, J. Vandenhaute, J. J. Letesson, and X. D. Bolle. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43:945-960. [DOI] [PubMed] [Google Scholar]
- 7.Bovallius, Å., and B. Zacharias. 1971. Variations in the metal content of some commercial media and their effect on microbial growth. Appl. Microbiol. 22:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers, L. M., E. B. Shapland, and K. R. Ryan. 2008. Who's in charge here? Regulating cell cycle regulators. Curr. Opin. Microbiol. 11:547-552. [DOI] [PubMed] [Google Scholar]
- 9.Brassinga, A. K. C., R. Siam, W. McSween, H. Winkler, D. Wood, and G. T. Marczynski. 2002. Conserved response regulator CtrA and IHF binding sites in the α-Proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 184:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callister, S. J., R. C. Barry, J. N. Adkins, E. T. Johnson, W. J. Qian, B. J. M. Webb-Robertson, R. D. Smith, and M. S. Lipton. 2006. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 5:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callister, S. J., C. D. Nicora, X. Zeng, J. H. Roh, M. A. Dominguez, C. L. Tavano, M. E. Monroe, S. Kaplan, T. J. Donohue, R. D. Smith, and M. S. Lipton. 2006. Comparison of aerobic and photosynthetic Rhodobacter sphaeroides 2.4.1 proteomes. J. Microbiol. Methods 67:424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper, R. 1963. The biosynthesis of coproporphyrinogen, magnesium protoporphyrin monomethyl ester and bacteriochlorophyll by Rhodopseudomonas capsulata. Biochem. J. 89:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig, R., and R. C. Beavis. 2004. TANDEM: matching proteins with mass spectra. Bioinformatics 20:1466-1467. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, P. J., D. Foster-Hartnett, K. K. Gabbert, and R. G. Kranz. 1994. Structure and expression of the alternative sigma factor, RpoN, in Rhodobacter capsulatus; physiological relevance of an autoactivated nifU2-rpoN superoperon. Mol. Microbiol. 11:51-65. [DOI] [PubMed] [Google Scholar]
- 15.DasSarma, S., and P. Arora. 1997. Genetic analysis of the gas vesicle gene cluster in haloarchaea. FEMS Microbiol. Lett. 153:1-10. [Google Scholar]
- 16.Emetz, D., and G. Klug. 1998. Cloning and characterization of the rpoH gene of Rhodobacter capsulatus. Mol. Gen. Genet. 260:212-217. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 18.Green, H. A., and T. J. Donohue. 2006. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J. Bacteriol. 188:5712-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta, R., and A. Mok. 2007. Phylogenomics and signature proteins for the alpha Proteobacteria and its main groups. BMC Microbiol. 7:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallez, R., A. F. Bellefontaine, J. J. Letesson, and X. De Bolle. 2004. Morphological and functional asymmetry in α-proteobacteria. Trends Microbiol. 12:361-365. [DOI] [PubMed] [Google Scholar]
- 21.Haselkorn, R., A. Lapidus, Y. Kogan, C. Vlcek, J. Paces, V. Paces, P. Ulbrich, T. Pecenkova, D. Rebrekov, A. Milgram, M. Mazur, R. Cox, N. Kyrpides, N. Ivanova, V. Kapatral, T. Los, A. Lykidis, N. Mikhailova, G. Reznik, O. Vasieva, and M. Fonstein. 2001. The Rhodobacter capsulatus genome. Photosynth. Res. 70:43-52. [DOI] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horne, I. M., J. M. Pemberton, and A. G. McEwan. 1998. Manganous ions suppress photosynthesis gene expression in Rhodobacter sphaeroides. Microbiology 144:2255-2261. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry, R., B. Bolstad, F. Collin, L. Cope, B. Hobbs, and T. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, R., and R. Haselkorn. 1989. The DNA sequence of the Rhodobacter capsulatus ntrA, ntrB and ntrC gene analogs required for nitrogen fixation. Mol. Gen. Genet. 215:507-516. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, R. T., J. S. Page, Q. Luo, R. J. Moore, D. J. Orton, K. Tang, and R. D. Smith. 2006. Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal. Chem. 78:7796-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiebel, G., K. Auberry, N. Jaitly, D. Clark, M. Monroe, E. Peterson, N. Tolić, G. Anderson, and R. Smith. 2006. PRISM: a data management system for high-throughput proteomics. Proteomics 6:1783-1790. [DOI] [PubMed] [Google Scholar]
- 28.Kyndt, J. A., J. K. Hurley, B. Devreese, T. E. Meyer, M. A. Cusanovich, G. Tollin, and J. J. Van Beeumen. 2004. Rhodobacter capsulatus photoactive yellow protein: genetic context, spectral and kinetics characterization, and mutagenesis. Biochemistry 43:1809-1820. [DOI] [PubMed] [Google Scholar]
- 29.Lang, A. S., and J. T. Beatty. 2002. A bacterial signal transduction system controls genetic exchange and motility. J. Bacteriol. 184:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, A. S., and J. T. Beatty. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. U. S. A. 97:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 33.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 34.Marrs, B. L. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. U. S. A. 71:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayampurath, A., N. Jaitly, S. Purvine, M. Monroe, K. Auberry, J. Adkins, and R. Smith. 2008. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics 24:1021-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, T. R., and R. Belas. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648-1659. [DOI] [PubMed] [Google Scholar]
- 37.Monroe, M., N. Tolić, N. Jaitly, J. Shaw, J. Adkins, and R. Smith. 2007. VIPER: an advanced software package to support high-throughput LC-MS peptide identification. Bioinformatics 23:2021-2023. [DOI] [PubMed] [Google Scholar]
- 38.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 39.Pasternak, C., W. Chen, C. Heck, and G. Klug. 1996. Cloning, nucleotide sequence and characterization of the rpoD gene encoding the primary sigma factor of Rhodobacter capsulatus. Gene 176:177-184. [DOI] [PubMed] [Google Scholar]
- 40.Polpitiya, A. D., W. J. Qian, N. Jaitly, V. A. Petyuk, J. N. Adkins, D. G. Camp II, G. A. Anderson, and R. D. Smith. 2008. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24:1556-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quon, K. C., G. T. Marczynski, and L. Shapiro. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83-93. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 43.Schaefer, A. L., T. A. Taylor, J. T. Beatty, and E. P. Greenberg. 2002. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 184:6515-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelswell, K. J., T. A. Taylor, and J. T. Beatty. 2005. Photoresponsive flagellum-independent motility of the purple phototrophic bacterium Rhodobacter capsulatus. J. Bacteriol. 187:5040-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skerker, J. M., and M. T. Laub. 2004. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2:325-337. [DOI] [PubMed] [Google Scholar]
- 46.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slightom, R. N., and A. Buchan. 2009. Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl. Environ. Microbiol. 75:6027-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, R. D., G. A. Anderson, M. S. Lipton, L. Pasa-Tolic, Y. Shen, T. P. Conrads, T. D. Veenstra, and H. R. Udseth. 2002. An accurate mass tag strategy for quantitative and high-throughput proteome measurements. Proteomics 2:513-523. [DOI] [PubMed] [Google Scholar]
- 49.Solioz, M., H. C. Yen, and B. Marrs. 1975. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 123:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer, W., R. Siam, M. C. Ouimet, D. P. Bastedo, and G. T. Marczynski. 2009. CtrA, a global response regulator, uses a distinct second category of weak DNA binding sites for cell cycle transcription control in Caulobacter crescentus. J. Bacteriol. 191:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staron, A., H. J. Sofia, S. Dietrich, L. E. Ulrich, H. Liesegang, and T. Mascher. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74:557-581. [DOI] [PubMed] [Google Scholar]
- 52.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363-375. [DOI] [PubMed] [Google Scholar]
- 53.Vlcek, C., V. Paces, N. Maltsev, J. Paces, R. Haselkorn, and M. Fonstein. 1997. Sequence of a 189-kb segment of the chromosome of Rhodobacter capsulatus SB1003. Proc. Natl. Acad. Sci. U. S. A. 94:9384-9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wall, J. D., P. F. Weaver, and H. Gest. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 105:217-224. [DOI] [PubMed] [Google Scholar]
- 55.Williams, K. P., B. W. Sobral, and A. W. Dickerman. 2007. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 189:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
- 57.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.