Abstract
Corynebacterineae is a specific suborder of Gram-positive bacteria that includes Mycobacterium tuberculosis and Corynebacterium glutamicum. The ultrastructure of the cell envelope is very atypical. It is composed of a heteropolymer of peptidoglycan and arabinogalactan (AG) covalently associated to an outer membrane. Five arabinosyltransferases are involved in the biosynthesis of AG in C. glutamicum. AftB catalyzes the transfer of Araf (arabinofuranosyl) onto the arabinan domain of the arabinogalactan to form terminal β(1 → 2)-linked Araf residues. Here we show that ΔaftB cells lack half of the arabinogalactan mycoloylation sites but are still able to assemble an outer membrane. In addition, we show that a ΔaftB mutant grown on a rich medium has a perturbed cell envelope and sheds a significant amount of membrane fragments in the external culture medium. These fragments contain mono- and dimycolate of trehalose and PorA/H, the major porin of C. glutamicum, but lack conventional phospholipids that typify the plasma membrane, suggesting that they are derived from the atypical mycolate outer membrane of the cell envelope. This is the first report of outer membrane destabilization in the Corynebacterineae, and it suggests that a strong interaction between the mycolate outer membrane and the underlying polymer is essential for cell envelope integrity. The presence of outer membrane-derived fragments (OMFs) in the external medium of the ΔaftB mutant is also a very promising tool for outer membrane characterization. Indeed, fingerprint analysis of major OMF-associated proteins has already led to the identification of 3 associated mycoloyltransferases and an unknown protein with a C-terminal hydrophobic anchoring domain reminiscent of that found for the S-layer protein PS2 of C. glutamicum.
Corynebacterineae is a specific suborder of Gram-positive bacteria that includes medically and economically important species, such as Mycobacterium tuberculosis, Mycobacterium leprae, and Corynebacterium glutamicum. The ultrastructure of cell envelopes in the Corynebacterineae has been intensively studied during recent decades and has revealed a completely unexpected scheme. Indeed, they are composed of a heteropolymer of peptidoglycan and arabinogalactan (AG) covalently associated to an outer membrane. This outer membrane is made up of mycolic acids that either esterify trehalose (free mycolates) or are terminal Araf residues of AG skeleton (bound mycolates) (38-40). The disclosure of this very atypical structure initially came from functional studies in which pore-forming proteins were identified in almost all members of the Corynebacterineae (41, 62), pointing out the presence of an unexpected additional hydrophobic barrier in the cell envelope of these Gram-positive bacteria. Its presence was recently clearly visualized by cryo-electron microscopy of vitreous sections (CEMOVIS) (29, 67), but the exact physiological properties of this barrier are poorly documented because only a few integral outer membrane proteins (OMPs) are yet characterized. In mycobacteria, two examples of integral OMPs are well documented: OmpA of M. tuberculosis and MspA of Mycobacterium smegmatis. MspA is a major porin involved in mycolate outer membrane permeability (58, 64) and has a very specific structure based on a β-barrel core reminiscent of that found in Gram-negative porins (25). In C. glutamicum, four different porins of very low molecular mass (PorA [36, 42], PorH [30], and PorB and PorC [18]) have been identified. Very recently it was shown that a heterooligomeric structure composed of PorA and PorH is needed to form the major cell wall channel of C. glutamicum (6). Although these proteins have been extensively characterized in vitro, their physiological importance remains elusive. Surprisingly, preliminary structural studies of PorB suggested that it could be organized in an α-helical structure, in contrast to all the “canonical” porins, and thus could represent a new class of outer membrane proteins (66).
Because Gram-negative outer membrane proteins and MspA from M. smegmatis are organized in β-barrel structures, two studies aimed to identify M. tuberculosis OMPs by a bioinformatic approach using β-barrel prediction algorithms (43, 57). Although very attractive, this approach has several limitations. Indeed, these algorithms generally produce a high number of false-positive proteins and are not able to predict amphipathic α-helical structures such as those expected for PorB. Nevertheless, this approach led to the identification of a new channel-forming outer membrane protein of M. tuberculosis, Rv1698, which is conserved among all members of the Corynebacterineae (55). This protein increases the susceptibility of bacteria to hydrophilic antibiotics and increases the rate of glucose uptake. Its structure has not yet been characterized.
Isolation of the outer membrane of mycobacteria or corynebacteria using standard biochemical methods is difficult due to the covalent links between mycolic acids and the underlying heteropolymer hampering the identification of new OMPs. In order to overcome this problem, an alternative approach could come from outer membrane-derived vesicles (OMVs), whose release in the medium is a conserved mechanism among Gram-negative bacteria and has been observed in many environments (34). Increased OMV release has been reported for mutants lacking either components of the Tol-Pal system (8, 63) or Lpp, the major lipoprotein involved in noncovalent interactions between the outer membrane and the peptidoglycan (8, 14, 15, 22, 31). This suggested that OMV production in Gram-negative bacteria is controlled through specific domains that promote outer membrane protein-peptidoglycan and outer membrane protein-inner membrane interactions (22). OMVs from Gram-negative bacteria are composed of lipopolysaccharide (LPS), phospholipids, and outer membrane proteins but lack inner membrane proteins. In some cases, elementary units of peptidoglycan and periplasmic proteins are also recovered in these vesicles (35, 37).
C. glutamicum is the prototype of amino acid producers and has been widely used in biotechnology for several decades. More recently it was also shown to be a very attractive model for depicting cell wall biosynthesis of the Corynebacterineae (45). Indeed, an extensive library of mutants involved in the different steps of mycolic acid and arabinogalactan synthesis is available for C. glutamicum but not for mycobacteria, where these components are strictly essential for viability. In this work, we wanted to screen different C. glutamicum cell wall mutants for their ability to release outer membrane-derived vesicles in the external medium. More particularly, we focused on mutants with altered covalent linkage between the mycolate outer membrane and the arabinogalactan. Accordingly, we constructed an arabinosyltransferase mutant unable to catalyze the transfer of Araf onto the arabinan domain of AG to form terminal β(1 → 2)-linked Araf residues. This abnormal AG structure results in both a decrease in mycoloylation sites on arabinogalactan and a concomitant appearance of outer membrane-derived fragments in the external medium.
MATERIALS AND METHODS
Strains and plasmids.
C. glutamicum strains CGL2005, ATCC 13032, and RES167 (24) were cultured on brain heart infusion (BHI) medium (Difco) at 30°C. For molecular biology experiments, we used the Escherichia coli DH5α strain in order to clone mytA-His, mytC-His, and cspB genes in the shuttle vector pCGL482.
Kanamycin (Km), chloramphenicol (Cm), and sucrose (Suc) were added when required at final concentrations of 25 μg ml−1, 6 μg ml−1 (for C. glutamicum), and 10% (wt/vol), respectively. Transformation of C. glutamicum by electroporation was performed as described by Bonamy et al. (12).
Primers were synthesized by Genosys and MWG. All DNA sequencing was carried out by Cogenics and MWG (52).
Construction of ΔaftB mutant.
In order to delete C. glutamicum aftB (cg-aftB) (NCgl2780), we used the strategy described by Schafer et al. (52). In brief, two DNA fragments overlapping the gene to be deleted at its 5′ and 3′ extremities were amplified by PCR from C. glutamicum total DNA by using the primers 2780-del1 (5′-TATTCTAGATCTCCGGATTCATGCTC-3′)/2780-del2 (5′-CTCGAGTCAAAACATCGG-3′) (464 bp) and 2780-del3 (5′-TATAGCCTCGAGTAAACACTTATTCGC-3′)/2780-del4 (5′-ATGTTGTTTGAGATCTAGAGGCGCG-3′) (519 bp) and cloned into the nonreplicative vector pK18mobSac. The resulting plasmid (pK18mobsacΔaftB) was sequenced and introduced into C. glutamicum RES167 by electroporation. Transformants in which the construct was integrated into the chromosome by single crossover were selected on BHI plates containing Km. The second crossover event was selected by plating Kmr clones on BHI plates containing sucrose. Kms and Sucr colonies were screened by PCR for the correct deletion of the gene using combinations of primers localized upstream and downstream of the NCgl2780 sequence. After verification of PCR products by sequencing, one strain carrying the NCgl2780 deletion (ΔaftB) was selected for further studies.
Construction of expression vectors.
An expression vector encoding cg-AftB was constructed using pCGL482 (44) as the cloning vector. We chose to clone this open reading frame (ORF) under the control of its own putative promoter (542 bp upstream of NCgl2780 ATG). The corresponding DNA fragment (2,639 bp) was amplified by PCR from C. glutamicum RES167 chromosomal DNA using the primer pair 2780del1/2780XB (5′-GACACTCGAGGGTAGTTATC-3′) and ligated with pCGL482 digested by SmaI to give pCGL482-AftB. Transformants were selected on Cm-containing plates.
An expression vector encoding PS2Δ (PS2 deleted for amino acids 345 to 350) was constructed using pCGL824, a vector that contains the cspB gene of ATCC 17965 (44). For that purpose, we used the overlap extension method (28). Two overlapping PCR products, corresponding to the regions up- and downstream of the deletion site, were synthesized separately using pCGL824 as a template and the primer pairs SD10 (5′-AGATCTCGAGGCCTGCAGCCC-3′)/SD21 (5′-CGCTGATCCCTAACGCAAACATCAGCG-3′) and SD22 (5′-CGCTGATGTTTGCGTTAGGGATCAGCG-3′)/SD5 (5′-AGATGGCTCTATCAACCCAGATAC-3′). These fragments were used as a template for a second PCR using the primer pair SD10/SD5. This second PCR resulted in a cspB gene fragment carrying the desired deletion and containing a PvuI site and a StuI site that are present in pCGL824. After digestion with StuI and PvuI, PCR fragments were ligated to pCGL824, digested with the same restriction endonucleases to give pES859. Transformants were selected on Cm-containing plates.
Expression vectors encoding MytA-His and MytC-His were constructed using pCGL482 (44) as the cloning vector. We chose to clone these ORFs under the control of their native promoters (414 and 432 bp upstream of the mytA and mytC ATGs, respectively). The corresponding DNA fragments (2,388 and 1,555 bp) were amplified by PCR from C. glutamicum ATCC 13032 chromosomal DNA using the primer pairs 336his-Bam (5′-AGTTTTTGGGCGGATCCTAGCATTTCTTAGACTCCAAAGG-3′)/336his-Xho (5′-GAGCTAGAGGCCCATCATCATCATCATCATTAGCTCGAGAT-3′) and 2777his-Bam (5′-CTGTTTTCCACACAACCGGATCCTCAAGGGGAAAACAAGG-3′)/2777his-Xho (5′-CTCTCGAGTTAATGATGATGATGATGATGGCGAGACTCCTC-3′) digested by BamHI and XhoI and ligated with pCGL482 to give pCGL482-MytAhis and pCGL482-MytChis. Transformants were selected on Cm-containing plates.
An expression vector encoding FbpA under its own promoter and its own sequence signal was obtained as described previously (51).
Cryo-transmission electron microscopy (cryo-TEM).
The purified fraction recovered from the gradient was diluted 10-fold with 10 mM HEPES (pH 7.5). An aliquot (4 μl) was adsorbed onto a holey carbon-coated grid (Quantifoil, Germany), blotted with Whatman 1 filter paper, and vitrified into liquid ethane at −178°C (23). Frozen grids were transferred onto a Philips CM200-FEG electron microscope using a Gatan 626 cryo-holder. Electron micrographs were recorded at an accelerating voltage of 200 kV and a magnification of ×50,000, using a low-dose system (20 e−/Å2) and keeping the sample at −175°C. Micrographs were recorded on Kodak SO-163 films and then digitized with a Heidelberg Primescan 7100 scanner at a 4-Å/pixel resolution at the specimen level.
Freeze fracture electron microscopy.
A small droplet of the purified fraction recovered from the gradient was placed on the copper holder and quenched in liquid propane cooled with nitrogen. The frozen sample was fractured at −125°C in vacuo at about 10−5 Pascals with a liquid nitrogen-cooled knife in a Balzers 300 freeze-etching unit as described previously (2). The fractured sample was replicated with a 1- to 1.5-nm deposit of platinum-carbon (Pt/C) and coated with a 20-nm carbon film. The Pt/C replica was cleaned with 2% SDS, washed with pure water, transferred onto a copper electron microscopy (EM) grid, and observed with a Philips CM100 electron microscope.
Purification of ΔaftB outer membrane fragments.
Outer membrane vesicles were purified from 300 ml ΔaftB BHI culture. After centrifugation at 7,000 (7K) × g, the supernatant was completely clarified on a 0.22-μm filter and then centrifuged at 35K × g for 2 h in a 45TI rotor. The pellet was carefully resuspended in 25 mM HEPES (pH 7.5) buffer containing 0.1 mg of Pefabloc and then layered on a sucrose step gradient ranging from 0 to 55% (wt/wt) for 16 h at 10°C in a SW65 rotor at 50K × g. After centrifugation, the band corresponding to membrane vesicles was recovered, diluted 10 times in HEPES buffer, and centrifuged for 30 min in a TLA 100.2 rotor at 65K × g. Finally, the pellet was resuspended in HEPES buffer with 0.1 mg/ml of Pefabloc and frozen at −20°C.
Extraction and quantification of corynomycolic acids from whole cells.
Lipids were first extracted from wet cells for 16 h with CHCl3-CH3OH (1:2 [vol/vol]) at room temperature and then reextracted with CHCl3-CH3OH (1:1 [vol/vol]) and finally with CHCl3-CH3OH (2:1 [vol/vol]) for 16 h. The three organic phases were pooled and concentrated by means of rotary evaporation. The crude lipid extracts were partitioned between the aqueous and organic phases arising from a mixture of CHCl3-H2O (1:1 [vol/vol]). The lower organic phases were collected and evaporated to dryness to yield the crude lipid extracts from each strain. Subsequently, they were comparatively examined using thin-layer chromatography (TLC) on silica gel-coated plates (Durasil-25, 0.25-mm thickness; Macherey-Nagel) developed with CHCl3-CH3OH-H2O (65:25:4 [vol/vol/vol]). Glycolipids were detected by spraying plates with 0.2% anthrone in concentrated H2SO4, followed by heating. The corynomycolate content of extractable lipids was determined in three independent experiments as follows: lipid extracts (100 mg) of the various strains were dried under vacuum, saponified (21), and then acidified with 20% H2SO4. The resulting fatty acids were extracted with diethyl ether, washed with water, converted to methyl esters with diazomethane, dried under vacuum, and weighed. The fatty acid methyl esters were separated from contamination on a silica gel column irrigated with different concentrations of diethyl ether in petroleum ether (0, 5, 10, 20%, and 100% [vol/vol]). Fractions were analyzed by TLC with development with CH2Cl2. Lipids were detected by spraying plates with rhodamine B, and fractions containing corynomycolates were pooled and weighed. The arabinogalactan-bound mycolic acids present in the delipidated cells, i.e., the material after the chloroform-methanol extraction (described above), were saponified with 40% KOH in 2-methoxyethanol (1:7 [vol/vol]) at 110°C for 3 h, and the solution was neutralized using 20% H2SO4. The released corynomycolic acids were extracted with diethyl ether and methylated with diazomethane. The resulting fatty acid methyl esters were examined using TLC on silica gel-coated plates developed as above. When significant amounts of fatty acids other than corynomycolic acids were present in the saponification products, corynomycolates were isolated by chromatography on a silica gel column as described above and weighed.
Lipid and sugar analysis of ΔaftB membrane fragments.
Lipids οf ΔaftB fragments were extracted by the method of Bligh and Dyer (10). Briefly, wet fragment pellet from 300 ml of BHI culture supernatant was incubated in a one-phase solvent system, CHCl3-CH3OH (1:2 [vol/vol]) for 16 h at room temperature. Then, CHCl3 and H2O (1:1 [vol/vol]) were added to obtain two phases: the organic lower phase, containing lipids, and the aqueous upper phase, containing sugars. The organic phase was dried under nitrogen, and lipids were resuspended in a minimal volume of CHCl3, deposited on silica gel Durasil-25 plates (0.25 mm, 10 by 20 cm; Nacherey-Nagel), which were developed in CHCl3-CH3OH-H2O (65/25/4 [vol/vol/vol]). Glycolipids were revealed by spraying the TLC plates with 0.2% anthrone (wt/vol) in concentrated H2SO4, followed by heating at 110°C. The corresponding aqueous phase was analyzed to determine the sugar composition of ΔaftB fragments. Dry fraction was subjected to hydrolysis with anhydrous 1.5 M CH3OH-HCl for 16 h at 80°C. The resulting methyl glycosides were then dried under nitrogen (traces of acids were removed by two coevaporations with methanol) and trimethylsilylated (59) before gas chromatography (GC) analysis. GC analysis was performed using a Hewlett-Packard 4890 chromatograph equipped with a fused silica OV1 capillary column (0.3 mm by 25 m). The temperature program was 60°C to 100°C with an increasing gradient of 20°C/min and then 100°C to 310°C with an increasing gradient of 5°C/min.
Arabinogalactan preparation and analysis.
Cell walls were prepared as previously described (19). The glycosyl composition of purified cell walls was determined by hydrolyzing an aliquot of mAGP (mycolyl-arabinogalactan-peptidoglycan) with 2 M trifluoroacetic acid (TFA) for 2 h at 110°C, followed by trimethylsilylation (59) and GC analysis of the resulting products.
For 13C nuclear magnetic resonance (NMR) analysis, arabinogalactans were solubilized in 2H2O (30 mg/0.5 ml) and analyzed at 150.9 MHz on a Bruker Avance 600-MHz instrument equipped with a TCI cryoprobe. 1H-decoupled 13C spectra were obtained with a DEPT 135 pulse sequence in a DQD acquisition mode.
Biochemical analysis.
Cell wall protein extracts were prepared as described by Joliff (32). Briefly, cells were recovered by centrifugation and resuspended in Laemmli buffer (100 optical density [OD] units per ml), and the sample was heated for 5 min. After progressive cooling, unbroken cells were discarded by centrifugation, and the supernatant was analyzed by SDS-PAGE or Western blotting.
SDS-PAGE and Western blotting were done according to standard methods. The transfer of proteins was realized during 1 h at 30 V on a Whatman Protran BA85 (0.45 μm) nitrocellulose membrane for FbpA analysis and 30 min at 30 V on a Pall Life Science Biotrace NT membrane (0.22 μm) for analysis of PorA. The membrane was probed with anti-PorA antibodies diluted at 1/1,000 (from R. Benz) or anti-FbpA diluted at 1/10 (from K. Huygen).
For proteinase K digestion, outer membrane fragments (100 μg of proteins) were incubated in 500 μl of 25 mM HEPES buffer at 30°C with 10 μg of proteinase K. The proteolysis reaction was stopped by precipitation of the fragments in 20% trichloroacetic acids (TCA). A control experiment was done by addition of TCA immediately after the addition of proteinase K. After acetone washing, the pellet was resuspended in Laemmli buffer and analyzed by SDS-PAGE. All samples were run in duplicate.
For urea treatment, outer membrane fragments were resuspended either in HEPES buffer (25 mM [pH 7.0]) or in the same buffer containing 4 M freshly dissolved urea. After 30 min of incubation at room temperature, outer membrane fragments were recovered by centrifugation at 65K × g for 30 min in a TLA 100.2 rotor at 10°C to avoid urea crystallization. The pellet was resuspended in Laemmli buffer before SDS-PAGE analysis.
For outer membrane fragment solubilization, outer membrane fragments were resuspended either in HEPES buffer (25 mM, pH 7.0) or in the same buffer containing 1% of octylglucoside, 2% of LDAO (lauryldimethylamine-oxide), or 2% of Triton X-100. After 30 min of incubation at room temperature, outer membrane fragments were recovered by centrifugation at 65K × g for 30 min in a TLA 100.2 rotor at 10°C. The supernatant was TCA precipitated and washed with cold acetone. Both pellets from the 65K × g centrifugation and TCA precipitation were resuspended in an identical volume of Laemmli buffer before SDS-PAGE analysis.
RESULTS
The ΔaftB mutant has a partially disrupted cell surface and sheds cell envelope fragments in the external medium.
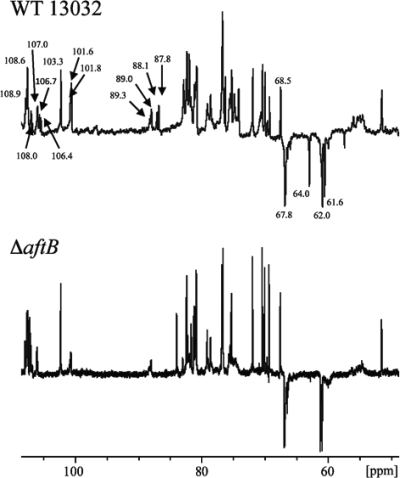
Arabinofuranosyltransferases enzymes, C. glutamicum Emb (Cg-Emb) (3), AftA (4), AftB (54), AftC (9), and AftD (56), play an important role in the biosynthesis of arabinogalactan in C. glutamicum. Inactivation of one of these enzymes is expected to impact the AG structure and dramatically affects the covalent link between the outer membrane and AG. We chose to delete the aftB gene in C. glutamicum ATCC 13032, and we characterized the resulting mutant (ΔaftB) using biochemical and electron microscopy approaches. Quantification of cell wall lipids of the ΔaftB mutant indicated a significant decrease (about 40%) in bound mycolates and a concomitant increase in free mycolates (1.7-fold) (Table 1), suggesting a defect either in arabinogalactan structure or in mycoloyltransferase activity. To decipher the structural features of the arabinogalactans from the parental and ΔaftB mutant strains, cell wall polysaccharides were recovered by means of alkaline hydrolysis (19, 20) and analyzed by 13C NMR spectroscopy (Fig. 1). Based on previous assignments deduced from the spectra of M. tuberculosis (19) and other members of the Corynebacterineae (20), resonances attributable to the C-1s of t-β-Araf and 2-linked-α-Araf of the nonreducing end of the arabinogalactan molecules were observed at 101 to 102 ppm and 106 to 107 ppm, respectively. The resonances of the C-2s of 2-linked-α-Araf were identified at δ 87 to 89, while that of the C-5 of t-β-Araf was seen at 64 ppm. The resonances of primary alcohol carbons of unsubstituted C-5s of α-Araf and C-6s of β-Galf were observed at δ 61 and 62, whereas those of 5- and 3,5-Araf and 6- and 5,6-Galf were seen at δ 67.8. The signal at 103.3 ppm was attributed to the resonance of t-Rhap, which is specific to the arabinogalactan of C. glutamicum (3). Compared to the wild-type (WT) strain, the spectrum from the ΔaftB mutant lacks four series of peaks attributed to C-1 of t-β-Araf and 2-linked-α-Araf, C-2 of 2-linked-α-Araf, and C-5 of t-β-Araf.
TABLE 1.
Quantification of corynomycolates from WT strain 13032 and the ΔaftB and ΔaftB(pCGL482-aftB) strains of C. glutamicum
| Parameter | Value for strain |
||
|---|---|---|---|
| WT 13032 | ΔaftB strain | ΔaftB(pCGL482-aftB) strain | |
| % of corynomycolates in extractible lipidsa | 30.75 ± 0.75 | 51.5 ± 4.5 | 36.5 ± 3.5 |
| % of cell wall-linked corynomycolatesb | 1.13 ± 0.13 | 0.58 ± 0.02 | 0.89 ± 0.19 |
Whole cells grown on BHI medium were extracted with organic solvents, and corynomycolates were determined by saponification followed by weighing of both fatty acid and corynomycolic methyl esters separated in silica gel chromatography. The amount of corynomycolates is expressed as a percentage of bacterial dry weight (mean ± SD).
The resulting delipidated cells were saponified, and the amount of corynomycolate was compared to the bacterial dry weight (mean ± SD).
FIG. 1.
Partial 13C NMR spectra of the cell wall arabinogalactan from strain ATCC 13032 and the ΔaftB mutant strain. The polysaccharides were solubilized in 2H2O (30 mg per ml) and analyzed at 150.9 MHz on a Bruker Avance 600-MHz NMR spectrometer equipped with a TCI cryoprobe. The anomeric carbon resonances of t-β-Araf and 2-linked-α-Araf are indicated.
As expected, complementation of C. glutamicum ΔaftB with pCGL482-AftB restored the normal amount of both cell wall-bound and free mycolates (Table 1). It was concluded that AftB catalyzes the transfer of Araf onto the arabinan domain of AG to form terminal β(1 → 2)-linked Araf residues, as previously shown by Seidel (54).
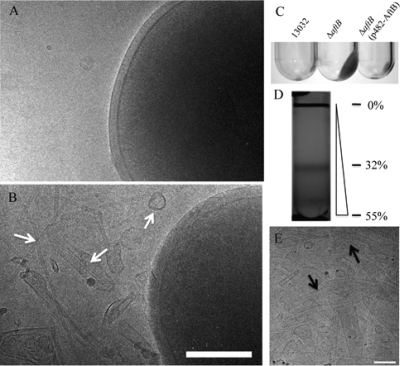
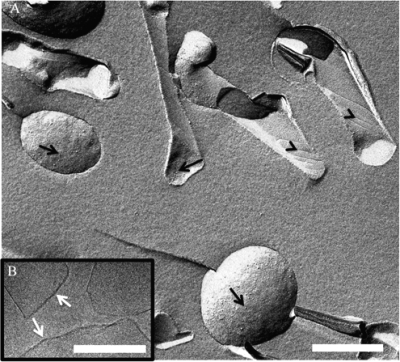
Despite these deleterious characteristics, the mutant strain is fully viable and grows normally except that it tends to aggregate in BHI rich medium, suggesting that the bacterial surface was altered (data not shown). Cryo-electron microscopy observations (Fig. 2A and B) revealed that large patches of the cell envelope seem to detach from the ΔaftB mutant but not from the wild type. In addition, membrane fragments and/or vesicles were always observed around mutant bacteria (Fig. 2B). Expression of aftB from a plasmid in trans restored membrane integrity, confirming that this phenotype was the result of the ΔaftB mutation (data not shown and Fig. 2C). In order to further characterize this material, we ultracentrifuged a filtered supernatant of a ΔaftB culture grown on rich medium to stationary phase. The pellet was then layered on a sucrose gradient as described in Methods in order to purify cell envelope fragments. After centrifugation for 16 h, an intense homogenous band was recovered at a concentration of 32% of sucrose (Fig. 2D) and analyzed by cryo-electron microscopy (Fig. 2E). Large membranous structures (from 20 to 200 nm or more) were observed. They are very diverse in size and form either round vesicles, tubes, or membrane fragments rolling up like cigarette paper. When observed by freeze fracture electron microscopy (Fig. 3A), they display a characteristic striated surface 〈arrowheads〉 resembling the fracture surface previously observed on whole C. glutamicum cells (16, 49). This ordered behavior may be attributed to the high transition temperature of cell wall mycolates compared to that of phospholipids (11). Interestingly, as indicated by the arrows in Fig. 3A, small particles are distributed all over the fracture surface and reveal the presence of integral proteins embedded in the bilayer. These particles are not very abundant compared to similar images obtained from inner membrane vesicles (data not shown) and suggest a low protein/lipid ratio. Indeed, the purified fragments released by the ΔaftB mutant are less dense than inner membrane vesicles obtained from either E. coli or C. glutamicum (data not shown). Frozen-hydrated fragments were also observed by cryo-electron microscopy. High-resolution images show clearly the bilayer structure of vesicles and fragments (Fig. 3B) with a thickness of 5 nm, in good agreement with the value deduced from cryo-electron microscopy of vitreous sections reports for the outer membrane of C. glutamicum.
FIG. 2.
Identification of membrane fragments in the culture medium of the ΔaftB mutant. (A and B) Cryo-electron microscopy images of C. glutamicum 13032 and ΔaftB cells grown on BHI medium, respectively. Culture supernatant from the wild type, the ΔaftB mutant, or the complemented strain was ultracentrifuged as described in Materials and Methods. An important pellet is clearly visible for the mutant strain but not for the wild type or the complemented strain (C). Membrane fragments released by the ΔaftB strain were then purified on a sucrose gradient (D) and were observed directly by cryo-electron microscopy (E). Fragments of different sizes and forms are indicated by arrows. The percentages of sucrose (wt/wt) at specific positions are indicated next to the gradient. Bars, 200 nm.
FIG. 3.
Characterization of the bilayer structure of ΔaftB membrane fragments. (A) Freeze fracture electron microscopy of purified ΔaftB membrane fragments. Arrows indicate the presence of protein particles, and arrowheads indicate striations on the fracture surface. Scale bar, 400 nm. (B) High-resolution image of frozen hydrated sample observed by cryo-EM reveals the bilayer structure of the fragments (arrows). Scale bar, 200 nm.
ΔaftB cell envelope fragments contain all known specific markers of the outer membrane.
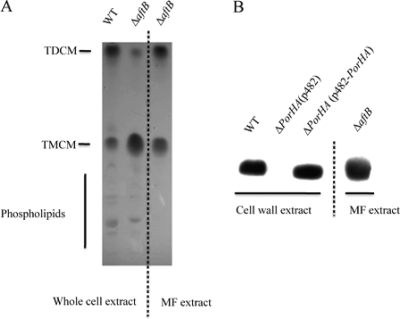
Equivalent amounts (100 μg) of total cell lipid extract from strain ATCC 13032 and from its isogenic ΔaftB mutant were analyzed by thin layer chromatography (TLC). (In the case of the ΔaftB mutant, this extract did not contain the released membranous material). Glycolipids were revealed with anthrone in concentrated sulfuric acid as described in Methods. As expected (47), two main spots were detected for trehalose dicorynomycolate (TDCM) and trehalose monocorynomycolate (TMCM), while several minor spots were visible for more hydrophilic lipids, corresponding mainly to phospholipids and phosphatidylinositol mannosides (PIMs) (Fig. 4A, lanes 1 and 2). Analysis of 100 μg of lipids extracted from highly purified ΔaftB cell envelope fragments clearly revealed the presence of 2 intense spots corresponding to TDCM and TMCM (Fig. 4A, lane 3). No phospholipids were detected. The ratio between TDCM and TMCM in ΔaftB cell envelope fragments is close to 1 (Fig. 4A, lane 3), while TMCM clearly accumulates in ΔaftB total lipid extract (Fig. 4A, lane 2). This observation may indicate that a significant amount of TMCM accumulates in the cell wall of this mutant but is not released with membrane fragments. Extracted lipids from ΔaftB cell envelope fragments were also subjected to mass spectrometry analysis, and two sets of peaks corresponding to sodium adducts of TMCM and TDCM were detected (data not shown). The different peaks differ by 26 Da and thus correspond to C32:0, C34:1, and C36:2 chains bound to trehalose. Again, phospholipids were not detected in this experiment.
FIG. 4.
Lipid and protein characterization of membrane fragments (MF) released by the ΔaftB mutant. (A) TLC analysis of lipids from whole cells (13032 and ΔaftB strains) or from purified membrane fragments. (B) SDS protein extract from membrane fragments or cell wall protein extract from strains 13032, 13032ΔPorHA, and 13032ΔPorHA (pCGL-PorHA) were analyzed by SDS-PAGE followed by Western blotting with anti-PorA polyclonal antibodies.
We then checked the presence of sugars in this material after complete acid hydrolysis. Trace amounts of glucose, mannose, and arabinose but not galactose were detected by gas chromatography (data not shown), indicating that the cell wall arabinogalactan does not contaminate the membrane fragment preparation. Finally, we checked for the presence of the well-characterized pore-forming protein PorA in these fragments. PorA is the major porin of C. glutamicum and thus has been proposed to be localized in the outer membrane. PorA has also been detected at the cell surface by specific antibodies labeled with gold particles (36). As shown in Fig. 4B, a very intense spot migrating at the expected position for PorA was detected in ΔaftB cell envelope fragments. Taken together, these results strongly suggest that membranous fragments released by the ΔaftB mutant derived from the mycolic acid-containing outer membrane; thus, they will here be referred to as outer membrane-derived fragments (OMFs).
Mycoloyltransferases are major proteins associated to ΔaftB OMFs.
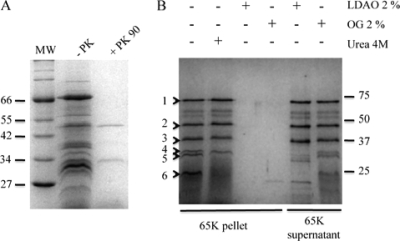
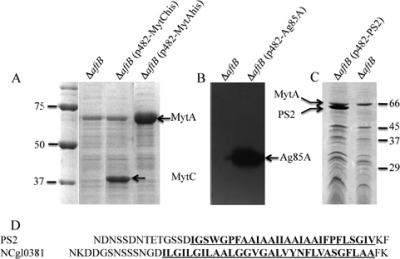
Purified OMFs were analyzed by SDS-PAGE in order to characterize their associated proteins. As shown in Fig. 5B (lane 1), several discrete bands (numbered from 1 to 6) were detected in this fraction after Coomassie blue staining of the gel. Silver nitrate staining revealed a lot of additional minor spots (data not shown). Most of these proteins are firmly associated to the fragments, since a high ionic strength (NaCl, 1 M; data not shown) or a harsh treatment with 4 M urea (Fig. 5B, lane 2) did not break this interaction. These proteins could be solubilized only by detergent, LDAO (0.4%) (Fig. 5B lane 3), octylglucoside (2%) (Fig. 5B, lane 4), or Triton X-100 (1%) (data not shown) or degraded by proteinase K (Fig. 5A, lane 2). This result indicates that most OMF-associated proteins are accessible to the external medium (not trapped inside membrane vesicles) and tightly associated to the membrane by hydrophobic interactions. In order to identify these proteins, we performed a fingerprint analysis of the 6 major bands that were consistently detected after Coomassie blue staining of the gel. The results (Table 2) clearly identified 3 mycoloyltransferases, MytA (NCgl2777; band 1), MytC (NCgl0336; band 2), and MytB (NCgl2779; bands 4 and 5) and one hypothetical protein, NCgl0381 (band 2). The protein corresponding to band 6 could not be determined. Because strain ATCC 13032 encodes 6 mycoloyltransferases, the presence of 3 of them in OMFs is a very strong indication that their presence is not the result of a contamination or an artifact. However, in order to confirm their location in the OMFs, we cloned two of them, i.e., MytC and MytA, with a His tag extension at the C terminus. MytC and MytA are homologous, but MytA has an additional specific C-terminal domain that is not involved in mycoloytransferase activity (46) and has been proposed to be involved in specific interactions with the cell wall or in cell shape (1, 13). Both His-tagged mycoloyltransferases were detected in large amounts in OMFs released by the ΔaftB strain (Fig. 6A). To date, mycoloyltransferases have always been described as soluble proteins, and their association with outer membrane-derived vesicles was quite unexpected. In order to know if this particular behavior of MytC and MytA is conserved in the mycoloyltransferase family, we examined the ability of a mycobacterial mycoloyltransferase to associate with OMFs. To this end, we expressed FbpA from M. tuberculosis in the ATCC 13032 and ΔaftB strains. FbpA was detected in the cell wall of strain 13032 (data not shown), in agreement with previous results that showed that FbpA could be expressed and localized correctly in C. glutamicum strain 2005 (51). When expressed in the ΔaftB mutant, FbpA was detected in the OMF fraction (Fig. 6B), strongly suggesting that FbpA is also associated with the mycolate layer. This important finding demonstrates that, more generally, mycoloyltransferases are major outer membrane-associated proteins of Corynebacterineae.
FIG. 5.
Effects of proteinase K, detergents, and urea on OMF-associated proteins. (A) SDS protein extract from OMFs was analyzed by SDS-PAGE before or after proteinase K treatment. (B) SDS protein extract from OMFs was analyzed by SDS-PAGE after treatment with 4 M urea, 2% LDAO, or 1% octylglucoside and centrifugation at 65K × g. Proteins solubilized by LDAO or octylglucoside were precipitated from the 65K supernatant before analysis. Molecular masses are given in kDa. Bands numbered from 1 to 6 were subjected to fingerprint analysis.
TABLE 2.
Major OMF-associated proteins
| Band no. | Protein name and locus taga | MOWSE scoreb | Peptide match | % of sequence recovery |
|---|---|---|---|---|
| 1 | Mycoloyltransferase NCgl2777 (MytA, Cop1) | 182 | 18/25 | 31 |
| 2 | Hypothetical protein NCgl0381 | 101 | 8/15 | 21 |
| 3 | Mycoloyltransferase NCgl0336 (MytC, Cmt1) | 122 | 12/19 | 31 |
| 4, 5 | Mycoloyltransferase NCgl02779 (MytB, Cmt2) | 72, 63 | 5/6, 4/5 | 19, 13 |
| 6 | Not identified |
Major bands (1 to 6) observed in the total protein extract of OMFs (see Fig. 5B) were excised from the gel and digested with trypsin, and peptides were analyzed by mass spectrometry. Identified proteins are listed according to locus tag nomenclature. Common alternative names are given in parentheses.
MOWSE score, molecular weight search score.
FIG. 6.
MytAhis, MytChis, Ag85A, and PS2 are localized in OMFs. Total SDS extract of OMFs purified from the ΔaftB, ΔaftB (pCGl482-MytChis), and ΔaftB (pCGl482-MytAhis) (A), ΔaftB and ΔaftB (pCGl482-Ag85A) (B), or ΔaftB (pCGl482-PS2) and ΔaftB (C) strains are shown. Panels A and C show Coomassie blue SDS-PAGE gels, while panel B is a Western blot revealed with anti-Ag85A monoclonal antibodies. In panel D, the C-terminal sequences of NCgl0381 and PS2Δ are aligned. Molecular masses are given in kDa.
The S-layer protein PS2 is anchored in ΔaftB OMFs.
NCgl0381, identified as band 3 on the SDS-PAGE gel presented in Fig. 5, is an unknown protein. Nevertheless, careful examination of its sequence revealed the presence of a long (27-amino-acid [aa]) C-terminal hydrophobic domain. Moreover, as shown in Fig. 6D, the hydrophobic sequence is delimited upstream by a serine/aspartate/asparagine-rich region and downstream by a very short C terminal end including a basic and an aromatic amino acid (K and F). These features are remarkably similar to those of the well-characterized S-layer PS2 protein of C. glutamicum. This protein, encoded by cspB, forms a crystalline surface layer (S layer) covering the whole surface of strain CGL 2005 and several other C. glutamicum strains (but not ATCC 13032) (27, 44). Its C-terminal hydrophobic sequence was shown to be essential for cell wall anchoring (17) and thus was proposed to be the insert in the outer membrane. The similarity of the C-terminal domains between NCgl0381 and PS2 suggests that both proteins are physically associated to the outer membrane of C. glutamicum.
In order to test this hypothesis, we expressed PS2 in strain ATCC 13032. Because the presence of a crystalline S layer could alter the formation of OMFs in the ΔaftB mutant, we chose to express a variant of PS2 deleted for amino acids 345 to 350 (PS2Δ). Indeed, we have previously shown that PS2Δ is localized at the cell surface of strains CGL 2005 and ATCC 13032 but is unable to form a two-dimensional (2D) crystal (C. Houssin, M. Chami, and N. Bayan, unpublished data). As visualized in SDS-PAGE (Fig. 6C), PS2Δ is highly expressed in the ΔaftB mutant and is clearly recovered in OMFs. Moreover, association of PS2Δ with OMFs is very strong since it is not relieved by 4 M urea, although complete solubilization of the protein is achieved by nondenaturing detergents (data not shown). This result is a clear indication that the S-layer protein PS2 is tightly anchored to ΔaftB mycolate fragments and definitely demonstrates that PS2 is physically associated to the outer membrane of C. glutamicum.
DISCUSSION
In this article, we showed that the ΔaftB mutant grown on a rich medium has a highly perturbed cell envelope and sheds a significant amount of membrane vesicles and/or membrane fragments (OMFs) in the external culture medium. These fragments contain mono- and dicorynomycolate of trehalose and PorA, the major porin of C. glutamicum, but lack the hallmark lipids typifying the plasma membrane (phospholipids). These characteristics strongly suggested that they are derived from the atypical mycolate outer membrane of the cell envelope. The biochemical characterization of the ΔaftB mutant previously reported by Seidel (54) and confirmed by the data presented in this work clearly indicates that ΔaftB cells lack half of the arabinogalactan mycoloylation sites but are still able to produce a mycomembrane, as shown by our electron microscopy images. We can assume that the loss of “bound” mycolates has been balanced by “free” mycolates, i.e., TDCM or more abundantly TMCM, resulting in a mycomembrane only loosely associated with the underlying AG heteropolymer. This weak association may explain the release of large patches of outer membrane in the culture medium. Interestingly, this situation may be compared to that of Gram-negative bacteria, where it has been shown that the formation of membrane vesicles was the result of the absence of a physical link between the outer membrane and the underlying peptidoglycan skeleton (22). The absence of such a link can occur either all around the cell during growth elongation or at the septum region during growth division. This also happens for mutants lacking Lpp, OmpA, or Pal, which are all known to interact simultaneously with peptidoglycan and the outer membrane, thus contributing to the tight association between the two structures. If such a mechanism is involved in C. glutamicum, it implies that in the ΔaftB mutant, there is a particular arrangement of arabinogalactan mycoloylation sites, such as relatively large areas of the outer membrane, that are completely devoid of covalent links to the AG. These domains could be easily destabilized when outer membrane growth overpasses that of the cell wall.
An alternative model comes from the observation that the ΔaftB mutant accumulates TMCM. This accumulation is likely the result of a decreased number of acceptor sites on the arabinogalactan. The unusually large amounts of this amphipathic molecule in the inner leaflet of the outer membrane may act like cup-former molecules or lysolipids (53, 65). This may in turn lead to the lateral expansion of the inner leaflet and finally induce tension in the outer membrane. This may finally lead to the destabilization of large membrane patches in the external medium exhibiting isolated cell wall-bound mycolates esters at the cell surface. Exposure of these lipids pointing out of the bacterium will make the cell surface hydrophobic and may explain bacterial aggregation observed in liquid media. In this hypothesis, the limited AG mycoloylation sites in the ΔaftB mutant may facilitate the detachment of membrane patches but is not the initial triggering factor.
Whatever mechanism is involved, the formation and the characteristics of OMVs in Gram-negative bacteria and OMFs in C. glutamicum differ in some different ways. First, we were not able to detect any vesicles or fragments released by the wild-type ATCC 13032 strain even after very careful examination of septum sites by cryo-electron microscopy. This may indicate that during normal growth, arabinogalactan and outer membrane synthesis are very efficiently coordinated. Second, the material that is released by the ΔaftB mutant is very atypical and does not resemble either inner or outer membrane vesicles of Gram-negative bacteria. It is highly heterogeneous in size, along with a high variety of morphologies, and displays a specific striated surface. Third, in the case of the ΔaftB mutant, the isolated fragments are devoid of arabinogalactan, peptidoglycan, or periplasmic proteins. Indeed, no sugar was detected, and no protein was found trapped inside these atypical structures.
As observed on a sucrose density gradient, isolated OMFs have a lower density than inner membrane vesicles. This observation also correlates with the presence of a limited number of protein imprints on fracture surfaces of OMFs visualized by electron microscopy. SDS-PAGE analysis confirmed that OMFs contain a discrete number of proteins as detected by Coomassie blue staining. Three of them correspond to mycoloyltransferases that are very important proteins involved in mycolate outer membrane synthesis. In vitro, they catalyze the transfer of mycolate from one TMCM to another TMCM, leading to the formation of TDCM (7). In vivo, converging data from different groups indicated that they are also involved in the transfer of mycolate from TDCM to β terminal arabinofuranosyl residues of arabinogalactan (13, 33, 46). These proteins have been largely studied in C. glutamicum. Six mycoloyltransferases have been found in ATCC 13032, and all seem to have redundant functions (13, 33). For mycobacteria, 3 functional mycoloyltransferases (FbpA, FbpB, and FbpC2) have been described (48). The three-dimensional structures of these mycobacterial proteins (expressed in E. coli) revealed an α/β-fold without any obvious membranous domain (50). Surprisingly, when expressed in the C. glutamicum ΔaftB mutant, FbpA was found tightly associated to OMFs, exactly like corynebacterial mycoloyltransferases, excluding that the latter group may have an additional specific membrane anchor domain that has not been observed in mycobacterial enzymes. Mycoloyltransferases are found associated to the cell envelope but are also secreted in the external medium. The nature of the interaction with the cell envelope is not known, but careful observation of the three-dimensional structure of FbpB in the presence of trehalose led Anderson and collaborators (5) to propose that the protein could be tethered to the membrane and may function at the interface of the outer membrane by a “scooting” mechanism. In the trehalose-bound FbpB crystal structure, trehalose was found in two binding sites. One is oriented so that the meromycolate chain of TMCM could fit in a long hydrophobic groove while the α-chain of TMCM could anchor FbpB in the mycolate outer membrane. The second trehalose binding site could simply help membrane interaction by the sugar moiety of TMCM, while both mycolate chains are embedded in the bilayer. This attractive model, which is consistent with our findings, would allow a very tight association of mycoloyltransferases with the mycomembrane and an efficient catalysis of mycolate transfer without releasing the hydrophobic mycolate chain in the aqueous medium.
Besides mycoloyltransferases, we showed that two proteins (PS2 and NCgl0381) with similar C-terminal domains were localized in ΔaftB OMFs. These C-terminal domains include a hydrophobic stretch of about 30 amino acids preceded by an S/D/N-rich region and followed by aromatic and basic amino acids. PS2 is an S-layer protein that forms a 2D crystal network at the cell surface (16, 44). Its C-terminal hydrophobic domain has been described to be essential to its association with the cell envelope (17). The presence of PS2 in ΔaftB OMFs is in good agreement with all previous published data and suggests that the C-terminal stretch of hydrophobic amino acids provides a physical anchor into the mycolate outer membrane. The discovery of another protein (NCgl0381), sharing with PS2 the same type of association with the outer membrane, may suggest the existence of a more general mycolate outer membrane protein anchoring system in the Corynebacterineae. In Gram-negative bacteria, outer membrane proteins are essentially β-barrel proteins except for lipoproteins and for a newly described protein that has been shown to be tethered to the external surface of the outer membrane by an uncleaved TAT sequence signal (26). In Gram-positive bacteria, a large number of cell wall proteins are covalently linked to the peptidoglycan by a sortase-mediated mechanism involving an LPXTG motif upstream of a C-terminal hydrophobic sequence (60). In PS2 and NCgl0381, the LPXTG motif is not present and the hydrophobic region contains a much higher number of hydrophobic amino acids. In C. glutamicum (a Gram-positive bacterium with a mycolate outer membrane), the anchoring system for PS2 and NCgl0381 may have evolved from the sortase-mediated system, which, surprisingly, is present in Corynebacterium diphtheriae and Corynebacterium efficiens (61) but seems to be lacking in C. glutamicum.
The presence of mycolate outer membrane-derived vesicles in the external medium has been described for a high number of Gram-negative bacteria, but this is the first report concerning the Corynebacterineae, which are Gram-positive bacteria. This discovery adds significant knowledge of the highly complex cell envelope of C. glutamicum and more generally of the cell envelope of the Corynebacterineae. It is also an important milestone on the way toward the biochemical characterization of the mycomembrane, which is still widely undocumented. Indeed, the ΔaftB mutant strain could be a very powerful tool for identifying new mycolate outer membrane proteins in C. glutamicum but also, as shown for FbpA, for determining the localization of mycobacterial cell envelope proteins.
Acknowledgments
We are very grateful to Pierre Le Maréchal, Paulette de Cottignies, and Christophe Marchand for their valuable help in fingerprint analysis and also Elodie Point and Christelle Lémy for their excellent technical assistance. Anne Lemassu is greatly acknowledged for her experience in NMR. We thank Roland Benz and Chris Huygens for their generous gift of the anti-PorA polyclonal antibodies and anti-FbpA monoclonal antibodies, respectively. We are also very grateful to Muriel Masi for careful correction of the manuscript and Andreas Engel (University of Basel) for his support.
This work has received financial support from the “Agence Nationale pour la Recherche” grant ANR-07-BLAN-0363, the CNRS, and the University of Paris Sud-XI.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Adindla, S., K. K. Inampudi, K. Guruprasad, and L. Guruprasad. 2004. Identification and analysis of novel tandem repeats in the cell surface proteins of archaeal and bacterial genomes using computational tools. Comp. Funct. Genomics 5:2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggerbeck, L. P., and T. Gulik-Krzywicki. 1986. Studies of lipoproteins by freeze-fracture and etching electron microscopy. Methods Enzymol. 128:457-472. [DOI] [PubMed] [Google Scholar]
- 3.Alderwick, L. J., E. Radmacher, M. Seidel, R. Gande, P. G. Hitchen, H. R. Morris, A. Dell, H. Sahm, L. Eggeling, and G. S. Besra. 2005. Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280:32362-32371. [DOI] [PubMed] [Google Scholar]
- 4.Alderwick, L. J., M. Seidel, H. Sahm, G. S. Besra, and L. Eggeling. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281:15653-15661. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, D. H., G. Harth, M. A. Horwitz, and D. Eisenberg. 2001. An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (antigen 85B), a mycolyl transferase. J. Mol. Biol. 307:671-681. [DOI] [PubMed] [Google Scholar]
- 6.Barth, E., M. A. Barcelo, C. Klackta, and R. Benz. 2010. Reconstitution experiments and gene deletions reveal the existence of two-component major cell wall channels in the genus Corynebacterium. J. Bacteriol. 192:786-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belisle, J. T., V. D. Vissa, T. Sievert, K. Takayama, P. J. Brennan, and G. S. Besra. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420-1422. [DOI] [PubMed] [Google Scholar]
- 8.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubes. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birch, H. L., L. J. Alderwick, A. Bhatt, D. Rittmann, K. Krumbach, A. Singh, Y. Bai, T. L. Lowary, L. Eggeling, and G. S. Besra. 2008. Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1-→3) arabinofuranosyltransferase. Mol. Microbiol. 69:1191-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 11.Bokas, D., D. Uy, F. Grattepanche, G. Duportail, E. Guedon, S. Delaunay, and J. L. Goergen. 2007. Cell envelope fluidity modification for an effective glutamate excretion in Corynebacterium glutamicum 2262. Appl. Microbiol. Biotechnol. 76:773-781. [DOI] [PubMed] [Google Scholar]
- 12.Bonamy, C., A. Guyonvarch, O. Reyes, F. David, and G. Leblon. 1990. Interspecies electro-transformation in Corynebacteria. FEMS Microbiol. Lett. 54:263-269. [DOI] [PubMed] [Google Scholar]
- 13.Brand, S., K. Niehaus, A. Puhler, and J. Kalinowski. 2003. Identification and functional analysis of six mycolyltransferase genes of Corynebacterium glutamicum ATCC 13032: the genes cop1, cmt1, and cmt2 can replace each other in the synthesis of trehalose dicorynomycolate, a component of the mycolic acid layer of the cell envelope. Arch. Microbiol. 180:33-44. [DOI] [PubMed] [Google Scholar]
- 14.Braun, V., and U. Sieglin. 1970. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur. J. Biochem. 13:336-346. [DOI] [PubMed] [Google Scholar]
- 15.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubes. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chami, M., N. Bayan, J. Dedieu, G. Leblon, E. Shechter, and T. Gulik-Krzywicki. 1995. Organization of the outer layers of the cell envelope of Corynebacterium glutamicum: a combined freeze-etch electron microscopy and biochemical study. Biol. Cell 83:219-229. [DOI] [PubMed] [Google Scholar]
- 17.Chami, M., N. Bayan, J. L. Peyret, T. Gulik-Krzywicki, G. Leblon, and E. Shechter. 1997. The S-layer protein of Corynebacterium glutamicum is anchored to the cell wall by its C-terminal hydrophobic domain. Mol. Microbiol. 23:483-492. [DOI] [PubMed] [Google Scholar]
- 18.Costa-Riu, N., E. Maier, A. Burkovski, R. Kramer, F. Lottspeich, and R. Benz. 2003. Identification of an anion-specific channel in the cell wall of the Gram-positive bacterium Corynebacterium glutamicum. Mol. Microbiol. 50:1295-1308. [DOI] [PubMed] [Google Scholar]
- 19.Daffe, M., P. J. Brennan, and M. McNeil. 1990. Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J. Biol. Chem. 265:6734-6743. [PubMed] [Google Scholar]
- 20.Daffe, M., M. McNeil, and P. J. Brennan. 1993. Major structural features of the cell wall arabinogalactans of Mycobacterium, Rhodococcus, and Nocardia spp. Carbohydr. Res. 249:383-398. [DOI] [PubMed] [Google Scholar]
- 21.Daffe, M. M. 1983. Structure of the lipid constituents of Mycobacterium leprae. Comparison with various species of mycobacteria. Acta Leprol. 1:205-209. (In French.) [PubMed] [Google Scholar]
- 22.Deatherage, B. L., J. C. Lara, T. Bergsbaken, S. L. Rassoulian Barrett, S. Lara, and B. T. Cookson. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72:1395-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21:129-228. [DOI] [PubMed] [Google Scholar]
- 24.Dusch, N., A. Puhler, and J. Kalinowski. 1999. Expression of the Corynebacterium glutamicum panD gene encoding L-aspartate-alpha-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 65:1530-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faller, M., M. Niederweis, and G. E. Schulz. 2004. The structure of a mycobacterial outer-membrane channel. Science 303:1189-1192. [DOI] [PubMed] [Google Scholar]
- 26.Ferrandez, Y., and G. Condemine. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol. Microbiol. 69:1349-1357. [DOI] [PubMed] [Google Scholar]
- 27.Hansmeier, N., A. Albersmeier, A. Tauch, T. Damberg, R. Ros, D. Anselmetti, A. Puhler, and J. Kalinowski. 2006. The surface (S)-layer gene cspB of Corynebacterium glutamicum is transcriptionally activated by a LuxR-type regulator and located on a 6 kb genomic island absent from the type strain ATCC 13032. Microbiology 152:923-935. [DOI] [PubMed] [Google Scholar]
- 28.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunten, P., B. Schiffler, F. Lottspeich, and R. Benz. 2005. PorH, a new channel-forming protein present in the cell wall of Corynebacterium efficiens and Corynebacterium callunae. Microbiology 151:2429-2438. [DOI] [PubMed] [Google Scholar]
- 31.Inouye, M., J. Shaw, and C. Shen. 1972. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J. Biol. Chem. 247:8154-8159. [PubMed] [Google Scholar]
- 32.Joliff, G., L. Mathieu, V. Hahn, N. Bayan, F. Duchiron, M. Renaud, E. Schechter, and G. Leblon. 1992. Cloning and nucleotide sequence of the csp1 gene encoding PS1, one of the two major secreted proteins of Corynebacterium glutamicum: the deduced N-terminal region of PS1 is similar to the Mycobacterium antigen 85 complex. Mol. Microbiol. 6:2349-2362. [DOI] [PubMed] [Google Scholar]
- 33.Kacem, R., C. De Sousa-D'Auria, M. Tropis, M. Chami, P. Gounon, G. Leblon, C. Houssin, and M. Daffe. 2004. Importance of mycoloyltransferases on the physiology of Corynebacterium glutamicum. Microbiology 150:73-84. [DOI] [PubMed] [Google Scholar]
- 34.Kuehn, M. J., and N. C. Kesty. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645-2655. [DOI] [PubMed] [Google Scholar]
- 35.Lee, E. Y., J. Y. Bang, G. W. Park, D. S. Choi, J. S. Kang, H. J. Kim, K. S. Park, J. O. Lee, Y. K. Kim, K. H. Kwon, K. P. Kim, and Y. S. Gho. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143-3153. [DOI] [PubMed] [Google Scholar]
- 36.Lichtinger, T., F. G. Riess, A. Burkovski, F. Engelbrecht, D. Hesse, H. D. Kratzin, R. Kramer, and R. Benz. 2001. The low-molecular-mass subunit of the cell wall channel of the Gram-positive Corynebacterium glutamicum. Immunological localization, cloning and sequencing of its gene porA. Eur. J. Biochem. 268:462-469. [DOI] [PubMed] [Google Scholar]
- 37.Mashburn-Warren, L. M., and M. Whiteley. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61:839-846. [DOI] [PubMed] [Google Scholar]
- 38.McNeil, M., M. Daffe, and P. J. Brennan. 1990. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 265:18200-18206. [PubMed] [Google Scholar]
- 39.McNeil, M., M. Daffe, and P. J. Brennan. 1991. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J. Biol. Chem. 266:13217-13223. [PubMed] [Google Scholar]
- 40.Minnikin, D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, p. 95-184. In C. R. J. L. Stanford (ed.), The biology of Mycobacteria, vol. 1. Academic press, London, United Kingdom. [Google Scholar]
- 41.Niederweis, M. 2003. Mycobacterial porins—new channel proteins in unique outer membranes. Mol. Microbiol. 49:1167-1177. [DOI] [PubMed] [Google Scholar]
- 42.Niederweis, M., E. Maier, T. Lichtinger, R. Benz, and R. Kramer. 1995. Identification of channel-forming activity in the cell wall of Corynebacterium glutamicum. J. Bacteriol. 177:5716-5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajon, R., D. Yero, A. Lage, A. Llanes, and C. J. Borroto. 2006. Computational identification of beta-barrel outer-membrane proteins in Mycobacterium tuberculosis predicted proteomes as putative vaccine candidates. Tuberculosis (Edinb.) 86:290-302. [DOI] [PubMed] [Google Scholar]
- 44.Peyret, J. L., N. Bayan, G. Joliff, T. Gulik-Krzywicki, L. Mathieu, E. Schechter, and G. Leblon. 1993. Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum. Mol. Microbiol. 9:97-109. [DOI] [PubMed] [Google Scholar]
- 45.Portevin, D., C. De Sousa-D'Auria, C. Houssin, C. Grimaldi, M. Chami, M. Daffe, and C. Guilhot. 2004. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. U. S. A. 101:314-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puech, V., N. Bayan, K. Salim, G. Leblon, and M. Daffe. 2000. Characterization of the in vivo acceptors of the mycoloyl residues transferred by the corynebacterial PS1 and the related mycobacterial antigens 85. Mol. Microbiol. 35:1026-1041. [DOI] [PubMed] [Google Scholar]
- 47.Puech, V., M. Chami, A. Lemassu, M. A. Laneelle, B. Schiffler, P. Gounon, N. Bayan, R. Benz, and M. Daffe. 2001. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147:1365-1382. [DOI] [PubMed] [Google Scholar]
- 48.Puech, V., C. Guilhot, E. Perez, M. Tropis, L. Y. Armitige, B. Gicquel, and M. Daffe. 2002. Evidence for a partial redundancy of the fibronectin-binding proteins for the transfer of mycoloyl residues onto the cell wall arabinogalactan termini of Mycobacterium tuberculosis. Mol. Microbiol. 44:1109-1122. [DOI] [PubMed] [Google Scholar]
- 49.Richter, W., F. Hanel, and M. Hilliger. 1985. Freeze-fracture observations of Corynebacterium glutamicum: the occurrence of an outer membrane-like structure and the influence of temperature on the cytoplasmic membrane. J. Basic Microbiol. 25:527-536. [DOI] [PubMed] [Google Scholar]
- 50.Ronning, D. R., T. Klabunde, G. S. Besra, V. D. Vissa, J. T. Belisle, and J. C. Sacchettini. 2000. Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines. Nat. Struct. Biol. 7:141-146. [DOI] [PubMed] [Google Scholar]
- 51.Salim, K., V. Haedens, J. Content, G. Leblon, and K. Huygen. 1997. Heterologous expression of the Mycobacterium tuberculosis gene encoding antigen 85A in Corynebacterium glutamicum. Appl. Environ. Microbiol. 63:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 53.Seeman, P., and S. Roth. 1972. General anesthetics expand cell membranes at surgical concentrations. Biochim. Biophys. Acta 255:171-177. [DOI] [PubMed] [Google Scholar]
- 54.Seidel, M., L. J. Alderwick, H. L. Birch, H. Sahm, L. Eggeling, and G. S. Besra. 2007. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 282:14729-14740. [DOI] [PubMed] [Google Scholar]
- 55.Siroy, A., C. Mailaender, D. Harder, S. Koerber, F. Wolschendorf, O. Danilchanka, Y. Wang, C. Heinz, and M. Niederweis. 2008. Rv1698 of Mycobacterium tuberculosis represents a new class of channel-forming outer membrane proteins. J. Biol. Chem. 283:17827-17837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skovierova, H., G. Larrouy-Maumus, J. Zhang, D. Kaur, N. Barilone, J. Kordulakova, M. Gilleron, S. Guadagnini, M. Belanova, M. C. Prevost, B. Gicquel, G. Puzo, D. Chatterjee, P. J. Brennan, J. Nigou, and M. Jackson. 2009. AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19:1235-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, H., R. Sandie, Y. Wang, M. A. Andrade-Navarro, and M. Niederweis. 2008. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb.) 88:526-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailander, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58:714-730. [DOI] [PubMed] [Google Scholar]
- 59.Sweeley, C. C. 1963. Purification and partial characterization of sphingomyelin from human plasma. J. Lipid Res. 4:402-406. [PubMed] [Google Scholar]
- 60.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 61.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12:228-234. [DOI] [PubMed] [Google Scholar]
- 62.Trias, J., V. Jarlier, and R. Benz. 1992. Porins in the cell wall of mycobacteria. Science 258:1479-1481. [DOI] [PubMed] [Google Scholar]
- 63.Webster, R. E. 1991. The tol gene products and the import of macromolecules into Escherichia coli. Mol. Microbiol. 5:1005-1011. [DOI] [PubMed] [Google Scholar]
- 64.Wolschendorf, F., M. Mahfoud, and M. Niederweis. 2007. Porins are required for uptake of phosphates by Mycobacterium smegmatis. J. Bacteriol. 189:2435-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoo, J., and Q. Cui. 2009. Curvature generation and pressure profile modulation in membrane by lysolipids: insights from coarse-grained simulations. Biophys. J. 97:2267-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziegler, K., R. Benz, and G. E. Schulz. 2008. A putative alpha-helical porin from Corynebacterium glutamicum. J. Mol. Biol. 379:482-491. [DOI] [PubMed] [Google Scholar]
- 67.Zuber, B., M. Chami, C. Houssin, J. Dubochet, G. Griffiths, and M. Daffe. 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190:5672-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]