Abstract
The open reading frame Rv2228c from Mycobacterium tuberculosis is predicted to encode a protein composed of two domains, each with individual functions, annotated through sequence similarity searches. The N-terminal domain is homologous with prokaryotic and eukaryotic RNase H domains and the C-terminal domain with α-ribazole phosphatase (CobC). The N-terminal domain of Rv2228c (Rv2228c/N) and the full-length protein were expressed as fusions with maltose binding protein (MBP). Rv2228c/N was shown to have RNase H activity with a hybrid RNA/DNA substrate as well as double-stranded RNase activity. The full-length protein was shown to have additional CobC activity. The crystal structure of the MBP-Rv2228c/N fusion protein was solved by molecular replacement and refined at 2.25-Å resolution (R = 0.182; Rfree = 0.238). The protein is monomeric in solution but associates in the crystal to form a dimer. The Rv2228c/N domain has the classic RNase H fold and catalytic machinery but lacks several surface features that play important roles in the cleavage of RNA/DNA hybrids by other RNases H. The absence of either the basic protrusion of some RNases H or the hybrid binding domain of others appears to be compensated by the C-terminal CobC domain in full-length Rv2228c. The double-stranded-RNase activity of Rv2228c/N contrasts with classical RNases H and is attributed to the absence in Rv2228c/N of a key phosphate binding pocket.
The bacterium Mycobacterium tuberculosis is the causative agent of the disease tuberculosis (TB), which kills 2 million to 3 million people worldwide every year. One-third of the world's population has latent infection, and 10% of these will develop the active form of the disease. The evolution of multidrug-resistant strains and the increase in HIV-related immunocompromisation have led to serious reemergence of the disease. The sequencing and annotation of the M. tuberculosis genome (9) have enabled a fuller evaluation of the biology of this important human pathogen and the identification of new potential targets for anti-TB drug discovery, although annotations are potentially compromised by the absence of direct structural or functional data (5). Some examples of misannotations have already been noted (6, 20, 46).
An area of direct relevance to the emergence of drug-resistant strains of M. tuberculosis is that of DNA replication and repair (3). Although many genes homologous to the DNA repair machinery of other organisms can be recognized, some apparent absences have been noted (29). Here, we focus on an unusual gene product, Rv2228c, which is annotated as a bifunctional, two-domain protein, comprising an N-terminal RNase H domain and a C-terminal domain homologous with α-ribazole phosphatase (CobC), presumed to act in vitamin B12 biosynthesis.
The RNases H are a family of endonucleases that specifically degrade the RNA of RNA/DNA hybrids (43). These enzymes are found in eukaryotes, bacteria, archaea, and retroviruses, where they have essential roles in DNA replication and repair (11, 17, 19, 22, 32). They are highly variable in size, sequence, and specificity, making classification difficult. Most commonly, they are divided into two classes: type 1 and type 2. The classical type 1 RNase H enzymes are encoded by the rnhA gene and are typically less than 20 kDa in size, although N-terminal and C-terminal extensions frequently provide additional domains that modulate function (8, 44). Eukaryotic RNase HI enzymes, for example, have N-terminal hybrid binding domains that precede the C-terminal catalytic domain (7). The type 2 RNase H enzymes, encoded by the rnhB or rnhC gene, are typically larger and more diverse in sequence but nevertheless have in common a similar RNase H catalytic domain (7).
The M. tuberculosis genome contains no classical rnhA gene, although one rnhB gene, encoding Rv2902c, is present. BLAST searches do, however, identify the N-terminal domain of the open reading frame Rv2228c (Rv2228c/N) as having 31% sequence identity with RNase HI from Escherichia coli (EcRNaseH) and 23% identity with human RNase HI (HsRnaseH). This leads to the hypothesis that this domain provides the essential RNase HI activity in M. tuberculosis. The C-terminal domain of Rv2228c presents a puzzle, however. It has 34% sequence identity with the α-ribazole phosphatase CobC of Synechococcus sp., but it is also homologous with PhoE from Bacillus subtilis (34% identity) and Rv3214 from M. tuberculosis (28% identity), both of which have acid phosphatase activity (39, 46). Bifunctional proteins similar to Rv2228c are encoded by the genomes of other Actinomycetales bacteria, including those of the Mycobacterium, Streptomyces, Corynebacterium, and Nocardia genera, and one of these bifunctional proteins, SCO2299 from Streptomyces coelicolor, has RNase HI activity in its N-terminal domain and acid phosphatase activity in its C-terminal domain (34).
We undertook the structural and functional characterization of Rv2228c/N in order to establish the function of this domain and the possible significance of its associated C-terminal domain. The crystal structure of Rv2228c/N, determined at 2.25-Å resolution as a maltose binding protein (MBP) fusion protein, reveals a classic RNase H fold, but with structural and functional characteristics that make it most like the archaeal RNase H from Sulfolobus tokodaii and differentiate it from classical RNases H. Functional studies confirm the RNase H activity of Rv2228c/N and show that the C-terminal domain has both acid phosphatase and CobC activity, together with a role in enhancing the RNase H activity of the N-terminal domain.
MATERIALS AND METHODS
Protein expression and purification.
The N-terminal domain of Rv2228c (Rv2228c/N), on its own and as a fusion protein with MBP, was produced and purified as described previously (45). The open reading frame corresponding to Rv2228c/N was amplified and cloned into the MBP fusion vector pMAL-C2X, giving a construct comprising an N-terminal MBP moiety joined to Rv2228c/N by a 20-residue linker that incorporates a Factor Xa cleavage site. The resulting MBP fusion protein was then overexpressed in E. coli and purified using amylose affinity chromatography and gel filtration. Factor Xa cleavage was used to also give the Rv2228c/N domain on its own, and this protein was purified similarly. The full-length Rv2228c protein was cloned into the Gateway vector pDEST-566 and expressed as a maltose binding protein fusion. It was purified as for the Rv2228c/N domain fusion (45). The MBP tag was then removed by digestion with recombinant tobacco etch virus protease, and the full-length Rv2228c protein was isolated and purified by amylose affinity chromatography and size exclusion chromatography.
The CobC and CobT genes from E. coli were cloned from E. coli K-12 genomic DNA into the Gateway vector pDEST-17. They were expressed in Luria-Bertani medium at 37°C, transferred to 18°C, and induced with 1 mM isopropylthiogalactoside at an optical density at 600 nm (OD600) of 0.6. Purification was carried out by nickel affinity chromatography as described previously (45).
Crystallization and data collection.
Crystals were grown as described previously (45), by vapor diffusion, using a Honeybee nanoliter dispensing robot to prepare sitting drops (100 nl protein mixed with 100 nl reservoir solution) in 96-well Intelliplates (Art Robbins Instruments). Crystals were grown at 291 K using 20% polyethylene glycol 2000 (PEG 2000), 0.2 M ammonium tartrate as the reservoir solution and were cryoprotected with 20% ethylene glycol before being flash frozen in liquid nitrogen. The crystals were monoclinic, of space group P21, with unit cell dimensions of 73.63 Å for a, 101.38 Å for b, 76.09 Å for c, and 109.01° for β. Two fusion protein molecules of 57,417 Da were present in each asymmetric unit, consistent with a Matthews's coefficient, Vm, of 2.31 Å3 Da−1 and a solvent content in the crystals of 46.8%. Diffraction data to 2.25-Å resolution were collected at 113 K at a wavelength of 0.98397 Å on beam line 9-2 at the Stanford Synchrotron Radiation Laboratory, CA. The data were processed using MOSFLM (25) and SCALA (10), giving the statistics shown in Table 1.
TABLE 1.
Data collection, refinement, and model details
| Characteristic | Valuea |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | a = 73.63; b = 101.38; c = 76.09 Å; β = 109.01° |
| Wavelength (Å) | 0.983970 |
| Resolution range (Å) | 41.45-2.25 |
| No. of observed reflections | 187,273 (27,345) |
| No. of unique reflections | 50,220 (7,326) |
| % completeness | 99.99 (100.0) |
| <I/σ(I)> | 11.5 (3.7) |
| Multiplicity | 3.7 (3.7) |
| Rmergeb | 0.121 (0.462) |
| Refinement and model details | |
| Resolution (Å) | 41.45-2.25 |
| No. of reflections (work/test) | 47,615/2,575 |
| Rworkc/Rfreed (%) | 18.2/23.8 |
| No. of atoms | |
| Protein | 7,780 |
| Water molecules | 329 |
| Maltotriol/tartrate/ethylene glycol | 2/1/3 |
| Avg B factor (Å2) | 18.3 |
| rms deviations from ideal geometry | |
| Bond length (Å) | 0.022 |
| Bond angle (°) | 1.9 |
| Ramachandran plot | |
| % residues in most favored region | 96.0 |
| No. of outliers | 0 |
The figure for the outermost shell is shown in parentheses.
Rmerge = Σ|Ii − <I>/Σ Ii.
R = Σh||Fobs(h)|−|Fcalc(h)||/Σh|Fobs(h)|.
Rfree = Σh||Fobs(h)|−|Fcalc(h)||/Σh|Fobs(h)| for 5% of reflections not used in refinement.
Structure determination and refinement.
The Rv2228c/N domain shares 31% sequence identity with the E. coli RNase HI domain (Protein Data Bank [PDB] code 1RBS), and the MBP component of the fusion protein is identical to that used in the MBP-60S ribosomal protein fusion construct (8) (PDB code 1NMU). A hybrid search model was therefore created using CHAINSAW (10, 42) to combine the models for the two components of the fusion protein. All solvent molecules and ligands were removed from the model structures, after which molecular replacement and model building were carried out using Phaser (28). The complete maltose binding protein structure and approximately 60% of the RNase H domain structure were auto-built by Phaser. Further building was carried out with COOT (14), from 2Fo-Fc and Fo-Fc maps, with refinement in PHENIX-REFINE (1, 2). Maximum likelihood refinement was initially carried out using strict noncrystallographic symmetry restraints in cycles of simulated annealing and energy minimization. Later refinement cycles were carried out using TLS refinement. Water molecules were added, checked, and edited so that only those that made appropriate hydrogen-bonding contacts and had spherical densities above 1σ and 3σ in the 2Fo-Fc and Fo-Fc maps, respectively, were included in the model. Two maltotriol molecules were modeled into large peaks of positive density in equivalent positions in the MBP moiety of the fusion protein. Ethylene glycol and tartrate molecules were also modeled into areas of positive electron density. Full refinement statistics are in Table 1.
Enzyme assays.
RNase HI activity was assayed by monitoring the increase in fluorescence that results from degradation of a DNA/RNA duplex (35, 36). The 5′-fluorescein-labeled RNA strand and the 3′-dabcyl-labeled DNA strand, custom synthesized by Bioneer, were resuspended to a concentration of 100 μM in 50 mM Tris-Cl, 60 mM KCl solution, treated with diethyl pyrocarbonate (DEPC), and autoclaved to remove any contaminating RNases. Equal quantities of these two oligonucleotides were mixed to give a 50 μM solution, heated at 95°C for 5 min, and allowed to cool slowly to room temperature. The hybrid substrate was stored at −20°C. The assay was carried out at an enzyme concentration of 0.5 nM, with substrate concentrations ranging from 2.5 to 400 nM. The increase in fluorescence was monitored every minute for 30 min at 25°C. Double-stranded RNase (dsRNase) activity was assayed using the same method, with an RNA/RNA substrate including 5 mM MnCl2 (final concentration) in the reaction mixture. Bovine pancreatic RNase A (Sigma) was used as a positive control. Reaction rates were measured using an Envision plate reader (PerkinElmer).
Phosphatase activity was assayed using p-nitrophenyl phosphate as previously described (46) to determine the pH optima for all Rv2228c constructs and their Michaelis-Menten kinetic constants. α-Ribazole-5-phosphate (CobC) activity was assayed using a variation of the method of Ohtani et al. (34). In this case, the production of phosphate by the second stage of the reaction was monitored using the malachite green assay (15). Acid phosphatase from potato (Sigma) was used as a positive control. Kinetic constants were determined using 2 mM 5,6-dimethylbenzimidazole and varying the nicotinate mononucleotide concentration between 0 and 10 mM.
Protein structure accession number.
Atomic coordinates and structure factors have been deposited in the PDB with accession code 3HST.
RESULTS
Structure determination and final model.
The N-terminal domain of Rv2228c was expressed and crystallized as a fusion protein with E. coli maltose binding protein (MBP) (45) (Fig. 1) after unsuccessful attempts to crystallize the full-length Rv2228c protein and the Rv2228c/N domain on its own; without the MBP moiety, Rv2228c/N was too unstable for crystallization. The crystals contained two MBP-Rv2228c/N fusion molecules in the asymmetric unit, allowing structure solution by molecular replacement using a complete MBP model and a partial model of E. coli RNase HI as search models. The structure was refined at 2.25-Å resolution to final values of 0.182 for R and 0.238 for Rfree (Table 1).
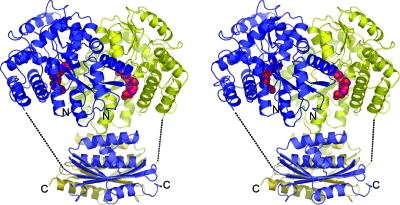
FIG. 1.
Stereo diagram showing the Rv2228c/N-maltose binding protein (MBP) fusion protein, with the two molecules of the asymmetric unit shown in blue and yellow. The MBP moieties (upper) are in the closed conformation with maltotriol (magenta) in the binding cleft. The Rv2228c/N domains are below.
The final model comprises 7,780 protein atoms from two MBP molecules, comprising residues 14 to 374 of molecule A and residues 9 to 374 of molecule B, and two Rv2228c/N domains, for which residues 1 to 138 and 1 to 131, respectively, have been modeled. The C-terminal residue of MBP is joined to the N-terminal Val1 residue of Rv2228c/N by a 20-residue linker, but for both molecules, this is disordered except for the first 4 residues. Each molecule of MBP was found to be in a closed conformation (37), with a maltotriol molecule bound (Fig. 1), and the overall model was completed by one tartrate ion, two ethylene glycols, and 329 water molecules. A Ramachandran plot showed that 96.0% of residues occupied the most-favored regions, as defined by MolProbity (13).
Structure of the Rv2228c/N domain.
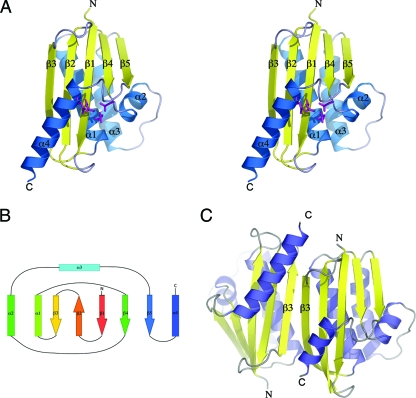
The Rv2228c/N domain has the classic RNase H fold, consisting of a central 5-stranded β-sheet against which are packed four α-helices; α1, α2, and α3 pack against one face, and the C-terminal helix α4 against the other face (Fig. 2). The β-sheet comprises four parallel β-strands and one antiparallel strand (β2), with the connectivity 3-2-1-4-5. The same core fold is found for all the type 1 RNases H that have been characterized structurally to date: the enzymes from E. coli, Bacillus halodurans, Sulfolobus tokodaii, Thermus thermophilus, Moloney murine leukemia virus, the HIV reverse transcriptase (HIV-RT), and human (12, 18, 21, 26, 30, 31, 48). The active site is situated on the α4 side of the molecule at a point where strands β1 and β4 diverge from each other, with the critical divalent metal ion-binding residues contributed by strand β1 (Asp8), helix α1 (Glu49), strand β4 (Asp73), and helix α4 (Asp123).
FIG. 2.
(A) Stereo diagram for the monomer unit of the Rv2228c RNase HI domain, showing β-strands in yellow and α-helices in blue. The four acidic residues at the active site are shown in stick representation, in magenta. (B) Topology of the Rv2228c/N domain, characteristic of the classical RNase H fold. (C) Crystal dimer formed by the Rv2228c/N domain through antiparallel pairing of the two “edge” β3 strands. The C-terminal helices (α4) are of different lengths due to lack of electron density for seven C-terminal residues of molecule B (shown on the right).
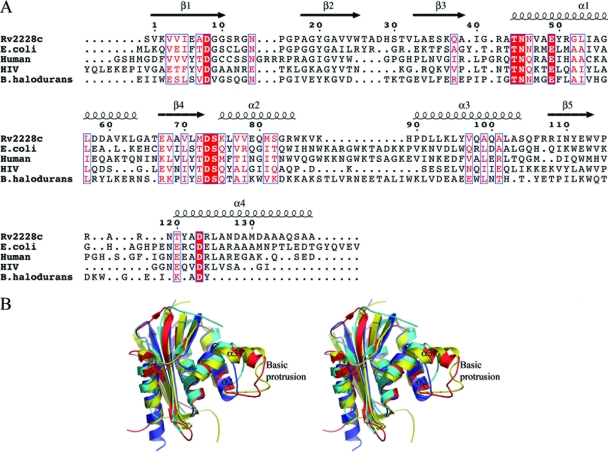
Pairwise superpositions of Rv2228c/N onto other RNases H show that its closest structural homolog is the archaeal RNase H from S. tokodaii (StRNaseH) (49), with which it shares 38% sequence identity and a root-mean-square (rms) difference in Cα atom positions of 1.24 Å over 120 residues. A lower level of similarity is seen with the E. coli RNase HI (EcRNaseH), with 31% sequence identity and an rms difference of 1.53 Å over 114 equivalent Cα atom positions, and human RNase HI (HsRNaseH), with 23% sequence identity and an rms difference of 1.34 Å for 109 equivalent Cα atoms. Insertions and deletions occur in the connecting loops between secondary structures, but the major differences are at the N and C termini and in the region between helices α2 and α3 (Fig. 3). In Rv2228c/N, the HIV-RT RNase H, and StRNaseH, helices α2 and α3 are connected by a short linker of 3 or 4 residues. In the E. coli, B. halodurans, and human enzymes, however, this is the site of a large insertion, which in EcRNaseH and HsRNaseH forms a prominent “basic protrusion” (Fig. 3B). This basic protrusion contacts the RNA/DNA substrate in the complex formed by HsRNase H (33) and has been shown to be critical for substrate binding by the E. coli enzyme; deletion of 18 residues from this region in the latter led to a 40-fold reduction in its affinity for the RNA/DNA hybrid and essentially inactivated the enzyme (16).
FIG. 3.
Comparison of Rv2228c/N with structural homologs. (A) Structure-based sequence alignment comparing the sequence of the Rv2228c/N domain with the RNases H from S. tokodaii, E. coli, human, HIV-RT, and B. halodurans. Residue numbering and the location of secondary structural elements are shown for Rv2228c. The absence of the basic protrusion between helices α2 and α3 in the Rv2228c and HIV-RT structures can clearly be seen. (B) Stereo diagram showing a structural superposition of the RNase HI domain of Rv2228c (dark blue) on to the RNase H domains from S. tokodaii (gray), HIV-RT (cyan), human (red), and E. coli (yellow), showing the additional basic protrusion between helices α2 and α3 in the human and E. coli enzymes.
Unlike all other RNases H that have been structurally characterized to date, Rv2228c/N forms a dimer in the crystal structure. The dimer is formed by the antiparallel association of the β3 strands of two adjacent molecules, with main chain hydrogen bonds between them, generating a continuous 10-stranded β-sheet that extends over the two molecules (Fig. 2C). This brings the MBP moieties into close proximity, but without any significant interaction between them. Analysis of the monomer-monomer interaction using the PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (23) indicates, however, that the contacts most likely result from crystal packing and that the solution state of the Rv2228c/N domain is monomeric. This would be consistent with other prokaryotic RNases HI, all of which exist as monomers in solution (21, 30, 31), and was confirmed by analytical gel filtration (data not shown).
Active site.
The active site of Rv2228c/N is characterized by four acidic residues, Asp8, Glu49, Asp73, and Asp 123, which correspond to the conserved DEDD metal-binding quartet of residues that are found across the whole RNase H family (Fig. 3A). These four residues serve to bind two divalent metal ions, which may be Mg2+ or Mn2+, and which are essential for catalysis. Stable metal binding appears to be seen only in the presence of substrate, however (30, 31). Consistent with this, no metal ion is present in the Rv2228c/N structure; several water molecules are bound in the vicinity of the acidic residues, but none corresponds with an expected metal ion position and none has the appropriate coordination geometry for a magnesium ion. The apo form of B. halodurans RNase H (BhRNaseH) likewise has no bound metal ion, although two Mg2+ ions are bound in its complexes with RNA/DNA substrates (30). Similarly, the substrate complex of the human RNase H domain contains two bound Ca2+ ions occupying the same sites; one (metal B) is coordinated by the residues equivalent to Asp8, Glu49, and Asp73 (Rv2228c numbering), two water molecules, and an oxygen atom from the scissile phosphate of the substrate RNA strand, and the other (metal A) is coordinated by the equivalents of Asp8 and Asp123, two water molecules, and the nonbridging oxygen atom of the scissile phosphate (31). Despite the absence of bound metal ions, the four acidic residues in Rv2228c/N are oriented as in the human structure, indicating that this is a prepared site, ready for metal ion binding and catalysis, and that it is probably not perturbed by subsequent substrate binding.
Apart from the lack of the “basic protrusion,” described above, the surface topography of Rv2228c/N appears generally similar to that of other RNases H. Two shallow grooves which could accept the RNA and DNA strands are present, as in the substrate complexes of the human and B. halodurans enzymes (30, 31). These grooves are separated by a ridge carrying Asn14, Asn45, and Asn46, which fit into the minor groove; the equivalent residues are Asn151, Asn182, and Gln 183 in HsRNaseH and Asn77, Asn105, and Asn106 in BhRNaseH. The residues that impart specificity for the RNA strand by hydrogen bonding to the 2′-OH groups of four consecutive ribonucleotides, two on each side of the scissile bond, are also conserved in Rv2228c/N.
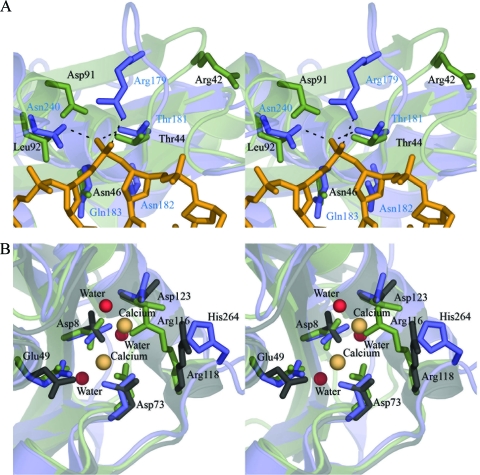
There are two notable differences in Rv2228c/N, however. First, a phosphate binding pocket, present in HsRNaseH and BhRNaseH, is believed to confer specificity for DNA by requiring a nucleotide conformation that is accessible only to DNA (27). This pocket is disrupted in Rv2228c/N, however (Fig. 4A). A threonine residue that is a conserved feature of the phosphate binding sites of HsRNaseH and BhRNaseH is present as Thr44 in Rv2228c/N, but the other residues that would be required to complete a binding cavity are missing. Thus, the β3-α1 loop, which contributes Arg179 to the phosphate pocket of HsRNaseH, has a 1-residue deletion in Rv2228c and adopts a very different conformation. As a result, Arg42 of Rv2228c/N, nominally equivalent to Arg179, is oriented in the opposite direction, with its guanidinium group ∼12 Å away from the putative phosphate site. Another residue that hydrogen bonds to the phosphate in HsRNaseH, Asn240, is replaced by Leu in Rv2228c, and an acidic residue Asp91 further disfavors phosphate binding. The archaeal StRNaseH also lacks this phosphate binding pocket, and it has been suggested that its absence accounts for the loss of RNA/DNA specificity in this enzyme and its ability to degrade both RNA/DNA and double-stranded RNA (dsRNA) substrates (48).
FIG. 4.
Comparisons of the Rv2228c/N domain with other RNase HI enzymes. (A) Stereo diagram of the DNA phosphate binding site in human RNase HI superimposed on the equivalent region of Rv2228c/N. The human structure is in blue, with blue residue labeling, with the bound DNA strand in gold, and with hydrogen bonds with the DNA phosphate shown by broken lines. The Rv2228c/N structure is in green, with residues labeled in black. Key differences are described in the text. (B) Stereo diagram showing the active site region of Rv2228c/N (green) superimposed onto the equivalent regions of the human (blue) and S. tokodaii (gray) enzymes. The four conserved acidic residues are shown in stick mode, labeled according to Rv2228c/N numbering, together with the residue that projects into the active site and is implicated in product release (His264 in HsRNaseH, Arg116 in Rv2228c/N, and Arg118 in StRNaseH). Spheres represent water molecules (red) in Rv2228c/N and the two Ca2+ ions (beige) in HsRNaseH.
Second, product release is thought to be facilitated by a residue on a mobile loop following strand β5; in BhRNase H, this is Glu188, and in HsRNase H, it is His264. These residues point into the active site, clashing with the cleaved product phosphate group (30, 31). The corresponding loop is drastically shortened in Rv2228c/N, as in StRNaseH, through a deletion of 3 or 4 residues, with only 3 residues separating the end of β5 from the start of the C-terminal helix α4. Nevertheless, an arginine residue, Arg116, corresponds spatially to His264 and actually projects further into the active site (Fig. 4B), with its guanidinium group occupying the site of one of the Mg-coordinating waters in the human structure. StRNaseH also has an arginine residue (Arg118) in this position, where it is shown to play a significant role in catalysis, possibly again in substrate release (48).
Activity assays.
Assays for the putative RNase H activity of Rv2228c/N were carried out using a fluorescence-based protocol as opposed to the more conventional radioisotope-based assay (4). This assay confirmed that Rv2228c/N is indeed an RNase H. We then compared the activity of this domain on its own with the activities of its fusion protein with MBP, and of the full-length Rv2228c construct, in order to investigate the impact of the additional N- or C-terminal domains of these forms. All of these proteins were active, with broadly similar Km values that ranged from 53 nM for the MBP fusion protein to 173 nM for the full-length Rv2228c construct (Table 2). This indicated that the substrate binding affinity is largely unaffected by the presence or absence of the additional N- or C-terminal domains. On the other hand, the value of Vmax for full-length Rv2228c, with its additional C-terminal acid-phosphatase (CobC) domain, is almost 100-fold higher than those of the Rv2228c/N domain alone or its MBP fusion.
TABLE 2.
Kinetic data for RNase H activity of Rv2228c constructs
| Construct | Km (nM) | Vmax (M−1 s−1) |
|---|---|---|
| RNase HI domain | 161 ± 4 | 3.05 × 10−15 ± 2.4 × 10−16 |
| RNase HI-MBP fusion | 53 ± 3 | 1.12 × 10−15 ± 2.2 × 10−16 |
| Rv2228c full-length | 173 ± 4 | 1.55 × 10−13 ± 2.4 × 10−14 |
| E. coli RNase H (control) | 307 ± 5 | 1.23 × 10−14 ± 3.7 × 10−15 |
Comparisons with the E. coli RNase HI protein and the HIV-RT RNase H domain (Table 2) are also instructive. The E. coli protein possesses a basic protrusion that is evidently necessary for full activity (16). The HIV-RT RNase H domain is completely inactive on its own, requiring the presence of the polymerase domain for activity; the latter apparently plays a role similar to that of the binding domains or basic protrusions of the other proteins. Loss of the C-terminal domain of Rv2228c does not have such drastic effects, as is shown by the activity of the isolated Rv2228c/N domain and its MBP fusion, although this activity is significantly reduced from that of the full-length protein.
Given the similarity of Rv2228c/N to StRNaseH (48), in the lack of the phosphate pocket that selects for a DNA strand, and in the location of Arg116 in the active site, we also assayed for dsRNase activity, using the same assay as for RNA/DNA, but with an RNA/RNA substrate (Table 3). This showed that Rv2228c/N does indeed have dsRNase activity, at a level similar to that of bovine pancreatic RNase A, taken as a positive control. As in the case of the RNase H activity, substrate binding is not significantly affected by the absence of the C-terminal domain, as judged by the Km value, but again, Vmax increases 100-fold when the C-terminal domain is present.
TABLE 3.
Kinetic constants for dsRNase activity for Rv2228c constructs
| Construct | Km (nM) | Vmax (M−1 s−1) |
|---|---|---|
| RNase HI domain | 10.0 ± 3 | 1.27 × 10−15 ± 8 × 10−16 |
| RNase HI-MBP fusion | 282.7 ± 6 | 1.43 × 10−14 ± 5 × 10−16 |
| Rv2228c full length | 172.6 ± 4 | 1.55 × 10−13 ± 4 × 10−15 |
| Bovine pancreatic RNase A (control) | 307 ± 2 | 1.23 × 10−14 ± 4 × 10−16 |
We also sought to assay for the predicted activity of the Rv2228c C-terminal domain itself. Generic phosphatase activity assays carried out using p-nitrophenol phosphate as a substrate (46) showed that the full-length Rv2228c protein has phosphatase activity that is unaffected by the presence or absence of the maltose binding protein moiety (Table 4). The pH optimum of 4.0 defines this as an acid phosphatase activity. Neither the RNase H domain alone nor its MBP fusion showed any discernible phosphatase activity. The C-terminal domain is also predicted to have α-ribazole-5-phosphatase, CobC, activity on the basis of sequence similarities with authentic CobC proteins. This was tested using an assay coupled to the preceding enzyme in the vitamin B12 biosynthesis pathway, CobT. Activity was detected using the Malachite green method of inorganic phosphate detection. This showed that the C-terminal domain does indeed have CobC activity, with a Km of 0.15 mM and a Vmax of 59.2 pmol min−1. This is in a range similar to those observed for the activity and substrate binding affinities of the E. coli CobC enzyme (Table 5) and is in direct contrast to the demonstrated inactivity of the protein SCO2299 (34). In this case, the C-terminal domain, though predicted to have CobC activity, showed only broad-specificity phosphatase activity.
TABLE 4.
Kinetic constants for generic phosphatase activity for Rv2228c constructs
| Construct | Vmax (M−1s−1) | Km (mM) |
|---|---|---|
| RNase HI domain | No activity | No activity |
| RNase HI-MBP fusion | No activity | No activity |
| Rv2228c full length | 8.04 × 10−12 ± 1.3 × 10−13 | 5.5 ± 1 |
| Rv2228c full length MBP fusion | 6.5 × 10−12 ± 1.1 × 10−13 | 13.6 ± 2 |
| Acid phosphatase from potato (control) | 2.7 × 10−10 ± 1.8 × 10−11 | 2.2 ± 1 |
TABLE 5.
Kinetic constants for CobC activity
| Construct | Vmax (M−1s−1) | Km (mM) |
|---|---|---|
| Rv2228c full-length | 4.45 × 10−11 ± 8.2 × 10−13 | 1.07 ± 1 |
| CobC E. coli | 1.38 × 10−11 ± 7.6 × 10−13 | 2.03 ± 1 |
DISCUSSION
RNase H activity, defined as the ability to cleave the RNA strand of RNA/DNA hybrids, appears to be common to all domains of life, with recognizable RNase H enzymes present in eukaryotes, prokaryotes, and archaea (33). The RNase H enzymes themselves are highly variable, however. Some type 1 enzymes, such as those from E. coli (21) and S. tokodaii (48), consist of a single domain only, which carries all the elements necessary for activity. In others, however, the RNase H domain is combined with a hybrid binding domain, as for the human and B. halodurans enzymes (30, 31), or is incorporated into larger multidomain proteins, such as the HIV reverse transcriptase (12). The one type 1 putative RNase H encoded by the M. tuberculosis genome, Rv2228c, is representative of a novel class of RNase H enzymes in which an RNase H domain is combined with a second domain having quite distinct activity to create a bifunctional protein. Only one member of this subfamily has been characterized so far, the product of the SCO2299 gene from S. coelicolor (34), but no structural information is available on any RNase H of this class.
The crystal structure of the N-terminal domain of Rv2228c, reported here, confirms that this domain shares the archetypal RNase H fold, which is conserved across the entire family of currently characterized RNases HI (30), and our functional assays further confirm that the Rv2228c/N domain is indeed a functional RNase H. Structurally, all the components necessary for catalysis appear to be present. The four acidic residues (DEDD) which characterize the typical RNase H active site (4) are conserved in Rv2228c/N and have spatial arrangements similar to those of the corresponding residues in other RNases H. Even though no metal ions are bound in the present crystal structure, it is apparent that this is a prepared metal binding site, ready for the binding of the two essential Mg2+ or Mn2+ ions (47). The important substrate binding residues identified from the substrate complexes of HsRNaseH and BhRNaseH are generally conserved. The catalytically important residue from the β5-α4 loop, His264 in HsRNaseH and Glu188 in BhRNaseH (30), is replaced in Rv2228c/N by Arg116 but may perform the same role in regulating substrate entry or product release; an equivalent Arg is present in StRNaseH (48).
Two major features of the Rv2228c/N domain differentiate it from most of the RNase H enzymes characterized previously. The first is the disruption of the pocket that in BhRNaseH and HsRNaseH binds a DNA phosphate group 2 residues prior to the scissile bond (30, 31). This site evidently has high affinity for phosphate (it is occupied by a tightly bound sulfate ion in the BhRNaseH apoenzyme) and defines the specificity for RNA/DNA hybrid substrates. Classical RNase H enzymes such as these can bind dsRNA, but evidently not in a productive mode for catalysis; these enzymes do not have dsRNase activity (30, 31). Disruption of the phosphate binding pocket, as seen in the S. tokodaii enzyme (48) and now also in Rv2228c/N, places these enzymes in a different functional class that has both RNase H and dsRNase activity. The Km and Vmax values for the dsRNase activity of Rv2228c/N are in a range similar to those determined for RNase H activity. A similar 100-fold increase is seen in Vmax for this activity in the presence of the C-terminal domain, indicating that stabilization of this domain is still critical to product release from the active site.
The key macroscale difference in Rv2228c/N, in comparison with other RNases H, is in the absence of the basic protrusion or of a typical substrate binding domain, such as is found in eukaryotic RNases H (7) and in the B. halodurans enzyme (44). Our functional data strongly suggest that the C-terminal CobC domain serves this role in full-length Rv2228c. The presence of this domain increases RNase H and dsRNase activity to levels similar to those of E. coli RNase H and bovine dsRNase, respectively. The fact that Km values for the full-length Rv2228c protein are not greatly different from those for the N-terminal domain alone suggests that its role is not primarily related to the affinity of binding. Rather, the 100-fold increase in Vmax for both activities in the full-length protein indicates a decrease in the energy difference between the transition state and product release. This suggests that the C-terminal domain, perhaps aided by Arg116, may play a role in changing the conformation of the active site and thus increasing the efficiency of product release.
The influence of the C-terminal domain of Rv2228c in enhancing the RNase H and dsRNase activities of the N-terminal RNase H domain is in marked contrast to the homologous SCO2299 bifunctional protein from S. coelicolor. In the latter, the activities of the two domains, RNase H and acid phosphatase, are completely independent (34). Significantly, however, the interdomain linker is much longer in SCO2299 than in Rv2228c, as determined by comparison of ∼120 residues, with only 10 in the M. tuberculosis enzyme. This suggests that it is the short linker of the latter that enables its C-terminal domain to modulate the activity of the N-terminal domain. In this respect, Rv2228c may be similar to the HIV reverse transcriptase, where the RNase H domain also lacks the basic protrusion, but where the N-terminal DNA polymerase domain may substitute for the basic protrusion (12), as it occupies a similar position relative to the RNase H domain.
As the only authenticated RNase HI in M. tuberculosis, Rv2228c seems likely to play an important role in the physiology of this organism. Whether it is essential for in vitro or in vivo growth has not yet been proven, since the rv2228c gene was not included in the library used by Sassetti et al. in their genome-wide transposon mutagenesis analyses (38, 40, 41). Similarly, in the study by Lamichhane et al. (24), transposon insertion occurred in the ATG end codon and would not have compromised Rv2228c function. A pointer to the important physiological role of this protein, however, lies in the presence of rv2228c homologs in the genomes of all pathogenic mycobacterial species, including the attenuated species Mycobacterium leprae, as the only candidate genes for an RNase HI enzyme. In contrast, nonpathogenic species, such as Mycobacterium smegmatis, contain an additional rnhA gene homologous with that for EcRNaseH, suggesting that Rv2228c represents the ancestral mycobacterial RNase HI enzyme.
Beyond its role as the M. tuberculosis RNase HI, there is a potentially wider significance to the combination of three biochemical activities in the two domains of this protein. The infected macrophage is a phosphate-deprived environment, as is illustrated by the upregulation of proteins associated with phosphate transport (39). This would suggest that any function leading to release or recycling of phosphate would be advantageous to the bacterium within the macrophage. A potentially coupled generation of phosphate from the RNase H and dsRNase reaction products and the CobC reaction would greatly increase the available pool of inorganic phosphate and thus improve the environment inside the macrophage for the organism. The absence of the full complement of B12 biosynthetic genes from the M. tuberculosis genome raises questions about the in vivo role of the CobC domain of Rv2228c. Even if the latter does have the alternative function as an adenosylcobalamin phosphatase that was demonstrated for Salmonella enterica CobC (49), and a possible role in B12 recycling, the fact remains that the CobC domain of Rv2228c is unequivocally a phosphatase. This suggests that phosphate generation may be a primary role of the C-terminal domain of Rv2228c.
The full-length Rv2228c protein appears to have 3 explicit functions in a two-domain protein. This could be explained as an energy saving mechanism for the M. tuberculosis bacterium in the low-nutrient, low-energy environment of the infected macrophage. The combination of both the RNase H and the dsRNase functions of Rv2228c/N could cover a large spectrum of RNA-related functions while also combining with the C-terminal domain, not only for enhanced RNase function but also for phosphate generation.
Acknowledgments
This work was supported by the Health Research Council of New Zealand.
We gratefully acknowledge Tom Caradoc-Davies, Victoria Money, and Richard Bunker for data collection and help with crystallographic aspects of the work and Stephanie Dawes for many helpful suggestions. We also thank the following institutions for funding for purchase of the Envision plate reader: NZ Lottery Grants Board (Health), the Maurice & Phyllis Paykel Trust, and the Allan Wilson Centre for Molecular Ecology and Evolution.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L.-W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Afonine, P. V., R. W. Grosse-Kunstleve, P. D. Adams, V. Y. Lunin, and A. Urzhumtsev. 2007. On macromolecular refinement at subatomic resolution with interatomic scatterers. Acta Crystallogr. D 63:1194-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff, H. I. M., M. B. Reed, C. E. I. Barry, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 4.Busen, W., and P. Hausen. 1975. Distinct ribonuclease H activities in calf thymus. Eur. J. Biochem. 52:179-190. [DOI] [PubMed] [Google Scholar]
- 5.Camus, J. C., M. J. Pryor, C. Medigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148:2967-2973. [DOI] [PubMed] [Google Scholar]
- 6.Card, G. L., N. A. Peterson, C. A. Smith, B. Rupp, B. M. Schick, and E. N. Baker. 2005. The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. J. Biol. Chem. 280:13978-13986. [DOI] [PubMed] [Google Scholar]
- 7.Cerritelli, S. M., and R. J. Crouch. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, J. A., G. S. Prasad, S. A. White, C. D. Stout, and J. R. Williamson. 2003. Inherent protein structural flexibility at the RNA-binding interface of L30e. J. Mol. Biol. 326:999-1004. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta, S., H. Masukata, and J. Tomizawa. 1987. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell 51:1113-1122. [DOI] [PubMed] [Google Scholar]
- 12.Davies, J. F., II, Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 13.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Geladopoulos, T. P., T. G. Sotiroudis, and A. E. Evangelopoulos. 1991. A malachite green colorimetric assay for protein phosphatase activity. Anal. Biochem. 192:112-116. [DOI] [PubMed] [Google Scholar]
- 16.Haruki, M., E. Noguchi, S. Kanaya, and R. J. Crouch. 1997. Kinetic and stoichiometric analysis for the binding of Escherichia coli ribonuclease HI to RNA-DNA hybrids using surface plasmon resonance. J. Biol. Chem. 272:22015-22022. [DOI] [PubMed] [Google Scholar]
- 17.Horiuchi, T., H. Maki, and M. Sekiguchi. 1984. RNase H-defective mutants of Escherichia coli: a possible discriminatory role of RNase H in initiation of DNA replication. Mol. Gen. Genet. 195:17-22. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, K., M. Okumura, K. Katayanagi, S. Kimura, S. Kanaya, H. Nakamura, and K. Morikawa. 1993. Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution. J. Mol. Biol. 230:529-542. [DOI] [PubMed] [Google Scholar]
- 19.Itoh, T., and J. Tomizawa. 1980. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. U. S. A. 77:2450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, J. M., V. L. Arcus, C. J. Morton, M. W. Parker, and E. N. Baker. 2003. Crystal structure of a putative methyltransferase from Mycobacterium tuberculosis: misannotation of a genome clarified by protein structural analysis. J. Bacteriol. 185:4057-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayanagi, K., M. Miyagawa, M. Matsushima, M. Ishikawa, S. Kanaya, M. Ikehara, T. Matsuzaki, and K. Morikawa. 1990. Three-dimensional structure of ribonuclease H from E. coli. Nature 347:306-309. [DOI] [PubMed] [Google Scholar]
- 22.Kogoma, T., N. L. Subia, and K. von Meyenburg. 1985. Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol. Gen. Genet. 200:103-109. [DOI] [PubMed] [Google Scholar]
- 23.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 24.Lamichhane, G., M. Zignol, N. J. Blades, D. E. Geiman, A. Dougherty, J. Grosset, K. W. Broman, and W. R. Bishai. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 100:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie, A. G. 1999. Integration of macromolecular diffraction data. Acta Crystallogr. D 55:1696-1702. [DOI] [PubMed] [Google Scholar]
- 26.Lim, D., G. G. Gregorio, C. Bingman, E. Martinez-Hackert, W. A. Hendrickson, and S. P. Goff. 2006. Crystal structure of the moloney murine leukemia virus RNase H domain. J. Virol. 80:8379-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima, W. F., J. B. Rose, J. G. Nichols, H. Wu, M. T. Migawa, T. K. Wyrzykiewicz, A. M. Siwkowski, and S. T. Crooke. 2007. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 71:83-91. [DOI] [PubMed] [Google Scholar]
- 28.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizrahi, V., and S. J. Anderson. 1998. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 29:1331-1339. [DOI] [PubMed] [Google Scholar]
- 30.Nowotny, M., S. A. Gaidamakov, R. J. Crouch, and W. Yang. 2005. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121:1005-1016. [DOI] [PubMed] [Google Scholar]
- 31.Nowotny, M., S. A. Gaidamakov, R. Ghirlando, S. M. Cerritelli, R. J. Crouch, and W. Yang. 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28:264-276. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa, T., G. G. Pickett, T. Kogoma, and A. Kornberg. 1984. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 81:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtani, N., M. Haruki, M. Morikawa, and S. Kanaya. 1999. Molecular diversities of RNases H. J. Biosci. Bioeng. 88:12-19. [DOI] [PubMed] [Google Scholar]
- 34.Ohtani, N., N. Saito, M. Tomita, M. Itaya, and A. Itoh. 2005. The SCO2299 gene from Streptomyces coelicolor A3(2) encodes a bifunctional enzyme consisting of an RNase H domain and an acid phosphatase domain. FEBS J. 272:2828-2837. [DOI] [PubMed] [Google Scholar]
- 35.Parniak, M. A., K. L. Min, S. R. Budihas, S. F. Le Grice, and J. A. Beutler. 2003. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 322:33-39. [DOI] [PubMed] [Google Scholar]
- 36.Potenza, N., L. De Colibus, and A. Russo. 2005. Gel-based assay for ribonuclease H activity toward unlabeled poly(A)-poly(dT). Anal. Biochem. 337:167-169. [DOI] [PubMed] [Google Scholar]
- 37.Quiocho, F. A., J. C. Spurlino, and L. E. Rodseth. 1997. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure 5:997-1015. [DOI] [PubMed] [Google Scholar]
- 38.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:8327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigden, D. J., L. V. Mello, P. Setlow, and M. J. Jedrzejas. 2002. Structure and mechanism of action of a cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus with broad specificity phosphatase activity. J. Mol. Biol. 315:1129-1143. [DOI] [PubMed] [Google Scholar]
- 40.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 41.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarzenbacher, R., A. Godzik, S. K. Grzechnik, and L. Jaroszewski. 2004. The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D 60:1229-1236. [DOI] [PubMed] [Google Scholar]
- 43.Stein, H., and P. Hausen. 1969. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science 166:393-395. [DOI] [PubMed] [Google Scholar]
- 44.Tadokoro, T., and S. Kanaya. 2009. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 276:1482-1493. [DOI] [PubMed] [Google Scholar]
- 45.Watkins, H. A., and E. N. Baker. 2008. Cloning, expression, purification and preliminary crystallographic analysis of the RNase HI domain of the Mycobacterium tuberculosis protein Rv2228c as a maltose-binding protein fusion. Acta Crystallogr. F 64:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watkins, H. A., and E. N. Baker. 2006. Structural and functional analysis of Rv3214 from Mycobacterium tuberculosis, a protein with conflicting functional annotations, leads to its characterization as a phosphatase. J. Bacteriol. 188:3589-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, H., W. F. Lima, and S. T. Crooke. 2001. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 276:23547-23553. [DOI] [PubMed] [Google Scholar]
- 48.You, D. J., H. Chon, Y. Koga, K. Takano, and S. Kanaya. 2007. Crystal structure of type 1 ribonuclease H from hyperthermophilic archaeon Sulfolobus tokodaii: role of arginine 118 and C-terminal anchoring. Biochemistry 46:11494-11503. [DOI] [PubMed] [Google Scholar]
- 49.Zayas, C. L., and J. C. Escalante-Semerena. 2007. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: evidence that dephosphorylation of adenosyl-5′-phosphate by the CobC phosphatase is the last step of the pathway. J. Bacteriol. 189:2210-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]