Mutations in the Sex comb on midleg (Scm) gene in Drosophila cause strong derepression of homeotic genes, the hallmark phenotype of mutations in Polycomb group (PcG) genes. The SCM protein is conserved between Drosophila and mammals, but its role in PcG repression has remained elusive. In this issue, Wang et al. (14) provide evidence that SCM plays a role in recruitment of the well-characterized PcG protein complexes PRC1 and PRC2. Not only does this paper provide insight into the role of SCM in PcG repression, it helps to explain why Polycomb response elements (PREs), the DNA elements that bind PcG proteins in Drosophila, are so complex.
PcG genes encode a diverse group of proteins that act together to repress gene expression (for recent reviews, see references 2 and 12). Originally discovered in Drosophila as important for maintenance of the spatially restricted expression patterns of homeotic genes, PcG proteins bind to hundreds of sites in the Drosophila genome and are now thought to regulate hundreds of genes. Similarly, mammalian PcG proteins are bound to hundreds of genes and are implicated in stem cell maintenance, as well as differentiation. Levels of PcG proteins are also altered in some cancers, where their misexpression may alter the expression levels of critical genes. At the Drosophila homeotic genes, PcG proteins are epigenetic silencers of gene expression, while at other loci, they may modulate expression levels. The regulation of gene expression by PcG proteins is an area of active exploration.
Many PcG proteins act in protein complexes to modify chromatin. The best-characterized complexes, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2), are conserved between Drosophila and mammals. Here we describe only the Drosophila complexes; the mammalian genome contains multiple paralogs of many of the Drosophila PcG genes. The core components of PRC1 in Drosophila are Polycomb (PC), Polyhomeotic (PH) (there are two homologs, PH-D and PH-P), RING (also called Sex combs extra [SCE]), and Posterior sex combs (PSC) [or its homolog SU(Z)2]. Biochemical activities attributed to PRC1 in vitro include inhibition of chromatin remodeling, inhibition of transcriptional elongation, and chromatin compaction. In addition, RING/SCE is an H2A ubiquitin ligase, but this biochemical activity is more likely mediated through another recently identified complex in Drosophila, RAF, which includes RING/SCE, PSC, and the demethylase KDM2 (encoded by the CG11033 gene), coupling histone H2A ubiquitylation to histone H3 demethylation (4). Thus, RING/SCE and PSC are present in two PcG complexes, and their activities must be evaluated in that light. PRC2 consists of the PcG proteins E(Z), ESC (or its homolog ESCL), SU(Z)12, and the histone-binding protein CAF1 (also known as NURF55). PRC2 methylates histone H3 at lysine 27, and H3K27me3 modified chromatin is a hallmark of PcG-regulated genes. Finally, the DNA-binding PcG protein Pleiohomeotic (PHO) (or its homolog PHOL) is in a complex called PhoRC (for Pho repressive complex) with the PcG protein SFMBT. All of these protein complexes are bound to PREs, but exactly how this occurs is unknown.
How does SCM fit into this scheme? SCM copurifies with PRC1 in substoichiometric amounts. Further, SCM can interact with the PRC1 component PH in vitro and can even be incorporated into a PRC1 complex when coexpressed in the baculoviral system (8). Despite this, the bulk of SCM in Drosophila embryos is in an uncharacterized protein complex that is distinct from PRC1 (8). More recently, SCM has been shown to interact both physically and genetically with SFMBT (1). A discussion of the domains present in SCM, SFMBT, and PH is useful at this point in making sense of this information.
The conserved protein domains in SCM provide some insight into function. The SCM proteins contain two copies of the mbt domain, a protein domain of about 100 amino acids that binds methylated histones. First identified in the Drosophila l(3)mbt [lethal (3) malignant brain tumor] protein, mbt domains appear to be present only in metazoans. In addition to SCM and l(3)mbt, Drosophila has a third mbt domain-containing protein, SFMBT (Scm-related gene containing four mbt domains). Urochordates and vertebrates have homologs of all three Drosophila mbt-containing proteins. The nematode Caenorhabditis elegans has only two MBT proteins (LIN61 and MBTR-1), both of which contain four mbt domains. Sequence comparisons also suggest that the nematode proteins are more related to SFMBT homologs than to SCM or l(3)mbt homologs. mbt domains appear to occur in tandem arrays with from two to four domains per protein, but for the Drosophila proteins, only the most carboxy-terminal mbt domain of each array appears to have a binding pocket that can accommodate methyl-lysine residues (1). The functions of the remaining mbt domains are not clear.
There are also Zn finger motifs in all three Drosophila MBT proteins, as well as in the PRC1 component PH. The Drosophila SFMBT and SCM proteins interact physically through N-terminal fragments that include the Zn finger motifs (1). Finally, a C-terminal putative protein interaction domain, an SPM-type SAM (sterile alpha motif) domain, is present in SFMBT, SCM, and PH. The SCM SPM domain mediates interactions with the PH SPM domain, as well as with itself (8). Mutations in the SPM domain disrupt SCM function both in the endogenous gene and in an artificial repression system where SCM is tethered to DNA by a heterologous DNA-binding protein (10). Tethered SCM repression is dependent on PH, suggesting that SCM can recruit PH (and presumably PRC1) to DNA. Overexpression of the SCM SPM domain disrupts PcG repression by an unknown mechanism (8). It is worth noting that SAM domains may also be able to bind RNA, which could add to the complexity of PcG-induced silencing (3).
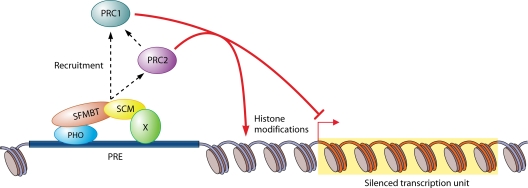
How do all of these protein complexes (PhoRC, PRC1, PRC2, and the 500-kDa uncharacterized SCM complex) get recruited to the target genes? As stated above, PhoRC contains the DNA-binding protein PHO, and PHO-binding sites are required for the activity of all PREs tested so far. Some genome-wide studies suggest that PHO is bound at nearly all sites where PRC1 and/or PRC2 is bound, (11), whereas other studies suggest that it is bound at only 50% of these sites (cited in reference 12). Nevertheless, there is no doubt that PHO binding is an important component of many PREs. In experiments utilizing RNA interference (RNAi) and mutations to knock down various PcG proteins (approaches also used in the work described by Wang et al. [14]), PHO binding was required for the localization of both PRC1 and PRC2 to a PRE (13). Further, PRC1 binding was dependent on PRC2. The model that emerged from these studies is that PhoRC recruits PRC2, which then trimethylates histone H3K27. The PRC1 subunit PC then binds to H3K27me3, which could mediate PRC1 recruitment. In vitro, PHO interacts with the PRC2 subunits ESC and E(Z) as well as the PRC1 subunits PC and PH (6), suggesting that it could directly recruit both complexes. Enter SCM. Wang et al. (14) used RNAi and mutations to knock down expression levels of various PcG proteins and show that the binding of PRC1 and PRC2, but not PhoRC, to the PRE is SCM dependent. They further suggest that SCM might be in a complex with an unknown DNA-binding protein, protein X, and that the SCM-protein X complex could cooperate with PhoRC to recruit PRC1 and PRC2 (Fig. 1). This attractive model provides not only a role for the SCM protein but also a potential role for one of the myriad of other DNA-binding sites required for PRE function.
FIG. 1.
Model for the role of SCM in Polycomb-induced gene silencing. PHO-SFMBT and SCM-protein X are recruited independently to the PRE. SFMBT and SCM physically interact, and both facilitate either recruitment or retention of PRC2 and PRC1 at the PRE. PRC2 also facilitates recruitment or retention of PRC1 at the PRE. Silencing of the target gene by PRC1 and PRC2 involves histone modifications as well as other mechanisms.
PREs are complex elements made up of binding sites for many different proteins (reviewed in references 7 and 9), including PHO, GAGA factor (GAF), Pipsqueak (PSQ) (which binds the same site as GAF), the Sp1/KLF family of proteins, DSP1, ZESTE, and Grainyhead (GRH). While the role of PHO sites in PRE function is well established, the roles of the other protein binding sites are less clear. Numerous studies have shown that GAF/PSQ-binding sites are important for PRE function, and in vitro studies suggest that binding of GAF may make the DNA more accessible to PHO binding (5). Genome-wide chromatin immunoprecipitation (ChIP) studies show that only a subset of locations that bind PRC1 and/or PRC2 have GAF bound (11). It is not known whether PSQ associates with PREs in vivo. Likewise, DSP1 and ZESTE are bound only to a subset of PREs. There are 9 or 10 Sp1/KLF family members in Drosophila, and it is not known which, if any, are PRE associated. Thus, the current understanding of which proteins are necessary for PRE function is poor. Nevertheless, it is absolutely clear that PHO sites alone are not sufficient for PRE function, that PREs contain binding sites for at least three different proteins, and that mutation of any one binding site disrupts PRE function.
Could one of the known PRE-binding proteins be protein X? Characterization of the proteins present in the 500-kDa embryonic SCM complex may provide the answer. Another approach is to look at whether SCM binds to PRE transgenes that contain mutations in the DNA-binding sites necessary for PRE activity. The model predicts that mutation of the site that binds protein X will cause a loss of SCM binding. This interesting paper (14) has brought us much closer to an understanding of PRE function and PcG recruitment.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Grimm, C., R. Matos, N. Ly-Hartig, U. Steuerwald, D. Lindner, V. Rybin, J. Müller, and C. W. Muller. 2009. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28:1965-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerppola, T. K. 2009. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 19:692-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, C. A., and J. U. Bowie. 2003. SAM domains: uniform structure, diversity of function. Trends Biochem. Sci. 28:625-628. [DOI] [PubMed] [Google Scholar]
- 4.Lagarou, A., A. Mohd-Sarip, Y. M. Moshkin, G. E. Chalkley, K. Bezstarosti, J. A. A. Demmers, and C. P. Verrijzer. 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 22:2799-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoudi, T., L. M. P. Zuijderduijn, A. Mohd-Sarip, and C. P. Verrijzer. 2003. GAGA facilitates binding of Pleiohomeotic to a chromatinized Polycomb response element. Nucleic Acids Res. 31:4147-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller, J., and J. A. Kassis. 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16:476-484. [DOI] [PubMed] [Google Scholar]
- 8.Peterson, A. J., D. R. Mallin, N. J. Francis, C. S. Ketel, J. Stamm, R. K. Weller, R. E. Kingston, and J. A. Simon. 2004. Requirement for Sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167:1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ringrose, L., and R. Paro. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134:223-232. [DOI] [PubMed] [Google Scholar]
- 10.Roseman, R. R., K. Morgan, D. R. Mallin, R. Roberson, T. J. Parnell, D. J. Bornemann, J. A. Simon, and P. K. Geyer. 2001. Long-range repression by multiple Polycomb group (PcG) proteins targeted by fusion to a defined DNA-binding domain in Drosophila. Genetics 158:291-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuettengruber, B., M. Ganapathi, B. Leblanc, M. Portoso, R. Jaschek, B. Tolhuis, M. Van Lohuizen, A. Tanay, and G. Cavalli. 2009. Functional anatomy of Polycomb and Trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 7:146-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon, J. A., and R. E. Kingston. 2009. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10:697-708. [DOI] [PubMed] [Google Scholar]
- 13.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of Polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 14.Wang, L., N. Jahren, E. L. Miller, C. S. Ketel, D. R. Mallin, and J. A. Simon. 2010. Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other Polycomb group repressors at a Drosophila Hox gene. Mol. Cell. Biol. 30:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]