Abstract
Imprinted gene expression corresponds to parental allele-specific DNA CpG methylation and chromatin composition. Histone tail covalent modifications have been extensively studied, but it is not known whether modifications in the histone globular domains can also discriminate between the parental alleles. Using multiplex chromatin immunoprecipitation-single nucleotide primer extension (ChIP-SNuPE) assays, we measured the allele-specific enrichment of H3K79 methylation and H4K91 acetylation along the H19/Igf2 imprinted domain. Whereas H3K79me1, H3K79me2, and H4K91ac displayed a paternal-specific enrichment at the paternally expressed Igf2 locus, H3K79me3 was paternally biased at the maternally expressed H19 locus, including the paternally methylated imprinting control region (ICR). We found that these allele-specific differences depended on CTCF binding in the maternal ICR allele. We analyzed an additional 11 differentially methylated regions (DMRs) and found that, in general, H3K79me3 was associated with the CpG-methylated alleles, whereas H3K79me1, H3K79me2, and H4K91ac enrichment was specific to the unmethylated alleles. Our data suggest that allele-specific differences in the globular histone domains may constitute a layer of the “histone code” at imprinted genes.
Imprinted genes are defined by the characteristic monoallelic silencing of either the paternally or maternally inherited allele. Most imprinted genes exist in imprinted gene clusters (10), and these clusters are usually associated with one or more differentially methylated regions (DMRs) (27, 65). DNA methylation at DMRs is essential for the allele-specific expression of most imprinted genes (31). Maternal or paternal allele-specific DNA methylation of a subset of DMRs (germ line DMRs) is gamete specific (27, 39). These maternal or paternal methylation differences are established during oogenesis or spermatogenesis, respectively, by the de novo DNA methyltransferases Dnmt3a and Dnmt3b together with Dnmt3L (5, 26, 48). The gamete-specific methylation differences set the stage for the parental allele-specific action of germ line DMRs, some of which have been shown to control the monoallelic expression of the associated genes in the respective domains (11, 34, 36, 53, 66, 71-73, 77). These DMRs are called imprinting control regions (ICRs).
Two recurring themes have been reported for ICR action. ICRs can function as DNA methylation-regulated promoters of a noncoding RNA or as methylation-regulated insulators. Recent evidence suggests that both of these mechanisms involve chromatin organization by either the noncoding RNA (45, 50) or the CTCF insulator protein (17, 32) along the respective imprinted domains. The CTCF insulator binds in the unmethylated maternal allele of the H19/Igf2 ICR and blocks the access of the Igf2 promoters to the shared downstream enhancers. CTCF cannot bind in the methylated paternal ICR allele; hence, here the Igf2 promoters have access to the enhancers (4, 18, 24, 25, 62). When CTCF binding is abolished in the ICR of the maternal allele, Igf2 expression becomes biallelic, and H19 expression is missing from both alleles (17, 52, 58, 63). Importantly, CTCF is the single major organizer of the allele-specific chromatin along the H19/Igf2 imprinted domain (17). Significantly, CTCF recruits, at a distance, Polycomb-mediated H3K27me3 repressive marks at the Igf2 promoter and at the Igf2 DMRs (17, 32).
A role for chromatin composition is suggested in the parental allele-specific expression of imprinted genes. Repressive histone tail covalent modifications, such as H3K9me2 H3K9me3, H4K20me3, H3K27me3, and the symmetrically methylated H4R3me2 marks, are generally associated with the methylated DMR alleles, while activating histone tail covalent modifications, such as acetylated histone tails and also H3K4me2 and H3K4me3, are characteristic of the unmethylated alleles (7-9, 12-15, 17, 21, 33, 35, 43, 44, 51, 55, 56, 67, 69, 74, 75). Importantly, the maintenance of imprinted gene expression depends on the allele-specific chromatin differences. ICR-dependent H3K9me2 and H3K27me3 enrichment in the paternal allele (67) is required for paternal repression of a set of imprinted genes along the Kcnq1 imprinted domain in the placenta (30). Imprinted Cdkn1c and Cd81 expression depends on H3K27 methyltransferase Ezh2 activity in the extraembryonic ectoderm (64). Similarly, H3K9 methyltransferase Ehmt2 is required for parental allele-specific expression of a number of imprinted genes, including Osbpl5, Cd81, Ascl2, Tfpi2, and Slc22a3 in the placenta (44, 45, 70).
There is increasing evidence that covalent modifications, not only in the histone tails but also in the histone globular domains, carry essential information for development and gene regulation. The H3K79 methyltransferase gene is essential for development in Drosophila (60) and in mice (22). H3K79 methylation is required for telomeric heterochromatin silencing in Drosophila (60), Saccharomyces cerevisiae (47, 68), and mice (22). The H4K91 residue regulates nucleosome assembly (76). Whereas mutations at single acetylation sites in the histone tails have only minor consequences, mutation of the H4K91 site in the histone H4 globular domain causes severe defects in silent chromatin formation and DNA repair in yeast (37, 42, 76).
Contrary to the abundant information that exists regarding the allele-specific chromatin composition at DMRs of imprinted genes, no information is available about the parental allele-specific marking in the histone globular domains at the DMRs. We hypothesized that chromatin marks in the globular domains of histones also distinguish the parental alleles of germ line DMRs. In order to demonstrate this, we measured the allele-specific enrichment of H3K79me1, H3K79me2, H3K79me3, and H4K91ac at 11 mouse DMRs using quantitative multiplex chromatin immunoprecipitation-single nucleotide primer extension (ChIP-SNuPE) assays. In general, H3K79me3 was associated with the methylated allele at most DMRs, whereas the unmethylated allele showed enrichment for H3K79me1, H3K79me2, and H4K91ac. These results are consistent with the possibility that allele-specific differences in the globular domains of histones contribute to the “histone code” at DMRs.
MATERIALS AND METHODS
Chromatin immunoprecipitation.
Mouse embryonic fibroblasts (MEFs) were derived from 13.5-days-postcoitum (dpc) embryos. 129S1 (129) and JF1 inbred mice (28) were purchased from the Jackson Laboratory. The CAST/Ei (CS) line FVB/NJ.CAST/Ei(N7) is a distal chromosome 7 partial congenic strain (63). The CTCF site mutant (CTCFm) mouse line carries point mutations at for ICR-CTCF sites (63). Chromatin preparation from 129 × CS, CS × 129, CTCFm × CS, 129 × JF1, and JF1 × 129 primary MEFs was done as described earlier (17). Briefly, the chromatin was cross-linked with formaldehyde. After sonication in lysis buffer, an aliquot of the chromatin was reverse cross-linked and quantitated by optical density (OD), and the efficiency of sonication was verified on agarose gel. The chromatin was then diluted to a 0.4-mg/ml concentration and snap-frozen in small aliquots. Each aliquot was thawed only once on the day that ChIP was performed. The following antibodies, purchased from Abcam, were used in the chromatin immunoprecipitation (ChIP) assays: anti-monomethyl-histone H3 (Lys79), ab2886; anti-trimethyl-histone H3 (Lys79), ab2621; anti-acetyl-histone H4 (Lys91), ab4627; and anti-acetyl-histone H4 (Lys16), ab61240. Anti-dimethyl-histone H3 (Lys79), 07-366; anti-dimethyl-histone H3 (Lys4), 07-030; anti-trimethyl-histone H3 (Lys9), 17-625; anti-trimethyl-histone H3 (Lys27), 07-449; and anti-acetyl-histone H3 (Lys9), 07-352, were purchased from Millipore. The globular histone covalent modifications were detected using cross-linking and ChIP (X-ChIP) conditions. Native ChIP (N-ChIP) chromatin was used for detecting the histone tail modifications. The chromatin immunoprecipitation was performed as described previously (17), with minor modifications. Preblocked A/G beads obtained from Santa Cruz (sc-2003) were used. Four micrograms of DNA equivalent chromatin was used for ChIP.
Real-time PCR.
Real-time PCR was performed to measure the region-specific overall ChIP enrichment levels at the H19-Igf2 domain as described previously (17). Equal aliquots (3 μl out of 100 μl) of ChIP elution DNA were amplified with region-specific primers. A dilution series of known amounts of genomic DNA was used for quantitating copy numbers from ChIP and input samples. The input DNAs were from the exact chromatin aliquots used for the ChIP. These were used for fine adjustment of the ChIP intensities. PCR and extension primers for the 11 DMRs and control regions can be found in Table S1 in the supplemental material.
Analysis of allele-specific histone enrichment.
To measure allele-specific chromatin differences, we used the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) allelotyping analysis method obtained from Sequenom. This method uses mass spectrometry quantification of the extended SNuPE primers based on the differences in molecular mass between alleles (19, 23). Single nucleotide polymorphisms (SNPs) for the H19-Igf2 region were obtained by DNA sequencing of inbred 129S1 (129) and CAST/Ei (CS) at specific regions of interest as described previously (17). The SNP for the Rasgrf1 DMR was obtained by sequencing inbred 129S1 and JF1 mouse DNA. For the other DMRs, we used known JF1 polymorphisms (20, 27, 38) that we had confirmed by DNA sequencing. PCR and extension primers (see Table S1 in the supplemental material) for multiplex assays (H19-Igf2, 7-plex; DMRs, 16-plex) were designed using MassArray assay design software version 3.1. Six-microliter PCR mixtures contained 1 pmol of each of the corresponding PCR primer pairs, 25 ng genomic DNA or 10 μl of the chromatin immunoprecipitated DNA sample, and hot-start reaction mix (Qiagen). PCR conditions were as follows: 94°C for 15 min, followed by 40 cycles of 94°C (20 s), 56°C (30 s), and 72°C (60 s), and a final extension of 72°C for 3 min. Amplification of the H19 promoter was performed separately in a 10-μl reaction with Roche long-range buffer 1 and a mixture of Taq DNA polymerase (Roche) and Taq Vent (New England Biolabs) and utilized the following PCR conditions: 95°C for 2 min, followed by 37 cycles of 95°C (30 s), 65°C (45 s), 72°C (45 s), and a final extension of 72°C for 5 min. Amplified samples were spotted onto a 384 SpectroChip array using a Nanodispenser and analyzed in a MassArray compact mass spectrometer (Sequenom). Automated spectra acquisition was performed using SpectroAcquire (Sequenom). Samples were analyzed with the MassArray Typer version 3.4. Allelotyping was performed by first generating an allele skew correction file using a heterozygote DNA sample to correct for any allelic imbalance that may be present in the allele mass products of true heterozygote crosses. All samples were then exported from Typer version 3.4 and, in the process, were applied to the skew correction file in order to normalize any existing allelic imbalance in the SNP allele products. The final allelotype data report contained the ratio present of each allele product at that given SNP. Serial dilutions (see Fig. S1 and S2 in the supplemental material) were included in every experiment for quality control. Samples were run in duplicate.
RNA knockdown using shRNA.
To generate the pseudoviral particles, 293T cells were cotransfected by calcium phosphate with 15 μg of each small hairpin RNA (shRNA) plasmid (Sigma, St. Louis, MO) and 10 μg of pPACK packaging plasmid mix (SBI, Mountain View, CA) at a cell density of 4 × 106 per 10-cm culture dish. The culture medium was replaced with fresh medium after 6 h. The supernatant was collected 24 h and 48 h after transfection. To determine the vector titers, 105 HT1080 cells were seeded in a six-well plate and transduced with various dilutions of the vector in the presence of 4 μg of Polybrene/ml (Sigma, St. Louis, MO). The culture medium was replaced 48 h later with fresh medium containing puromycin (Sigma, St. Louis, MO) at a concentration of 1.5 μg/ml, and puromycin-resistant colonies were counted 10 days after transduction. 129 × JF1 MEFs were transduced with Mission pseudoviral particles TRCN0000125099, TRCN0000125100, TRCN0000125101, TRCN0000125102, and TRCN0000125103 against mouse Dot1L (Sigma, St. Louis, MO) at an optimal multiplicity of infection (MOI) of 5 and collected 10 days later for RNA analysis. The experiment was repeated, and similar results were obtained at day 16 and day 21.
RNA isolation and RT-PCR.
RNA was isolated from MEFs or from the Dot1L+/+ and Dot1L1lox/1lox embryos (22) at 8.5 dpc using RNA-Bee, according to manufacturer's instructions (Tel-Test). The pellet was dissolved in diethyl pyrocarbonate (DEPC) water containing RNasin (Promega) and 10 mM dithiothreitol (DTT). Contaminating DNA was removed with the DNA-free kit (Ambion). Reverse transcription was performed on equal amounts of RNA with random hexamers using the SuperScript III random primer synthesis kit for RT-PCR (Invitrogen), according to manufacturer's instructions. A total of 3 μl of first-strand cDNA was used for real-time quantitative PCR. 129 × JF1 MEF cDNA was used for standard curve dilutions. RT-PCR primers are listed in Table S1 in the supplemental material.
RESULTS
Multiplex quantitative ChIP-SNuPE assays for measuring allele-specific chromatin composition at 11 DMRs and at the H19/Igf2 imprinted domain.
Allele-specific chromatin analysis has been described for individual imprinted genes or small sets of DMRs in a limited number of cell types. To gain an understanding of how different chromatin modifications may influence the allele-specific expression of imprinted genes, we designed assays that allow a comparative and comprehensive chromatin analysis of a large set of germ line DMRs. To achieve a high-throughput allele-specific chromatin analysis at imprinted domains, we developed nonradioactive multiplex ChIP-SNuPE assays based on the Sequenom allelotyping platform (23). These assays distinguish allele-specific incorporation of dideoxynucleoside triphosphates (ddNTPs) into the SNuPE primer based on differences in molecular mass. Our previous singleplex ChIP-SNuPE assays were based on radionucleotide incorporation at sites of single nucleotide polymorphisms between 129 and CS mouse genomic sequences along the H19/Igf2 imprinted domain. Our first 7-plex Sequenom assay used the same SNPs (Fig. 1) at the Igf2 DMR1, Igf2 P2 promoter, Igf2 DMR2, two halves of the ICR (−3 kb and −4 kb), the H19 gene body (+2 kb), and an intermediary region −8 kb from the H19 transcriptional start site. Each assay in the 7-plex assay and also in the H19 promoter assay, run separately, were rigorously quantitative, as shown by DNA mixing experiments (see Fig. S1 in the supplemental material).
FIG. 1.
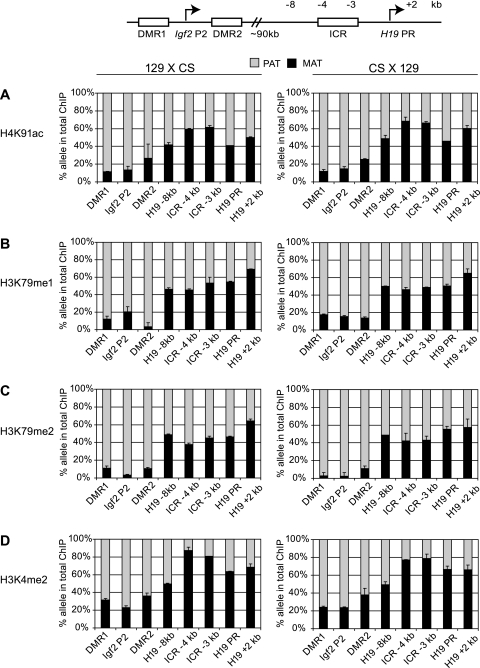
Activating chromatin composition along the H19/Igf2 imprinted domain. Allele-specific activating chromatin was measured by quantitative ChIP-SNuPE assays at the H19/Igf2 imprinted domain, using the 7-plex assay (see Fig. S1 in the supplemental material) and the H19 promoter assay. The regions of interest are depicted in the schematic drawing and indicated under each column. ChIP was done in duplicate, using antibodies against specific histone modifications (indicated on the left side of each row of charts) to precipitate chromatin from 129 mother × CS father MEFs or reciprocal CS mother × 129 father MEFs (indicated at the top). The ratio of an allele-specific histone modification at a specific region was expressed as a percentage of maternal (MAT) or paternal (PAT) allele in the total (maternal plus paternal, or 100%) immunoprecipitation. Standard deviations are indicated as error bars. Active chromatin histone globular domain modifications H4K91ac (A), H3K79me1 (B), and H3K79me2 (C) and the control histone tail modification H3K4me2 (D) clearly distinguished the paternal alleles at the Igf2 regions. These modifications were slightly biased or not biased toward the maternal alleles at the H19 regions. No allele-specific chromatin differences existed at a “neutral” intermediary region −8 kb upstream of the H19 promoter (PR). Reciprocal mouse crosses had very similar allele-specific chromatin composition.
To extend our high-throughput allele-specific chromatin analysis to a comprehensive set of imprinted regions, we also designed a 16-plex ChIP-SNuPE reaction. This allows for measuring allelic ratios of chromatin at 11 different DMRs based on SNPs between 129 and JF1 inbred strains. Four of these DMRs were represented in the assay with two or three alternative SNPs along their sequences (see Fig. S2 in the supplemental material). These multiplex assays again were rigorously quantitative (see Fig. S2 and S3 in the supplemental material). Most of these assays showed a linear response, except Gnas1A DMR, which required a simple curve-fitting calculation step. The parental allele specificity of histone modifications at each of these DMRs was very similar between ChIP replicates, between alternative SNPs in the same DMRs, between the reciprocal mouse crosses, and (for the ICR) between 129 × CS and 129 × JF1 crosses (Fig. 1 and 2; see Fig. 6 to 8).
FIG. 2.
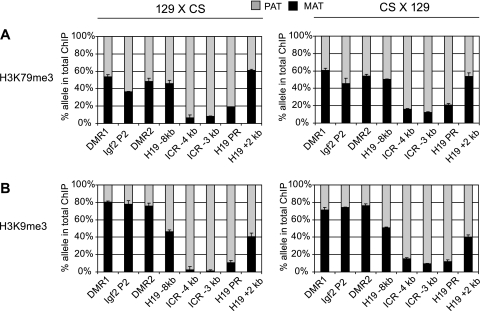
Repressive chromatin composition along the H19/Igf2 imprinted domain. Repressive globular domain histone modification H3K79me3 (A) and the control histone tail modification H3K9me3 (B) localized to the paternal allele at the H19 ICR and H19 promoter sequences. H3K9me3 but not H3K79me3 distinguished the maternal allele at the Igf2 regions. No allele-specific chromatin differences existed at −8 kb upstream of the H19 promoter. Other details are provided in Fig. 1.
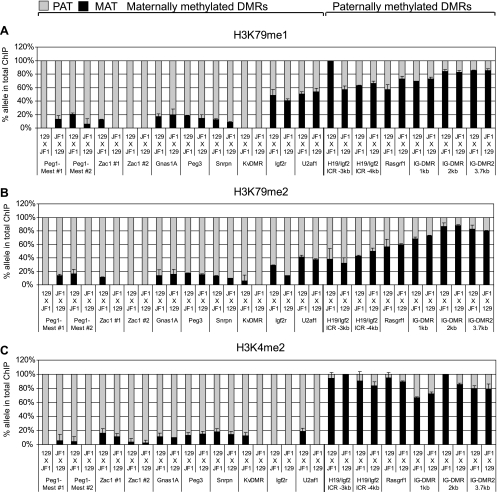
FIG. 6.
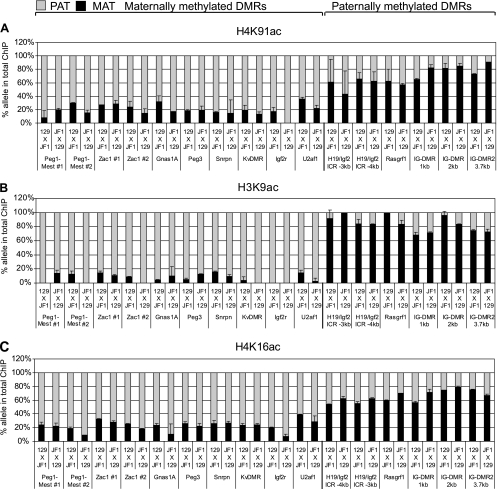
Histone acetylation marks distinguish the CpG-unmethylated alleles of maternally and paternally methylated DMRs. Allele-specific chromatin composition was determined at single nucleotide polymorphisms (SNPs) within maternally methylated (Peg1, Zac1, Gnas1A, Peg3, Snrpn, Igf2r, U2af1, DMRs, and KvDMR) and paternally methylated (H19/Igf2, Rasgrf1, DMRs, and IG-DMR) DMRs by quantitative multiplex assays (see Fig. S2 and S3 in the supplemental material) and represented as shown in Fig. 1. Alternative SNPs are included for the H19/Igf2 ICR (−3 kb and −2 kb from the transcription start site of H19), IG-DMR (at 1, 2, or 3.7 kb along the DMR), Peg1-Mest (no. 1 and 2 along the DMR), and Zac1 (no. 1 and 2) DMRs. ChIP was performed in duplicate using antibodies against specific modified histones (indicated on the top of each row of charts) from 129 × JF1 MEFs or the reciprocal JF1 × 129 MEFs (indicated under each column, maternal allele comes first). (A) The allele specificity of the H4K91ac globular histone modification is similar to the H3K9ac (B) and H4K16ac (C) histone tail modifications.
FIG. 8.
Repressive chromatin marks distinguish the parental alleles of maternally and paternally methylated DMRs. The globular histone mark H3K79me3 (A) showed allele-specific distribution at DMRS similar to that of other repressive histone tail modifications H3K9me3 (B) and H3K27me3 (C). The paternally methylated DMRs exhibited a paternal-allele specific bias, whereas some of the maternally methylated DMRs showed a maternal allele-specific bias for H3K79me3. Other details are shown in Fig. 6.
Assessment of the specificity of the antibodies.
To gain a better understanding of the function of globular domain histone modifications, we decided to test the allele-specific enrichment of different methylated forms of H3K79 at DMRs of imprinted genes. The H3K79me1, H3K79me2, H3K79me3, and control H3K9me3 antibodies recognized the corresponding peptides in immuno-dot blot assays with high specificity (see Fig. S4 in the supplemental material).
Allele-specific chromatin composition at the H19/Igf2 imprinted region.
We first investigated the parental allele-specific enrichment of histone covalent modifications in the globular domains of H3 and H4 at the H19/Igf2 imprinted domain using the 7-plex Sequenom assay (see Fig. S1 in the supplemental material). We found that the H4K91ac, H3K79me1, and H3K79me2 marks were strongly paternal allele specific at the Igf2 DMR1, Igf2 P2 promoter, and Igf2 DMR2 sequences (Fig. 1A to C). This pattern was similar to the pattern of an activating histone tail modification, H3K4me2 (Fig. 1D). The H19/Igf2 ICR, the H19 promoter, and the H19 gene body showed no or only slight bias toward the maternal alleles for the H4K91ac, H3K79me1, and H3K79me2 marks (Fig. 1A to C). This pattern was different from the maternally biased pattern of H3K4me2 (Fig. 1D).
H3K79me3 enrichment was strongly paternal allele specific at the H19/Igf2 ICR and at the H19 promoter (Fig. 2A), similar to the pattern of H3K9me3 at these sequences (Fig. 2B). The 7-plex SNuPE assay was more sensitive than our previous manual ChIP-SNuPE assays (17) for the H19/Igf2 domain. With the multiplex assay, we were able to measure the H3K9me3 allelic enrichment at the Igf2 DMR1 and Igf2 P2 promoter and also measure more precisely the H3K9me3 allelic bias at the Igf2 DMR2. Whereas H3K9me3 was biased toward the repressed maternal allele throughout the Igf2 locus (Fig. 2B), these regions did not exhibit allele-specific enrichment for H3K79me3 (Fig. 2A). The control intermediary region at −8 kb did not exhibit allele-specific differences for any of the histone modifications examined.
In summary, the histone globular domain covalent modifications we examined exhibited a strong allelic bias toward only the paternally inherited chromosome along the H19/Igf2 imprinted domain.
Effects of the ICR-CTCF site mutations on the allele-specific chromatin composition of histone globular domain modifications at the H19/Igf2 imprinted domain.
CTCF binding in the H19/Igf2 ICR is required for organization of the maternal allele's chromatin composition, with regard to histone tail modifications (17). CTCF binding in the maternal allele recruited active chromatin to the H19 locus and repressive chromatin to the Igf2 locus and also excluded repressive chromatin from the H19 locus and active chromatin from the Igf2 locus. We asked whether CTCF binding in the H19/Igf2 ICR has also an effect on the allele specificity of histone globular domain covalent modifications. We compared normal 129 × CS and mutant CTCFm × CS MEF chromatin along the domain (Fig. 3). The latter cells carried point mutations at each of the four CTCF binding sites in the ICR (17, 63) and, as a consequence, lacked in vivo ICR-CTCF binding (17). Lack of insulation resulted in biallelic Igf2 expression and lack of H19 expression in CTCFm × CS fetal kidneys and livers (63) and in CTCFm × CS MEFs (17). We measured the amount of immunoprecipitated DNA from equal amounts of chromatin in normal versus mutant cells using real-time PCR (Fig. 3) and determined the parental allele specificity of chromatin in the mutant cells using ChIP-SNuPE (Fig. 4).
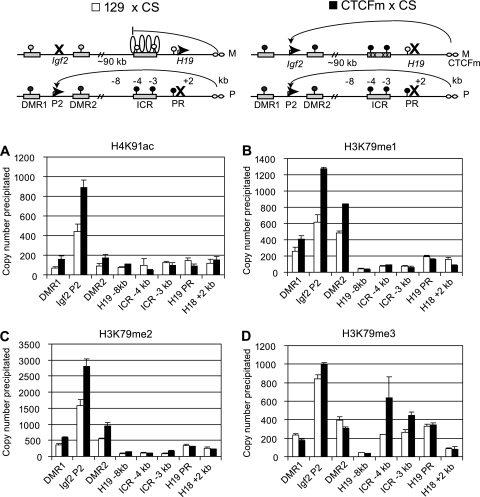
FIG. 3.
CTCF is responsible for region-specific enrichment of chromatin components at the H19 and Igf2 loci. The overall enrichment for specific chromatin modifications was compared between 129 × CS MEFs (white bars) and CTCFm × CS MEFs (black bars) by ChIP and real-time PCR. The schematic drawings at the top depict the expressed versus silenced status (horizontal arrow versus X) and methylation of the H19 and Igf2 imprinted genes in normal 129 × CS MEFs and mutant CTCFm × CS MEFs (17). In normal cells, CTCF protein (vertical oval) binding in the ICR (rectangle) in the unmethylated (white lollipop) maternal allele (M) but not in the methylated (black lollipop) paternal allele (P) insulates the Igf2 promoter from the downstream enhancers (small horizontal ovals). In the mutant cells, CTCF binding is abolished in the maternal ICR by point mutations (x) resulting in lack of insulation and, hence, biallelic Igf2 expression. The levels of active chromatin marks H4K91ac (A), H3K79me1 (B), and H3K79me2 (C) greatly increased in the mutant cells at the DMR1, the DMR2, and the Igf2 P2 promoter. The repressive H3K79me3 signal (D) greatly increased at the H19 ICR in CTCFm × CS MEFs compared to that in normal cells. There was no change at the −8-kb region. Average precipitation values are expressed in copy numbers and are shown with standard deviations.
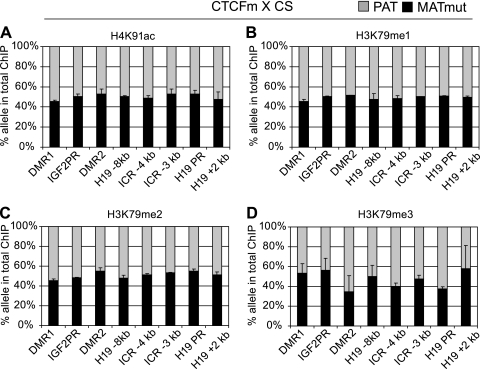
FIG. 4.
CTCF is required for allele-specific chromatin composition locally and at a distance. Quantitative analyses of chromatin composition reveal the consequences of ICR CTCF site mutations. Allele-specific enrichment is no longer apparent: the activating globular domain histone marks H4K91ac (A), H3K79me1 (B), and H3K79me2 (C) have shifted toward the maternal allele at the Igf2 locus. (D) H3K79me3 has shifted toward the maternal allele at the H19 locus. Chromatin was precipitated from CTCFm × CS MEFs in duplicate with the specific antibodies indicated on top of each chart. Allele-specific histone modification at a specific region was expressed as a percentage of the maternal mutant (MATmut) or paternal wild type (PAT) allele in the total immunoprecipitate.
Remarkably, the ICR CTCF site point mutations caused a 2-fold increase in the heterochromatin mark H3K79me3 at the ICR sequences (Fig. 3D), where it was strongly paternal allele specific in normal cells (Fig. 2A). H3K79me3 became biallelic in the mutant cells at the ICR (Fig. 4D), providing evidence that CTCF is required in the ICR for excluding H3K79me3 from the maternal allele. H3K79me3 occupancy at the H19 promoter similarly switched from paternal to biallelic (Fig. 4D), but this was not accompanied by a change in the level of H3K79me3 (Fig. 3D). The ICR CTCF site point mutations also caused a 2-fold increase in the H4K91ac, H3K79me1, and H3K79me2 levels at the Igf2 P2 promoter and Igf2 DMRs in CTCFm × CS MEFs compared to wild-type 129 × CS MEFs (Fig. 3A to C), and these paternal allele-specific (Fig. 1A to C) activating chromatin marks became biallelic in the mutant cells at the Igf2 P2 promoter and Igf2 DMRs (Fig. 4A to C).
H4K91ac levels were low at the H19 locus (Fig. 3A) and were only slightly biased at the ICR toward the maternal allele in normal cells (Fig. 1A) but became unbiased in the mutant cells (Fig. 4A). These data suggest that CTCF has a slight effect on H4K91ac recruitment at the H19 locus. H3K79me1 and H3K79me2 levels were low in abundance (Fig. 3A to C) and showed biallelic enrichment in normal cells at the H19 locus (Fig. 1A to C), but the CTCF site mutations did not change these features (Fig. 3 and 4A to C), suggesting that CTCF does not regulate H3K79me1 and H3K79me2 enrichment at the H19 locus. At the Igf2 P2 promoter and at the Igf2 DMRs, H3K79me3 levels were relatively high compared to those of other sequences in the H19/Igf2 domain but did not change significantly in response to the CTCF site mutations (Fig. 3D). Also, H3K79me3 was biallelically enriched at the Igf2 locus in normal and mutant cells (Fig. 2A and 4D), indicating that CTCF-ICR binding is not responsible for including K79me3-modified H3 in the maternal allele at the Igf2 locus. Taken together, the ICR CTCF site mutations have caused the paternalization of the maternal allele's chromatin composition along the H19/Igf2 imprinted domain by exclusion. CTCF binding in the H19 ICR was required in the maternal allele at the H19 locus for excluding H3K79me3 and, at a distance, for excluding H3K79me1, HeK79me2, and H4K91ac at the Igf2 locus. CTCF did not significantly contribute to recruiting globular histone domain modifications (Fig. 5).
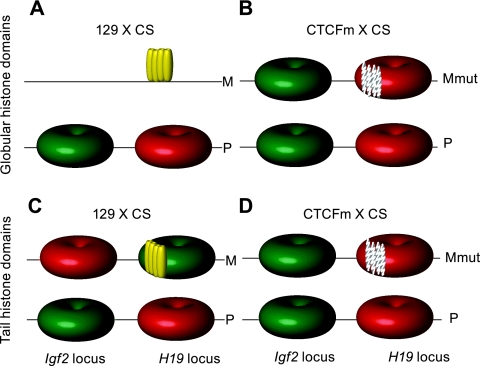
FIG. 5.
Comparison of the examined histone globular domain and histone tail modifications at the H19/Igf2 imprinted domain. (A) Allele-specific differences in repressive (red) and activating (green) covalent modifications in the globular domain of histones are specific to the paternal (P) but not the maternal (M) allele at the H19 and Igf2 loci in normal MEFs where CTCF binding (yellow ovals) in the ICR is maternal allele specific. (B) The globular domain histone composition of the maternally inherited CTCFm chromosome (Mmut) becomes similar to that of the normal paternal chromosome due to the ICR CTCF site mutations (white speckled ovals). (C) Allele-specific differences in repressive and activating covalent histone tail modifications distinguish the paternal (P) and maternal (M) alleles at the H19 and Igf2 loci in normal MEFs. (D) CTCF site mutations cause the “paternalization” of the maternal allele's chromatin composition in the histone tail modifications along the H19/Igf2 imprinted region (17). CTCF is responsible for defining the maternal allele's identity at the level of chromatin along the H19/Igf2 imprinted domain by recruiting and excluding histone tail modifications (C and D) and by excluding histone globular domain modifications (A and B).
Allele-specific histone modifications in the globular domains of H3 and H4 at eight maternally methylated DMRs.
To investigate whether it is a general phenomenon that histone globular domain residues H4K91ac, H3K79me1, H3K79me2, and H3K79me3 exhibit parental allele-specific enrichment at imprinted regions, we extended our analysis to 10 additional (8 maternally and 2 paternally methylated) germ line DMRs. We precipitated chromatin in primary MEFs derived from 129 × JF1 and the reciprocal JF1 × 129 mouse crosses using the specific antibodies for these modified residues and also with control antibodies H3K9ac, H4K16ac, H3K4me2, H3K9me3, and H3K27me3 recognizing histone tail modifications. Real-time PCR quantitation of the precipitated DNA at each DMR revealed that every single specific antibody precipitated chromatin at much higher level than nonspecific IgG (Table 1). As expected, we observed background levels of precipitation with antibodies against H4K91ac, H3K4me2, and H3K79me2 at heterochromatin control regions such as intracisternal A particles and major satellites, but these antibodies very strongly precipitated a euchromatin control region at the c-myc promoter. We then subjected the precipitated chromatin preparations to allele-specific multiplex DMR ChIP-SNuPE assays (see Fig. S2 and S3 in the supplemental material).
TABLE 1.
Real-time PCR quantitation of immunoprecipitated chromatin at DMRsa
| Histone modification | Copy no. precipitated | Region of interest |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peg1-Mest | Zac1 | Gnas1A | Peg3 | Snrpn | KvDMR1 | Igf2r | U2af1 | H19 ICR | Rasgrf1 | IG DMR | c-myc | IAP | Maj. sat. | ||
| H3K9ac | Avg | 5,749 | 181 | 10,543 | 123 | 77 | 267 | 893 | 784 | 64 | 929 | 74 | 3,405 | 9 | 6 |
| SD | 943 | 34 | 1,999 | 32 | 15 | 73 | 229 | 33 | 15 | 119 | 17 | 261 | 13 | 1 | |
| H4K91ac | Avg | 334 | 90 | 1,870 | 48 | 12 | 85 | 261 | 148 | 15 | 299 | 20 | 684 | 18 | 1 |
| SD | 41 | 18 | 659 | 17 | 2 | 6 | 42 | 60 | 11 | 1 | 8 | 53 | 5 | 0 | |
| H3K4me2 | Avg | 17,666 | 989 | 29,108 | 309 | 302 | 367 | 1,875 | 5,217 | 310 | 10,082 | 369 | 7,804 | 41 | 54 |
| SD | 5,261 | 3 | 2,725 | 9 | 28 | 48 | 114 | 41 | 32 | 392 | 13 | 1,275 | 36 | 14 | |
| H3K79me1 | Avg | 21,866 | 2,798 | 58,070 | 387 | 240 | 667 | 5,902 | 36,233 | 67 | 1,919 | 203 | 5,023 | 119 | 24 |
| SD | 4,408 | 129 | 7,959 | 92 | 66 | 17 | 708 | 2,776 | 11 | 293 | 12 | 159 | 29 | 12 | |
| H3K79me2 | Avg | 6,319 | 771 | 14,060 | 137 | 61 | 467 | 2,790 | 11,476 | 24 | 209 | 42 | 1,144 | 111 | 12 |
| SD | 1,098 | 20 | 52 | 5 | 12 | 9 | 11 | 1,111 | 8 | 41 | 11 | 439 | 10 | 4 | |
| H3K79me3 | Avg | 619 | 84 | 464 | 33 | 42 | 246 | 426 | 1,018 | 40 | 2,280 | 51 | 50 | 39 | 45 |
| SD | 81 | 15 | 131 | 13 | 9 | 66 | 24 | 622 | 0 | 1,137 | 9 | 31 | 24 | 2 | |
| H3K27me3 | Avg | 331 | 93 | 840 | 34 | 41 | 19 | 143 | 160 | 70 | 10,790 | 96 | 19 | 46 | 21 |
| SD | 2 | 10 | 245 | 14 | 0 | 2 | 35 | 46 | 1 | 429 | 13 | 13 | 22 | 0 | |
| H3K9me3 | Avg | 4,438 | 473 | 1,903 | 316 | 232 | 253 | 1,159 | 1,246 | 205 | 11,056 | 247 | 138 | 253 | 243 |
| SD | 354 | 29 | 269 | 26 | 27 | 55 | 158 | 323 | 15 | 2,216 | 18 | 6 | 19 | 7 | |
| IgG (N-ChIP) | Avg | 4 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 3 | 6 |
| IgG (X-ChIP) | Avg | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 0 | 1 | 2 | 5 |
The numbers are averages and standard deviations (SD) of DNA copies precipitated at each DMR and at the control genomic regions with the antibodies indicated on the left. Maj. sat., major satellites.
Each of the DMRs (27) with maternal allele-specific CpG methylation (Peg1-Mest, Zac1, Gnas1A, Peg3, Snrpn, KvDMR, Igf2r DMR2, and U2af1) exhibited a strong acetylation bias toward the paternal allele at H4K91 (Fig. 6A). This strong paternal allele-specific H4K91ac bias was very similar to that of H3K9ac and H4K16ac (Fig. 6B and C).
Monomethylation of H3K79 showed a strong paternal allele-specific bias at the Peg1-Mest, Zac1, Gnas1A, Peg3, and Snrpn DMRs and at the KvDMR (Fig. 7A) but did not exhibit allelic bias at the Igf2r DMR2 and U2af1 DMR. Dimethylation of H3K79 was strongly paternal allele specific at the Peg1-Mest, Zac1, Gnas1A, Peg3, and Snrpn DMRs and at the KvDMR, was weakly paternal allele specific at the Igf2r DMR2, and was not allele specific at the U2af1 DMR (Fig. 7B). The pattern of mono- and dimethylation at the H3K79 globular histone residue was similar to the pattern of dimethylation of H3K4 in the histone tail (Fig. 7C).
FIG. 7.
Active histone methylation marks distinguish the parental alleles of maternally and paternally methylated DMRs. H3K79me1 (A) and H3K79me2 (B) globular domain modifications are comparable to the H3K4me2 histone tail mark (C). Reciprocal mouse crosses had nearly identical allele-specific chromatin composition. The maternally methylated DMRs exhibited a paternal allele-specific bias for H3K79me1, H3K79me2, and H4K91ac histone marks, whereas the paternally methylated DMRs were more maternally biased for these modifications. Other details are shown in Fig. 6.
Trimethylation of H3K79 showed a maternal allele-specific bias at the Peg1-Mest, Zac1, Gnas1A, Peg3, and Snrpn DMRs but was not allele specific at the KvDMR, Igf2r DMR2, and U2af1 DMR (Fig. 8A). The pattern of trimethylation at the H3K79 globular histone residue was similar to the patterns of trimethylation of H3K9 and H3K27 in the H3 histone tail (Fig. 8B and C), with the H3K9me3 differences being the most polarized.
Allele-specific histone modifications in the globular domains of H3 and H4 at three paternally methylated DMRs.
The H4K91ac enrichment at the three paternally methylated DMRs, H19/Igf2 ICR, Rasgrf1 DMR, and IG-DMR (Fig. 6A), was different than what we had found at the maternally methylated DMRs: it was not paternal allele specific. H4K91ac exhibited a maternal-specific bias at the IG-DMR, like H3K9ac (Fig. 6B). H4K91ac, however, was biallelic at the H19/Igf2 ICR and the Rasgrf1 DMR similar to H3K16ac but unlike H3K9ac (Fig. 6B and C).
Similar to the bias of H3K4me2 (Fig. 7C), the H3K79me1 and H3K79me2 marks were strongly biased toward the maternal allele at the IG-DMR (Fig. 7A and B), but unlike H3K4me2, they did not exhibit allele specificity at the H19/Igf2 ICR and at the Rasgrf1 DMR (Fig. 7A and B).
H3K79 trimethylation was strongly biased toward the paternal allele at the H19/Igf2 ICR and less biased toward the paternal allele at the Rasgrf1 DMR and at the IG-DMR (Fig. 8A). The allele-specific H3K79me3 pattern was almost identical to the H3K9me3 pattern (Fig. 8B) at all three paternally methylated DMRs, but it was not similar to the H3K27me3 pattern, which showed a clear maternal allele-specific bias at the Rasgrf1 DMR (Fig. 8C). The latter finding was in agreement with the antagonistic roles of H3K27me3 and DNA methylation at the Rasgrf1 DMR (35).
Testing the function of H3K79 methyltransferase Dot1L in imprinting.
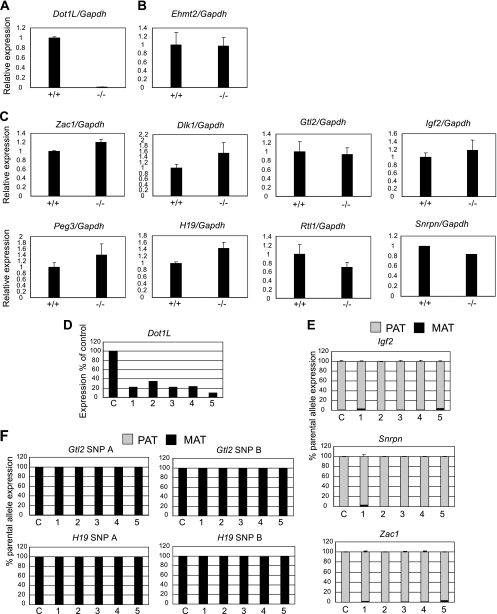
In general, H3K79me3 was associated with the CpG-methylated allele at most DMRs, while the unmethylated allele showed enrichment for H3K79me1 and H3K79me2. We asked whether the overall H3K79 methylation levels have a role in the expression of imprinted genes. We measured the levels of expression for a set of imprinted genes in 8.5-dpc Dot1L mutant (22) embryos using real-time RT-PCR (Fig. 9A to C). Dot1L expression was abolished (Fig. 9A), whereas the control histone methyltransferase Ehmt2 transcript level was unaffected in the Dot1L−/− embryos (Fig. 9B). We did not detect significant changes in the expression levels of imprinted genes (Fig. 9C). It is not possible to directly compare the 129 × CS and 129 × JF1 MEF chromatin data with the Dot1L−/− embryo data, because these mutant embryos die between 9.5 and 10.5 dpc before MEFs can be derived (at 13.5 dpc). The Dot1L mutant mouse line also did not allow for the assessment of allele-specific gene expression. To reveal whether allele-specific changes may occur in response to Dot1L downregulation but may be masked at the level of overall expression, we knocked down Dot1L in 129 × JF1 MEFs using five different small hairpin RNAs against Dot1L (Fig. 9D). More than 99% of the 129 × JF1 MEFs died after 2 weeks of shRNA treatment. The extremely low cell numbers precluded the analysis of chromatin. However, we were able to analyze RNA from surviving cells 10 days after treatment. The Dot1L RNA level was reduced in the shRNA-treated MEFs by an average of 80% and up to 90%, as measured by real-time RT-PCR (Fig. 9D), compared to that of the vector control sample. The knockdown efficiency was similar in an independent study of embryonic stem (ES) cells with the same shRNA constructs (2). Those authors verified by RT-PCR that Dot1L expression was reduced by 80% and by Western blotting that H3K79 methylation was absent (2). Whereas cell viability in undifferentiated ES cells was not affected, the Dot1L shRNAs caused a great reduction of cell viability upon differentiation (2). These results suggest that loss of viability of 129 × JF1 MEFs most likely resulted from demethylation of H3K79 in the near absence of Dot1L after shRNA knockdown. We measured the allele-specific expression of H19, Igf2, Gtl2, Snrpn, and Zac1 imprinted genes using multiplex RNA SNuPE Sequenom assays (see Fig. S5 in the supplemental material) and found that each of these imprinted genes was strictly expressed from only the correct parental allele (Fig. 9E and F). Taken together, the two independent functional assays suggest that Dot1L-dependent overall H3K79 methylation levels are not essential for maintaining imprinted gene expression in the mouse embryo.
FIG. 9.
Dot1L is not required for parental allele-specific expression of imprinted genes in the embryo. (A) Expression of Dot1L is reduced in Dot1L mutant (−/−) embryos compared to that of wild-type Dot1L (+/+) littermates. Real-time RT-PCR with oligonucleotides that recognize the catalytic domain is shown. (B) The transcript levels did not change for a control histone methyltransferase gene, Ehmt2. (C) Imprinted genes exhibit no significant changes in transcript levels in Dot1L mutant embryos. Real-time RT-PCR using duplicate embryos is shown. (D) Dot1L is downregulated by shRNA. The transcript level of Dot1L in the MEFs with different shRNA constructs (1-5) is presented as a percentage of the Dot1L transcript levels in MEFs with control (c) vector transduction. Real-time RT-PCR results are shown. (E and F) Imprinted genes exhibit normal allele-specific transcription in 129 × JF1 MEFs after Dot1L knockdown. Allele-specific expression of paternally (E) and maternally (F) expressed imprinted genes was measured using RNA SNuPE multiplex assays after Dot1L knockdown. The average percentage of maternal (MAT) or paternal (PAT) alleles in the total 100% expression is shown in black and gray, respectively, from duplicate measurements with standard deviations.
DISCUSSION
In this study we provide a comprehensive map of the allele-specific chromatin composition of 9 different histone modifications at 11 DMRs in reciprocal mouse crosses. Our results reveal that H4K91 acetylation and H3K79 methylation allele specifically mark the germ line DMRs in the globular domains of histones H3 and H4. We provide evidence that along the H19/Igf2 imprinted domain, CTCF insulator binding controls the globular domain marks by excluding them from the maternal allele. We show that H3K79me2 and H3K79me3 are biased toward functionally opposite and epigenetically distinct alleles of the DMRs. Therefore, a single methyl group specifies H3K79me2 and H3K79me3 association with euchromatin and heterochromatin, respectively.
Globular domain histone modifications exclusively mark the paternal allele at the H19/Igf2 imprinted region in a CTCF insulator-dependent fashion.
In normal cells, we found paternal allele-specific bias for H3K79me1, H3K79me2, and H4K91ac at the paternally expressed Igf2 P2 promoter and at Igf2 DMR1 and DMR2 and a paternal allele-specific bias for H3K79me3 at the H19/Igf2 ICR and H19 promoter. In CTCFm × CS cells, H3K79me1, H3K79me2, and H4K91ac became biallelic at the biallelically expressed Igf2 P2 promoter, and H3K79me3 became biallelic at the biallelically silent H19 promoter. These findings suggested that H3K79me1 and -2/H4K91ac and H3K79me3 may have activating and repressing regulatory roles at the Igf2 and H19 loci, respectively. On the other hand, H3K79me3 was enriched in both alleles at the Igf2 P2 promoter in normal MEFs and also in CTCFm × CS MEFs, and the expression of H19 and Igf2 did not change in Dot1L mutant embryos and after Dot1L knockdown, arguing that H3K79 methylation has no regulatory role at the H19/Igf2 imprinted domain in the embryo. Alternatively, the H3K79 methylation marks may be redundant with other epigenetic marks in the embryo at this imprinted domain.
The present data expand our previous finding that CTCF is the master organizer of chromatin at the H19/Igf2 imprinted domain (17). CTCF is required for specifying the maternal allele's chromatin by recruiting certain histone tail marks to the maternal allele and for excluding other tail marks from the maternal allele. CTCF directly recruits Suz12-mediated H3K27 trimethylation to the Igf2 locus in the maternal allele (32). On the other hand, CTCF controls histone tail modifications at the H19 promoter indirectly by setting the activity state of the promoter (69). In the case of globular histone marks, CTCF was responsible for the chromatin composition of the maternal allele by excluding H4K91ac, H3K79me1, and H3K79me2 at the Igf2 locus and by excluding H3K79me3 at the H19 locus from the maternal allele, but it did not recruit any of these globular marks to the maternal allele (Fig. 5). CTCF may directly or indirectly exclude H3K79me3 at the ICR and H3K79me2 at the Igf2 locus from the maternal allele. We cannot distinguish between these possibilities at the Igf2 locus because lack of CTCF binding in the ICR invariably results in biallelic Igf2 expression. If CTCF has a direct role in excluding globular marks, it may do so by excluding the H3K79 methyltransferase from the maternal allele or by recruiting H3K79 demethylase activities to the maternal allele.
CTCF may control chromatin composition by controlling DNA methylation (17, 52, 58, 63) at the ICR and distantly at the Igf2 DMRs. The H19 promoter, however, is unmethylated in CTCFm × CS MEFs (17), at the same time undergoes heterochromatinization with regard to histone tail modifications, and also attains biallelic H3K79me3. Therefore, CTCF-mediated chromatin regulation can be independent of CTCF-dependent maintenance of DNA hypomethylation.
Whereas CTCF binding in the ICR exhibited a regulatory role for histone tail and globular domain modifications at the H19/Igf2 imprinted domain, further work is required to find out whether CTCF organizes chromatin at other DMRs and imprinted domains.
Allele-specific enrichment of globular domain histone modifications at DMRs.
Our systematic analysis determined that all of the germ line DMRs examined exhibited allele-specific enrichment for one or more globular domain modifications. H4K91ac, similar to H3K9ac, showed a paternal allele-specific bias at each of the maternally methylated DMRs (Peg1-Mest, Zac1, Gnas1A, Peg3, Snrpn, KvDMR, Igf2r DMR2, and U2af1), whereas H3K79me3, similar to H3K9me3, exhibited a paternal allele-specific bias at each of the paternally methylated DMRs (H19/Igf2 ICR, Rasgrf1 DMR, and IG-DMR). Unlike H3K9ac, however, H4K91ac was not maternal allele specific at each of the paternally methylated DMRs (H19/Igf2 ICR and Rasgrf1 DMR were biallelic). Unlike H3K9me3, H3K79me3 was not maternal specific at each of the maternally methylated DMRs (KvDMR, Igf2r DMR2, and U2af1 DMR were biallelic). These differences suggest a level of independence between the specific enzymes modifying the respective tail and globular domain residues.
A histone cross talk model has been proposed previously in yeast: acetylation of H4K16 excludes Sir3 and allows Dot1 to bind to the H4 histone tail and dimethylate K79 in the H3 globular domain (1). The allele-specific enrichment of H4K16 acetylation at 11 DMRs in the mouse (Fig. 6C) shows a general correlation with H3K79me2 (Fig. 7B). This is consistent with the possibility that the asymmetry of H3K79me2 and H3K79me3 allelic enrichment could be a response to H4K16 acetylation in the CpG-hypomethylated allele.
The globular domain histone modifications correlated with the DNA methylation status at the DMRs. In each case, when allelic bias existed at a DMR, the allele specificity was consistent, as follows: H4K91ac, H3K79me1, and H3K79me2 were biased toward the unmethylated allele, whereas H3K79me3 was biased toward the CpG-methylated allele. This pattern suggests that globular histone marks may have functional relevance at DMRs. Hypoacetylated H4K91 in the methylated allele may stabilize nucleosomes due to the positive charge at this residue when the acetyl group is lacking (76), whereas acetylated H4K91 in the unmethylated allele may destabilize nucleosomes, leading to more accessible chromatin. H3K79me3 marks may reinforce repressive chromatin in the CpG-methylated DMR alleles. The findings that expression of imprinted genes was unaffected in 8.5-dpc Dot1L mutant embryos and in MEFs where Dot1L was knocked down suggest that H3K79 methylation marks must be functionally redundant with other epigenetic marks, such as DNA methylation and histone tail modifications in the embryo. Nevertheless, H3K79 methylation may have a role in the regulation of imprinted genes in a tissue-specific manner, for example, in the placenta where the role of DNA methylation is less important for imprinted gene expression (30, 67). The H3K27 methyltransferase Ezh2 is required for imprinted expression of Cdkn1c and Cd81 in the extraembryonic ectoderm (64), and the H3K9 methyltransferase Ehmt2 is required for monoallelic expression of the Osbpl5, Cd81, Ascl2, Tfpi2, and Slc22a3 imprinted genes in the placenta (44, 45, 70).
One methyl group between H3K79me2 and H3K79me3 distinguishes euchromatin from heterochromatin.
The literature has contradicting reports on whether H3K79 methylation is an active or repressing chromatin mark. It was originally considered a hallmark of euchromatin (46, 68), but in the early studies, only H3K79me2 enrichment was assessed. Genome-wide ChIP analyses later found H3K79me2 enrichment at active regions in Drosophila (41, 59) but not in human (3) cells. Association was found between H3K4/K79me2 and preengaged transcription (16). H3K79me2 levels increase in promoters after Myc induction (40). H3K79me2 is dynamically induced upon gene activation but is destabilized in highly transcribed promoters at the globin loci (57). H3K79me3 is enriched within the transcribed regions of genes in yeast (54) and mammalian (61) cells. H3K79me3 colocalizes with repressed promoters in human cells (3) and with DNase I-hypersensitive sites proximally of silent genes (6). In fibroblasts and oocytes, H3K79me2 was observed throughout the genome by immunocytochemistry, whereas H3K79me3 was localized in the pericentromeric heterochromatin regions (49).
The maternally and paternally inherited alleles of DMRs exist in opposite epigenetic states and provide a well-defined experimental system to assess whether a particular histone component is specific to euchromatin or heterochromatin. Association of H3K79me3 with H3K9me3 at the CpG-methylated alleles of most DMRs suggests that H3K79me3 is a component of heterochromatin, whereas association of H3K79me2 with H3K4me2 and H3K9ac at the unmethylated DMR alleles implies that H3K79me2 is a component of euchromatin.
It will be interesting to reveal whether H3K79me2 and H3K79me3 distinguish euchromatin or heterochromatin in organisms where gene regulation is achieved without DNA methylation and whether the role of chromatin marks, therefore, is less redundant. In yeast, the H3K79 methyltransferase Dot1 is essential not only for heterochromatin-mediated silencing but also for blocking heterochromatin spread into euchromatin (1, 29, 47, 60, 68). Subtelomeric heterochromatin was characterized with low levels of H3K79 methylation, but only H3K79me2 levels were measured, and H3K79me3 was not assessed (46). In view of our findings, lack of H3K79me2 would be consistent with high levels of H3K79me3 in subtelomeric heterochromatin. The H3K79 methyltransferase mutant grappa exhibited striking contradicting Polycomb and Trithorax phenotypes in Drosophila (60). Our data suggest that H3K79me1 and -2 and H3K79me3 might be involved in maintaining the levels of euchromatin and heterochromatin, respectively, at the regulatory domains of homeotic selector genes, and both active and repressive chromatin would be affected in the grappa mutants.
Dot1L is required for mono-, di-, and trimethylation of H3K79 in the mouse (22). The enzyme that removes these methyl groups is unknown. The addition/removal of one methyl group determines the association of H3K79me2 and H3K79me3 with heterochromatin and euchromatin, respectively. It will be interesting to find out what triggers the H3K79me2-to-H3K79me3 transition and its reversal, if this one methyl group may function to regulate gene activation/repression in the imprinted genes of mammal tissues specifically, and if it affects heterochromatin-euchromatin distribution in different organisms globally.
Supplementary Material
Acknowledgments
We thank Hiroyuki Sasaki for sharing JF1 SNP information with us. We thank Gerd Pfeifer and Nathan Oates for their comments on the manuscript. We thank Hui Su, Shirley Tsai, and Claudia Kowolik for technical assistance.
This work was supported by Public Health Service grant GM064378 from the National Institute of General Medical Sciences. Thomas B. Nicholson and Taiping Chen are employees of the Novartis Institutes for Biomedical Research.
Footnotes
Published ahead of print on 29 March 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28:1002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, E. R., W. Krueger, C. M. Jakuba, E. Veilleux, D. J. Ambrosi, C. E. Nelson, and T. P. Rasmussen. 2009. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 27:1538-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823-837. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 5.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, A. P., S. Davis, H. P. Shulha, P. Meltzer, E. H. Margulies, Z. Weng, T. S. Furey, and G. E. Crawford. 2008. High-resolution mapping and characterization of open chromatin across the genome. Cell 132:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, M. S., A. Yevtodiyenko, C. L. Schmidt, and J. V. Schmidt. 2007. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 89:280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, J. H., J. D. Kim, J. H. Chung, L. Stubbs, and J. Kim. 2006. Allele-specific deposition of macroH2A1 in imprinting control regions. Hum. Mol. Genet. 15:717-724. [DOI] [PubMed] [Google Scholar]
- 9.Delaval, K., J. Govin, F. Cerqueira, S. Rousseaux, S. Khochbin, and R. Feil. 2007. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 26:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, C. A., and A. C. Ferguson-Smith. 2007. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19:281-289. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick, G. V., P. D. Soloway, and M. J. Higgins. 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32:426-431. [DOI] [PubMed] [Google Scholar]
- 12.Fournier, C., Y. Goto, E. Ballestar, K. Delaval, A. M. Hever, M. Esteller, and R. Feil. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandjean, V., L. O'Neill, T. Sado, B. Turner, and A. Ferguson-Smith. 2001. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 488:165-169. [DOI] [PubMed] [Google Scholar]
- 14.Gregory, R. I., L. P. O'Neill, T. E. Randall, C. Fournier, S. Khosla, B. M. Turner, and R. Feil. 2002. Inhibition of histone deacetylases alters allelic chromatin conformation at the imprinted U2af1-rs1 locus in mouse embryonic stem cells. J. Biol. Chem. 277:11728-11734. [DOI] [PubMed] [Google Scholar]
- 15.Gregory, R. I., T. E. Randall, C. A. Johnson, S. Khosla, I. Hatada, L. P. O'Neill, B. M. Turner, and R. Feil. 2001. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell. Biol. 21:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guccione, E., F. Martinato, G. Finocchiaro, L. Luzi, L. Tizzoni, V. Dall’ Olio, G. Zardo, C. Nervi, L. Bernard, and B. Amati. 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 8:764-770. [DOI] [PubMed] [Google Scholar]
- 17.Han, L., D. H. Lee, and P. E. Szabó. 2008. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol. Cell. Biol. 28:1124-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 19.Haun, W. J., and N. M. Springer. 2008. Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J. 56:903-912. [DOI] [PubMed] [Google Scholar]
- 20.Hiura, H., J. Komiyama, M. Shirai, Y. Obata, H. Ogawa, and T. Kono. 2007. DNA methylation imprints on the IG-DMR of the Dlk1-Gtl2 domain in mouse male germline. FEBS Lett. 581:1255-1260. [DOI] [PubMed] [Google Scholar]
- 21.Hu, J. F., J. Pham, I. Dey, T. Li, T. H. Vu, and A. R. Hoffman. 2000. Allele-specific histone acetylation accompanies genomic imprinting of the insulin-like growth factor II receptor gene. Endocrinology 141:4428-4435. [DOI] [PubMed] [Google Scholar]
- 22.Jones, B., H. Su, A. Bhat, H. Lei, J. Bajko, S. Hevi, G. A. Baltus, S. Kadam, H. Zhai, R. Valdez, S. Gonzalo, Y. Zhang, E. Li, and T. Chen. 2008. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4:e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurinke, C., M. F. Denissenko, P. Oeth, M. Ehrich, D. van den Boom, and C. R. Cantor. 2005. A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutat. Res. 573:83-95. [DOI] [PubMed] [Google Scholar]
- 24.Kaffer, C. R., M. Srivastava, K. Y. Park, E. Ives, S. Hsieh, J. Batlle, A. Grinberg, S. P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14:1908-1919. [PMC free article] [PubMed] [Google Scholar]
- 25.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 26.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoto, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429:900-903. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, H., C. Suda, T. Abe, Y. Kohara, T. Ikemura, and H. Sasaki. 2006. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 113:130-137. [DOI] [PubMed] [Google Scholar]
- 28.Koide, T., K. Moriwaki, K. Uchida, A. Mita, T. Sagai, H. Yonekawa, H. Katoh, N. Miyashita, K. Tsuchiya, T. J. Nielsen, and T. Shiroishi. 1998. A new inbred strain JF1 established from Japanese fancy mouse carrying the classic piebald allele. Mamm. Genome 9:15-19. [DOI] [PubMed] [Google Scholar]
- 29.Lacoste, N., R. T. Utley, J. M. Hunter, G. G. Poirier, and J. Cote. 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277:30421-30424. [DOI] [PubMed] [Google Scholar]
- 30.Lewis, A., K. Mitsuya, D. Umlauf, P. Smith, W. Dean, J. Walter, M. Higgins, R. Feil, and W. Reik. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36:1291-1295. [DOI] [PubMed] [Google Scholar]
- 31.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 32.Li, T., J. F. Hu, X. Qiu, J. Ling, H. Chen, S. Wang, A. Hou, T. H. Vu, and A. R. Hoffman. 2008. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol. Cell. Biol. 28:6473-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, T., T. H. Vu, G. A. Ulaner, Y. Yang, J. F. Hu, and A. R. Hoffman. 2004. Activating and silencing histone modifications form independent allelic switch regions in the imprinted Gnas gene. Hum. Mol. Genet. 13:741-750. [DOI] [PubMed] [Google Scholar]
- 34.Lin, S. P., N. Youngson, S. Takada, H. Seitz, W. Reik, M. Paulsen, J. Cavaille, and A. C. Ferguson-Smith. 2003. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35:97-102. [DOI] [PubMed] [Google Scholar]
- 35.Lindroth, A. M., Y. J. Park, C. M. McLean, G. A. Dokshin, J. M. Persson, H. Herman, D. Pasini, X. Miro, M. E. Donohoe, J. T. Lee, K. Helin, and P. D. Soloway. 2008. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet. 4:e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, J., M. Chen, C. Deng, D. Bourc'his, J. G. Nealon, B. Erlichman, T. H. Bestor, and L. S. Weinstein. 2005. Identification of the control region for tissue-specific imprinting of the stimulatory G protein alpha-subunit. Proc. Natl. Acad. Sci. U. S. A. 102:5513-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, X. J., J. Wu, B. A. Altheim, M. C. Schultz, and M. Grunstein. 1998. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. U. S. A. 95:6693-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mager, J., N. D. Montgomery, F. P. de Villena, and T. Magnuson. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33:502-507. [DOI] [PubMed] [Google Scholar]
- 39.Mann, J. R., P. E. Szabó, M. R. Reed, and J. Singer-Sam. 2000. Methylated DNA sequences in genomic imprinting. Crit. Rev. Eukaryot Gene Expr. 10:241-257. [DOI] [PubMed] [Google Scholar]
- 40.Martinato, F., M. Cesaroni, B. Amati, and E. Guccione. 2008. Analysis of Myc-induced histone modifications on target chromatin. PLoS One 3:e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. U. S. A. 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Megee, P. C., B. A. Morgan, B. A. Mittman, and M. M. Smith. 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247:841-845. [DOI] [PubMed] [Google Scholar]
- 43.Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman, G. Giannoukos, P. Alvarez, W. Brockman, T. K. Kim, R. P. Koche, W. Lee, E. Mendenhall, A. O'Donovan, A. Presser, C. Russ, X. Xie, A. Meissner, M. Wernig, R. Jaenisch, C. Nusbaum, E. S. Lander, and B. E. Bernstein. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monk, D., A. Wagschal, P. Arnaud, P. S. Muller, L. Parker-Katiraee, D. Bourc'his, S. W. Scherer, R. Feil, P. Stanier, and G. E. Moore. 2008. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 18:1270-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagano, T., J. A. Mitchell, L. A. Sanz, F. M. Pauler, A. C. Ferguson-Smith, R. Feil, and P. Fraser. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717-1720. [DOI] [PubMed] [Google Scholar]
- 46.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U. S. A. 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng, H. H., Q. Feng, H. Wang, H. Erdjument-Bromage, P. Tempst, Y. Zhang, and K. Struhl. 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 49.Ooga, M., A. Inoue, S. Kageyama, T. Akiyama, M. Nagata, and F. Aoki. 2008. Changes in H3K79 methylation during preimplantation development in mice. Biol. Reprod. 78:413-424. [DOI] [PubMed] [Google Scholar]
- 50.Pandey, R. R., T. Mondal, F. Mohammad, S. Enroth, L. Redrup, J. Komorowski, T. Nagano, D. Mancini-Dinardo, and C. Kanduri. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32:232-246. [DOI] [PubMed] [Google Scholar]
- 51.Pannetier, M., E. Julien, G. Schotta, M. Tardat, C. Sardet, T. Jenuwein, and R. Feil. 2008. PR-SET7 and SUV4-20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep. 9:998-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pant, V., P. Mariano, C. Kanduri, A. Mattsson, V. Lobanenkov, R. Heuchel, and R. Ohlsson. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedone, P. V., M. J. Pikaart, F. Cerrato, M. Vernucci, P. Ungaro, C. B. Bruni, and A. Riccio. 1999. Role of histone acetylation and DNA methylation in the maintenance of the imprinted expression of the H19 and Igf2 genes. FEBS Lett. 458:45-50. [DOI] [PubMed] [Google Scholar]
- 54.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 55.Regha, K., M. A. Sloane, R. Huang, F. M. Pauler, K. E. Warczok, B. Melikant, M. Radolf, J. H. Martens, G. Schotta, T. Jenuwein, and D. P. Barlow. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamoto, A., J. Liu, A. Greene, M. Chen, and L. S. Weinstein. 2004. Tissue-specific imprinting of the G protein Gsalpha is associated with tissue-specific differences in histone methylation. Hum. Mol. Genet. 13:819-828. [DOI] [PubMed] [Google Scholar]
- 57.Sawado, T., J. Halow, H. Im, T. Ragoczy, E. H. Bresnick, M. A. Bender, and M. Groudine. 2008. H3 K79 dimethylation marks developmental activation of the beta-globin gene but is reduced upon LCR-mediated high-level transcription. Blood 112:406-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 59.Schubeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shanower, G. A., M. Muller, J. L. Blanton, V. Honti, H. Gyurkovics, and P. Schedl. 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steger, D. J., M. I. Lefterova, L. Ying, A. J. Stonestrom, M. Schupp, D. Zhuo, A. L. Vakoc, J. E. Kim, J. Chen, M. A. Lazar, G. A. Blobel, and C. R. Vakoc. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28:2825-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szabó, P., S. H. Tang, A. Rentsendorj, G. P. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607-610. [DOI] [PubMed] [Google Scholar]
- 63.Szabó, P. E., S. H. Tang, F. J. Silva, W. M. Tsark, and J. R. Mann. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol. Cell. Biol. 24:4791-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terranova, R., S. Yokobayashi, M. B. Stadler, A. P. Otte, M. van Lohuizen, S. H. Orkin, and A. H. Peters. 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 15:668-679. [DOI] [PubMed] [Google Scholar]
- 65.Thorvaldsen, J. L., and M. S. Bartolomei. 2007. SnapShot: imprinted gene clusters. Cell 130:958. [DOI] [PubMed] [Google Scholar]
- 66.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umlauf, D., Y. Goto, R. Cao, F. Cerqueira, A. Wagschal, Y. Zhang, and R. Feil. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36:1296-1300. [DOI] [PubMed] [Google Scholar]
- 68.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 69.Verona, R. I., J. L. Thorvaldsen, K. J. Reese, and M. S. Bartolomei. 2008. The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol. Cell. Biol. 28:71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagschal, A., H. G. Sutherland, K. Woodfine, A. Henckel, K. Chebli, R. Schulz, R. J. Oakey, W. A. Bickmore, and R. Feil. 2008. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell. Biol. 28:1104-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williamson, C. M., S. T. Ball, W. T. Nottingham, J. A. Skinner, A. Plagge, M. D. Turner, N. Powles, T. Hough, D. Papworth, W. D. Fraser, M. Maconochie, and J. Peters. 2004. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat. Genet. 36:894-899. [DOI] [PubMed] [Google Scholar]
- 72.Williamson, C. M., M. D. Turner, S. T. Ball, W. T. Nottingham, P. Glenister, M. Fray, Z. Tymowska-Lalanne, A. Plagge, N. Powles-Glover, G. Kelsey, M. Maconochie, and J. Peters. 2006. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 38:350-355. [DOI] [PubMed] [Google Scholar]
- 73.Wutz, A., O. W. Smrzka, N. Schweifer, K. Schellander, E. F. Wagner, and D. P. Barlow. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389:745-749. [DOI] [PubMed] [Google Scholar]
- 74.Yang, Y., J. F. Hu, G. A. Ulaner, T. Li, X. Yao, T. H. Vu, and A. R. Hoffman. 2003. Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites. J. Cell. Biochem. 90:1038-1055. [DOI] [PubMed] [Google Scholar]
- 75.Yang, Y., T. Li, T. H. Vu, G. A. Ulaner, J. F. Hu, and A. R. Hoffman. 2003. The histone code regulating expression of the imprinted mouse Igf2r gene. Endocrinology 144:5658-5670. [DOI] [PubMed] [Google Scholar]
- 76.Ye, J., X. Ai, E. E. Eugeni, L. Zhang, L. R. Carpenter, M. A. Jelinek, M. A. Freitas, and M. R. Parthun. 2005. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell 18:123-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoon, B. J., H. Herman, A. Sikora, L. T. Smith, C. Plass, and P. D. Soloway. 2002. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.