Abstract
Sex Comb on Midleg (SCM) is a transcriptional repressor in the Polycomb group (PcG), but its molecular role in PcG silencing is not known. Although SCM can interact with Polycomb repressive complex 1 (PRC1) in vitro, biochemical studies have indicated that SCM is not a core constituent of PRC1 or PRC2. Nevertheless, SCM is just as critical for Drosophila Hox gene silencing as canonical subunits of these well-characterized PcG complexes. To address functional relationships between SCM and other PcG components, we have performed chromatin immunoprecipitation studies using cultured Drosophila Schneider line 2 (S2) cells and larval imaginal discs. We find that SCM associates with a Polycomb response element (PRE) upstream of the Ubx gene which also binds PRC1, PRC2, and the DNA-binding PcG protein Pleiohomeotic (PHO). However, SCM is retained at this Ubx PRE despite genetic disruption or knockdown of PHO, PRC1, or PRC2, suggesting that SCM chromatin targeting does not require prior association of these other PcG components. Chromatin immunoprecipitations (IPs) to test the consequences of SCM genetic disruption or knockdown revealed that PHO association is unaffected, but reduced levels of PRE-bound PRC2 and PRC1 were observed. We discuss these results in light of current models for recruitment of PcG complexes to chromatin targets.
The Polycomb group (PcG) proteins are a set of conserved chromatin regulators that work together to execute gene silencing during development (see references 45, 46, 73, 81, and 83 for reviews). Most PcG proteins were initially identified in Drosophila based upon their roles in Hox gene silencing along the anterior-posterior body axis (7, 27, 28, 40, 79, 86). Subsequently, genome-wide studies revealed that PcG proteins collaborate to control hundreds of target genes in both flies and mammals (4, 5, 38, 47, 72, 84, 88). In mammalian systems, PcG proteins play key regulatory roles in X-chromosome inactivation (58, 78, 99) and in maintenance of both embryonic and lineage-restricted stem cells (4, 38, 54, 57, 83). In addition, human PcG proteins have been linked to oncogenesis in many tissue types (see references 62, 82, and 83 for reviews), suggesting that they contribute to abnormal chromatin states in cancer cells (51, 69, 87, 98). Thus, there is great interest in determining mechanisms by which PcG proteins alter chromatin and control gene expression in both normal development and disease.
Biochemical studies have defined subunit compositions and molecular activities of several PcG complexes purified from Drosophila embryos or from human cells (13, 16, 35, 39, 44, 75). Three Drosophila PcG complexes have been purified and characterized so far; these are termed Polycomb repressive complex 1 (PRC1), PRC2, and PHO-RC (16, 21, 34, 44, 75). PHO-RC contains two subunits, Pleiohomeotic (PHO) and SFMBT (34). PHO is the best-characterized sequence-specific DNA-binding protein in PcG silencing (10, 11, 22, 77), and it plays a key role in recognizing PcG target genes and recruiting other PcG complexes to these loci (34, 36, 42, 52, 71, 93).
Fly PRC2 contains four core subunits, E(Z), ESC, SU(Z)12, and NURF55 (16, 44). PRC2 possesses histone methyltransferase (HMTase) activity, which is provided by the SET domain of E(Z). Contributions from the ESC and SU(Z)12 subunits are also required for robust enzyme function (31, 49). The PRC2 HMTase methylates histone H3 on lysine 27 (K27) (13, 44), and this histone modification is commonly associated with PcG-silenced genes in vivo (18, 53, 72). In vitro studies show that methylation of H3-K27 creates a binding site for the PRC1 subunit PC (19, 41), which may help recruit PRC1 and/or influence its interactions with local chromatin (73, 81). Besides PC, PRC1 contains three other core subunits, PSC, PH, and dRING1 (21, 75). Among these, PSC is a central contributor to transcriptional silencing by PRC1 in vitro (32, 33), and dRING1 supplies ubiquitin ligase activity that can modify histone H2A on K119 (12, 92). In addition to histone ubiquitylation, previous studies have implicated PRC1 in inhibition of nucleosome remodeling (21, 75), compaction of nucleosome arrays (20), blockage of RNA polymerase II initiation (17), and/or arrested transcription elongation (85). The subunit compositions and biochemical activities of PRC1 and PRC2 are conserved between the fly and human versions. There are also mammalian homologs of both PHO-RC subunits (76, 90), although a mammalian PHO-RC complex has not yet been described.
The PcG protein Sex Comb on Midleg (SCM) is just as critical as any subunit of PHO-RC, PRC1, or PRC2 for Hox gene silencing in fly embryos (2, 7). However, the biochemical role of SCM in the context of these well-characterized PcG complexes has not been determined. In vitro interactions between SCM and a PRC1 subunit, PH, initially suggested that SCM might function as a component of PRC1 (55). Indeed, SCM can assemble into a recombinant PRC1 complex via this PH interaction (56). However, biochemical tests on fly embryo extracts revealed that SCM does not behave as a core subunit of PRC1. First, gel filtration chromatography yields separable peaks for SCM and PRC1 (56). Second, coimmunoprecipitations detect little or no SCM association with PRC1 subunits (34, 56). Finally, purified preparations of PRC1 contain some SCM but consistently in substoichiometric quantities (39, 67, 75). Thus, the sum of biochemical tests so far indicates that SCM is not a core subunit of PRC1, PRC2, or PHO-RC in vivo (34, 50, 56, 67).
Based upon its domain content (Fig. 1, top), SCM appears to lack intrinsic enzymatic or DNA recognition functions. However, the SCM domain organization does offer functional clues and intriguing similarities with other PcG components. In particular, SCM shares a C-terminal domain, called the SPM domain, with the PRC1 subunit PH and with the PHO-RC subunit SFMBT (Fig. 1). This domain is a subtype within the larger family of SAM domains (59), and, like other family members, it is a multihelix bundle that mediates homotypic and heterotypic protein interactions (61). Indeed, this domain mediates the in vitro SCM-PH binding described previously (55, 56). Thus, although SCM is not stably associated with PRC1 or PHO-RC in fly embryo extracts, this shared domain provides the capacity, or at least the potential, to interact with these other PcG complexes.
FIG. 1.
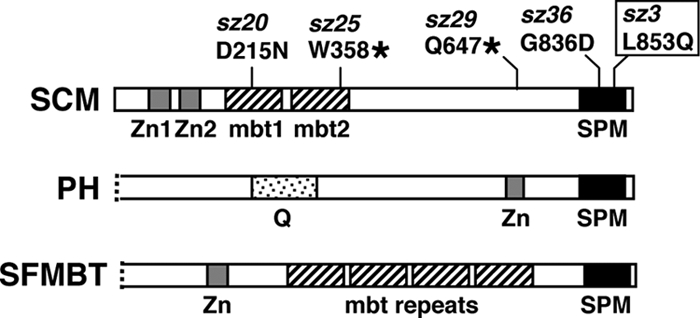
Domain organizations of SCM and two related PcG proteins. PH is a subunit of PRC1 (21, 75), and SFMBT is a subunit of PHO-RC (34). All three proteins share a C-terminal SPM domain (solid black) and copies of a Cys2-Cys2 zinc finger (gray). SCM and SFMBT also share multiple mbt repeats (hatched). “Q” represents a glutamine-rich domain. N-terminal portions of PH and SFMBT lacking homology domains are not shown. The mutant lesions of five newly characterized Scm alleles are displayed. Two are nonsense mutations (sz25 and sz29), and three are missense mutations (sz3, sz20, and sz36). The sz3 allele (boxed) is a pupal-lethal hypomorphic mutation used for wing disc ChIPs in this study. The sz20 missense change is identical to the independently isolated ScmSu(z)302 allele (2, 97).
We reasoned that a chromatin context, which is lacking in tests using soluble nuclear extracts, may be required to fully assess the functional relationship of SCM to other PcG components. To address this, we employed chromatin immunoprecipitation (ChIP) to directly test SCM in the context of a bona fide chromatin target in vivo. Using both Drosophila Schneider line 2 (S2) cells and larval wing discs, we found that SCM colocalizes with components of PHO-RC, PRC2, and PRC1 on a well-characterized Polycomb response element (PRE) located upstream of the Hox gene Ultrabithorax (Ubx). To investigate functional relationships between SCM and these PcG complexes, we performed ChIP analyses to interrogate chromatin from cells and/or tissues where levels of SCM, PHO, E(Z), or PC have been reduced by RNA interference (RNAi). We also tested PcG associations in chromatin from wing discs bearing a newly described Scm loss-of-function allele. Our results provide further evidence that SCM is not an integral subunit in any of the three fly PcG complexes defined so far. Furthermore, they suggest that SCM targeting to chromatin sites can occur, to a significant extent, independently of these PcG complexes.
MATERIALS AND METHODS
Fly stocks and crosses.
The Scmsz3 mutant was kindly provided by Henrik Gyurkovics (Biological Research Center, Szeged, Hungary). ScmH1 is a null allele that converts W248 to a stop codon (2). Scmsz3/ScmH1 mutant larvae were selected as non-Tb progeny from a cross between Scmsz3/TM6B Tb males and ScmH1/TM6B Tb females. The fly stock Pc-R4 (stock number 32443R-4; http://www.shigen.nig.ac.jp/fly/nigfly) contains a transgene to express a short hairpin RNA (shRNA), covering approximately 400 bp near the 5′ end of the PC-coding region, under upstream activating sequence (UAS) control. Expression of the Pc shRNA was driven by GAL4 activator provided from a second construct in trans (6). For phenotypic analysis in adults (see Fig. 6B), homozygous Pc-R4 males were crossed to females homozygous for the A9-GAL4 wing disc driver (25), and the progeny were reared at 25°C. To produce wing discs for analysis by Western blotting, reverse transcription-PCR (RT-PCR), and ChIPs (see Fig. 6C to E), homozygous Pc-R4 males were crossed to females homozygous for the ubiquitous da-GAL4 driver (96), and the progeny were raised at 29°C prior to harvesting of wing discs from third-instar larvae. Consistent with temperature sensitivity of GAL4-UAS expression in Drosophila (6), the extent of PC loss from the Ubx PRE was slightly greater if this cross was performed at 29°C rather than 25°C. A y Df(1)w67c2 fly stock was used routinely as an essentially wild-type control in Western blotting, immunostaining, RT-PCR, and ChIP assays. This stock was raised at 25°C, except when used as a control in experiments with Pc-R4 (see Fig. 6C to E), when it was raised at 29°C.
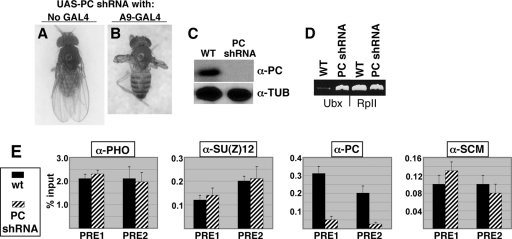
FIG. 6.
Consequences of PC knockdown in wing discs. (A and B) Phenotypes of adult flies bearing the UAS-Pc shRNA construct without a GAL4 driver (A) or combined with the A9-GAL4 wing disc driver (B). (C) Western blots to detect PC or α-tubulin (loading control) in wing discs from wild-type (WT) or UAS-Pc shRNA; da-GAL4 larvae. (D) RT-PCR analysis of Ubx and RpII140 mRNA levels in wing discs isolated from wild-type or UAS-Pc shRNA; da-GAL4 larvae. (E) Bar graphs depict Q-PCR ChIP signals obtained using antibodies against the indicated four PcG proteins and wing disc chromatin samples from wild-type (solid bars) or UAS-Pc shRNA; da-GAL4 (hatched bars) larvae. Error bars show standard deviations from the mean determined using a single ChIP for each PcG protein and six independent Q-PCRs.
Antibodies.
Primary antibodies used to detect PcG proteins in Western blots and chromatin immunoprecipitations were the following rabbit polyclonal antibodies described previously: anti-PHO (22), anti-E(Z) (14), anti-PC (93), anti-SU(Z)12 (44), and anti-SCM (2).
Western blots.
Protein extracts from whole larvae were prepared as described previously (2) using 5 to 10 μl of 2× SDS sample buffer per larva supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 μg/ml leupeptin. Wing disc protein samples were prepared using approximately 50 discs disrupted in 15 μl of 2× SDS sample buffer. Protein extracts from S2 cells were prepared as described previously (93). Proteins were resolved by SDS-PAGE on 10% gels and transferred to Protran (Whatman) membranes. Blots were blocked in 5% nonfat dry milk for 1 h before being incubated in primary antibody overnight at 4°C. Antibodies against PHO, PC, and SCM were each used at 1:500 dilutions. Antibody against E(Z) was used at 1:1,000. A monoclonal antibody against α-tubulin (Sigma), used to gauge equivalence of lane loadings, was diluted 1:2,000. After washing, blots were incubated with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Jackson) at 1:5,000, except for the antitubulin blots, which were incubated with an HRP-conjugated goat anti-mouse secondary antibody at 1:10,000. Secondary antibody incubations were at room temperature for 1 h, and signals were developed using an ECL chemiluminescence detection kit (Amersham).
Detection of Hox gene expression.
Reverse transcription-PCR (RT-PCR) to detect Ubx mRNA levels, and control RpII140 mRNA levels was performed as described previously (93), using total RNA extracted from S2 cells or larval wing discs with Trizol reagent (Invitrogen). Immunostaining to detect ABD-A in embryos was performed as described previously (79) using a rabbit polyclonal antibody (30). Immunostaining to detect UBX in larval discs was performed essentially as described previously (9) using a mouse monoclonal primary antibody (FP.3.38 [95]). For initial analyses of Scm mutant discs, anti-UBX was diluted 1:1,000 and immunodetection was performed using biotinylated goat anti-mouse secondary antibody (1:1,000) and the VectaStain ABC kit (Vector Laboratories). For the immunostainings shown in Fig. 4A, anti-UBX was diluted 1:500 and a goat anti-mouse secondary antibody conjugated to Alexa568 (Invitrogen) was used at 1:2,000.
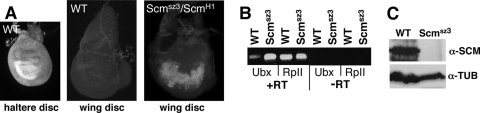
FIG. 4.
Ubx desilencing in Scmsz3 mutant larvae. (A) Patterns of UBX accumulation revealed by immunostaining of a wild-type (WT) haltere disc (left), a wild-type wing disc (middle), and an Scmsz3/ScmH1 wing disc (right). (B) RT-PCR analysis of Ubx and RpII140 mRNA levels in wing discs isolated from wild-type or Scmsz3/ScmH1 mutant larvae. (C) Western blots to detect SCM or α-tubulin (loading control) in extracts from wild-type or Scmsz3/ScmH1 mutant larvae.
RNA interference.
Double-stranded RNAs specific for PHO, E(Z), PC, and green fluorescent protein (GFP) were produced as described previously (13, 93), using cloned cDNAs as initial templates. For SCM knockdown, an Scm cDNA was used as template to generate a 578-bp double-stranded RNA (dsRNA) extending from 37 bp upstream of the ATG to 541 bp downstream. Double-stranded RNAs were synthesized and transfected into S2 cells as described previously (13).
ChIP assays.
Formaldehyde-cross-linked chromatin was prepared from fly S2 cells or wing imaginal discs (typically ∼200 per batch), and immunoprecipitations were performed as described previously (93) using 5 μl of antiserum against PHO, SU(Z)12, PC, or SCM per chromatin immunoprecipitation (ChIP). Amplification of Ubx PRE fragments and a control RpII140 fragment by endpoint PCR was performed as described previously (93). Amplification of Ubx PRE fragments from wing disc ChIPs by real-time quantitative PCR (Q-PCR) used the primer pairs 5′-CGC ACT CAA AAT CCG AAA AT-3′ and 5′-CGC ACG TCA GAC TTG GAA TA-3′ for fragment “PRE1” and 5′-GGG CTA TTC CAA GTC TGA CG-3′ and 5′-GGC CAT TAC GAA CGA CAG TT-3′ for fragment “PRE2.” Q-PCR was performed using Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) according to the manufacturer's instructions, except that the total reaction volume was 15 μl. Q-PCR was performed using 45 cycles consisting of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s on a Mastercycler RealPlex 2S (Eppendorf).
RESULTS
SCM colocalizes with other PcG components at the Ubx PRE.
One of the best-characterized Polycomb response elements in Drosophila is located approximately 25 kb upstream of the Hox gene Ubx, within the bxd regulatory region. This PRE can confer PcG silencing of reporters in vivo (15, 80), and it has been delimited to an approximately 0.5-kb region (22, 26). Furthermore, the associations of PcG proteins with this Ubx PRE have been mapped via chromatin immunoprecipitation (ChIP) assays on cultured fly cells, embryos, or larval imaginal discs (13, 29, 34, 48, 53, 68, 93). Thus, it is well established that subunits of PHO-RC, PRC2, and PRC1 associate with this Ubx PRE region in vivo, with peak association detected on the b4 and b5 fragments (93), as depicted in Fig. 2A.
FIG. 2.
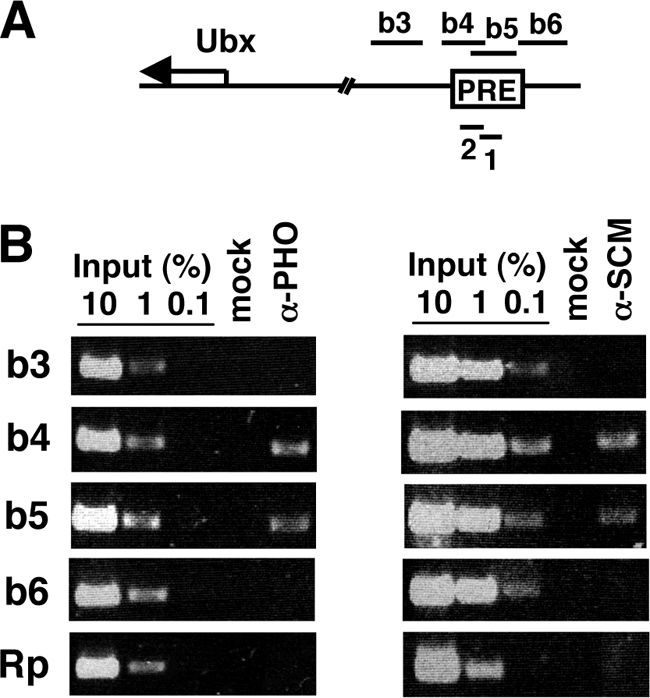
SCM associates with a Polycomb response element (PRE) upstream of Ubx. (A) The map depicts the Ubx transcription start region and a PRE located ∼25 kb upstream within the bxd regulatory region. Numbered fragments above the map have been described previously (13, 93, 94) and were used in ChIP assays with signals obtained by endpoint PCR. Fragments 1 and 2, below the map, were used in ChIP assays employing Q-PCR signal detection. (B) ChIPs to detect distributions of PHO (left panel) and SCM (right panel) on the Ubx PRE in Drosophila S2 cells. Antibodies used for immunoprecipitations are indicated at the top, and amplified fragments are indicated at the left. “Mock” indicates control immunoprecipitation with protein A-agarose beads alone, and “Rp” indicates a control fragment from the RpII140 locus.
To determine if SCM tracks with these other PcG components on the Ubx PRE, we performed ChIP assays using Drosophila Schneider line 2 (S2) cells. Figure 2B compares the distributions of SCM (right panel) and PHO (left panel) across four fragments (b3 to b6) from the Ubx PRE region and on a negative-control fragment (Rp) derived from the gene encoding an RNA polymerase II subunit. This analysis shows that the SCM distribution coincides with those of PHO (Fig. 2B) and PRC2 and PRC1 (13, 93) within this Ubx PRE region.
SCM functions in Ubx gene silencing in fly S2 cells.
Previous studies have shown that Ubx mRNA levels are elevated in fly S2 cells when subunits of PHO-RC, PRC2, or PRC1 are depleted by RNA interference (8, 13, 93). To determine if SCM is also functionally engaged in Ubx gene silencing in S2 cells, we transfected short double-stranded RNAs to deplete SCM and assayed for changes in Ubx mRNA levels by RT-PCR. Figure 3A (bottom) shows that this RNA interference (RNAi) yields greater than an 8-fold reduction in the level of SCM. Separate RNAi treatments produce similar degrees of knockdown in the levels of PHO, E(Z), or PC (Fig. 3A). Figure 3B shows that each one of these RNAi knockdowns, including SCM knockdown (lane 5), causes Ubx desilencing as measured relative to a control sample treated with a nonspecific (GFP) double-stranded RNA. This degree of Ubx desilencing is similar to that observed previously after RNAi depletion of PHO, E(Z), or PC in S2 cells (93). These results demonstrate that, like other PcG proteins, SCM functions in Ubx gene silencing in S2 cells. Thus, fly S2 cells provide a valid platform for investigating functional interdependence among SCM and the previously defined PcG complexes.
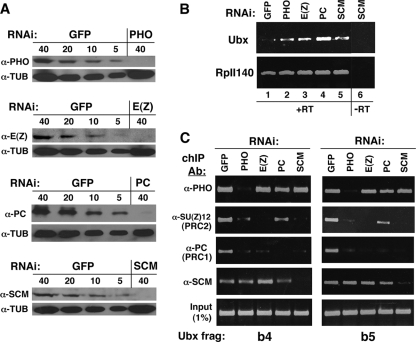
FIG. 3.
Consequences of SCM depletion and other PcG protein depletions in S2 cells. (A) Western blots to detect levels of individual PcG proteins after depletion by RNA interference. Labels at the left denote antibodies to detect the indicated PcG proteins or α-tubulin, which was used as a loading control. Labels at the top of each panel identify samples from mock-treated (GFP) or PcG-depleted S2 cells containing the indicated amounts of protein in μg. (B) RT-PCR analysis of Ubx and RpII140 expression after mock treatment (GFP) or treatment with the indicated PcG double-stranded RNAs. “+RT” indicates reverse transcriptase added, and “−RT” indicates a control with reverse transcriptase omitted from the reaction mixture. (C) ChIP analyses to detect associations of the indicated PcG proteins with Ubx PRE fragments b4 (left panel) and b5 (right panel). Labels at the left indicate antibodies (Ab) used in each chromatin immunoprecipitation, and labels at the top identify samples with individual PcG proteins depleted by RNAi. Lanes labeled “GFP” identify mock-depleted samples. PHO is a subunit of PHO-RC, SU(Z)12 is a subunit of PRC2, and PC is a subunit of PRC1.
Chromatin associations after depletion of PHO-RC, PRC2, and PRC1 subunits.
ChIP assays were performed using cross-linked chromatin from S2 cells after RNAi depletion of PHO, E(Z), or PC. For comparison, parallel mock-depleted samples were obtained after treatment with nonspecific GFP dsRNA. Besides Western blotting (Fig. 3A), RNAi depletions were verified by loss of corresponding ChIP signals on the b4 (Fig. 3C, left panel) and b5 (Fig. 3C, right panel) fragments. For example, the PHO ChIP signals are substantially reduced in the PHO RNAi samples (Fig. 3C, top row). PRC2 chromatin association was gauged by ChIP using an antibody against the SU(Z)12 subunit, and PRC1 association was tracked using an antibody to PC.
As reported previously (93), association of both PRC2 and PRC1 with the Ubx PRE is reduced after PHO depletion, consistent with the proposed role of PHO-RC in targeting of other PcG complexes. However, PHO depletion has little or no effect upon the association of SCM, which is retained at both the b4 and b5 fragments (Fig. 3C, fourth row, second column). Similarly, E(Z) knockdown (third column) dislodges PRC2 and PRC1 but not SCM, and PC knockdown (fourth column) dislodges PRC1 but not SCM. These results suggest that none of the three characterized fly PcG complexes is critically required for SCM association with the Ubx PRE in S2 cells.
Chromatin associations after SCM knockdown.
ChIP assays were also performed on S2 cells with SCM depleted. SCM chromatin association was significantly reduced, as expected, after SCM RNAi (Fig. 3C, fifth column, fourth row in both panels). This result verifies the specificity of the SCM antibody in generating the SCM ChIP signal.
To assess whether SCM plays a role in chromatin association of other PcG components, the SCM-depleted samples were tested by ChIP for PHO, SU(Z)12, and PC (Fig. 3C, fifth columns). PHO association with b4 and b5 appears to be unaffected by SCM loss, whereas associations of both PRC2 and PRC1 appear to be compromised. Thus, this matrix of ChIP assays suggests parallels between PHO and SCM in their effects upon PcG complex recruitment; both PHO and SCM can associate with the Ubx PRE independently of other PcG components, and loss of either PHO or SCM appears to reduce or dislodge PRC2 and PRC1.
Characterization of a pupal-lethal Scm allele.
In addition to performing S2 cell experiments, we wished to extend our analysis of SCM by interrogating chromatin from fly tissues in vivo. Several studies have successfully exploited Drosophila wing imaginal discs to track PcG chromatin associations (13, 53, 68, 93). A key feature of wing discs is that Ubx is kept off in this tissue due to PcG silencing. Thus, in contrast to intact embryos, where Ubx is expressed in about half the cells and silenced in the other half, larval wing discs enable Ubx ChIP studies where PcG silencing greatly predominates.
A key challenge to testing Scm loss of function in larval wing discs is that Scm null mutants die as embryos (2, 7). Thus, we sought a hypomorphic Scm mutant that survives to late larval or pupal stages and that nevertheless displays substantial Ubx desilencing in wing discs. Hypomorphic pupal-lethal Scm alleles have been described, but their relatively mild phenotypes and retained SCM targeting to Hox loci (2) suggested that these alleles were too weak for our envisioned ChIP analysis. Consequently, we investigated a set of 19 novel Scm fly mutants to identify at least one that might supply wing discs with robust desilencing of Ubx. These 19 new mutants, kindly provided by Henrik Gyurkovics (Biological Research Center, Szeged, Hungary), were isolated in a screen for modifiers of miniwhite silencing by linked PREs. This screen has previously been reported to yield PcG mutations (74, 91).
The 19 Scm alleles were first categorized as putative null alleles versus hypomorphic alleles using two criteria: determination of lethal phase and spatial expression of the ABD-A HOX protein in embryos. Scm null mutants die by late embryogenesis with robust ectopic accumulation of ABD-A, whereas hypomorphic mutants typically survive to larval or pupal stages and display little or no ABD-A misexpression in embryos (2, 3, 7). These tests identified seven putative hypomorphic Scm mutants that survive to pupal stages and show normal ABD-A patterns in embryos. These hypomorphic mutants were then tested for Ubx desilencing in larval imaginal discs. Each mutant was crossed to an Scm null mutant, ScmH1 (2), and tissues dissected from the Scmhypomorph/ScmH1 larval progeny were immunostained to reveal patterns of UBX accumulation. As shown in Fig. 4A, UBX normally accumulates in the haltere disc but is absent in wild-type wing discs, where it is subject to PcG silencing (1). Of the seven new Scm hypomorphs, most showed only subtle UBX misexpression consisting of isolated cells or small patches of cells. In contrast, a single mutant, Scmsz3, displayed substantial ectopic UBX in the wing disc (Fig. 4A, right panel). The UBX pattern varied from disc to disc, but it typically encompassed >30% of the disc territory with preferential accumulation in the wing pouch area. Similarly, RT-PCR analysis revealed Ubx desilencing at the mRNA level in Scmsz3/ScmH1 mutant wing discs (Fig. 4B).
By these criteria, Scmsz3 was the most useful Scm mutant for ChIP analysis of wing discs. Furthermore, Western blot analyses of larval extracts indicated substantially reduced levels of SCM in the Scmsz3 mutant (Fig. 4C). In contrast, SCM accumulates at levels comparable to wild type in previously characterized Scm hypomorphs, such as ScmSu(z)302 and ScmR5-13 (2). Intriguingly, sequence analysis revealed that sz3 is a missense mutation (L853Q) within the SPM domain (Fig. 1, top). Presumably, this alteration within the conserved protein interaction domain destabilizes SCM, perhaps through a reduced ability to bind partners, or to oligomerize, in vivo. Indeed, we note that the same sz3 missense change was independently isolated in a targeted two-hybrid screen for binding-defective SPM domain mutants (L48Q in reference 56). The map locations of the sz3 allele, plus one other hypomorphic allele (sz20) and three other null alleles (sz25, sz29, and sz36) determined in this study, are displayed in Fig. 1.
PcG chromatin associations in Scm mutant wing discs.
Wing discs were harvested from Scmsz3/ScmH1 larvae and were subjected to ChIP analysis in parallel with control discs from wild-type larvae. Figure 5A (left panel) shows that in wild-type discs, PHO, SU(Z)12, PC, and SCM are all detected in association with the b4 and b5 fragments of the Ubx PRE but not on the flanking b6 fragment or on the control fragment from the RpII140 locus. Thus, the normal wing disc distributions of these PcG proteins, including SCM, mirror their distributions in S2 cells (Fig. 2 and 3C) (24). Figure 5A (right panel) shows ChIP results from the Scmsz3 mutant. In agreement with Western blot analysis (Fig. 4C), little or no SCM is detected on the Ubx PRE in this mutant (rightmost column). The remaining ChIP samples show that PHO is retained on the Ubx PRE but that the signals for SU(Z)12 and PC are reduced. Thus, PHO association appears to be unaffected by SCM loss of function, whereas PRE binding by both PRC2 and PRC1 is diminished. These results are in close agreement with PcG associations detected after SCM knockdown in S2 cells (Fig. 3C).
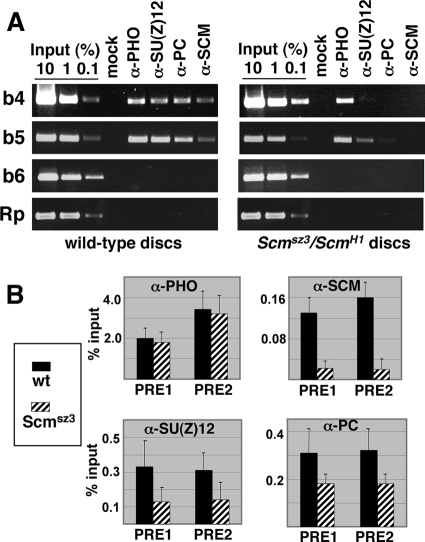
FIG. 5.
PcG chromatin associations at the Ubx PRE in Scm mutant wing discs. ChIP assays were used to detect PRE binding of PHO, SU(Z)12, PC, and SCM in wild-type or Scmsz3/ScmH1 wing discs as revealed by endpoint PCR (A) and quantitative (real-time) PCR (B). (A) The left panel shows ChIPs using wild-type wing discs, and the right panel shows ChIPs of Scmsz3/ScmH1 wing discs. Antibodies used for immunoprecipitations are indicated at the top, and amplified fragments are indicated at the left. “Mock” indicates control immunoprecipitation with protein A-agarose beads alone. “Rp” indicates a control fragment from the RpII140 locus. (B) Bar graphs depict Q-PCR ChIP signals obtained using antibodies against the indicated PcG proteins and chromatin samples from wild-type (solid bars) or Scmsz3/ScmH1 (hatched bars) wing discs. Error bars show standard deviations from the mean determined using at least two independent ChIP samples for each PcG protein and at least six separate Q-PCRs. For the SU(Z)12 and PC ChIP data (bottom panels), Student's t test yields a P value of ≤0.02 for all comparisons of Scm mutant versus wild type. “PRE1” and “PRE2” correspond to fragments 1 and 2, respectively, in Fig. 2A.
In addition to endpoint PCR analysis of the wing disc samples (Fig. 5A), we also performed real-time PCR assays to obtain more quantitative readouts (Fig. 5B). These quantitative PCR (Q-PCR) assays relied upon amplification of Ubx PRE fragments 1 and 2, depicted in Fig. 2A, which are subfragments of the b5 and b4 fragments, respectively. The Q-PCR results confirm that there is substantial loss of SCM signal in the Scm mutant discs and that PHO is unaffected by this SCM disruption (Fig. 5B, top panels). In addition, the Q-PCR assays detected reduced SU(Z)12 and PC signals, but they do not appear to be eliminated (Fig. 5B, bottom panels). We conclude that PHO is retained and there is partial loss of PRC2 and PRC1 from the Ubx PRE when Scm function is compromised in vivo.
PcG chromatin associations in wing discs after PC knockdown.
In order to conduct similar ChIP analyses on wing discs bearing loss of PRC1 function, we investigated depletion of the PC subunit via an RNA interference approach. Specifically, we tested whether conditional expression of a short hairpin RNA (shRNA) targeted against Pc mRNA could produce Pc loss of function in wing discs. A similar shRNA approach has been used to create in vivo knockdown of the PcG protein PCL (68). We obtained a transgenic fly line, denoted Pc-R4, from among a collection of fly lines (www.shigen.nig.ac.jp/fly/nigfly) that express individual shRNAs under the control of the GAL4 upstream activating sequence (UAS). Crosses were performed to generate progeny bearing both the UAS-Pc shRNA construct and a “driver” construct that expresses GAL4. As shown in Fig. 6B, wing-to-haltere transformations were produced when the A9-GAL4 wing disc driver (25) was used. Thus, targeted expression of Pc shRNA produced the canonical phenotype expected for Pc loss of function in wing discs. When the ubiquitous daughterless-GAL4 driver (96) was used instead, the consequence was pupal lethality, presumably reflecting more widespread PC knockdown. Western blot analysis showed that PC levels are dramatically reduced in wing discs dissected from these da-GAL4/+; UAS-Pc shRNA/+ larvae (Fig. 6C). Likewise, RT-PCR revealed Ubx desilencing in these wing discs (Fig. 6D), verifying that this genotype disrupts PcG silencing at the Ubx target locus.
Figure 6E shows the results of ChIP assays performed using wing discs harvested from da-GAL4/+; UAS-Pc shRNA/+ larvae. Q-PCR analysis of the ChIP samples revealed that this shRNA approach yields 6- to 8-fold reductions of PC bound to the Ubx PRE compared to wild-type (third panel). In contrast, levels of PHO, SU(Z)12, and SCM bound to the PRE appear to be unaltered (Fig. 6E). Thus, SCM chromatin association is retained at this PRE in vivo despite substantial removal of a core PRC1 subunit. This result is consistent with our findings using S2 cells (Fig. 3C). The retention of PHO and SU(Z)12 despite PC knockdown (Fig. 6E) is also consistent with previously described tests on PcG complex recruitment (93).
DISCUSSION
SCM and PcG complex recruitment to the Ubx PRE.
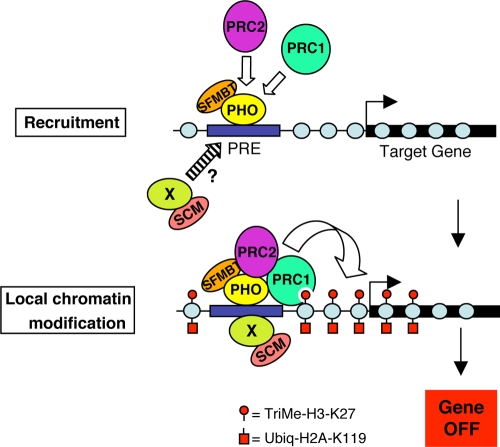
Chromatin immunoprecipitation studies using fly S2 cells, embryos, and wing discs indicate that PHO binds PREs in vivo and plays a key role in recruiting other PcG components to target sites (34, 36, 52, 53, 71, 93). The mechanisms of PRC2 and PRC1 recruitment are not fully understood but could involve physical interactions between PHO and subunits of these complexes (42, 43, 93). Thus, most models for recruitment of fly PcG complexes to target chromatin feature an early step involving PHO binding to PRE DNA, as depicted in Fig. 7. Since PHO exists in a stable heterodimer with the PcG protein SFMBT, which colocalizes with PHO at PREs in vivo, the PRE-bound species is the two-subunit PHO-RC complex (34, 52).
FIG. 7.
Model for chromatin association of PcG complexes. A PcG target gene with an upstream Polycomb response element (PRE). In the first step, PHO-RC, consisting of subunits PHO and SFMBT (34), binds to the PRE via DNA-binding activity of PHO. PRC2 is then recruited, possibly through contact with PHO (93), which methylates local nucleosomes on H3-K27. PRC1 associates with the target gene via interactions with methylated H3-K27 and/or with PHO (42, 43) or PRC2 (60). Red circles depict trimethyl-H3-K27 deposited by PRC2, and red squares depict ubiquitylated H2A on K119 created by PRC1 and/or the PRC1-related dRAF complex (12, 37, 92). The present ChIP data suggest that SCM is recruited independently of PRC2 and PRC1, possibly in parallel to PHO-RC. The protein labeled “X” (green) represents a hypothetical DNA-binding protein that could partner with SCM, akin to PHO partnership with SFMBT. Gray circles represent nucleosomes, and the curved open arrow depicts deposition of histone modifications. The arrow pointing rightward represents the transcription start site.
Once PRC2 has arrived at target chromatin, it can trimethylate H3-K27 on local nucleosomes (13, 16, 31, 35, 44). Since the PRC1 subunit PC can bind trimethylated H3-K27 via its chromodomain (19, 41), it has been hypothesized that this interaction helps recruit PRC1 to target loci. Indeed, the loss of PRC1 from target loci following knockdown or inactivation of PRC2 subunits in either Drosophila or mammalian cells is consistent with this view (4, 12, 13, 93). However, the overall contribution of the PC/triMe-K27 interaction to PRC1 targeting in vivo is yet to be determined. As an alternative to initial targeting, binding of PRC1 to trimethyl-H3-K27 could promote intralocus looping to bring PRE-bound PcG complexes into contact with the body of the gene to be silenced (45, 73).
How might SCM fit in molecularly with the other PcG components depicted in Fig. 7? Although in vitro associations of SCM with PRC1 subunits have been described (55, 56, 67, 75), the ChIP analyses here indicate that SCM can associate with the Ubx PRE despite the loss of PRC1 (Fig. 3C and 6E). Similarly, although SCM can bind to the PHO-RC subunit SFMBT in a pairwise assay (24), SCM localization at the PRE does not appear to be dependent on PHO (Fig. 3C). Taken together, these ChIP results are consistent with biochemical studies that reveal SCM separability from PHO-RC, PRC1, and PRC2 in fly embryo extracts (34, 50, 56).
An intriguing finding from our matrix of molecular epistasis tests is that SCM exhibits recruitment properties very similar to those of PHO. Specifically, both SCM and PHO can localize to the Ubx PRE independent of all other PcG components tested, and loss of either SCM or PHO diminishes PRC2 and PRC1 association with the PRE (Fig. 3C, 5A and B, and 6E). This similarity suggests that SCM may function, like PHO-RC, at an early step in PcG recruitment (Fig. 7A, top). In this context, it is worth emphasizing the striking overall similarities between SCM and the PHO-RC subunit SFMBT (Fig. 1). Perhaps SCM partners with a yet-to-be identified PcG DNA-binding protein, akin to the functional partnership of SFMBT with PHO (34). Indeed, since PHO-binding sites are insufficient for PRE function in vivo (22, 77) and many other DNA-binding proteins have been implicated in Drosophila PcG silencing (see references 45, 63, and 70 for reviews), there is abundant evidence that PRE recognition involves more than just PHO-RC. The common view is that many PREs contain a composite of PHO sites plus additional types of factor-binding motifs (45, 63, 70). At present, little is known about the nature of SCM-containing complexes beyond the detection of an approximately 500-kDa moiety in fly embryo extracts (56). It will be informative to characterize stably associated SCM partner proteins and evaluate their potential roles in binding to PRE DNA.
Although the ChIP assays presented here, together with previous biochemical tests (34, 50, 56), emphasize SCM separability from other PcG components, SCM must still integrate with its PcG cohorts to achieve gene silencing. This interdependence is highlighted by in vivo assays where robust silencing of a miniwhite reporter by a tethered form of SCM is disrupted if the PRC1 subunit PH is compromised by mutation (64). Despite advances in understanding biochemical activities of individual PcG complexes, it is not yet clear how their multiple functions are integrated to achieve gene silencing (81). Further studies will be needed to determine how SCM functions in concert with other PcG components at target chromatin.
Functional domains shared in SCM and SFMBT.
Ultimately, a precise understanding of SCM function requires deciphering the mechanistic contributions of each of its three identified domains. As depicted in Fig. 1, SCM contains a C-terminal SPM domain, two mbt repeats, and two Cys2-Cys2 zinc fingers. Strikingly, each of these domains is also present in SFMBT (Fig. 1), suggesting that the overall biochemical roles of these two PcG components may be very similar. Indeed, a recent study provides evidence of functional synergy between SCM and SFMBT (24). In addition, the PH PcG protein possesses two of these three homology domains (Fig. 1). This presents the curious situation of three different PcG proteins related by shared domains yet with none appearing to reside in a stable common complex in nuclear extracts (34, 50, 56).
There are currently in vitro and in vivo data on roles of the SPM domain and mbt repeats but little knowledge yet about the zinc fingers. The SPM domain is a subtype within the broader category of SAM domains that mediate protein interactions (3, 59, 61). The SCM version of this domain is capable of robust self-binding and cross-binding to the PH version in vitro (55). The importance of SPM domain interactions in vivo is emphasized by PcG phenotypes observed after overexpressing a dominant-negative isolated SPM domain in developing flies (56). However, it remains unclear precisely what SPM interactions contribute to the PcG silencing mechanism. The simple idea that they constitutively glue PcG complex subunits together is at odds with the biochemical separabilities in embryo extracts. Perhaps SPM interactions function primarily directly at chromatin targets, where they could sponsor contacts among different PcG complexes rather than among subunits within the same complex. Such chromatin-specific interactions could contribute to intralocus loops, which have been hypothesized to exist at PcG silenced loci (45, 73).
The functional significance of the SCM mbt repeats is reflected by partial loss-of-function alleles that alter the first repeat (2) and by Hox gene silencing defects observed after disruption of the second repeat (23). Structural determinations and in vitro binding studies have revealed that mbt repeats are modules for binding to methylated lysines (23, 34, 65, 66, 89). Since trimethylated H3-K27 is a prominent feature of PcG-silenced chromatin, the mbt repeats could, at first glance, play a role akin to that of the PC chromodomain. However, there are important differences between the substrate-binding properties of these mbt repeats and the PC chromodomain. First, the mbt repeats prefer mono-and dimethylated lysines, whereas the chromodomain prefers the trimethylated form (19, 23, 34, 41, 65, 89). An intriguing hypothesis is that this mono/di preference could reflect a “grappling hook” function whereby hypomethylated nucleosomes are recognized and brought into proximity for trimethylation by PRC2 (34, 45). Another distinction is that the binding mode of mbt repeats is not much influenced by peptide sequence context, whereas chromodomain binding features extensive contact with residues flanking the methylated lysine (19, 23, 65). Consistent with this, the SCM mbt repeats lack binding preference for any particular histone tail lysines (23, 65). Thus, mbt repeats provide a pocket for methyl-lysine binding, but it is not yet clear if the relevant substrate for SCM is a particular methylated histone residue or even a nonhistone protein. Certainly, the in vitro binding preferences could be modified by additional associated factors in vivo.
Figure 8 shows a sequence alignment of the Cys2-Cys2 fingers present in SCM, SFMBT and PH. This zinc finger is a distinct subtype that adheres to the consensus sequence CXXCG-Xn-K/R-X-F/Y-CSXXC. These fingers do not appear to function by binding DNA, since sequence-specific binding is not observed in vitro for any of them (2, 3, 34). Thus, their molecular role is unknown, but their common inclusion in these related fly PcG proteins suggests some key contribution to PcG chromatin function. Curiously, both the SCM and SFMBT human homologs appear to have lost their Cys2-Cys2 fingers, whereas all three human PH homologs have retained them (Fig. 8). Thus, if these zinc fingers are critical in PcG silencing, then they apparently can be supplied from different combinations of PcG proteins in flies and in mammals. It will be important to test the genetic requirement for the SCM zinc fingers in Drosophila and to further define the mechanistic contributions of all three SCM functional domains to PcG chromatin silencing.
FIG. 8.

Alignment of Cys2-Cys2 zinc fingers in SCM, SFMBT, and PH. Amino acid residues are from the two zinc fingers in Drosophila SCM, the single zinc fingers in Drosophila SFMBT and PH, and the single zinc fingers in the three human PH homologs. This type of zinc finger, termed an FCS zinc finger (IPR012313), is also found in the l(3)mbt [lethal (3) malignant brain tumor] protein, which shares homology domains but is not known to function with PcG proteins. Two human SCM homologs and two human SFMBT homologs apparently lack these FCS zinc fingers. Presumed zinc-coordinating cysteines are shown in bold. The residues shown from each protein are indicated at the right. A consensus sequence is shown at the bottom.
Acknowledgments
We are grateful to Henrik Gyurkovics for providing new Scm alleles. We thank Rick Jones, Judy Kassis, Jürg Müller, and Mike O'Connor for antibody reagents and the National Institute of Genetics Fly Stock Center (Japan) for shRNA-expressing fly stocks.
This work was supported by National Institutes of Health grant GM49850 to J.A.S. N.J. and C.S.K. were supported in part by NIH training grant HD07480.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Beuchle, D., G. Struhl, and J. Muller. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993-1004. [DOI] [PubMed] [Google Scholar]
- 2.Bornemann, D., E. Miller, and J. Simon. 1998. Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics 150:675-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bornemann, D., E. Miller, and J. Simon. 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122:1621-1630. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349-353. [DOI] [PubMed] [Google Scholar]
- 5.Bracken, A. P., N. Dietrich, D. Pasini, K. H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 7.Breen, T. R., and I. M. Duncan. 1986. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 118:442-456. [DOI] [PubMed] [Google Scholar]
- 8.Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. [DOI] [PubMed] [Google Scholar]
- 9.Brower, D. L. 1987. Ultrabithorax gene expression in Drosophila imaginal discs and larval nervous system. Development 101:83-92. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. L., C. Fritsch, J. Mueller, and J. A. Kassis. 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130:285-294. [DOI] [PubMed] [Google Scholar]
- 11.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 12.Cao, R., Y. Tsukada, and Y. Zhang. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20:845-854. [DOI] [PubMed] [Google Scholar]
- 13.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 14.Carrington, E. A., and R. S. Jones. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development 122:4073-4083. [DOI] [PubMed] [Google Scholar]
- 15.Chan, C. S., L. Rastelli, and V. Pirrotta. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13:2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 17.Dellino, G. I., Y. B. Schwartz, G. Farkas, D. McCabe, S. C. Elgin, and V. Pirrotta. 2004. Polycomb silencing blocks transcription initiation. Mol. Cell 13:887-893. [DOI] [PubMed] [Google Scholar]
- 18.Ebert, A., G. Schotta, S. Lein, S. Kubicek, V. Krauss, T. Jenuwein, and G. Reuter. 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 18:2973-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis, N. J., R. E. Kingston, and C. L. Woodcock. 2004. Chromatin compaction by a polycomb group protein complex. Science 306:1574-1577. [DOI] [PubMed] [Google Scholar]
- 21.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 22.Fritsch, C., J. L. Brown, J. A. Kassis, and J. Muller. 1999. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126:3905-3913. [DOI] [PubMed] [Google Scholar]
- 23.Grimm, C., A. G. de Ayala Alonso, V. Rybin, U. Steuerwald, N. Ly-Hartig, W. Fischle, J. Muller, and C. W. Muller. 2007. Structural and functional analyses of methyl-lysine binding by the malignant brain tumour repeat protein Sex comb on midleg. EMBO Rep. 8:1031-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm, C., R. Matos, N. Ly-Hartig, U. Steuerwald, D. Lindner, V. Rybin, J. Muller, and C. W. Muller. 2009. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28:1965-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haerry, T. E., O. Khalsa, M. B. O'Connor, and K. A. Wharton. 1998. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development 125:3977-3987. [DOI] [PubMed] [Google Scholar]
- 26.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, R. S., and W. M. Gelbart. 1990. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics 126:185-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurgens, G. 1985. A group of genes controlling spatial expression of the bithorax complex in Drosophila. Nature 316:153-155. [Google Scholar]
- 29.Kahn, T. G., Y. B. Schwartz, G. I. Dellino, and V. Pirrotta. 2006. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 281:29064-29075. [DOI] [PubMed] [Google Scholar]
- 30.Karch, F., W. Bender, and B. Weiffenbach. 1990. abdA expression in Drosophila embryos. Genes Dev. 4:1573-1587. [DOI] [PubMed] [Google Scholar]
- 31.Ketel, C. S., E. F. Andersen, M. L. Vargas, J. Suh, S. Strome, and J. A. Simon. 2005. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell. Biol. 25:6857-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, I. F., R. B. Emmons, N. J. Francis, B. Wild, J. Muller, R. E. Kingston, and C. T. Wu. 2005. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol. Cell. Biol. 25:6578-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King, I. F., N. J. Francis, and R. E. Kingston. 2002. Native and recombinant polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol. Cell. Biol. 22:7919-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klymenko, T., B. Papp, W. Fischle, T. Kocher, M. Schelder, C. Fritsch, B. Wild, M. Wilm, and J. Muller. 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20:1110-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong, C., B. Adryan, I. Bell, L. Meadows, S. Russell, J. R. Manak, and R. White. 2008. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 4:e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagarou, A., A. Mohd-Sarip, Y. M. Moshkin, G. E. Chalkley, K. Bezstarosti, J. A. Demmers, and C. P. Verrijzer. 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 22:2799-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, T. I., R. G. Jenner, L. A. Boyer, M. G. Guenther, S. S. Levine, R. M. Kumar, B. Chevalier, S. E. Johnstone, M. F. Cole, K. Isono, H. Koseki, T. Fuchikami, K. Abe, H. L. Murray, J. P. Zucker, B. Yuan, G. W. Bell, E. Herbolsheimer, N. M. Hannett, K. Sun, D. T. Odom, A. P. Otte, T. L. Volkert, D. P. Bartel, D. A. Melton, D. K. Gifford, R. Jaenisch, and R. A. Young. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine, S. S., A. Weiss, H. Erdjument-Bromage, Z. Shao, P. Tempst, and R. E. Kingston. 2002. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 22:6070-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis, E. B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565-570. [DOI] [PubMed] [Google Scholar]
- 41.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohd-Sarip, A., F. Cleard, R. K. Mishra, F. Karch, and C. P. Verrijzer. 2005. Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex. Genes Dev. 19:1755-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 45.Muller, J., and J. A. Kassis. 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16:476-484. [DOI] [PubMed] [Google Scholar]
- 46.Muller, J., and P. Verrijzer. 2009. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 19:150-158. [DOI] [PubMed] [Google Scholar]
- 47.Negre, N., J. Hennetin, L. V. Sun, S. Lavrov, M. Bellis, K. P. White, and G. Cavalli. 2006. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nekrasov, M., T. Klymenko, S. Fraterman, B. Papp, K. Oktaba, T. Kocher, A. Cohen, H. G. Stunnenberg, M. Wilm, and J. Muller. 2007. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 26:4078-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nekrasov, M., B. Wild, and J. Muller. 2005. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 6:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohm, J. E., K. M. McGarvey, X. Yu, L. Cheng, K. E. Schuebel, L. Cope, H. P. Mohammad, W. Chen, V. C. Daniel, W. Yu, D. M. Berman, T. Jenuwein, K. Pruitt, S. J. Sharkis, D. N. Watkins, J. G. Herman, and S. B. Baylin. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 39:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oktaba, K., L. Gutierrez, J. Gagneur, C. Girardot, A. K. Sengupta, E. E. Furlong, and J. Muller. 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15:877-889. [DOI] [PubMed] [Google Scholar]
- 53.Papp, B., and J. Muller. 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20:2041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park, I. K., D. Qian, M. Kiel, M. W. Becker, M. Pihalja, I. L. Weissman, S. J. Morrison, and M. F. Clarke. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302-305. [DOI] [PubMed] [Google Scholar]
- 55.Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock, and J. Simon. 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17:6683-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson, A. J., D. R. Mallin, N. J. Francis, C. S. Ketel, J. Stamm, R. K. Voeller, R. E. Kingston, and J. A. Simon. 2004. Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167:1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pietersen, A. M., and M. van Lohuizen. 2008. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 20:201-207. [DOI] [PubMed] [Google Scholar]
- 58.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 59.Ponting, C. P. 1995. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4:1928-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiao, F., and J. U. Bowie. 2005. The many faces of SAM. Sci. STKE 2005:re7. [DOI] [PubMed] [Google Scholar]
- 62.Rajasekhar, V. K., and M. Begemann. 2007. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells 25:2498-2510. [DOI] [PubMed] [Google Scholar]
- 63.Ringrose, L., and R. Paro. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134:223-232. [DOI] [PubMed] [Google Scholar]
- 64.Roseman, R. R., K. Morgan, D. R. Mallin, R. Roberson, T. J. Parnell, D. J. Bornemann, J. A. Simon, and P. K. Geyer. 2001. Long-range repression by multiple polycomb group (PcG) proteins targeted by fusion to a defined DNA-binding domain in Drosophila. Genetics 158:291-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santiveri, C. M., B. C. Lechtenberg, M. D. Allen, A. Sathyamurthy, A. M. Jaulent, S. M. Freund, and M. Bycroft. 2008. The malignant brain tumor repeats of human SCML2 bind to peptides containing monomethylated lysine. J. Mol. Biol. 382:1107-1112. [DOI] [PubMed] [Google Scholar]
- 66.Sathyamurthy, A., M. D. Allen, A. G. Murzin, and M. Bycroft. 2003. Crystal structure of the malignant brain tumour (MBT) repeats in sex comb on midleg-like2 (SCML2). J. Biol. Chem. 278:46968-46973. [DOI] [PubMed] [Google Scholar]
- 67.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 68.Savla, U., J. Benes, J. Zhang, and R. S. Jones. 2008. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development 135:813-817. [DOI] [PubMed] [Google Scholar]
- 69.Schlesinger, Y., R. Straussman, I. Keshet, S. Farkash, M. Hecht, J. Zimmerman, E. Eden, Z. Yakhini, E. Ben-Shushan, B. E. Reubinoff, Y. Bergman, I. Simon, and H. Cedar. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39:232-236. [DOI] [PubMed] [Google Scholar]
- 70.Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc, and G. Cavalli. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128:735-745. [DOI] [PubMed] [Google Scholar]
- 71.Schuettengruber, B., M. Ganapathi, B. Leblanc, M. Portoso, R. Jaschek, B. Tolhuis, M. van Lohuizen, A. Tanay, and G. Cavalli. 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 7:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon, M. Biggin, and V. Pirrotta. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38:700-705. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz, Y. B., and V. Pirrotta. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 74.Shanower, G. A., M. Muller, J. L. Blanton, V. Honti, H. Gyurkovics, and P. Schedl. 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169:173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 76.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-66. [DOI] [PubMed] [Google Scholar]
- 77.Shimell, M. J., A. J. Peterson, J. Burr, J. A. Simon, and M. B. O'Connor. 2000. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 218:38-52. [DOI] [PubMed] [Google Scholar]
- 78.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4:481-495. [DOI] [PubMed] [Google Scholar]
- 79.Simon, J., A. Chiang, and W. Bender. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114:493-505. [DOI] [PubMed] [Google Scholar]
- 80.Simon, J., A. Chiang, W. Bender, M. J. Shimell, and M. O'Connor. 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158:131-144. [DOI] [PubMed] [Google Scholar]
- 81.Simon, J. A., and R. E. Kingston. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10:697-708. [DOI] [PubMed] [Google Scholar]
- 82.Simon, J. A., and C. A. Lange. 2008. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647:21-29. [DOI] [PubMed] [Google Scholar]
- 83.Sparmann, A., and M. van Lohuizen. 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6:846-856. [DOI] [PubMed] [Google Scholar]
- 84.Squazzo, S. L., H. O'Geen, V. M. Komashko, S. R. Krig, V. X. Jin, S. W. Jang, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stock, J. K., S. Giadrossi, M. Casanova, E. Brookes, M. Vidal, H. Koseki, N. Brockdorff, A. G. Fisher, and A. Pombo. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9:1428-1435. [DOI] [PubMed] [Google Scholar]
- 86.Struhl, G., and M. Akam. 1985. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4:3259-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ting, A. H., K. M. McGarvey, and S. B. Baylin. 2006. The cancer epigenome—components and functional correlates. Genes Dev. 20:3215-3231. [DOI] [PubMed] [Google Scholar]
- 88.Tolhuis, B., E. de Wit, I. Muijrers, H. Teunissen, W. Talhout, B. van Steensel, and M. van Lohuizen. 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38:694-699. [DOI] [PubMed] [Google Scholar]
- 89.Trojer, P., G. Li, R. J. Sims, 3rd, A. Vaquero, N. Kalakonda, P. Boccuni, D. Lee, H. Erdjument-Bromage, P. Tempst, S. D. Nimer, Y. H. Wang, and D. Reinberg. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129:915-928. [DOI] [PubMed] [Google Scholar]
- 90.Usui, H., T. Ichikawa, K. Kobayashi, and T. Kumanishi. 2000. Cloning of a novel murine gene Sfmbt, Scm-related gene containing four mbt domains, structurally belonging to the Polycomb group of genes. Gene. 248:127-135. [DOI] [PubMed] [Google Scholar]
- 91.Vazquez, J., G. Farkas, M. Gaszner, A. Udvardy, M. Muller, K. Hagstrom, H. Gyurkovics, L. Sipos, J. Gausz, M. Galloni, et al. 1993. Genetic and molecular analysis of chromatin domains. Cold Spring Harbor Symp. Quant. Biol. 58:45-54. [DOI] [PubMed] [Google Scholar]
- 92.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873-878. [DOI] [PubMed] [Google Scholar]
- 93.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 94.Wang, L., N. Jahren, M. L. Vargas, E. F. Andersen, J. Benes, J. Zhang, E. L. Miller, R. S. Jones, and J. A. Simon. 2006. Alternative ESC and ESC-like subunits of a polycomb group histone methyltransferase complex are differentially deployed during Drosophila development. Mol. Cell. Biol. 26:2637-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.White, R. A. H., and M. Wilcox. 1985. Distribution of Ultrabithorax proteins in Drosophila. EMBO J. 4:2035-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wodarz, A., U. Hinz, M. Engelbert, and E. Knust. 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82:67-76. [DOI] [PubMed] [Google Scholar]
- 97.Wu, C. T., R. S. Jones, P. F. Lasko, and W. M. Gelbart. 1989. Homeosis and the interaction of zeste and white in Drosophila. Mol. Gen. Genet. 218:559-564. [DOI] [PubMed] [Google Scholar]
- 98.Yu, J., D. R. Rhodes, S. A. Tomlins, X. Cao, G. Chen, R. Mehra, X. Wang, D. Ghosh, R. B. Shah, S. Varambally, K. J. Pienta, and A. M. Chinnaiyan. 2007. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 67:10657-10663. [DOI] [PubMed] [Google Scholar]
- 99.Zhao, J., B. K. Sun, J. A. Erwin, J. J. Song, and J. T. Lee. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750-756. [DOI] [PMC free article] [PubMed] [Google Scholar]