Abstract
The TIP60 histone acetyltransferase plays diverse roles in DNA damage responses, DNA double-strand break repair, and transcriptional regulation. TIP60 resides within a multisubunit complex that has been shown to be targeted by transcription factors and to be involved in histone acetylation and transcriptional activation. p400, an SWI2/SNF2-related ATPase that serves as an ATP-dependent chromatin remodeling enzyme, exists as an integral subunit of a TIP60 complex but also resides within a distinct complex that presumably lacks TIP60 and appears to be involved in the transcriptional repression of basal p53 target gene expression. Here, we describe a TIP60-containing p400 complex population in which the acetyltransferase activity of TIP60 is repressed by interactions with p400. We further show that an SWI3-ADA2-N-CoR-TFIIIB (SANT) domain of p400 binds directly to the histone acetyltransferase (HAT) domain of TIP60 and blocks both its enzymatic activity and its coactivator function in regulating basal p21 gene expression. Our results thus suggest that p400 represses basal p21 gene expression through dual mechanisms that include the direct inhibition of TIP60 enzymatic activity described here and the previously described ATP-dependent positioning of H2A.Z at the promoter.
The compaction of eukaryotic DNA within nucleosomal and higher-order chromatin structures allows intricate multilevel regulatory responses to diverse environmental signals, although these structures themselves provide physically repressive barriers to critical nuclear processes such as DNA replication, transcription, and repair (12). Two types of enzymes have evolved to regulate chromatin structure and thereby facilitate the transcriptional repression or activation of genes. The first group includes ATP-dependent chromatin remodelers that alter DNA-histone contacts without covalent bond breakage and result in changes in nucleosome positioning or composition (33). The second group includes diverse histone-modifying enzymes that add or remove posttranslational modifications on histones (32). Each type of enzyme generally forms a multisubunit complex with specialized subunits that may play roles in targeting to DNA-bound transcription factors, in direct binding to chromatin through unmodified or modified histones, or in interactions with basal transcription machineries. In Saccharomyces cerevisiae, the two enzyme types are usually found in distinct complexes that act together in either a concerted or a sequential manner to regulate transcription (11). This is generally the case in higher eukaryotes as well, but in some cases, enzyme complexes have converged into a combined complex that contains both an ATP-dependent chromatin remodeler and a histone modifier (13, 36). The TIP60 complex is one of a few complexes of this type in metazoans to mediate transcriptional regulation. Consistent with an evolutionary convergence, the mammalian TIP60 complex has been experimentally mimicked in yeast by assimilating yeast NuA4 histone acetyltransferase and SWR1 ATP-dependent chromatin remodeling complexes into a single complex (1).
TIP60 is a histone acetyltransferase (HAT) that acetylates H2A and H4 histones within chromatin, and its enzyme activity has been shown to be critical for multiple cellular processes that include DNA damage responses, double-strand break repair, and transcriptional activation (25). Besides the normal core histone substrates, TIP60 targets also include histone variants (H2A.Z and H2AX), the ATM kinase, and DNA-bound transcription factors such as p53 (2, 15, 27, 29). A complex (“TIP60 complex”) that was affinity purified through an epitope tag on TIP60 was shown to exhibit HAT activity and to contain multiple subunits that included p400 (5, 13). p400 is an ATPase in the SWI2/SNF2 class of ATP-dependent chromatin remodelers (8) and was first identified as an E1A-interacting protein crucial for E1A-dependent apoptosis and cellular transformation (24). p400 has also been implicated in p53 target gene regulation through a repressive effect on basal-level p21 transcription involving the localization of histone variant H2A.Z to the promoter (9, 31). However, while there is a close correlation between p21 transcriptional repression and the joint recruitment of p400 and H2A.Z to the promoter, a molecular mechanism for gene repression by H2A.Z remains unknown. Of relevance here, p400 contains a conserved SWI3-ADA2-N-CoR-TFIIIB (SANT) domain that has been shown to interact both with histone tails and with other proteins and is further implicated in both positive and negative chromatin-regulatory modifications (26, 35).
Importantly, a complex (“p400 complex”) that was affinity purified with an anti-p400 antibody appeared devoid of TIP60 and any associated HAT activity, indicating the existence of a complex that is distinct and separable from the p400-containing TIP60 complex (8, 13). However, especially in light of the nearly identical polypeptide compositions of the p400 and TIP60 complexes and the presence of HAT activity in the TIP60 complex, it has not been clear why TIP60 was not evident in the p400 complex while the p400-containing TIP60 complex exhibited HAT activity. One possible explanation is that the isolated TIP60 complex was heterogeneous, with a population that contained p400 and one that did not, and that the TIP60-p400 complex is dynamic. Moreover, because p400 was found to antagonize p53-dependent transcriptional activation mainly under basal (nonstressed) conditions, little is known about how TIP60 and p400 enzymes in the same complex might be modulated to achieve p53-dependent transcriptional activation upon genotoxic DNA damage stress. Since genotoxic stress has been reported to result in p400 dissociation from the p21 promoter followed by increased TIP60 recruitment, the activated TIP60 complex at the promoter might well lack the repressive p400 subunit (4, 9).
Here, we present evidence for a p400 complex subpopulation that contains an integral TIP60 in an inactive form as a result of a direct interaction between the TIP60 and p400 proteins. Remarkably, this interaction and the associated repression have been shown to involve the SANT domain, which binds to the HAT domain of TIP60 and suppresses HAT activity. We also demonstrate that this novel interaction between p400 and TIP60 closely correlates with a loss of the TIP60 coactivator function for basal p21 promoter activity.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, 293T, HCT116, and U2OS cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Sf9 cells were cultured in Hink's TNM-FH medium (Invitrogen) supplemented with 10% fetal bovine serum. For transfection, cells were plated at approximately 60 to 80% confluence and transfected with expression vectors, as indicated, by using Fugene 6 (Roche).
Expression plasmids.
The cDNAs for Flag-p400 and hemagglutinin (HA)-TIP60 were subcloned into CβS mammalian expression vectors (19) for transient expression assays. The p400 ATPase motif was mutagenized by PCR-mediated site-directed mutagenesis (Stratagene) on the BsaBI-SgrAI cDNA fragment and then reinserted into wild-type p400 cDNA. To prepare either HeLa or U2OS stable cell lines, lentiviruses encoding Flag-p400 proteins were generated and then used for lentiviral infections according to the manufacturer's protocol (System Biosciences). For baculoviral protein expression, Flag-p400 or HA-TIP60 cDNA was subcloned into pFAST-Bac1, and baculoviruses were then generated according to the manufacturer's protocol (Invitrogen). Glutathione S-transferase (GST) fusion and His-tagged constructs were made in pGEX4T-1 and 6HpET11d vectors, respectively.
Luciferase reporter assay.
The p21 promoter-driven firefly luciferase plasmid (pWWP-luc) (22) and the control pRL-CMV plasmid for Renilla luciferase were used for dual-luciferase reporter assays (Promega), and experimental firefly luciferase activities were normalized to pRL-CMV-driven Renilla luciferase activities.
HAT assays.
HA-TIP60 preparations were isolated from nuclear extracts either on anti-HA agarose (Sigma) for HA-TIP60 alone or on M2 agarose (Sigma) for HA-TIP60 bound to coexpressed Flag-p400 or its derivatives. HA-TIP60 preparations were released by the addition of excess HA or Flag peptides in HAT assay buffer (50 mM Tris at pH 8.0, 10% glycerol, 50 mM KCl, 0.1 mM EDTA, 10 mM butyric acid, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]). HeLa core histones (2 μg) and [3H]acetyl coenzyme A (CoA) were incubated with each eluate, and reaction mixtures were resolved on an SDS-PAGE gradient gel (4% to 15%). Proteins were transferred onto a nitrocellulose membrane and subjected to immunoblotting with the indicated antibodies. The bottom section of the gel below the 30-kDa molecular mass marker was subjected to autoradiography for HAT activities. For reconstituted HAT inhibition experiments, protein mixtures were preincubated at 30°C for 15 min, followed by an additional 30 min of incubation in the presence of [3H]acetyl-CoA.
GST pulldown assay.
Two micrograms of GST fusion proteins was bound to glutathione-Sepharose 4B beads, and 300 ng of purified His-tagged proteins was mixed in binding buffer (20 mM Tris-Cl [pH 7.5], 150 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.05% NP-40, 1 mg/ml bovine serum albumin [BSA], and 0.5 mM PMSF). After 3 h of incubation at 4°C, the beads were washed with binding buffer, and bound proteins were resolved by SDS-PAGE and immunoblot analysis.
Protein purification.
For the purification of p400 complexes from HeLa cells that stably express Flag-p400 and HA-TIP60, nuclear extracts were prepared by a modified Dignam procedure and directly applied onto a Sepharose CL6B gel filtration column to remove unincorporated free HA-TIP60 (23). Fractions corresponding to the p400 protein peak, as determined by immunoblotting, were combined and subjected to M2 agarose affinity purification. After being serially washed with BC300 and BC500 buffers (20 mM Tris-Cl [pH 7.5], 0.2 mM EDTA, 20% glycerol, and 0.5 mM PMSF with 300 and 500 mM KCl, respectively) containing 0.1% NP-40, captured Flag-p400 was eluted by the addition of 250 μg/ml Flag peptide at 4°C. To prepare recombinant p400 plus TIP60 from Sf9 cells, 3 × 107 cells were plated onto 150-mm plates and infected with recombinant Flag-p400 and HA-TIP60 baculoviruses at a multiplicity of infection (MOI) of 5 to 10 each. At 24 h postinfection, cells were harvested, and nuclear extracts were subjected to the same Sepharose CL6B gel filtration chromatography and M2 agarose affinity purification procedures as those described above.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were conducted as described previously (23). Briefly, 107 cells were cross-linked by the direct addition of formaldehyde (final concentration, 1%) to cells for 10 min at room temperature. Fixed cells were harvested and washed twice with swelling buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 8.0), 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml}. Nuclei were resuspended in sonication buffer (0.2% sodium dodecyl sulfate, 0.5 mM PMSF, 100 ng of leupeptin and aprotinin per ml) and sonicated on ice to obtain 500-bp DNA fragments. Normalized chromatin lysates were incubated overnight with the indicated antibodies (anti-GAL4[DBD] antibody from Santa Cruz, anti-p53 antibody [FL-393] from Santa Cruz, anti-hyperacetylated histone H4 from Millipore, anti-Flag antibody from Sigma, and anti-TIP60 antibody raised in this study), and 20 μl of protein A beads was then added and incubated for an additional 1 h at 4°C. Beads were washed and eluted, and following cross-link reversal, eluted material was subjected to real-time PCR amplification. Primers used for ChIP PCR are available upon request.
Histone variant exchange assay.
The 196-bp DNA fragment from plasmid pG5ML (14) was labeled with biotin-14-dATP and then assembled with recombinant histones into a mononucleosome. One milligram of Dynabead M-280 streptavidin (Invitrogen) was incubated with 700 ng of assembled mononucleosomes for 3 h at room temperature. Dynabead-bound mononucleosomes were washed with preincubation buffer (20 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mg/ml BSA, 0.5 mM DTT, 0.5 mM PMSF) and then used for the exchange assay. The exchange reaction was initiated by addition of 35 ng of mononucleosomes, 10 ng of purified p400, 1.5 mM ATP, and 300 ng of the Flag-H2A.Z plus H2B (Flag-H2A.Z+H2B) dimer in exchange buffer (25 mM HEPES [pH 7.6], 0.1 mM EDTA, 5 mM MgCl2, 10% glycerol, 0.02% NP-40, 1 mM DTT, 0.1 mg/ml BSA, 70 mM KCl) for 1 h at 30°C. Dynabead-bound mononucleosomes were washed with exchange buffer and resolved by SDS-PAGE and immunoblot analysis.
RESULTS
A TIP60-containing p400 complex with suppressed HAT activity.
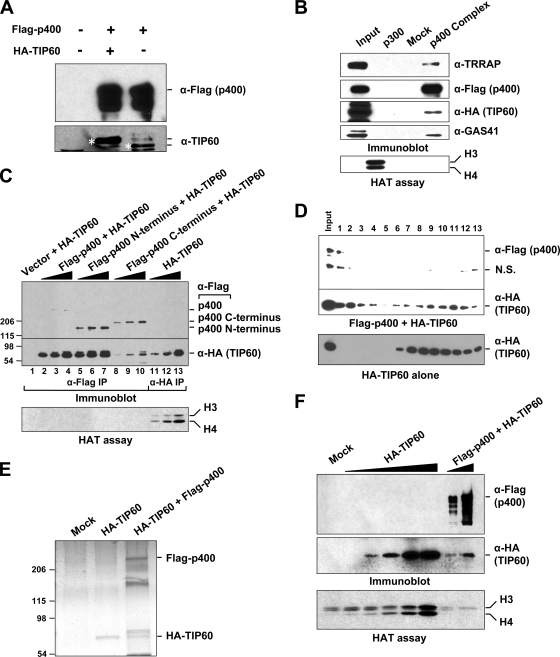
The TIP60 complex isolated through Flag-tagged TIP60 was previously reported to contain p400 as an integral subunit and exhibited a robust HAT activity (5, 13), whereas the p400 complex isolated through an anti-p400 antibody was reported previously to have neither TIP60 nor any significant HAT activity (8). Those previous results suggested possible heterogeneity, with respect to p400 occupancy, within isolated TIP60 complexes in addition to the possibility of a TIP60-free p400 complex. To see whether there might also be a TIP60-containing p400 complex lacking HAT activity, Flag-p400 was ectopically overexpressed in 293T cells either alone or together with HA-TIP60, and interactions were analyzed by coimmunoprecipitation and immunoblotting (Fig. 1A). The results indicate that Flag-p400 stably associates with both endogenous and ectopically expressed TIP60, thus arguing against the possibility of a p400 complex with little or no TIP60 protein. To test for TIP60-driven HAT activity from a TIP60-p400 complex, and in order to increase the population of complexes containing both p400 and TIP60, we established a HeLa cell line that stably expresses both Flag-p400 and HA-TIP60. The Flag-p400 complex that was affinity purified on M2 agarose contained stably associated TIP60 as well as other representative subunits, such as TRRAP and GAS41, but failed to show detectable HAT activity (Fig. 1B). Our results thus indicate that the lack of detectable HAT activity in a particular p400 complex may not be due simply to the normal absence of TIP60 or to an unstable TIP60 association.
FIG. 1.
Suppression of TIP60 HAT activity in the p400 complex. (A) Association of TIP60 with p400. Flag-p400 was ectopically expressed in 293T cells in the presence or absence of HA-TIP60 coexpression. Flag-p400 immunoprecipitates on M2 agarose were analyzed by immunoblotting with anti-Flag and anti-TIP60 antibodies. Coimmunoprecipitated HA-TIP60 and endogenous TIP60 are indicated by the upper and lower asterisks, respectively. (B) Analysis of the p400 complex containing TIP60. (Top) Immunoblot of polypeptides coimmunoprecipitated on M2 agarose with Flag-p400 from HeLa cells that stably coexpress Flag-p400 and HA-TIP60. (Bottom) Autoradiogram for HAT activities using free histones as a substrate. (C) Coimmunoprecipitation and HAT assays of p400-bound TIP60 that was ectopically coexpressed in 293T cells. HAT activities of HA-TIP60 that was either immunoprecipitated alone by anti-HA antibody (lanes 11 to 13) or coimmunoprecipitated by anti-Flag antibody with coexpressed full-length p400 (lanes 2 to 4), p400(1-1589) (lanes 5 to 7), or p400(1-108)+(1587-3122) (lanes 8 to 10) were compared. Input TIP60 amounts from immunoprecipitates (IP) on M2 agarose or anti-HA agarose were serially increased in both the immunoblot (top) and the HAT (bottom) assays. Relative amounts of Flag-p400 and HA-TIP60 polypeptides in the HAT reactions were determined by immunoblots with anti-Flag and anti-HA antibodies. (D) Immunoblotting of Flag-p400 and HA-TIP60 complexes that were resolved on Sepharose CL6B chromatography. (Top) Sf9 cells were coinfected with two baculoviruses expressing Flag-p400 and HA-TIP60, respectively, and Sf9 cell lysates were analyzed on Sepharose CL6B. (Bottom) As a control, the same analysis was conducted with an extract from Sf9 cells infected with a single baculovirus expressing HA-TIP60. Fractions were subjected to immunoblotting with the indicated antibodies. Higher fraction numbers represent lower molecular masses. N.S., not significant. (E) SDS-PAGE/silver stain analysis of mock control, HA-TIP60, and HA-TIP60+Flag-p400 preparations (from fractions 1 to 3) from the Sepharose CL6B gel filtration analysis described above (D) following affinity purification on anti-HA agarose (HA-TIP60) or M2 agarose (Flag-p400+HA-TIP60). (F) HAT activities of HA-TIP60 and Flag-p400+HA-TIP60. (Bottom) Increasing amounts of the purified materials used in E were analyzed for HAT activities using free core histones as a substrate. (Top) The relative amounts of HA-TIP60 and Flag-p400 in the HAT reaction mixtures were determined by immunoblotting with anti-HA and anti-Flag antibodies.
To understand the basis for the apparent suppression of HAT activity in the TIP60-containing p400 complex, we asked whether a direct one-to-one association between TIP60 and p400 results in TIP60 HAT inhibition. For this purpose, and as a control, we first showed that a preparation of HA-tagged TIP60, which was purified on anti-HA agarose following ectopic expression in 293T cells and (based on immunoblotting) deficient in p400, exhibited dose-dependent HAT activity with core histones as a substrate (Fig. 1C, lanes 11 to 13). We then coexpressed HA-TIP60 and either Flag-tagged p400 or p400 fragments and purified the corresponding HA-TIP60-containing Flag-p400/fragment complexes on M2 agarose. In contrast to purified HA-TIP60, and even when comparable amounts of HA-TIP60 (based on immunoblotting) were present in the HAT assay mixtures, the HA-TIP60-containing Flag-p400/fragment complexes showed no detectable HAT activity (Fig. 1C).
Since the TIP60-p400 association shown in a cell-based coimmunoprecipitation assay could be mediated indirectly through bridging proteins, we sought to determine whether there is a direct physical interaction between these two enzymes. For this purpose, Sf9 cells were coinfected with baculoviruses that independently express Flag-p400 and HA-TIP60, and cell lysates were analyzed by Sepharose CL6B gel filtration (Fig. 1D, top). As revealed by immunoblots with anti-Flag and anti-HA antibodies, Flag-p400 coeluted with a portion of HA-TIP60 in fractions (fractions 1 to ∼3) corresponding to higher molecular masses. In contrast, when expressed without Flag-p400 in Sf9 cells, HA-TIP60 was no longer detectable in high-molecular-mass fractions (Fig. 1D, bottom), suggesting that the molecular mass shift of TIP60 shown by gel filtration chromatography is mediated by a direct interaction with coexpressed p400 and not by related endogenous insect proteins.
From the gel filtration fractions containing Flag-p400 and HA-TIP60 (fractions 1 to ∼3) (Fig. 1D), we prepared a presumptive Flag-p400 and HA-TIP60 (Flag-p400+HA-TIP60) heterodimer by M2 agarose immunopurification. As a control, Sf9 cells that were infected with either mock or HA-TIP60 baculoviruses were used for mock and HA-TIP60 preparations, respectively (Fig. 1E). An SDS-PAGE analysis with silver staining revealed two prominent bands corresponding to Flag-p400 and HA-TIP60, in addition to several nonspecific bands, in the Flag-p400+HA-TIP60 preparation. This suggests that Flag-p400 is associated directly with HA-TIP60 in the Flag-p400+HA-TIP60 preparation. Next, the HAT activities of the baculovirus-expressed/affinity-purified proteins were compared. The HA-TIP60-only preparation displayed robust, dose-dependent HAT activity, whereas the Flag-p400+HA-TIP60 preparation failed to show detectable histone acetylation activity above the background level (Fig. 1F). Taken together, these results demonstrate a p400 complex subpopulation that contains TIP60 in an inactive form as a result of a direct interaction between the TIP60 and p400 proteins.
A SANT domain-containing p400 fragment interacts with TIP60 and reduces TIP60-enhanced basal p21 promoter activity.
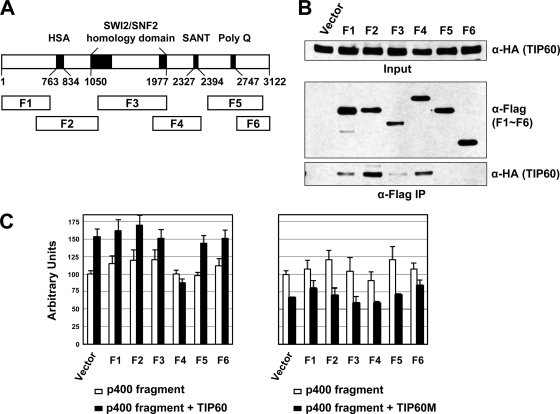
Because the TIP60 HAT activity was suppressed only when TIP60 was associated with p400, we sought to identify a specific p400 region that could inhibit the HAT activity of TIP60. To this end, vectors expressing six different Flag-tagged p400 fragments (fragment 1 [F1] to F6) were constructed for further analyses involving coexpression with HA-TIP60 in 293T cells followed by coimmunoprecipitation and HAT assays (Fig. 2A). Under conditions where comparable amounts of the six Flag-p400 fragments (scored by anti-Flag immunoblotting) were immunoprecipitated, the levels of coimmunoprecipitated HA-TIP60 (scored by anti-HA immunoblotting) indicated direct or indirect interactions with all three fragments (F1 to F3) spanning the N-terminal half of p400 (Fig. 2B). These results are consistent with previous findings showing that the p400 region spanning the helicase/SANT-associated (HSA) and SWI2/SNF2 homology domains (Fig. 2A) is critical for interactions with various subunits of the TIP60 complex (1, 8). Importantly, of the C-terminal p400 fragments, only fragment 4, containing the SANT domain, displayed a strong interaction with TIP60.
FIG. 2.
Identification of a p400 fragment that binds to TIP60 and suppresses TIP60-enhanced basal p21 promoter activity. (A) Schematic representation of the p400 polypeptide. The p400 fragments (F1 to F6) used for coimmunoprecipitation mapping and luciferase reporter assays are shown. (B) Association of p400 fragments with TIP60 in 293T cells. HA-TIP60 and the indicated Flag-p400 fragments were coexpressed in 293T cells, and after immunopurification on M2 agarose, bound Flag-p400 fragments and associated HA-TIP60 were monitored by immunoblots with anti-Flag (middle) and anti-HA (bottom) antibodies, respectively. (C) Effect of p400 fragments on TIP60-enhanced p21 promoter-driven luciferase activity. HCT116 cells were cotransfected with a p21 promoter-driven reporter plasmid (pWWP-luc) and plasmids encoding the indicated p400 fragments in the presence or absence of ectopic TIP60 expression. Plasmids expressing wild-type TIP60 (left) or HAT mutant TIP60 (right) were used for ectopic TIP60 expression. After normalization to an internal control reporter, the luciferase activity obtained from a vector control was set to 100, and relative values of each p400 fragment were then calculated.
Since p400 appears not to be required for DNA damage-induced p53 target gene activation, its repressive role in p53-dependent transcription is evident mainly under basal, nonstressed conditions (31). In addition, because the role of p400 in determining apoptotic versus growth arrest pathways depends on its ability to repress basal p21 gene expression, we chose the p21 promoter for studying the opposing functions of p400 and TIP60. To identify a p400 fragment that could prevent or reduce the coactivator function of TIP60, p21 promoter- driven luciferase assays were conducted by transiently coexpressing TIP60 and Flag-p400 fragments in HCT116 cells (Fig. 2C, left). The ectopic expression of TIP60 with an empty vector control induced a modest increase (∼50%) in basal (noninduced) p21 promoter activity. The ectopic expression of p400 fragments other than F4 did not significantly affect either basal p21 promoter activity or TIP60-enhanced p21 promoter activity. In contrast, the ectopic expression of F4 resulted in a loss of the ectopic TIP60 coactivator activity that consistently increases the basal p21 promoter activities in our cell-based reporter assay. As a control, the ectopic expression of a TIP60 HAT mutant resulted in a modest decrease, rather than an increase, in basal p21 promoter activity regardless of the p400 fragments that were coexpressed (Fig. 2C, right). Together with the coimmunoprecipitation results, these observations raised the possibility that the SANT domain in F4 might inhibit TIP60 coactivator function by a direct interaction.
The p400 SANT domain directly binds the TIP60 HAT domain and represses the HAT activity and associated coactivator function of TIP60.
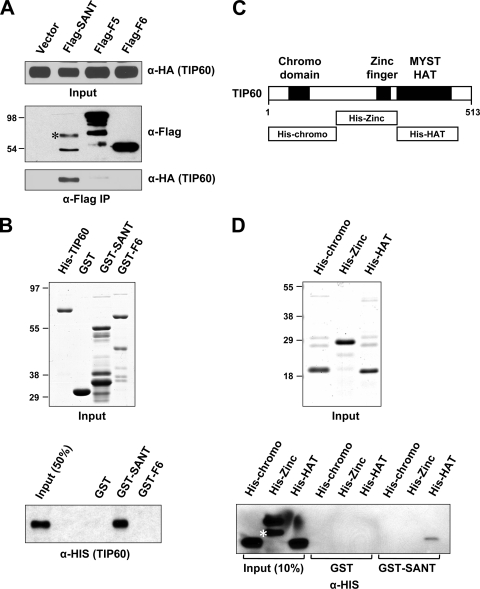
To further investigate the function of the p400 SANT domain, a smaller (311-residue) Flag-tagged SANT fragment (residues 2170 to 2480) was expressed in 293T cells and examined for interactions with coexpressed HA-TIP60 by a coimmunoprecipitation assay (Fig. 3A). This small fragment was expressed poorly relative to control F5 and F6 but nonetheless showed a strong and selective association with HA-TIP60. In a further analysis, His-tagged TIP60 and GST-SANT (residues 2260 to 2480) fusion proteins were purified from Escherichia coli cells (Fig. 3B, top) and employed in a GST pulldown assay. In contrast to the GST and GST-F6 control proteins, the GST-SANT polypeptide showed a robust binding of His-TIP60 (monitored by an anti-His immunoblotting) (Fig. 3B, bottom). These results indicate that the p400 SANT domain can bind directly to TIP60 in vitro without other bridging proteins.
FIG. 3.
Identification of the TIP60 HAT domain as a direct target of the p400 SANT domain. (A) Association of TIP60 with the p400 SANT domain in 293T cells. HA-TIP60 was coexpressed with Flag-p400 fragments (indicated at the top), and proteins bound to M2 agarose (anti-Flag immunoprecipitation) were analyzed by immunoblotting with anti-Flag and anti-HA antibodies. A nonspecific immunoblot signal is marked by an asterisk. (B) Direct binding of TIP60 to the p400 SANT domain. Recombinant His-tagged TIP60 and GST-p400 fragments were expressed in E. coli cells; purified through Ni-nitrilotriacetic acid (NTA) and glutathione-Sepharose 4B resins, respectively; analyzed by SDS-PAGE with Coomassie staining (top); and used in GST pulldown assays (bottom). His-TIP60 binding to GST or GST fusion proteins was scored by immunoblotting with an anti-His antibody. (C) Schematic representation of TIP60. Three TIP60 fragments corresponding to the chromodomain, the zinc finger, and the HAT domain are shown. (D) Binding of TIP60 fragments to the p400 SANT domain. The TIP60 fragments shown in C were purified from E. coli cells, analyzed by SDS-PAGE with Coomassie staining (top), and used in GST pulldown assays (bottom). TIP60 binding to GST-SANT was detected by immunoblotting with an anti-His antibody. A degraded His-chromodomain is indicated by an asterisk.
TIP60 contains three conserved domains corresponding to a chromodomain, a C2HC zinc finger, and a HAT domain (Fig. 3C) (34). To identify the TIP60 domain that binds to the p400 SANT domain, GST pulldown assays were conducted with recombinant GST-SANT (Fig. 3B, top) and His-tagged TIP60 fragments (Fig. 3D, top) that were purified from E. coli. Immobilized GST-SANT specifically bound the TIP60 HAT domain, whereas the TIP60 chromodomain- and zinc finger-containing fragments failed to show any interaction (Fig. 3D, bottom).
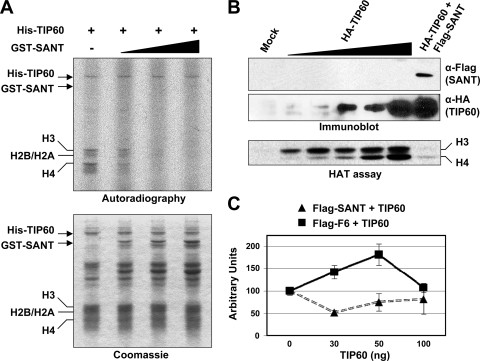
Given the demonstration of a direct interaction between the p400 SANT domain and the TIP60 HAT domain, we next asked whether this interaction affects the TIP60 HAT activity. To reconstitute the inhibitory activity of the SANT domain in vitro, purified His-tagged TIP60 and GST-SANT proteins (Fig. 3B, top, and 4A, bottom) were tested for HAT activity on core histones (Fig. 4A, top). Recombinant His-TIP60 alone acetylated free core histones, whereas the addition of the SANT domain to the HAT reactions significantly compromised TIP60-dependent HAT activity in a dose-dependent manner. Interestingly, the SANT domain did not inhibit TIP60 autoacetylation activity in vitro, suggesting that the inhibition by the SANT domain could be substrate specific (Fig. 4A, top). To validate the SANT domain-dependent inhibition of TIP60 HAT activity in vivo, 293T cells were transfected with vectors expressing either HA-TIP60 alone or both HA-TIP60 and Flag-SANT, and preparations of HA-TIP60 or HA-TIP60 with associated Flag-SANT were purified from cellular extracts on anti-HA agarose or M2 agarose, respectively, and subjected to comparative HAT assays. Quite significantly, the HA-TIP60-only preparation showed robust HAT activity with free core histones as a substrate, whereas the HA-TIP60 preparation with associated Flag-SANT, at a comparable TIP60 input, showed a drastically reduced level of HAT activity (Fig. 4B). Thus, the SANT domain efficiently binds to TIP60 and represses its intrinsic HAT activity.
FIG. 4.
p400 SANT domain-dependent inhibition of TIP60 HAT activity and coactivator function. (A) Reconstitution of SANT domain-mediated inhibition of TIP60 HAT activity with purified proteins. Recombinant His-TIP60 and GST-SANT proteins were purified as described in the legend of Fig. 3B. (Top) His-TIP60 was subjected to a HAT assay, with purified core histones as a substrate, in the presence of increasing amounts of GST-SANT. (Bottom) Amounts of recombinant proteins used in the assays are shown in the Coomassie-stained gel. (B) Inhibition of p400 SANT domain-associated TIP60 HAT activity. Ectopic HA-TIP60 was expressed either alone or with Flag-SANT in 293T cells, and the corresponding proteins were purified from cellular extracts on anti-HA agarose (HA-TIP60) or M2 agarose (HA-TIP60 plus Flag-SANT). The amounts of HA-TIP60 from the anti-HA immunoprecipitates were serially increased in the HAT and immunoblot assays for comparison with the HA-TIP60 that is associated with Flag-SANT. Input TIP60 amounts were determined by immunoblotting with an anti-HA antibody (middle), Flag-SANT was monitored by immunoblotting with an anti-Flag antibody (top), and HAT activities were scored by autoradiogram (bottom). (C) SANT domain inhibition of TIP60-mediated coactivator function on basal p21 promoter activity. HCT116 cells were cotransfected with either Flag-SANT or Flag-F6, the pWWP-luc reporter, and increasing amounts (as indicated) of the TIP60 expression plasmid. The luciferase activity obtained without ectopic TIP60 expression was set to 100.
Given the repressive effect of the SANT domain on TIP60 HAT activity, we next tested the ability of the ectopically expressed SANT domain to repress the TIP60-dependent enhancement of basal p21 promoter activity. To this end, HCT116 cells were cotransfected with the p21 promoter-driven luciferase reporter and plasmids expressing HA-TIP60 and either the Flag-SANT or Flag-F6 p400 fragment (Fig. 4C). The ectopic expression of TIP60 and the Flag-F6 control effected a nearly 2-fold increase in basal p21 promoter activity, whereas the coexpression of the SANT domain completely blocked the ectopic TIP60-dependent activity of the p21 promoter and further lowered the overall basal activity as well. Taken together, these results demonstrate that the p400 SANT domain binds directly to the TIP60 HAT domain and represses both the HAT activity and the associated coactivator function of TIP60 in basal p21 promoter activity.
ATPase-defective p400 derepresses basal p21 gene expression.
Since our results suggest a novel mechanism for the p400-mediated repression of basal p21 promoter activity, we sought to reconfirm data from previous reports showing that the ATPase-dependent function of p400 is also required for the repression of basal p21 promoter activity. Previous findings indicated a p400 requirement both for p53-dependent histone variant H2A.Z localization at the p21 promoter and for the repression of basal p53-dependent p21 gene transcription as well as a requirement of H2A.Z for p21 repression (9). However, these findings did not clearly demonstrate that the ATPase activity of p400 is required for p21 repression. To investigate requirements for the ATPase activity of p400, and based on the inactivation of the homologous SWR1 ATPase by the same mutation (21), a K1085G mutation was introduced into the ATP binding motif of the SWI2/SNF2 homology domain. The wild-type and ATPase-defective p400 proteins were expressed in Sf9 cells via baculovirus vectors, purified on M2 agarose, and tested for their ability to catalyze H2A.Z deposition into a mononucleosome. By incubating the wild-type Flag-p400 preparation with immobilized mononucleosomes, a Flag-H2A.Z+H2B heterodimer, and ATP, the transfer of Flag-H2A.Z to the nucleosome was detected by an anti-Flag immunoblot of the recovered mononucleosome (Fig. 5A). In contrast, no significant H2A.Z deposition was observed with the mock control or ATPase-defective p400 preparations. These results clearly indicate that the enzymatic activity of p400 is crucial for the catalysis of H2A.Z histone variant deposition into nucleosomes. On the other hand, an ATPase- defective p400 preparation that failed to catalyze H2A.Z deposition retained a strong helicase activity in vitro, suggesting that tightly associated contaminants, possibly insect-derived TIP49/TIP48 ATPases, play a dominant role in the observed helicase activity (data not shown) (8).
FIG. 5.
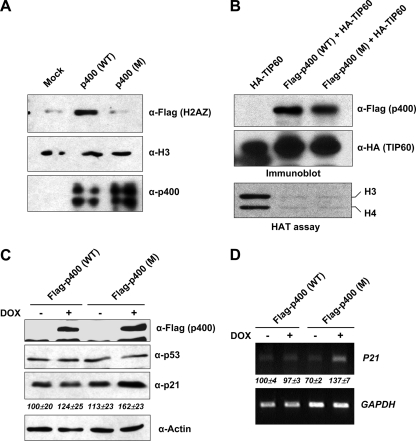
p400 ATPase function in basal p21 gene expression. (A) H2A.Z histone variant exchange assays. The assay, detailed in Materials and Methods, employed recombinant mononucleosomes and either wild-type (WT) or ATPase mutant (M) p400 proteins prepared from baculovirally infected Sf9 cells. Flag-H2A.Z incorporation into mononucleosomes was detected by immunoblotting with an anti-Flag antibody. (B) Comparison of TIP60 HAT activities associated with wild-type and mutant p400 proteins. (Bottom) A Flag-p400 preparation from 293T cells that coexpressed ectopic Flag-p400 (wild type or mutant) and HA-TIP60 was used for HAT assays. An HA-TIP60 preparation from 293T cells that expressed HA-TIP60 alone was used as a positive control. (Top) Relative amounts of p400 and TIP60 proteins in the HAT reactions were determined by immunoblots with anti-Flag and anti-HA antibodies. (C) Endogenous p21 protein expression. U2OS-TetOn cells were infected with lentivirus encoding wild-type (WT) or ATPase mutant (M) Flag-p400 proteins and selected with puromycin for 1 week. Flag-p400 expression was induced by doxycycline (DOX) (1 μg/ml) for 48 h, and cell extracts were analyzed by immunoblots with the indicated antibodies. Analyses of the expression levels of the p21 protein, normalized to actin, were carried out in triplicate, and average values are indicated. (D) Endogenous p21 mRNA expression. Total RNA from U2OS-TetOn cells used in C were analyzed by semiquantitative reverse transcription (RT)-PCR for p21 mRNA expression. Analyses of the expression levels of p21 mRNA, normalized to the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene, were carried out in triplicate, and average values are indicated.
The ATPase-defective p400 mutant retained the ability to bind TIP60 and was as effective as wild-type p400 in repressing the HAT activity of TIP60 in vitro (Fig. 5B). However, we were unable to establish a repressive effect of ectopically expressed p400, either wild type or mutant, on the basal promoter activity of a transfected p21 reporter (data not shown). Since reporter gene assays involving transient transfection may not be able to recapitulate chromatin structural features that impose a dependence upon the normal physiological functions of p400, we established U2OS cell lines that inducibly express either wild-type or ATPase mutant forms of p400 and then examined the effects on endogenous p21 gene expression. Following enrichment by antibiotic selection of U2OS cells expressing Flag-p400 proteins, the expression of wild-type and mutant Flag-p400 proteins was induced for 2 days with doxycycline. Both proteins were expressed at comparable levels, as shown by anti-Flag immunoblotting (Fig. 5C). However, as evidenced by both p21 protein (Fig. 5C) and mRNA (Fig. 5D) analyses, mutant Flag-p400 effected a modest upregulation of endogenous p21 gene expression, whereas wild-type p400 did not. The observed upregulation of basal p21 gene expression, presumably due to a dominant negative effect of mutant p400, is mediated by a transcriptional derepression mechanism since it was not accompanied by an increase in the p53 protein level (Fig. 5C). Our results thus indicate that the repression of basal p21 transcription by p400 requires both its ATPase activity and its TIP60-inhibitory activity.
p400 is evicted from the p21 promoter after DNA damage.
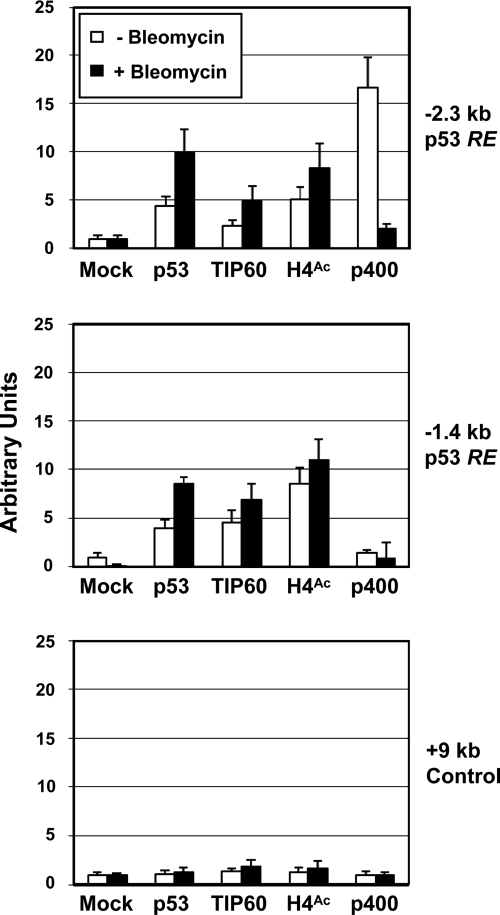
p400 and TIP60 play antagonistic roles in regulating p53 downstream target genes (31). It has been shown that in IMR-90 human fibroblast and U2OS osteosarcoma cells, p400 is prebound to the p53 binding sites in the p21 promoter during gene repression and released upon DNA damage-induced, p53-dependent transcriptional activation (4, 9). To confirm a reciprocal recruitment of p400 and TIP60 to p53 binding sites (kb −2.3 and −1.4) during p21 gene activation, chromatin immunoprecipitation assays were employed. Under normal growth conditions, U2OS cells have low basal levels of p53 binding to the p53 binding sites, but bleomycin-mediated DNA damage induced a significant increase in levels of p53 binding to both proximal and distal p53 binding sites (Fig. 6). DNA damage also effected a modest increase in TIP60 binding and histone H4 hyperacetylation at the p53 binding sites. p400 showed a higher level of binding to the distal p53 binding site during the normal cell cycle, but the level decreased to a basal level following DNA damage. A control region 9 kb downstream of the p21 promoter showed no significant binding by any of the analyzed proteins either before or after DNA damage. These results suggest a potential mechanism in which the dissociation of p400 leads not only to the relief of H2A.Z-dependent repression but also to an increase in TIP60 HAT activity at the promoter.
FIG. 6.
ChIP analysis of TIP60 and p400 binding during p21 gene activation. U2OS cells that stably express Flag-tagged p400 (polyclonal cells selected by puromycin) were used for chromatin immunoprecipitation analyses before and after bleomycin treatment. The values obtained with the mock antibody (anti-GAL4 antibody) in real-time PCR analyses were set to 1, and relative values were calculated for each of the other antibodies indicated at the bottom of each panel. The endogenous p21 gene regions monitored were the kb −2.3 p53 response element (top), the kb −1.4 p53 response element (RE) (middle), and a kb +9 control region (bottom).
DISCUSSION
TIP60, through its histone acetyltransferase activity, functions as a positive regulator of p53-dependent genes in the DNA damage response pathway, whereas p400, through an ATP-dependent chromatin remodeling activity that facilitates H2A.Z deposition, acts to repress specific genes (p21) in this pathway. Somewhat paradoxically, in view of these antagonistic roles, TIP60 and p400 have been reported to jointly reside in a large multisubunit complex that displays TIP60 activity. However, a report of the presence of p400 in a nearly identical complex lacking detectable TIP60 HAT activity suggested that the TIP60-p400 complex is dynamic. Here, in relation to the antagonistic effects of TIP60 and p400 on basal p21 promoter activity, we have demonstrated and functionally analyzed a TIP60-p400 complex in which the TIP60 HAT activity is repressed by p400 through the direct interaction of their respective HAT and SANT domains. These findings establish a new mechanism of transcriptional repression by p400 and have implications not only for the p53-dependent regulation of the p21 gene but also for gene- and cell-specific functions of TIP60 and p400 within the TIP60-p400 complex.
A direct p400 interaction inhibits TIP60 HAT and coactivator functions.
The previously reported antagonistic functions of p400 and TIP60 in p53 target gene transcription are consistent with, but not necessarily dependent upon, direct interactions of p400 and TIP60, especially since these factors were thought to act through distinct mechanisms (histone acetylation versus chromatin remodeling) (31). Importantly, however, we demonstrate here a direct physical interaction through these factors based on (i) the coelution of p400 and TIP60 upon gel filtration chromatography, (ii) copurification following coexpression from baculovirus vectors in Sf9 cells, and, most convincingly, (iii) the direct binding of TIP60 or the TIP60 HAT domain to the p400 SANT domain in studies with purified proteins. The functional consequences of these interactions are a repression of the in vitro TIP60 HAT activity, as assessed by in vitro HAT assays. This was evidenced both with purified recombinant TIP60-p400 complexes assembled in insect cells, where p400 alone was sufficient for repression, and with purified TIP60-p400 complexes assembled in 293T cells, where other common subunits of the TIP60/p400 complexes might have contributed to the inhibition. In this regard, previous reports implicated the TIP49 and GAS41 subunits in the repression of p21 transcription (4, 23), although it was not established whether this was related to an inhibition of the TIP60 HAT activity or to p400-mediated H2A.Z deposition. Importantly, the SANT domain of p400 proved sufficient, when assembled into a complex with TIP60 in vitro and in vivo, to completely inhibit TIP60 HAT activity (Fig. 4A and B). Note that a SANT motif in the SMRT corepressor was previously implicated in the inhibition of PCAF and CBP HAT activities but most likely through a different mechanism involving its histone tail binding activity (35). Thus, the present results establish a new p400 function, namely, the inhibition of TIP60 HAT activity through a direct interaction of the SANT domain with the TIP60 HAT domain. (Although not further analyzed here, the ability of the p400 N terminus, when assembled into a TIP60-containing complex in 293T cells, to repress the TIP60 HAT activity may reflect a secondary mechanism for the p400 inhibition of TIP60 HAT activity.)
Consistent with these results and with the previously established p53 coactivator function of TIP60 (2, 16, 31), the p400 SANT domain, when overexpressed, was shown to repress TIP60-enhanced basal p21 promoter activity. Importantly, these results establish a new p400-mediated repression mechanism that is complementary to, and may cooperate with, the previously established repression mechanism involving the deposition of H2A.Z (9). In further support of the latter mechanism, the present study also demonstrates an anticipated requirement for the ATPase activity of p400 in repressing basal p21 gene expression.
Dynamic TIP60 and p400 complexes and associated antagonistic functions of TIP60 and p400.
Our results, along with previous reports of a p400-containing TIP60 complex with HAT activity (3, 5), a p400-containing complex lacking TIP60 HAT activity (8), and dynamic changes in p400 and TIP60 associations during the activation of the p21 promoter (4, 9), are consistent with the possibility of dynamic p400 and TIP60 complexes. On the one hand, the TIP60 complex purified through an affinity tag on TIP60 (5) or on other subunits, including YL1 and MRGBP (3), could comprise a mixed population of HAT-inactive p400-containing complexes and HAT-active p400-deficient complexes. On the other hand, the HAT-deficient p400 complex(es) purified with an anti-p400 antibody (8) might contain both a TIP60-deficient complex and a TIP60 complex in which the HAT activity is repressed by the p400 association. Of special note, and whereas the p21 gene is characterized by a high p400-to-TIP60 ratio in the repressed basal state and a high TIP60-to-p400 ratio in the activated (daunorubicin-induced) state (9), our demonstration of a HAT-repressed TIP60-p400 complex allows the possibility of p400-repressed genes that, through the prebinding of a composite TIP60-p400 complex, are poised for rapid activation through a loss or inactivation of p400 and the consequent function of prebound TIP60.
The existence of a pool of p400-repressed TIP60 suggests the possibility of a stress-induced release of p400 with a consequent global increase in levels of TIP60 HAT activity. In this regard, it has been reported that the level of total cellular TIP60 HAT activity (including that due to ATM-associated TIP60) in HeLa cells is increased after exposure to bleomycin without any significant change in the non-ATM-associated TIP60 activity (27). Consistent with this observation, we failed to detect any significant increase in the level of the free non-ATM-associated TIP60 complex purified (through an affinity tag) from cells shortly after DNA damage (data not shown). This finding suggests that DNA damage may not increase the proportion of the active TIP60 complex pool that lacks the p400 module. In this regard, the p400-related yeast Eaf1 protein was shown previously to be critical for the assembly and stability of the corresponding HAT complex (1). Thus, analogous to the situation in yeast, a DNA damage-induced increase in the soluble p400-free TIP60 complex subpopulation may be precluded by the instability of this complex in the free (DNA-unbound) state such that an increased level of this complex may be evident mainly on induced, actively transcribed genes.
TIP60 acts at multiple steps during DNA damage response pathways (25), with documented functions of TIP60-mediated acetylation of (i) ATM in the DNA damage response, (ii) H2AX in the DNA repair process, and (iii) p53 and histones in p53-dependent transcriptional regulation (2, 15, 16, 27-29). The TIP60 level evidently must be tightly controlled during the normal cell cycle since levels that are too high or too low were shown previously to induce growth arrest/apoptosis or tumor formation, respectively (2, 10). Consistent with this notion and with previous and present demonstrations of antagonistic functions of TIP60 and p400, the p400-to-TIP60 ratio in colorectal cancer cells was found to determine cellular sensitivity to chemotherapy (18).
A model based on previous and present data for the reciprocal and antagonistic interaction of the TIP60 and p400 proteins in regulating p53 and the p21 target gene is shown in Fig. 7. This model emphasizes (i) the TIP60-mediated acetylation of p53 at lysine 120, which facilitates p53-mediated apoptosis over growth arrest (28, 29); (ii) the TIP60-dependent acetylation of histones at p53-activated promoters, which is correlated with gene activation (7, 25, 30); (iii) the p400-mediated deposition of H2A.Z on the p21 promoter, which contributes to the repression of p21 (9); (iv) based on findings reported for the yeast Esa1 homolog of TIP60 (20), a possible acetylation of H2A.Z at lysine 14 that relieves the inhibiting effect of H2A.Z; and (v) based on the present studies, a direct p400-mediated inhibition of TIP60 HAT and coactivator functions. This model reemphasizes the complementary and potentially cooperative functions of p400-mediated H2A.Z deposition and p400 inhibition of TIP60 in the repression of basal p21 gene expression.
FIG. 7.
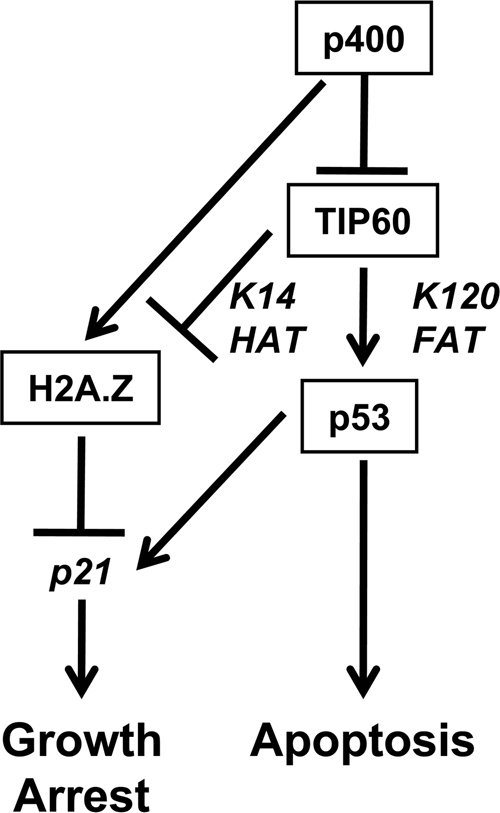
Dual-repression model of p400 in the regulation of p21 gene expression. HAT and FAT indicate histone (H2A.Z) and factor (p53) acetyltransferase activities of TIP60, respectively. For further details, see the text.
Promoter recruitment of TIP60 and p400 complexes with both gene- and cell-specific functions.
The existence of heterogeneous TIP60 and p400 complexes raises questions about which complexes are recruited to repressed versus activated genes as well as potential changes in promoter complexes during transitions from repressed to active states. In the case of the well-studied p21 promoter, the transition from a repressed to an activated state is typified by a loss of promoter-associated p400 and a gain of TIP60 (9). While clearly indicative of dynamic changes at the promoter, it is not yet clear whether this reflects dynamic changes in the same promoter-bound complex or the exchange of TIP60-deficient p400 complexes with p400-deficient TIP60 complexes. Previous results indicating that p400-containing TIP60 complexes represent a major subpopulation of the total pool of TIP60 complexes (5, 8) and our current demonstration of a TIP60-p400 complex in which TIP60 is repressed also suggest the possibility of at least a partial occupancy of the repressed p21 promoter by both TIP60 and p400, and other p400-repressed promoters (6, 31) might well show a greater occupancy by complete TIP60-p400 complexes in which p400 both represses the HAT activity and facilitates repression-associated H2AZ deposition. As mentioned above, this could facilitate a more rapid transition to an activated state dependent upon TIP60.
Another important question concerns the possible gene and cell type specificity of TIP60 and p400 functions. In relation to p53 target genes, the repression by p400 appears to be gene specific (31). Moreover, it remains to be determined whether the recruitment of the TIP60-deficient p400 complex and H2A.Z deposition on the repressed p21 promoter, with a subsequent transition to a p400-deficient TIP60 complex in the active state, can be generalized to other p400-repressed p53 target genes. Both gene-specific and cell-specific effects of p400 and TIP60 as well as variable p400 and TIP60 functions are also apparent from other studies indicating, for example, that p400 and TIP60 may be jointly required for either activated (17) or repressed (6) promoters. Such results indicating clear context effects suggest the possibility that the p400-mediated TIP60 inhibition found in the free TIP60-p400 complex pool described here could be relieved by unknown gene-specific mechanisms on promoter-bound complexes or that there may be an independent, stochastic recruitment of p400 and p400-deficient TIP60 complexes to the same promoter.
Acknowledgments
We thank Janice Ascano for critical reading of the manuscript, members of Roeder laboratory for fruitful discussions, Wei-Yi Chen for human H2A.Z cDNA, Jaehoon Kim for advice on baculovirus construction, and Michael Cole for CβS expression vectors.
This work was supported by grants from the NIH (CA129325 and DK071900), the Leukemia and Lymphoma Society (SCOR grant 7132-08), and the Starr Cancer Consortium (I2-A88) to R.G.R. and by a MURF grant to J.H.P.
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Auger, A., L. Galarneau, M. Altaf, A. Nourani, Y. Doyon, R. T. Utley, D. Cronier, S. Allard, and J. Cote. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell. Biol. 28:2257-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns, K., E. M. Hijmans, J. Mullenders, T. R. Brummelkamp, A. Velds, M. Heimerikx, R. M. Kerkhoven, M. Madiredjo, W. Nijkamp, B. Weigelt, R. Agami, W. Ge, G. Cavet, P. S. Linsley, R. L. Beijersbergen, and R. Bernards. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431-437. [DOI] [PubMed] [Google Scholar]
- 3.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665-13670. [DOI] [PubMed] [Google Scholar]
- 4.Chan, H. M., M. Narita, S. W. Lowe, and D. M. Livingston. 2005. The p400 E1A-associated protein is a novel component of the p53→p21 senescence pathway. Genes Dev. 19:196-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fazzio, T. G., J. T. Huff, and B. Panning. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134:162-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 9.Gevry, N., H. M. Chan, L. Laflamme, D. M. Livingston, and L. Gaudreau. 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21:1869-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorrini, C., M. Squatrito, C. Luise, N. Syed, D. Perna, L. Wark, F. Martinato, D. Sardella, A. Verrecchia, S. Bennett, S. Confalonieri, M. Cesaroni, F. Marchesi, M. Gasco, E. Scanziani, M. Capra, S. Mai, P. Nuciforo, T. Crook, J. Lough, and B. Amati. 2007. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448:1063-1067. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, P. D. 2001. Transcription and chromatin converge: lessons from yeast genetics. Curr. Opin. Genet. Dev. 11:142-147. [DOI] [PubMed] [Google Scholar]
- 12.Horn, P. J., and C. L. Peterson. 2002. Chromatin higher order folding—wrapping up transcription. Science 297:1824-1827. [DOI] [PubMed] [Google Scholar]
- 13.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 14.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 15.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 16.Legube, G., L. K. Linares, S. Tyteca, C. Caron, M. Scheffner, M. Chevillard-Briet, and D. Trouche. 2004. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 279:44825-44833. [DOI] [PubMed] [Google Scholar]
- 17.Martinato, F., M. Cesaroni, B. Amati, and E. Guccione. 2008. Analysis of Myc-induced histone modifications on target chromatin. PLoS One 3:e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattera, L., F. Escaffit, M. J. Pillaire, J. Selves, S. Tyteca, J. S. Hoffmann, P. A. Gourraud, M. Chevillard-Briet, C. Cazaux, and D. Trouche. 2009. The p400/Tip60 ratio is critical for colorectal cancer cell proliferation through DNA damage response pathways. Oncogene 28:1506-1517. [DOI] [PubMed] [Google Scholar]
- 19.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 20.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 22.Nakano, K., T. Mizuno, Y. Sowa, T. Orita, T. Yoshino, Y. Okuyama, T. Fujita, N. Ohtani-Fujita, Y. Matsukawa, T. Tokino, H. Yamagishi, T. Oka, H. Nomura, and T. Sakai. 1997. Butyrate activates the WAF1/Cip1 gene promoter through Sp1 sites in a p53-negative human colon cancer cell line. J. Biol. Chem. 272:22199-22206. [DOI] [PubMed] [Google Scholar]
- 23.Park, J. H., and R. G. Roeder. 2006. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol. Cell. Biol. 26:4006-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samuelson, A. V., M. Narita, H. M. Chan, J. Jin, E. de Stanchina, M. E. McCurrach, M. Fuchs, D. M. Livingston, and S. W. Lowe. 2005. p400 is required for E1A to promote apoptosis. J. Biol. Chem. 280:21915-21923. [DOI] [PubMed] [Google Scholar]
- 25.Squatrito, M., C. Gorrini, and B. Amati. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16:433-442. [DOI] [PubMed] [Google Scholar]
- 26.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 27.Sun, Y., X. Jiang, S. Chen, N. Fernandes, and B. D. Price. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U. S. A. 102:13182-13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sykes, S. M., H. S. Mellert, M. A. Holbert, K. Li, R. Marmorstein, W. S. Lane, and S. B. McMahon. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24:841-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, Y., J. Luo, W. Zhang, and W. Gu. 2006. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24:827-839. [DOI] [PubMed] [Google Scholar]
- 30.Taubert, S., C. Gorrini, S. R. Frank, T. Parisi, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 24:4546-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyteca, S., M. Vandromme, G. Legube, M. Chevillard-Briet, and D. Trouche. 2006. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 25:1680-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, G. G., C. D. Allis, and P. Chi. 2007. Chromatin remodeling and cancer, part I: covalent histone modifications. Trends Mol. Med. 13:363-372. [DOI] [PubMed] [Google Scholar]
- 33.Wang, G. G., C. D. Allis, and P. Chi. 2007. Chromatin remodeling and cancer, part II: ATP-dependent chromatin remodeling. Trends Mol. Med. 13:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, X. J. 2004. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32:959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J., Y. Li, T. Ishizuka, M. G. Guenther, and M. A. Lazar. 2003. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 22:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]