Abstract
Genetic programs that govern neural stem/progenitor cell (NSC) proliferation and differentiation are dependent on extracellular cues and a network of transcription factors, which can be regulated posttranslationally by phosphorylation. However, little is known about the kinase-dependent pathways regulating NSC maintenance and oligodendrocyte development. We used a conditional knockout approach to target the murine regulatory subunit (beta) of protein kinase casein kinase 2 (CK2β) in embryonic neural progenitors. Loss of CK2β leads to defects in proliferation and differentiation of embryonic NSCs. We establish CK2β as a key positive regulator for the development of oligodendrocyte precursor cells (OPCs), both in vivo and in vitro. We show that CK2β directly interacts with the basic helix-loop-helix (bHLH) transcription factor Olig2, a critical modulator of OPC development, and activates the CK2-dependent phosphorylation of its serine-threonine-rich (STR) domain. Finally, we reveal that the CK2-targeted STR domain is required for the oligodendroglial function of Olig2. These findings suggest that CK2 may control oligodendrogenesis, in part, by regulating the activity of the lineage-specific transcription factor Olig2. Thus, CK2β appears to play an essential and uncompensated role in central nervous system development.
Casein kinase 2 (CK2) is a conserved serine/threonine kinase with more than 300 substrates, mostly proteins related to transcription-directed signaling (27). CK2 is a heterotetrameric holoenzyme formed by two catalytic subunits, α and α′, that associate with a dimeric building block of regulatory β subunits (α2β2). CK2β modulates the substrate specificity of the CK2 enzymatic activity, and its architecture is consistent with its role as a docking partner for other interacting proteins (4). We previously demonstrated that disruption of CK2β function in mice results in postimplantation lethality. Moreover, many attempts to generate CK2β−/− embryonic stem (ES) cells failed, suggesting an essential role of mammalian CK2β for ES cell viability (2).
The function of CK2β for cell cycle progression has been investigated, but its precise role is largely unknown. The cell proliferation function of CK2β was first characterized in human fibroblasts, by use of antisense oligodeoxynucleotides and microinjection of specific antibodies (19, 26). Recently, a genome-wide survey of protein kinases required for cell progression into cultured Drosophila melanogaster S2 cells by double-stranded RNA demonstrated that CK2β is required for centrosomal normality (1). In the same vein, downregulation of CK2β by small interfering RNA (siRNA) results in delayed cell cycle progression in cultured mammalian cells (42). Finally, in a genetic screen for mutations affecting the central brain of Drosophila, a hypomorphic allele of D. melanogaster CK2β has been isolated, and that study suggested a role for CK2β in cell proliferation or cell survival during brain development (15).
In the developing mouse brain, neural stem/progenitor cells (NSCs), which reside in the ventricular zone (VZ), give rise to neurons and glial cells (astrocytes and oligodendrocytes) (23). Chronologically, neurons are generated first, and glial cells are generated subsequent to neurogenesis. Oligodendrocytes originate from ventral neural progenitors in the VZ, and once committed, they migrate laterally and dorsally as parenchymal precursors (oligodendrocyte precursor cells [OPCs]) to populate the entire embryonic telencephalon, where they terminally differentiate (reviewed in reference 29). Distinct genetic programs using combinations of transcription factors govern multiple spatiotemporal origins of oligodendroglial specification (reviewed in reference 28). The basic helix-loop-helix (bHLH) transcription factor Olig2 plays an essential role in the embryonic specification of neural progenitors into the oligodendrocyte lineage, and Olig2 gene function is absolutely required for OPC development (17, 21, 36, 43). Accordingly, Olig2 expression is detected first in ventral neural progenitors, precedes OPC-specific expression markers, and is maintained at all stages of oligodendrocyte development (29). It has been reported that the oligodendroglial function of Olig2 can be modulated by protein kinase AKT-mediated phosphorylation of its N-terminal domain (31). Interestingly, neural bHLH transcription factors have been described as being phosphorylated and regulated by the CK2 holoenzyme (9, 39). In the mouse brain, CK2 subunits (α, α′, and β) are expressed in all neural cells (A. Kolding and B. Boldyreff, unpublished data) in which the CK2β-to-CK2α-plus-CK2α′ ratio is largely elevated compared to that in other organs (41), suggesting a higher-threshold requirement in the amount of the holoenzyme in the central nervous system (CNS). All together, these observations suggest that CK2β may modulate neural homeostasis.
In this study, we have used Cre/loxP-mediated recombination to generate mice with a CK2β null allele in NSCs of the developing brain. Disruption of CK2β results in inhibition of NSC proliferation and in a severe deficiency for progenitors to specify OPCs. In addition, we identified, in vitro, the serine-threonine-rich (STR) domain of Olig2 as a strict CK2β-dependent, CK2-targeted substrate. We also show that the STR domain of Olig2 is a modulator of its oligodendroglial activity when tested in neurosphere assays, suggesting that CK2β could mediate its effect via Olig2 oligodendroglial activity. Hence, these findings provide insight into the genetic basis underlying neural progenitor proliferation and oligodendrocyte development.
MATERIALS AND METHODS
Mouse breeding.
Mice were of a C57BL/6 background. Nestin-cre transgenic mice (37) kept as heterozygotes were bred with CK2βloxP/loxP mice (2). Double-heterozygous mice (CK2βloxP/wt; Nestin-cre) were bred back to CK2βloxP/loxP mice to produce the genotypes used in this study, (i) CK2βloxP/wt, (ii) CK2βloxP/loxP, (iii) CK2βloxP/wt, Nestin-cre, and (iv) mutant CK2βloxP/loxP, Nestin-cre (referred to here as CK2β−). CK2βloxP/wt, CK2βloxP/loxP, and CK2βloxP/wt, Nestin-cre embryonic day 18.5 (E18.5) embryo phenotypes were identical and are referred to here as wild type (WT). Live-born mice, postnatal day 0 (P0) pups, and E18.5 embryos were genotyped by tail DNA PCR (2). Animal treatment was performed in accordance with the ethics committee (ComEth) of Grenoble, France.
Western blot analysis.
Olfactory bulb, cerebellum, and meningeal cells were removed from dissected brains. Forebrain protein extracts were obtained after homogenization and sonication in 25 mM Tris (pH 8.5)-1 mM dithiothreitol (DTT)-200 mM NaCl-5 mM EDTA. Cells were lysed in 50 mM Tris (pH 7.5)-5 mM EDTA-500 mM NaCl-1% Triton X-100. Extractions were done with protease and phosphatase inhibitor cocktails (Sigma). Samples (40 μg) were analyzed by 12% SDS-PAGE and processed for Western blot analysis. Antibodies used were rabbit anti-CK2β (1/500, directed against the C terminus of CK2β), rabbit anti-CK2α (1/1,000; Calbiochem), mouse anti-β-tubulin class III isoform (1/1,000; Chemicon), rabbit anti-poly(ADP-ribose) polymerase (anti-PARP) antibody (1/1,000; Cell Signaling), and mouse antihemagglutinin (anti-HA) antibody (clone 12CA5, 1/1,000; Roche).
Histological, immunochemical, and in situ hybridization analyses.
Embryos were perfused intracardially with 4% paraformaldehyde (PFA). Dissected brains were postfixed overnight at 4°C and processed for paraffin or OCT embedding. Paraffin samples were cut at 5 μm and used for hematoxylin and eosin (H&E) or for immunochemical analysis. In order to detect proliferating cells in S phase (bromodeoxyuridine [BrdU] labeling), a 1-h short pulse was performed as described previously (2). Paraffin-embedded sections were incubated with an anti-BrdU rat monoclonal antibody (1/75; Harlan, Indianapolis, IN). Mitotic cell activities were detected with a rabbit antibody, anti-Ser10 phospho-H3 (PH3, 1/2,000; Upstate). Frozen sections (16 μm) were used for immunostaining. Cryosections were incubated in a blocking solution (phosphate-buffered saline [PBS]-0.2% Triton X-100-5% normal goat serum) for 20 min and with a mouse IgG blocking reagent (Vector) for 1 h. The primary antibodies used were mouse anti-RC2 (1/10; Hybridoma Bank, University of Iowa), rat anti-GFAP (1/500; U.S. Biological), rabbit anti-NG2 antibody (1/100; Chemicon), rat anti-PECAM antibody (1/2; gift from A. Vecchi, Milan, Italy), rabbit anti-Olig2 antibody (1/20,000; gift from C. Stiles, Boston, MA), and mouse anti-Ki67 (clone MM1, 1/100; Novocastra). Primary antibodies were diluted in PBS-1% normal goat serum, followed by appropriate cyanin-3 dye-conjugated, cyanin-2 dye-conjugated (1/500; Jackson ImmunoResearch Laboratories), Alexa488 dye-conjugated (1/500; Molecular Probe), or horseradish peroxidase (HRP) 3′,3′-diaminobenzidine staining system secondary antibodies. Cells were stained with Hoechst dye 33342 (2 μg/ml) to visualize nuclei. Images were acquired using a Zeiss fluorescent microscope (Axiovert 200 M) with 16× and 40× objectives. Images were combined for figures by using Adobe Photoshop 8.0. In situ hybridization using antisense, digoxigenin-labeled Pdgfra riboprobe was performed as described previously (20).
Neurosphere culture.
E18.5 forebrains from diverse CK2β genetic backgrounds were subjected to trypsin digestion for 15 min at 37°C. Tissues were mechanically dissociated into single-cell suspensions, filtered through a 70-μm cell strainer, and further treated with 10 μg DNase I (Roche) for 10 min at room temperature. Cells from individual embryos, 2 × 106 cells/10-cm-diameter uncoated dish, were cultured for primary neurosphere assays in Dulbecco's modified Eagle's medium (DMEM)-F12 medium with B27 complement (D/F-B27) containing 10 ng/ml epidermal growth factor (EGF; PeproTech) and 10 ng/ml fibroblast growth factor-2 (FGF; gift from G. Bouche, Toulouse, France). Numbers of generated primary spheres were counted after 4 days of in vitro culture (4DIV). To assess for proliferation, 7DIV primary spheres were mechanically dissociated and individual viable cells were counted. These cells (6 × 105 cells/10-cm-diameter uncoated dish) were also used to assess for self-renewal in secondary neurosphere assays. Olig neurospheres were generated from neural stem/progenitor cells (NSCs) isolated from ganglionic eminences of Olig2+/− or Olig2−/− E14.5 embryos (18). For rescue experiments, Olig2−/− NSCs were subsequently infected with HA-tagged Olig2-encoding viruses (see below) and selected with blasticidin (2 μg/ml) 48 h after infection. Expression of HA-Olig2 proteins was confirmed by immunocytochemistry. For NSC differentiation assays, 4DIV neurosphere cultures were plated on poly-l-lysine-coated glass coverslips (at a density of ∼50 neurospheres/cm2) and shifted into D/F-B27 medium in the presence of 3.5% fetal bovine serum (FBS; Biowest) (differentiating medium) (11). 3DIV after plating, differentiated cultures were fixed with the PFA solution and processed for immunocytochemistry. Additional primary antibodies were mouse anti-β-tubulin III (1/500; Babco), mouse anti-O4 (1/10; gift from C. Soula, Toulouse, France), rat anti-Pdgf receptor a chain (1/100; BD PharMingen), and rat anti-HA (clone 3F10, 1/500; Roche).
Embryonic stem cell culture.
CK2βloxP/wt and CK2βloxP/− ES cell lines cultured in the presence of 103 units/ml leukemia inhibitory factor (LIF; Chemicon) and Cre-pMSCV-puro viral supernatant have been described previously (2). The PCR product corresponding to HA-tagged CK2βwt coding regions was amplified using oligonucleotide DNA primers (5′ primer A [5′-CGGAATTCGCCGCCACCATGTATCCTTATGATGTTCCTGATTATGCTATGAGCAGCTCCGAG GAGGTG-3′] and 3′ primer B [5′-GAAGATCTTCAGCGGATGGTCTTCACG-3′]) and cloned into EcoRI/BglII pMSCV-neo viral vector sites (Clontech). CK2β viral supernatants were generated by transfection of BOSC23 cells. Viral HA-CK2β-pMSCV-neo supernatants were first used to infect 2.5 × 104/cm2 CK2βloxP/− ES cells. To avoid chimera colonies, one day after infection, ES cells were subjected to trypsin treatment and individual cells were plated. Neomycin (Neo) selection (350 μg/ml) was started 48 h after infection, and Neor clones were expanded and screened for expression of exogenous the HA-CK2βwt protein by Western blot analysis using mouse anti-HA antibody. Selected Neor clones were further infected with Cre-pMSCV-puro supernatant. (CK2βloxP/wt ES cells were used as positive controls.) One day later, individualized ES cells were plated, and puromycin (puro) selection (1.5 μg/ml) was started 48 h postinfection for 6DIV or 9DIV. Individual clones were expanded and analyzed by PCR analysis and Southern blot analysis for the absence of the CK2βloxP allele (2) and by Western blot analysis for the absence of the endogenous wild-type CK2β protein. Neor puro-resistant CK2β−/− ES cells expressing only the exogenous HA-CK2βwt protein were seeded in the absence of LIF (3 × 106 cells/10-cm-diameter uncoated plate/10 ml) to form floating embryoid bodies (EBs). In order to stimulate neural differentiation, 5 μM retinoic acid was added after 4DIV of EB cultures and left for additional 4 days. EBs were further dissociated with trypsin, and individual cells (3 × 106 cells/10-cm-diameter uncoated plate/10 ml) were used in neurosphere assays.
CK2 activity.
Crude embryonic E18.5 forebrain extracts (adjusted to 5 mg/ml of protein concentration) were assayed for CK2β-dependent CK2 kinase activity at 22°C for 10 min using a 3 mM concentration of the CK2β-specific synthetic peptide RRREEETEEE (13) and [γ-32P]ATP. One unit of CK2 activity is defined as the amount of activity necessary to transfer 1 fmol of phosphate per minute into the synthetic peptide. Chromatographic fractions (5 μl) were assayed for 3 min with a 150 μM concentration of the CK2 consensus synthetic peptide RRREDEESDDEE (32) or with a purified fusion protein fragment consisting of glutathione S-transferase (GST) and Olig2 amino acids 1 to 177 [Olig2(1-177)]. The PCR product corresponding to the mouse Olig2(1-177) coding region was amplified from the Olig2 CMV2 template (34) by using the Expand high-fidelity PCR system (Roche) with oligonucleotide DNA primers (5′ primer C [5′-TTGAATTCATATGGACTCGGACGCCAGCCT-3′] and 3′ primer D [5′-AACTCGAGTCAGTAGATCTCGCTCACCAGTC-3′]) and subcloned into EcoRI/XhoI pGEX4T2 vector sites (Amersham Biosciences) in order to generate the GST-Olig2(1-177) fusion protein. Purified GST-Olig2 proteins (20 μg) were incubated at 22°C for 0 to 30 min with 25 μM [γ-32P]ATP and 12 mM MgCl2 in the presence of 60 to 240 ng of CK2α subunit with increasing amounts of equivalent CK2β subunit (0 to 230 ng). Phosphorylated proteins were analyzed by SDS electrophoresis and autoradiography.
Phosphopeptide mapping and automated Edman degradation.
The phosphorylated GST-Olig2(1-177) fusion protein was excised from the nitrocellulose membrane and digested in situ with trypsin (modified sequencing grade; Promega). Two-dimensional phosphopeptide mapping was done using thin-layer electrophoresis (HTLE-7000; CBS Scientific). First-dimension electrophoresis was performed in pH 1.9 buffer (formic acid, glacial acetic acid, and water at 50:156:1,794, vol/vol/vol) for 30 min at 2,000 V, and chromatography in the second dimension was performed in isobutyric acid-n-butyl alcohol-pyridine-glacial acetic acid-water (1,250:38:96:58:558, vol/vol/vol/vol/vol). After exposure, phosphopeptides were eluted from the plates in pH 1.9 buffer and lyophilized; aliquots of the samples were coupled to Sequelon-AA membrane (Millipore Corp.) by use of carbodiimide coupling and subjected to automated Edman degradation by using an Applied Biosystems sequencer (model 477A). Released phenylthiohydantoin derivates from each cycle were spotted onto thin-layer chromatography plates. The radioactivity in each spot was revealed by exposure on a FujiX Bio-imager.
GST pull-down assay.
Additional PCR products corresponding to coding regions for mouse Olig2 N-terminal amino acids 1 to 108 [N-ter(1-108)] and Olig2 bHLH amino acids 109 to 177 [bHLH(109-177)] were additionally amplified and subcloned into the pGEX4T2 vector as described above using 5′ primer C with 3′ primer E (5′-AACTCGAGTCACTGCTGCAGCTCGGGCTCAG-3′) and using 5′ primer F 5′-TTGAATTCATATGCTGCGCCTGAAGATCAACAGCCGCGA-3′ with 3′ primer D, respectively. Interaction assays were performed using prebound GST or GST-Olig2 glutathione Sepharose beads (Amersham Biosciences) equilibrated in PBS-0.05% Tween 20-1 mg/ml bovine serum albumin (BSA) and incubated in the presence of [35S]methionine-labeled CK2β (2 × 105 cpm) at 22°C for 90 min. Bound CK2β subunits were analyzed by SDS electrophoresis and autoradiography.
Construction of HA-tagged mouse Olig2 vectors.
A PCR product corresponding to the HA-tagged mouse Olig2(1-177) coding region was amplified using 5′ primer G (5′-TTGAATTCTGGGCCATGTACCCATACGATGTTCCAGATTACGCTATGGACTCGGACGCCAGCCT-3′) and 3′ primer H (5′-GAAGATCTCGCTCACCAGTCGCT-3′) and subcloned into EcoRI/BglII sites of the Olig2 CMV2 vector (34) to generate HA-tagged full-length mouse Olig2wt(1-323). The CMV2 vector with HA-Olig2 STR amino acids 77 to 94 (STR77-94) deleted (HA-Olig2ΔSTR) was generated by site directed mutagenesis using sense primer I (5′-TGGGCGGCGGTGGCTTCAAGAAGAAAGACAAGAAGCAGAT-3′) and antisense primer J (5′-ATCTGCTTCTTGTCTTTCTTCTTGAAGCCACCGCCGCCCA-3′). Construction of pWZL-BLAST viral vectors encoding HA-Olig2wt and HA-Olig2ΔSTR proteins were generated after subcloning EcoRI/XhoI PCR fragments obtained after amplification with respective CMV2 templates by using a GC-rich DNA polymerase (Invitrogen) with 5′ primer G and 3′ primer K (5′-AACTCGAGTCACTTGGCGTCGGAGGTGAG-3′).
Cos7 cell transfection and coimmunoprecipitation assay.
For coimmunoprecipitations, a Flag-tagged CK2β construct was produced after PCR amplification using 5′ primer L (5′-TTGGATCCGCCGCCACCATGGACTACAAGGACGACGACGACAAGATGAGCAGCTCCGAGG-3′) and 3′ primer M (5′-GAAGATCTTCAGCGGATGGTCTTCACG-3′) and cloned into BamHI/BglII pSG5 vector sites. The Flag-CK2β PSG5 and the HA-tagged full-length mouse Olig2wt(1-323) CMV2 constructs were transiently Lipofectamine-transfected into Cos7 cells cultured in DMEM-10% fetal calf serum (FCS). Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (48 h posttransfection) in the presence of protease inhibitors, and for each extract, protein expression levels were analyzed by Western blot analysis. Five micrograms of a monoclonal antibody against the Flag epitope (Sigma) were added in precleared lysates for 1 h at 4°C. Next, 50 μl of RIPA-equilibrated “mouse true blot” beads (eBioscience) was added and incubated under constant rotation for 1 h. After the beads were washed with RIPA buffer, the precipitated proteins were analyzed by SDS electrophoresis and Western blotting.
Statistical measures.
For kinase activities, three forebrain-derived extracts per genotype were tested in quadruplicate. For telencephalon cell counts, at least three embryos per genotype were used for each day postcoitum (dpc) indicated. Cells were counted on three or more nonadjacent sections per telencephalon. For in vitro CK2β neurosphere assays, sphere and cell counts were performed from three or six embryo-derived cultures per genotype. For in vitro neurosphere differentiation assays, cell counts were performed from four or more embryo-derived cultures per genotype with six glass coverslips per culture (∼50 neurospheres per coverslip). To evaluate the proliferation rate of neural stem cell-derived embryonic stem cells, cell counts were performed from seven differentiation cultures. Data are presented as averages ± standard errors. Statistical comparisons were established using Student's t test.
RESULTS
CK2β ablation in the central nervous system causes telencephalon defects during late development.
To gain insight into functions of CK2β in the developing central nervous system (CNS), we ablated CK2β in embryonic neural stem/progenitor cells (NSCs), since loss of CK2β is lethal to mouse embryos (2). We crossed CK2βloxP/loxP mice with hemizygous Nestin-cre transgenic mice (37) to generate conditional mutant CK2βloxP/loxP, Nestin-cre mice (referred to here as CK2β−). CK2β− mutant pups were born with a Mendelian ratio (Table 1) but did not feed and died shortly after birth. Therefore, the phenotype of embryos was analyzed. As expected, the CK2β protein was absent in CK2β− mutant E18.5 forebrain extracts (Fig. 1A). However, the CK2β protein was still detected in CK2β− mutant E16.5 extracts, although Cre-mediated recombination occurs at E9.5 to E10.5 (10). This observation is in agreement with the stability of the CK2β protein integrated in the structure of tetrameric CK2 holoenzymes (22) and could explain the late CK2β− mutant phenotype. Importantly, the expression of the CK2α catalytic subunit was unchanged upon CK2β ablation. We also measured the CK2β-dependent CK2 activity in E18.5 forebrain extracts by using phosphorylation of the CK2β-specific synthetic peptide RRREEETEE (13). In line with ablation of CK2β in CK2β− mutant forebrains, CK2β-dependent CK2 activity was significantly reduced (∼60%) in CK2β− mutant extracts compared to wild-type controls (Fig. 1B). The remaining activity in mutant extracts could be related to holoenzyme molecules derived from blood vessel cells present in dissected forebrains, in which Cre recombinase is not active (10). We conclude that expression of CK2β and, thus, the activity of CK2β-dependent phosphorylation are effectively impaired in CK2β− E18.5 forebrains.
TABLE 1.
Genotypes of live-born mice and embryos from CK2βloxP/wt, Nestin-cre × CK2βloxP/loxP crossingsa
| Offspring or embryo | No. with genotype: |
|||
|---|---|---|---|---|
| CK2βloxP/wt (WT) | CK2βloxP/loxP (WT) | CK2βloxP/wt, Nestin-cre (WT) | CK2βloxP/loxP, Nestin-cre (CK2β−) | |
| Live-born mice | 22 | 18 | 15 | 0 |
| P0 | 3 | 5 | 4 | 5 |
| E14.5/E16.5/E18.5 | 142 | 146 | 148 | 154 |
For details, see Materials and Methods.
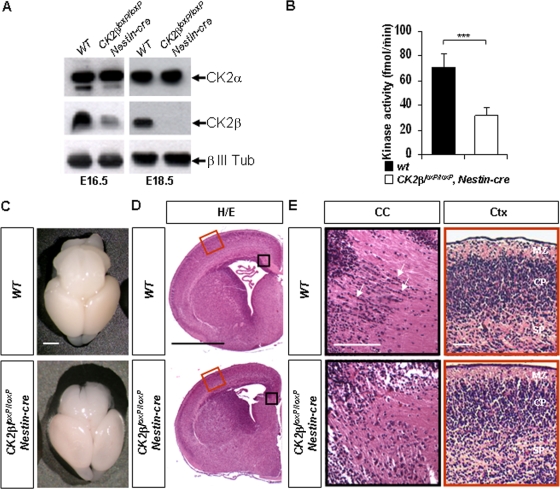
FIG. 1.
CK2β loss in the central nervous system impairs telencephalon development. (A) Western blot analysis of CK2β and CK2α expressions in E16.5 and E18.5 forebrain extracts. A specific neuronal (class III β-tubulin [β III Tub]) antibody was used to control protein normalization. (B) CK2β-dependent CK2 activity in E18.5 CK2βloxP/loxP, Nestin-cre forebrain extracts was significantly reduced (***, P = 2.7 × 10−9 [3 extracts in quadruplicate per genotype]) compared to that in wild-type controls. (C) Brain morphology of E18.5 CK2βloxP/loxP, Nestin-cre embryos compared to that of the wild type. (D) Representative H&E staining of E18.5 anterior forebrains. Black and red boxed areas are shown at higher magnification in panel E. (E) At the level of the corpus callosum (CC; black boxes), linear arrays of cells (arrows) detected in wild-type embryos were absent in the mutants. In contrast, laminar patterns appeared similar in the cortex (Ctx; red boxes). MZ, mantle zone; CP, cortical plate zone; SP, subplate zone. Scale bars, 1 mm (C and D) and 100 μm (E).
To investigate the effects of loss of the CK2β regulatory subunit on the development of the central nervous system, we analyzed the morphologies of E18.5 mutant brains. Gross morphologies of wild-type and CK2β− E18.5 brains appeared similar (Fig. 1C). However, further histological analyses showed defects in CK2β− telencephalons: an absence at the corpus callosum (CC) level of cells that formed linear, pearls-on-a-string arrays characteristic of oligodendroglial cells (Fig. 1E) (33) and an absence of the emerging hippocampal dentate gyrus (data not shown). In contrast, laminar patterns and cellular density were not affected in the cortex (Ctx; Fig. 1D and E), suggesting that cortical neurogenesis occurred normally in CK2β− mutants. Thus, CK2β loss in embryonic NSCs leads to morphology defects during late development of the brain.
CK2β disruption compromises forebrain stem/progenitor cell proliferation.
To understand the causes of the defects observed in mutant brains, we studied NSCs. In mice, NSCs in the VZs of developing brains possess radial glia morphology and express RC2. During the late embryonic (E18.5)/neonatal (P0) transition, radial glia gives rise to adult NSCs and progressively loose RC2 expression in favor of GFAP (23). In CK2β-ablated E18.5 embryos, neither RC2+ radial glia nor GFAP+ NSCs were substantially altered (Fig. 2A).
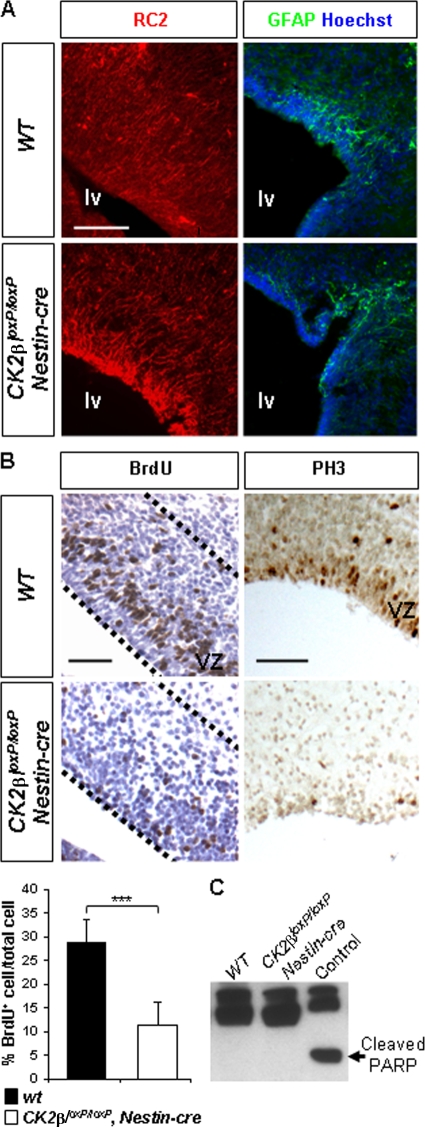
FIG. 2.
CK2β positively controls forebrain NSC proliferation but did not interfere with NSC specification. (A) Analysis of RC2 (red) and GFAP (green) expressions by immunohistochemistry (IHC) in E18.5 wild-type and CK2βloxP/loxP, Nestin-cre telencephalons. Cell nuclei were stained with Hoechst dye 33342 (blue). (B) There was a significant reduction (***, P = 2.7 × 10−12 [3 embryos per genotype and 3 nonadjacent sections per ventricular zone {VZ}]) of BrdU+ S-phase NSCs in CK2βloxP/loxP, Nestin-cre VZs (outlined with a dotted line) of lateral ventricles compared to the number in wild-type controls, as well as a near absence of PH3-positive mitotic cells. (C) Caspase-3-dependent apoptosis analysis in E18.5 forebrains. Western blot analysis of cleaved PARP in E18.5 forebrain extracts. The control represents the staurosporine-treated (1 μM, 3 h) NIH 3T3 positive extract; cleaved PARP (arrow) serves as a marker of cells undergoing caspase-3-dependent apoptosis. Scale bars, 100 μm (A) and 50 μm (B).
Previous results indicated that during mouse embryogenesis, CK2β is mainly involved in proliferation rather than in the prevention of apoptosis (2). Therefore, we examined at E18.5 the incorporation of a short pulse of BrdU in the germinal compartment of the telencephalon, the VZ of lateral walls (lw) of lateral ventricles (lv). CK2β− VZs displayed a significant reduction (∼60%) of NSCs in S phase as determined by the extent of BrdU-labeled nuclei compared to wild-type controls (Fig. 2B). In addition, the BrdU+ mutant nuclei showed a rounded morphology instead of an elongated morphology. To further address the effects of CK2β loss on the cell cycle, sections were stained with the mitosis marker phospho-histone H3. Only few CK2β− NSCs were found phospho-histone H3 positive (Fig. 2B). It is possible that the reduced NSC number represents a dysregulation of the cell cycle or an increase in cell death. We did not detect substantial apoptosis by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay staining in CK2β− E18.5 forebrains (data not shown). Accordingly, cleaved PARP, a hallmark of caspase-3-dependent apoptosis in vivo, was undetectable in CK2β− E18.5 forebrain protein extracts (Fig. 2C). Taken together, these results suggest that CK2β is important for the proliferation, but not the genesis, of forebrain NSCs.
CK2β loss in NSCs impairs oligodendroglial differentiation in developing telencephalons.
Next, we examined whether CK2β regulates oligodendrogenesis, as suggested by disorganization of the corpus callosum of CK2β− brains (Fig. 1E). We first analyzed the expression of the oligodendroglial precursor (OPC) marker NG2 (25) by immunohistochemistry (IHC). At E18.5, parenchymal NG2+ cells with OPC morphology were absent in CK2β− telencephalons, and only NG2+ pericytes associated with blood vessel were detected (Fig. 3B). However, in CK2β− brains, we were able to detect some NG2+ OPCs in a ventral location (Fig. 3C, E18.5 panel). In the mouse telencephalon, early OPCs (E12.5 to E14.5) are of ventral origin (reviewed in references 28 and 29). Thus, it was possible that the remaining E18.5 ventral OPCs in CK2β− brains were generated earlier during early embryogenesis, a period when the CK2β protein is not completely ablated (Fig. 1A). Figure 3C shows that early ventral NG2+ OPCs were generated in CK2β− mutant E14.5 telencephalons, but in contrast to wild-type controls, they did not expand during the late embryonic period (E16.5 to E18.5). Because NG2 cytoplasmic immunostaining is unsuitable for quantification, we analyzed the expression of another OPC marker, Pdgfra (12, 25), by in situ hybridization. At E14.5, the numbers of Pdgfra+ OPCs in wild-type and CK2β− brains were similar (Fig. 3D). However, there was a sharp decrease (∼75%) in the number of Pdgfra+ OPCs in E16.5 mutant telencephalons, despite incomplete inactivation of the CK2β protein. By E18.5, this difference was more pronounced (∼90%).
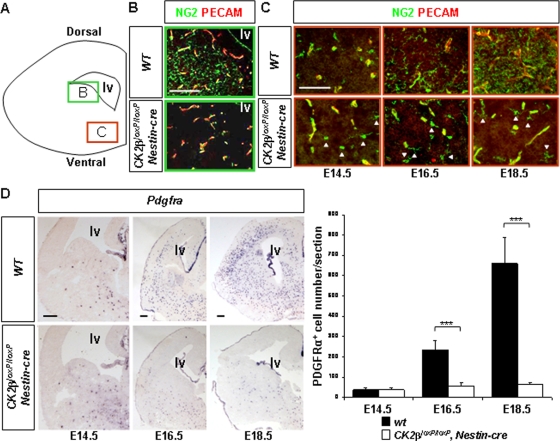
FIG. 3.
Defective OPC development in CK2βloxP/loxP, Nestin-cre telencephalons. (A) Schematic of a coronal telencephalon section. The green and red boxes indicate the regions analyzed for the detection of parenchymal OPCs in panel B and of ventral OPCs in panel C, respectively. (B) Analysis of NG2 (green) and PECAM (red) expressions by immunohistochemistry (IHC) in E18.5 wild-type and CK2βloxP/loxP, Nestin-cre telencephalons. Note the normal appearance of multiprocessed NG2+ parenchymal OPCs in the wild type and their absence in CK2βloxP/loxP, Nestin-cre embryos. The pattern of NG2 staining in vessel pericytes parallels PECAM endothelial cell labeling (yellow) and was unaffected. (C) NG2 and PECAM expression analysis (IHC) revealed the presence of ventral NG2+ OPCs (arrowheads) in CK2βloxP/loxP, Nestin-cre fetal (E14.5) and late embryonic (E16.5 to E18.5) telencephalons. (D) In situ hybridization (ISH) analysis of Pdgfra expression in fetal (E14.5) and late embryonic (E16.5 to E18.5) telencephalons. There was a significant reduction (***, P values of 2.5 × 10−10 at E16.5 and 1.0 × 10−6 at E18.5 [4 embryos per dpc and per genotype and 3 nonadjacent sections per telencephalon]) of Pdgfra+ OPCs in CK2βloxP/loxP, Nestin-cre telencephalons compared to the number in wild-type controls. lv, lateral ventricle. Scale bars, 100 μm (B and C) and 200 μm (D).
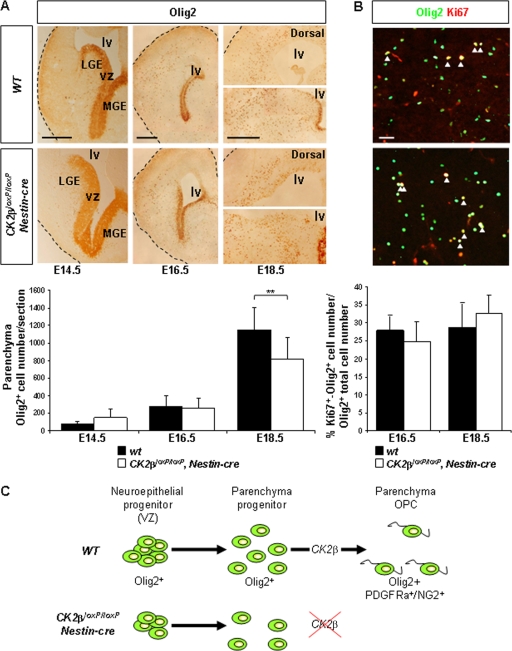
We also investigated the expression of Olig2, a basic helix-loop-helix (bHLH) transcription factor required for OPC development and maintained at all stages of oligodendrocyte development (17, 21, 29, 36, 43). In contrast to the defect observed for NG2+ and Pdgfra+ OPCs, analysis of Olig2 expression revealed identical numbers of parenchymal Olig2+ cells in CK2β− telencephalons and wild-type controls at E14.5 and E16.5 (Fig. 4A). However, by E18.5, a small decrease (∼30%) in the number of parenchymal Olig2+ cells was observed in CK2β− telencephalons. Because (i) Olig2 identifies both neural progenitors and OPCs (27, 28) and (ii) the decrease in the number of parenchymal Olig2+ cells is lower than the decrease in the number of Pdgfra+ OPCs in CK2β− brains (0% versus ∼75% at E16.5 and ∼30% versus ∼90% at E18.5), parenchymal Olig2+ cells in CK2β− brains may represent stalled neural progenitors unable to progress along the oligodendrocyte lineage. Alternatively, these Olig2+ cells may represent abnormal glia. To investigate whether the decrease in OPC numbers was due to defective proliferation of parenchymal Olig2+ progenitors as described for NSCs (Fig. 2B), we determined the ratio of Ki67+ Olig2+ cells to total Olig2+ cells in the parenchyma of wild-type and CK2β− telencephalons at E16.5 and E18.5 (Fig. 4B). Proliferation of parenchymal Olig2+ cells was unchanged in CK2β− brains and is therefore unlikely to account for OPC deficiency. Because parenchymal Olig2+ progenitors originate from Olig2+ neuroepithelial cells in the VZ (28, 29), the decrease in the number of Olig2+ cells in the parenchyma of CK2β− brains may be a consequence of a defective proliferation of Olig2+ cells in the VZ (Fig. 4C). The discrepancy observed between the effects of CK2β disruption on the proliferation of NSCs in the VZ and that of Olig2+ progenitors in the parenchyma may reflect different mechanisms involved in the control of the proliferation of progenitors downstream of the stem compartment. Collectively, these data suggest that CK2β loss leads to a defect in OPC specification of Olig2+ progenitors during telencephalon development (Fig. 4C).
FIG. 4.
Olig2+ progenitor analysis in fetal and late embryonic telencephalons. (A) IHC analysis of Olig2 expression in embryonic telencephalons. Parenchymal Olig2+ progenitors were detected in CK2βloxP/loxP, Nestin-cre E14.5, E16.5, and E18.5 telencephalons. lv, lateral ventricle; vz, ventricular zone; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence. There was a small decrease (**, P = 3.4 × 10−3 [4 embryos per dpc and per genotype and 3 nonadjacent sections per telencephalon]) in parenchymal Olig2+ progenitor numbers in CK2βloxP/loxP, Nestin-cre E18.5 telencephalons compared to the number in wild-type controls. (B) Proliferation analysis of parenchymal Olig2+ progenitors/OPCs by double labeling for Ki67 (red) and Olig2 (green). The ratios (percentages) of proliferating cells (Ki67+) coexpressing Olig2 in the parenchyma (yellow and arrowheads) to total parenchymal Olig2+ cells were not different between CK2βloxP/loxP, Nestin-cre and wild-type E16.5 and E18.5 telencephalons (3 embryos per dpc and per genotype and 4 nonadjacent sections per telencephalon). (C) CK2β regulates OPC specification of parenchymal Olig2+ progenitors. Scale bars, 100 μm (A) and 50 μm (B).
CK2β is required for neural stem cell proliferation and oligodendroglial differentiation in neurosphere culture.
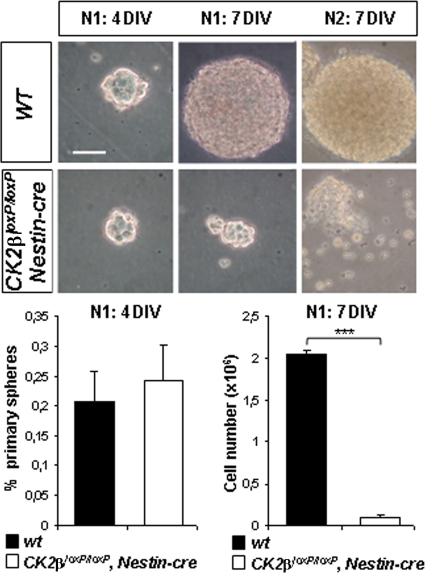
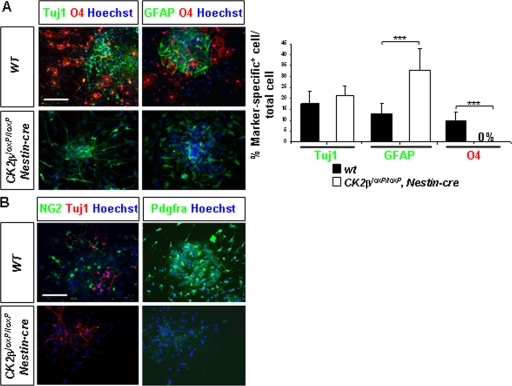
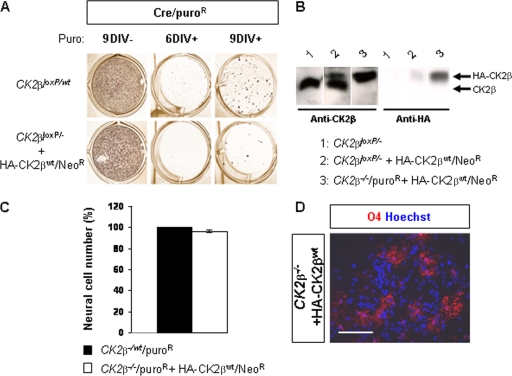
Embryonic NSCs located in the telencephalic VZ have been identified by their ability in vitro to generate neurospheres in response to FGF2 and EGF stimulation (38). In the absence of growth factors, neurosphere cells differentiate into neurons, astrocytes, and oligodendrocytes. To confirm whether CK2β loss affects NSC maintenance as shown in vivo, we cultured neurospheres derived from CK2β− E18.5 forebrains and induced their differentiation. There was no significant difference in the numbers of primary neurospheres generated from E18.5 wild-type and CK2β− forebrains in response to FGF2-EGF after 4 days of in vitro culture (4DIV) (Fig. 5). As observed in 7DIV cultures, wild-type cells formed large neurospheres, whereas CK2β− neurospheres did not expand. This was reflected by a significant reduction (∼95%) in the number of cells obtained from dissociated CK2β− neurospheres. In addition, we were unable to generate CK2β− secondary neurospheres, suggesting that the self-renewing potency of CK2β− NSCs was severely compromised. After differentiation, CK2β− neurosphere cells formed β-tubulin III-positive neurons and GFAP+ astrocytes but failed to generate O4+ differentiating OPCs (Fig. 6A). To confirm whether this defect was the result of a failure of OPC specification as observed in vivo, we examined NG2 and Pdgfra expression in differentiated neurosphere cultures. In striking contrast to control cultures, CK2β− neurospheres did not generate any NG2+ or Pdgfra+ OPCs (Fig. 6B). To further address the requirement of CK2β in NSC proliferation and in oligodendrogenesis, we expressed an HA-tagged CK2β protein in CK2β−/− NSC-derived embryonic stem (ES) cells (Fig. 7) (2). Such CK2β−/− NSCs expressing HA-CK2β were able to proliferate and to generate O4+ differentiating OPCs, excluding a genetic side effect associated with targeting of the CK2β locus. Taken together, these results suggest that the cell-autonomous defect(s) caused by CK2β disruption results in deficient NSC proliferation and OPC specification.
FIG. 5.
CK2β regulates NSC proliferation in neurosphere cultures. Primary neurospheres (N1; 4DIV and 7DIV) derived from E18.5 forebrain NSCs are shown. Compared to wild-type controls, CK2βloxP/loxP, Nestin-cre primary neurospheres in 7DIV cultures became smaller and did not generate secondary neurospheres (N2) after dissociation (7DIV). The ratio (percentage) of generated primary neurospheres to E18.5 forebrain-dissociated cells (6 primary neurosphere cultures per genotype) in CK2βloxP/loxP, Nestin-cre 4DIV cultures was not different from that of wild-type controls. The number of cells obtained from dissociated CK2βloxP/loxP, Nestin-cre primary neurospheres in 7DIV cultures was significantly reduced compared to that from wild-type controls (***, P = 4.3 × 10−7 [3 primary neurosphere cultures per genotype]). Scale bar, 100 μm.
FIG. 6.
CK2β disruption impairs OPC specification in neurosphere cultures. (A) Primary neurospheres derived from E18.5 forebrains were tested for their multipotency after differentiation. Cells were immunolabeled with the neuronal marker Tuj1 (green), the astrocytic marker GFAP (green), and the oligodendroglial marker O4 (red). The ratio (percentage) of cells expressing lineage-specific markers to the total cell number evaluated by Hoechst-stained nuclei (blue) showed an absence of O4+ cells (***, P = 1.3 × 10−8 [12 neurosphere cultures per genotype]) concomitant with a significant increase of GFAP+ astrocytes (***, P = 2.9 × 10−4) in CK2βloxP/loxP, Nestin-cre cultures compared to the number in wild-type controls. The percentages of Tuj1+ neurons did not differ between wild-type and mutant cultures. (B) CK2β loss altered the specification of NG2+ (green) and Pdgfra+ (red) OPCs in differentiation cultures. Scale bars, 100 μm.
FIG. 7.
Exogenous CK2β expression in NSC-derived CK2β−/− embryonic stem (ES) cells promotes proliferation and oligodendroglial differentiation. (A) Clonal selection of CK2β−/−, Cre-pMSCV-puro-cultured (Cre/puroR) ES cells expressing exogenous HA-CK2βwt/Neor protein. CK2β−/wt, Cre/puroR ES cell clones were generated in parallel and served as positive controls. (B) Western blot analysis with anti-CK2β (left panel) and anti-HA (right panel) antibodies of CK2βloxP/− ES cells, CK2βloxP/− and HA-CK2βwt/Neor ES cells, and CK2β−/−, Cre/puroR, and HA-CK2βwt/Neor ES cells. Note the complete absence of the endogenous CK2β protein in the CK2β−/− ES cell extract. (C) Proliferation analysis of NSC-derived ES cells. The ratio (percentage) of the number of cells present in neurosphere-derived CK2β−/− ES cells expressing the exogenous HA-CK2βwt protein to the number of cells present in control neurosphere-derived CK2β−/wt ES cells was unchanged when normalized to the CK2β−/wt ES cell line (7 differentiation cultures). (D) O4+ differentiating OPCs were identified from neurosphere-derived CK2β−/− ES cells expressing the exogenous HA-CK2βwt protein and allowed to differentiate. Scale bar, 100 μm.
Olig2 is an interacting partner of CK2β subunit and a strict CK2β-dependent substrate in vitro.
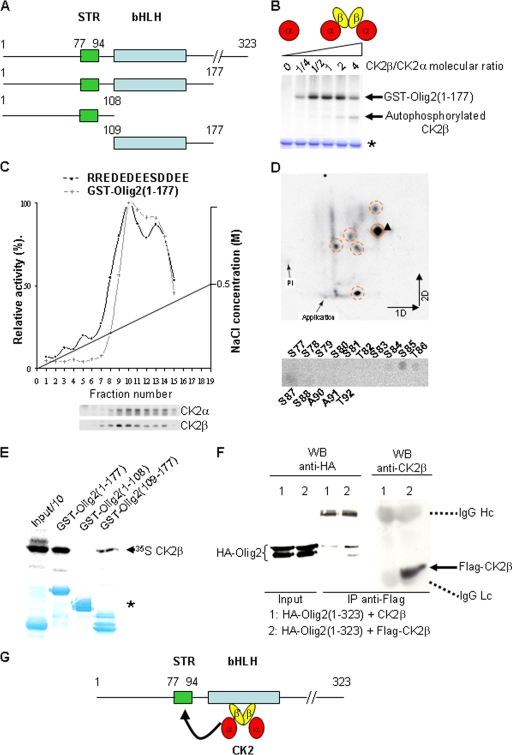
Given the essential roles of Olig2 and CK2β for OPC specification in fetal and late embryonic development (17, 21, 36, 43; this study) and the regulation of the oligodendroglial activity of Olig2 by phosphorylation (31), we investigated whether Olig2 may be a potential CK2 substrate. Amino acid sequence analysis of mouse Olig2 revealed that most putative CK2 phosphorylation sites (S/T; E/D/S n − 1, n + 1, n + 2, and/or n + 3) are present within an N-terminal fragment encompassing M1 to Y177. We designed a GST-Olig2(1-177) fusion protein that includes mouse Olig2 amino acids 1 to 177 (Fig. 8A) and determined the CK2-dependent incorporation of [γ-32P]ATP into the purified fragment. As shown in Fig. 8B, the Olig2(1-177) fragment was directly phosphorylated by CK2 only in the presence of the regulatory subunit CK2β. We further investigated CK2β-dependent phosphorylation of Olig2 by analyzing CK2 and Olig2 phosphorylation activities in fractions obtained after ion-exchange chromatography of wild-type E18.5 forebrain extracts (Fig. 8C). Phosphorylation experiments and Western blot analysis revealed coelution of CK2 and Olig2-dependent kinase. This coelution occurred in fractions containing the highest CK2β/CK2α subunits ratio. Moreover, no coelution of CK2 and Olig2-dependent kinase was observed when the chromatography was performed with CK2β− forebrain extracts (data not shown). In addition, Olig2 phosphorylation was abolished in the presence of 10 μM TBB, a specific in vitro CK2 inhibitor (data not shown). Taken together, these results suggest that the CK2 tetrameric holoenzyme (α2β2) is an Olig2 kinase.
FIG. 8.
CK2β positively modulates CK2-dependent phosphorylation of the Olig2(1-177) fragment in vitro. (A) Primary structure of mouse Olig2 and schematic representation of the Olig2 fragments used in this study. Represented are the serine-threonine-rich domain sequence (STR, amino acids 77 to 94), and the basic helix-loop-helix sequence (bHLH, amino acids 109 to 164). (B) Autoradiography of the purified recombinant GST-Olig2(1-177) fusion fragment after in vitro phosphorylation by the CK2 catalytic subunit (α) alone or in combination with increasing amounts of the CK2 regulatory subunit (β), which generate increased amounts of the holoenzyme (α2β2). Note that CK2β, an autosubstrate of the catalytic α subunit, competed in the reaction for the CK2 phosphorylation of Olig2 when present in excess. Protein normalization was controlled by Coomassie blue staining (*). (C) DEAE-Sephacel chromatography of crude wild-type E18.5 forebrain protein extracts. Each fraction was analyzed for consensus relative CK2 activity with the RRREDEESDDEE synthetic substrate/peptide and for relative CK2β-dependent Olig2(1-177) kinase activity. Activity profiles were normalized to the activity detected in the peak fractions. In parallel, a sample of each fraction was immunoblotted to detect the presence of CK2α and CK2β proteins (lower panels). (D) Two-dimensional (2D) phosphopeptide mapping analysis of CK2β-dependent Olig2(1-177) phosphorylation. The major phosphopeptide (arrowhead; upper panel) was subjected to automated Edman degradation and released derivates from each cycle were spotted onto plates and autoradiographed (lower panel). Deduced amino acids in the Olig2 sequence are indicated above or below the spots. (E) Recombinant GST fusion proteins containing different Olig2 fragments (1 to 177, 1 to 108, and 109 to 177) were incubated with [35S]methionine-labeled CK2β. The ability of the GST fusion proteins to pull down the CK2β subunit was detected by autoradiography after electrophoresis. On the left, one-tenth of the input is shown. Loading controls were checked after Coomassie blue staining of the gel (*). (F) Coimmunoprecipitation of Flag-CK2β and HA-Olig2(1-323) in Cos7 cells. Flag-CK2β was immunoprecipitated (IP) with anti-Flag antibody, and the immunoprecipitated complexes were analyzed by Western blotting (WB) with anti-HA and anti-CK2β antibodies. Hc, heavy chain; Lc, light chain. (G) Schematic representation of the suggested CK2-Olig2 interaction.
To delineate which particular motif of the mouse Olig2(1-177) fragment was involved in the phosphorylation by the CK2 holoenzyme, we performed two-dimensional tryptic phosphopeptide mapping (Fig. 8D). Analysis of the maps of 32P-labeled phosphopeptides revealed multiple spots. A major spot was suitable for Edman degradation. The released detectable radioactivity was observed mainly after cycles 9 and 11. Only one Olig2(1-177) tryptic peptide had serine/threonine residues at positions 9 and 11, allowing the identification of major phosphorylated sites at Ser85 and Ser87 in a serine-threonine-rich domain (STR77-94, tryptic peptide 77SSSSSTSSSTSSAATSSTK95) highly conserved in the human sequence. It should be noted that minor phosphorylation sites were also detected at Ser84, Thr86, and Ser88. Thus, these results identify the Olig2 STR sequence as an unconventional major domain of multiple acceptors for phosphorylation by the CK2 holoenzyme.
To investigate the regulation of Olig2 phosphorylation by CK2β, we tested the capacity of CK2 to interact with Olig2. GST pull-down assays showed that CK2β interacts with the bHLH fragment of Olig2 [Olig2(109-177)] but fails to interact with the fragment containing the STR domain [Olig2(1-108)] (Fig. 8E). Surprisingly, this Olig2(1-108) fragment was not phosphorylated by the CK2 holoenzyme (data not shown). Preincubation of CK2β with CK2α subunits which generate the tetrameric holoenzyme did not prevent its interaction with the bHLH Olig2(109-177) fragment (data not shown). We confirmed that CK2β can interact with Olig2 by coimmunoprecipitation of Flag-tagged CK2β with HA-tagged full-length mouse Olig2(1-323) after transient expression in Cos7 cells (Fig. 8F). Together, these data suggest that the bHLH domain of Olig2 is a docking site to promote the recruitment of CK2 via physical interaction with its regulatory CK2β subunit for efficient Olig2 phosphorylation on its STR sequence (Fig. 8G).
The CK2-targeted STR domain is required for the oligodendroglial activity of Olig2 in neurosphere culture.
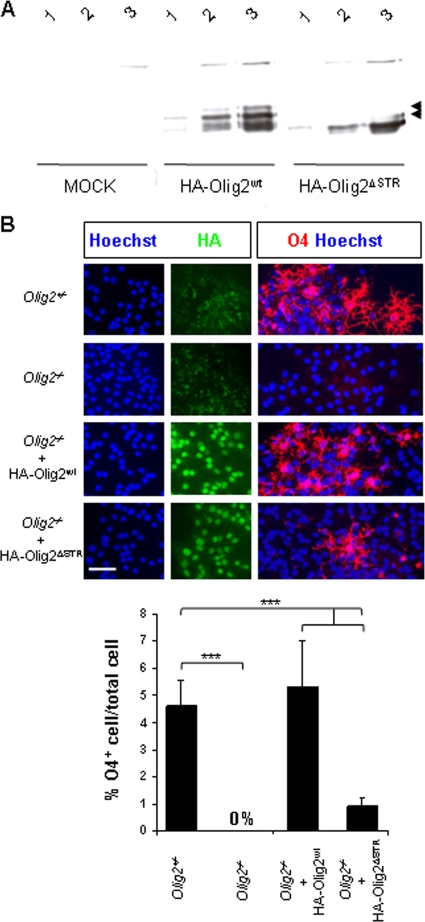
We next asked whether the CK2-phosphorylated STR domain is involved in mediating Olig2 biological function. Because of the many serine and threonine acceptor residues in the Olig2 STR that could potentially compensate for inactivation of the Ser85 and Ser87 major phosphorylation sites, we generated an Olig2 deletion mutant that lacks the entire STR77-94 domain (HA-Olig2ΔSTR). Transient transfection into Cos7 cells demonstrated identical levels of expression of HA-Olig2wt (amino acids 1 to 323; full-length) and HA-Olig2ΔSTR proteins (Fig. 9A). We infected Olig2−/− neurosphere cultures (18) with retroviruses encoding HA-Olig2wt or HA-Olig2ΔSTR and tested their potential to differentiate into the oligodendroglial lineage (Fig. 9B). As expected, expression of HA-Olig2wt in Olig2−/− NSCs restored the production of O4+ differentiating OPCs. In contrast, Olig2−/− NSCs transduced with HA-Olig2ΔSTR significantly failed to differentiate into O4+ cells. These findings demonstrate that, in vitro, the CK2-targeted STR domain is required for the oligodendroglial function of Olig2.
FIG. 9.
The Olig2 STR domain is required for NSC oligodendroglial potency in neurosphere cultures. (A) Transient expression analysis of the ΗΑOlig2ΔSTR mutant protein in Cos7 cells. Comparative pattern expressions of full-length (mouse; amino acids 1 to 323) HA-Olig2wt and HA-Olig2ΔSTR in transiently transfected Cos7 cells. Anti-HA Western blots revealed shifted-up bands within the HA-Olig2wt-transfected protein extracts (arrowheads), and these were not detected in the HA-Olig2ΔSTR version. This observation is reminiscent of the STR sequence as phosphorylated sites. “MOCK” indicates a control experiment performed with the empty vector. Amounts of protein are as follows: 1, 4 μg; 2, 8 μg; 3, 16 μg. (B) Olig2−/− NSCs were transduced with HA-Olig2wt(1-323) and HA-Olig2ΔSTR. NSC-derived neurospheres were allowed to differentiate and stained with HA (green) to detect exogenous Olig2 and with O4 (red) to assess for oligodendroglial potency. Olig2+/− and Olig2−/− assays are shown in parallel. The ratio (percentage) of cells expressing the O4 oligodendroglial marker to the total cell number evaluated by Hoechst-stained nuclei (blue) showed a significant deficiency (***, P = 5.6 × 10−7 [4 neurosphere cultures per genotype]) in the oligodendroglial activity of the HA-Olig2ΔSTR mutant compared to that of the HA-Olig2wt control. Scale bars, 50 μm.
DISCUSSION
Kinase-dependent pathways regulating NSC maintenance and fate specification are largely unknown. Here, we inactivated the protein kinase CK2 regulatory subunit (CK2β) gene in CNS embryonic neural progenitors and showed that CK2β is an important modulator of telencephalon development. Conditional CK2β mutant mice display impaired NSC proliferation, as well as defective OPC specification. We show that CK2β interacts physically with the bHLH transcription factor Olig2, an essential regulator of oligodendroglial differentiation, thereby allowing its serine-threonine-rich (STR) domain to be phosphorylated by CK2. Finally, we show that the Olig2 STR domain mediates the oligodendroglial activity of Olig2. Our results suggest that CK2β could mediate OPC specification through the modulation of the oligodendroglial activity of Olig2.
We present the first report of an in vivo role for CK2β in NSC proliferation in mammals. Loss of CK2β results in reduced numbers of dividing cells in the embryonic germinal zone, and our in vitro neurosphere assays show that CK2β is required for NSC proliferation and self-renewal. The altered nuclear morphology of BrdU+ CK2β− NSCs in the ventricular zone (Fig. 2B) is reminiscent of a cell cycle arrest event and suggests an interkinetic nuclear migration defect (40). During G1, the nuclei of NSCs ascend to the basal side of the VZ, where S phase is completed. Nuclei descend back toward the ventricular surface during G2, and this is where they stay during mitosis. The present study suggests that CK2β may control this process in the neural stem compartment, leading to a block in the G1/S phase in CK2β− NSCs. Therefore, CK2β− NSCs would not proceed through the G2 phase. Consequently, it would explain the near absence of PH3-positive mitotic CK2β− NSCs in the VZ (Fig. 2B) and the normal proliferation of the more downstream population of Olig2+ parenchymal progenitors in CK2β− embryos (Fig. 4B). Recently, it has been demonstrated that nuclear migration is governed by centrosomal specific microtubule-regulating proteins (40) and reported that CK2β is required for centrosomal normality (1). However, a strict CK2β-dependent CK2 substrate(s) involved in NSC proliferation has not been described.
We also show that CK2β− NSCs are specifically devoid of oligodendroglial potency. Our in vivo and in vitro studies demonstrate that depletion of the CK2β gene in embryonic neural progenitors blocks the production of NG2+ and Pdgfra+ OPCs. Several observations are in favor of a cell-intrinsic regulation by CK2β in OPC specification per se. (i) In E16.5 CK2β− telencephalons, the number of parenchymal Olig2+ progenitors was normal, despite a strong defect in NG2+ and Pdgfra+ OPCs, and by E18.5, whereas the number of Olig2+ progenitors increased, the number of OPCs in CK2β− telencephalons remained low compared to that in wild-type controls (Fig. 3 and 4). (ii) Olig2+ cell proliferation was unchanged in CK2β− brains (Fig. 4B). (iii) CK2β− neurosphere cultures recapitulated the deficiency in OPC specification observed in vivo (Fig. 6). (iv) CK2β− neurospheres cultured in the presence of other factors (PDGF-AA and insulin-like growth factor I [IGF-I]) were unable to differentiate into oligodendrocytes (data not shown), suggesting that a defect in a particular extracellular instruction for NSC specification to oligodendrogenesis cannot account for OPC deficiency (3, 5, 8, 14, 35). (v) CK2β is required to promote oligodendrogenesis in NSC-derived embryonic stem (ES) cells (Fig. 7). Thus, CK2β-dependent specification of OPCs may correspond to a molecular mechanism(s) shared by the multiple independent molecular, regional, and temporal origins of progenitors in the forebrain (reviewed in reference 28) and controlled by CK2β.
Genetic studies in mice demonstrated that during fetal and late embryogenesis, the bHLH transcription factor Olig2 is necessary for specification of progenitors into OPCs/oligodendrocytes (17, 21, 36, 43). Multiple regulations such as interactions (NFIA/B, SCL, and ID4) and phosphorylation (Akt/protein kinase B [PKB]) that inhibit Olig2 function dictate a loss into the oligodendroglial lineage (6, 24, 30, 31). The increase in astrocytogenesis observed in CK2β− neurosphere cultures after differentiation (Fig. 6A) is compatible with a glial switch following loss of the oligodendroglial function of Olig2 (6, 7, 21, 24, 30, 31, 36, 43). However, whether CK2β modulation of astrocytic instruction is direct or a consequence of a regulation of oligodendrocyte differentiation is unclear. We show that the Olig2(1-177) fragment is a strict CK2β-dependent CK2 substrate, probably through a CK2β-specific Olig2 recruitment by direct interaction with the bHLH domain, and that CK2 is an Olig2 kinase in forebrain extracts (Fig. 8). CK2-dependent major phosphorylation occurs in the STR domain. In neurosphere assays, we demonstrated that the CK2-targeted STR domain is required for the full oligodendroglial function of Olig2 (Fig. 9). Although we cannot exclude phosphorylation(s) of the STR domain by one or more other serine/threonine kinases, our results do not definitively prove, but do suggest, that CK2β-dependent CK2 activity may play a role as a determinant in the combinatorial code that trigger Olig2-dependent oligodendrocyte lineage progression. Alternatively, we cannot rule out other CK2β-dependent transcription factor substrates important for the oligodendroglial network which could act in synergy inside the same genetic pathway.
During CNS development, Olig2 is sequentially required for the generation of subsets of motor neurons (E9 to E10.5) and OPCs (E12.5 to E14.5) (21, 36, 43). If the assumption for a defect in Olig2 function in CK2β− progenitors is correct, then it is intriguing that the process of neurogenesis is not affected. However, in CK2β mutant brains, complete ablation of CK2β does not occur before E18.5, by which neurogenesis is largely completed. Additional studies will be needed to address the earlier role of CK2β in neuronal development.
The essential role of CK2β for cell viability initially detected in ES cells (2) precluded analysis of later roles for CK2 during development. Our conditional strategy revealed unexpected roles for CK2β in the CNS. We demonstrated that CK2β represents a unique signaling component in NSC proliferation and self-renewal and in the oligodendroglial lineage-restricted pathway. If CK2β-dependent CK2 activity is required for cancer stem cell proliferation and self-renewal as it is for NSCs, then inhibitors of interaction between the CK2β regulatory subunit and CK2 catalytic subunits (α and α′) could be useful chemotherapeutic agents. Conversely, because differentiation block of OPCs has been demonstrated to cause neurodegenerative diseases, such as chronic multiple sclerosis (16), activation of CK2β-dependent CK2 activity may be useful in promoting remyelination.
Acknowledgments
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Commissariat à l'Energie Atomique (CEA), the Ligue Nationale Contre le Cancer (Equipe Labelisée 2007), and the Institut National du Cancer (grant number 57).
We thank D. Rowitch for support, suggestions, and comments on the manuscript regarding in vivo analysis. We also thank I. Marechal and S. Bama for their efficiency with mouse breeding, C. Wernstedt for help with radiochemical sequencing, and I. Yakymovych for his competence in phosphopeptides analysis.
Footnotes
Published ahead of print on 5 April 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Bettencourt-Dias, M., R. Giet, R. Sinka, A. Mazumdar, W. G. Lock, F. Balloux, P. J. Zafiropoulos, S. Yamaguchi, S. Winter, R. W. Carthew, M. Cooper, D. Jones, L. Frenz, and D. M. Glover. 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432:980-987. [DOI] [PubMed] [Google Scholar]
- 2.Buchou, T., M. Vernet, O. Blond, H. H. Jensen, H. Pointu, B. B. Olsen, C. Cochet, O. G. Issinger, and B. Boldyreff. 2003. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 23:908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandran, S., H. Kato, D. Gerreli, A. Compston, C. N. Svendsen, and N. D. Allen. 2003. FGF-dependent generation of oligodendrocytes by a hedgehog-independent pathway. Development 130:6599-6609. [DOI] [PubMed] [Google Scholar]
- 4.Chantalat, L., D. Leroy, O. Filhol, A. Nueda, M. J. Benitez, E. M. Chambaz, C. Cochet, and O. Dideberg. 1999. Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J. 18:2930-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chojnacki, A., and S. Weiss. 2004. Isolation of a novel platelet-derived growth factor-responsive precursor from the embryonic ventral forebrain. J. Neurosci. 24:10888-10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deneen, B., R. Ho, A. Lukaszewicz, C. J. Hochstim, R. M. Gronostajski, and D. J. Anderson. 2006. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52:953-968. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda, S., T. Kondo, H. Takebayashi, and T. Taga. 2004. Negative regulatory effect of an oligodendrocytic bHLH factor OLIG2 on the astrocytic differentiation pathway. Cell Death Differ. 11:196-202. [DOI] [PubMed] [Google Scholar]
- 8.Gabay, L., S. Lowell, L. L. Rubin, and D. J. Anderson. 2003. Deregulation of dorsoventral patterning by FGF confers trilineage differentiation capacity on CNS stem cells in vitro. Neuron 40:485-499. [DOI] [PubMed] [Google Scholar]
- 9.Gratton, M. O., E. Torban, S. B. Jasmin, F. M. Theriault, M. S. German, and S. Stefani. 2003. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol. Cell Biol. 19:6922-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graus-Porta, D., S. Blaess, M. Senften, A. Littlewood-Evans, C. Damsky, Z. Huang, P. Orban, R. Klein, J. C. Schittny, and U. Muller. 2001. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron 31:367-379. [DOI] [PubMed] [Google Scholar]
- 11.Gritti, A., L. Bonfanti, F. Doetsch, I. Caille, A. Alvarez-Buylla, D. A. Lim, R. Galli, J. M. Verdugo, D. G. Herrera, and A. L. Vescovi. 2002. Multipotent neural stem cells reside into the rostral extension and olfactory bulb of adult rodents. J. Neurosci. 22:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, A., N. A. Giese, and W. D. Richardson. 1996. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development 122:4085-4094. [DOI] [PubMed] [Google Scholar]
- 13.Homma, M. K., D. Li, E. G. Krebs, Y. Yuasa, and Y. Homma. 2002. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. U. S. A. 99:5959-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh, J., J. B. Aimone, B. K. Kaspar, T. Kuwabara, K. Nakashima, and F. H. Gage. 2004. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J. Cell Biol. 164:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jauch, E., J. Melzig, M. Brkulj, and T. Raabe. 2002. In vivo functional analysis of Drosophila protein casein kinase 2 (CK2) β-subunit. Gene 298:29-39. [DOI] [PubMed] [Google Scholar]
- 16.Kuhlmann, T., V. Miron, Q. Cuo, C. Wegner, J. Antel, and W. Bruck. 2008. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain 131:1749-1758. [DOI] [PubMed] [Google Scholar]
- 17.Ligon, K. L., S. Kesari, M. Kitada, T. Sun, H. A. Arnett, J. A. Alberta, D. J. Anderson, C. D. Stiles, and D. H. Rowitch. 2006. Development of NG2 neural progenitor cells requires Olig gene function. Proc. Natl. Acad. Sci. U. S. A. 103:7853-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ligon, K. L., E. Huillard, S. Mehta, S. Kesari, H. Liu, J. A. Alberta, R. M. Bachoo, M. Kane, D. N. Louis, R. A. Depinho, D. J. Anderson, C. D. Stiles, and D. H. Rowitch. 2007. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 53:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenz, P., R. Pepperkok, W. Ansorge, and W. Pyerin. 1993. Cell biological studies with monoclonal and polyclonal antibodies against human casein kinase II subunit beta demonstrate participation of the kinase in mitogenic signaling. J. Biol. Chem. 268:2733-2739. [PubMed] [Google Scholar]
- 20.Lu, Q. R., D. Yuk, J. A. Alberta, Z. Zhu, I. Pawlitzky, J. Chan, A. P. McMahon, C. D. Stiles, and D. H. Rowitch. 2000. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25:317-329. [DOI] [PubMed] [Google Scholar]
- 21.Lu, Q. R., T. Sun, Z. Zhu, N. Ma, M. Garcia, C. D. Stiles, and D. H. Rowitch. 2002. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109:75-86. [DOI] [PubMed] [Google Scholar]
- 22.Lüscher, B., and D. W. Litchfield. 1994. Biosynthesis of casein kinase II in lymphoid cell lines. Eur. J. Biochem. 220:521-526. [DOI] [PubMed] [Google Scholar]
- 23.Merkle, F. T., A. D. Tramontin, J. M. Garcia-Verdugo, and A. Alvarez-Buylla. 2004. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. U. S. A. 101:17528-17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muroyama, Y., Y. Fujiwara, S. H. Orkin, and D. H. Rowitch. 2005. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature 438:360-363. [DOI] [PubMed] [Google Scholar]
- 25.Nishiyama, A., X. H. Lin, N. Giese, C. H. Heldin, and W. B. Stallcup. 1996. Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J. Neurosci. Res. 43:299-314. [DOI] [PubMed] [Google Scholar]
- 26.Pepperkok, R., P. Lorenz, R. Jakobi, W. Ansorge, and W. Pyerin. 1991. Cell growth stimulation by EGF: inhibition through antisense-oligodeoxynucleotides demonstrates important role of casein kinase II. Exp. Cell Res. 197:245-253. [DOI] [PubMed] [Google Scholar]
- 27.Pinna, L. A. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873-3878. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, W. D., N. Kessaris, and N. Pringle. 2006. Oligodendrocyte wars. Nat. Rev. Neurosci. 7:11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowitch, D. H. 2004. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 5:409-419. [DOI] [PubMed] [Google Scholar]
- 30.Samanta, J., and J. A. Kessler. 2004. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131:4131-4142. [DOI] [PubMed] [Google Scholar]
- 31.Setoguchi, T., and T. Kondo. 2004. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J. Cell Biol. 166:963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Songyang, Z., K. P. Lu, Y. T. Kwon, L. H. Tsai, O. Filhol, C. Cochet, D. A. Brickey, T. R. Soderling, C. Bartleson, D. J. Graves, A. J. DeMaggio, M. F. Hoekstra, J. Blenis, T. Hunter, and L. C. Cantley. 1996. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16:6486-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolt, C. C., S. Rehberg, M. Ader, P. Lommes, D. Riethmacher, M. Schachner, U. Bartsch, and M. Wegner. 2002. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, T., Y. Echelard, R. Lu, D. I. Yuk, S. Kaing, C. D. Stiles, and D. H. Rowitch. 2001. Olig bHLH proteins interact with homeodomain proteins to regulate cell fate acquisition in progenitors of the ventral neural tube. Curr. Biol. 11:1413-1420. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., S. K. Goderie, and S. Temple. 2005. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron 45:873-886. [DOI] [PubMed] [Google Scholar]
- 36.Takebayashi, H., Y. Nabeshima, S. Yoshida, O. Chisaka, K. Ikenaka, and Y. Nabeshima. 2002. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 12:1157-1163. [DOI] [PubMed] [Google Scholar]
- 37.Tronche, F., C. Kellendonk, O. Kretz, P. Gass, K. Anlag, P. C. Orban, R. Bock, R. Klein, and G. Schutz. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99-103. [DOI] [PubMed] [Google Scholar]
- 38.Tropepe, V., M. Sibilia, B. G. Ciruna, J. Rossant, E. F. Wagner, and D. van der Kooy. 1999. Distinct neural stem cells proliferate in response to EGF and FGF in the developing mouse telencephalon. Dev. Biol. 208:166-188. [DOI] [PubMed] [Google Scholar]
- 39.Viñals, F., J. Reiriz, S. Ambrosio, R. Bartrons, J. L. Rosa, and F. Ventura. 2004. BMP-2 decreases Mash1 stability by increasing Id1 expression. EMBO J. 23:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie, Z., L. Y. Moy, K. Sanada, Y. Zhou, J. Buchman, and L. H. Tsai. 2007. Cep 120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron 56:79-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu, X., P. A. Toselli, L. D. Russell, and D. C. Seldin. 1999. Globozoospermia in mice lacking the casein kinase II alpha' catalytic subunit. Nat. Genet. 23:118-121. [DOI] [PubMed] [Google Scholar]
- 42.Yde, C. W., B. B. Olsen, D. Meek, N. Watanabe, and B. Guerra. 2008. The regulatory beta-subunit of protein kinase CK2 regulates cell-cycle progression at the onset of mitosis. Oncogene 27:4986-4997. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, Q., and D. J. Anderson. 2002. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 109:61-73. [DOI] [PubMed] [Google Scholar]