Abstract
S100 proteins comprise a multigene family of EF-hand calcium binding proteins that engage in multiple functions in response to cellular stress. In one case, the S100B protein has been implicated in oligodendrocyte progenitor cell (OPC) regeneration in response to demyelinating insult. In this example, we report that the mitochondrial ATAD3A protein is a major, high-affinity, and calcium-dependent S100B target protein in OPC. In OPC, ATAD3A is required for cell growth and differentiation. Molecular characterization of the S100B binding domain on ATAD3A by nuclear magnetic resonance (NMR) spectroscopy techniques defined a consensus calcium-dependent S100B binding motif. This S100B binding motif is conserved in several other S100B target proteins, including the p53 protein. Cellular studies using a truncated ATAD3A mutant that is deficient for mitochondrial import revealed that S100B prevents cytoplasmic ATAD3A mutant aggregation and restored its mitochondrial localization. With these results in mind, we propose that S100B could assist the newly synthesized ATAD3A protein, which harbors the consensus S100B binding domain for proper folding and subcellular localization. Such a function for S100B might also help to explain the rescue of nuclear translocation and activation of the temperature-sensitive p53val135 mutant by S100B at nonpermissive temperatures.
The S100 proteins comprise a multigene family of low-molecular-weight EF-hand calcium binding and zinc binding proteins (5, 13, 16, 24, 33). To date, 19 different S100 proteins have been assigned to this protein family, and they show different degrees of similarity, ranging from 25 to 56% identity at the amino acid level. With S100B, S100P, and S100Z being the exceptions, the majority of the S100 genes are clustered on human chromosome 1q21 (33). Most S100 proteins serve as calcium sensor proteins that, upon activation, regulate the function and/or subcellular distribution of specific target proteins (13, 33, 47), and they are characterized by common structural motifs, including two low-affinity (KD [equilibrium dissociation constant] of ∼10 μM to 100 μM) helix-loop-helix calcium binding domains (EF hands) that are separated by a hinge region and flanked by amino- and carboxy-terminal domains. The carboxy-terminal domain is variable among S100 proteins, and it typically is the site that is responsible for the selective interaction of each individual S100 protein with specific target proteins (30). S100 proteins are often upregulated in cancers, in inflammation, and in response to cellular stress (14, 16), suggesting that they function in cell responses to stress situations. Consistent with this hypothesis, stress situations were necessary to reveal phenotypes associated with the S100 knockout in mice (11, 14, 33, 56). Moreover, recent observations revealed a new function for the S100 protein family that included their ability to assist and regulate multichaperone complex-ligand interactions (41, 50, 51).
One member of the S100 protein family, S100B, has attracted much interest in the past few years because, like other proteins implicated in neurodegeneration (e.g., amyloid, superoxide dismutase, and dual-specificity tyrosine phosphorylation-regulated kinase 1A), its gene is located within a segment of chromosome 21, which is trisomic in Down's syndrome (DS). Its expression in the brain of mammals coincides with defined periods of central nervous system (CNS) maturation and cell differentiation (43). In oligodendrocyte progenitor cells (OPC), S100B expression is associated with differentiation, and S100B contributes to OPC differentiation in response to demyelinating insult (11). To understand the contribution of S100B to OPC differentiation, we searched for high-affinity S100B target proteins in this cell type by using far-Western analysis. A major and highly specific S100B target protein was identified, the mitochondrial ATAD3A protein.
ATAD3A belongs to a new family of eukaryote-specific mitochondrial AAA+ ATPase proteins (17). In the human genome, two genes, Atad3A and Atad3B, are located in tandem on chromosome 1p36.33. The Atad3A gene is ubiquitous among multicellular organisms but absent in yeast. The Atad3B gene is specific to the human genome (27). ATAD3A is a mitochondrial protein anchored into the mitochondrial inner membrane (IM) at contact sites with the outer membrane (OM). Thanks to its simultaneous interaction with the two membranes, ATAD3A regulates mitochondrial dynamics at the interface between the inner and outer membranes and controls diverse cell responses ranging from mitochondrial metabolism, cell growth, and mitochondrial fission 20a, 25). The ATAD3A protein has also been identified as a mitochondrial DNA binding protein (23) and as a cell surface antigen in some human tumors (20, 21). The plasma membrane localization of ATAD3A in tumor cells is suggestive that ATAD3A mitochondrial routing can be compromised in pathological situations such as cancer. To understand the functional response resulting from the interaction between S100B and ATAD3A, we first characterized the minimal interaction domain on ATAD3A for S100B binding using thermodynamic studies of wild-type and ATAD3A variants as well as via nuclear magnetic resonance (NMR) spectroscopy techniques. These studies allowed us to further refine the consensus S100B binding motif, which is conserved in several other S100B target proteins, including the p53 protein and several newly discovered target proteins associated with the cell translational machinery. We next analyzed the cellular interaction of S100B with truncated ATAD3A mutants that harbor the S100B binding domain but that are deficient for mitochondrial import. These studies revealed that S100B could assist ATAD3A mutant proteins during cytoplasmic processing by preventing dysfunctional aggregation events. Our results are discussed in light of the possible function of S100B in assisting the cytoplasmic processing of proteins for proper folding and subcellular localization.
MATERIALS AND METHODS
Cells.
U373 cells were purchased from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM; GutaMAX; Gibco) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Rat OPC were obtained from newborn rat brain (10). OPC were grown on poly-l-lysine (Sigma)-coated plastic dishes in DMEM (1/3 dilution), F12 medium (1/3 dilution), B104 medium (1/3 dilution), decomplemented donor calf serum (DCS) (0.5%), N2 complement (1:100 dilution; Invitrogen), glucose (2%), recombinant human platelet-derived growth factor (PDGF) (10 ng/ml; Peprotech Inc.) and recombinant human basic fibroblast growth factor (bFGF) (10 ng/ml; G. Bouche, LBME, Toulouse, France). PDGF-AA and bFGF were added daily.
For differentiation studies, cells were shifted to new culture medium without PDGF-AA and bFGF but supplemented with thyroid hormone (50 ng/ml; Sigma) and forskolin (1 μM). Cells were incubated in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Antibodies.
Affinity-purified polyclonal antibodies to ATAD3A were raised in rabbit. N-terminal antibodies against the RPAPKDKWSNFDPTGC peptide and C-terminal antibodies against the CLKAEGPGRGDEPSPS peptide were obtained. Monoclonal anti-Myc IgG and anti-O4 IgM are homemade hybridoma supernatants. Mouse monoclonal anti-S100 antibody (S16) was a generous gift of M. Takahashi. The following commercial primary antibodies were used: mouse monoclonal anti-EF-1α (Upstate Biotechnology), rabbit anti-IQGAP1, goat anti-TCP-1η (Santa Cruz), and rabbit anti-S100B (Dako). Secondary antibodies conjugated to cyanin 3 or cyanin 5 were obtained from Jackson Immunoresearch Laboratories. Secondary antibodies conjugated to Alexa Fluor 488 were obtained from Molecular Probes Inc.
S100B protein preparation and purification.
Rat S100B was purified as described previously (46). Prior to storage at −80°C, buffer exchange was accomplished by using a G-25 gel filtration column preequilibrated with a solution containing 0.25 mM Tris and 25 mM NaCl (pH 7.2) and then dialyzed overnight in a chelexed solution containing 0.25 mM Tris (pH 7.2) followed by lyophilization. Such a preparation ensured the safe storage of the protein preparations as well as the removal of any trace dithiothreitol (DTT) or EDTA from our final protein sample, as necessary for isothermal titrating calorimetry (ITC) studies.
ATAD3A peptide preparation.
The 21-amino-acid peptide corresponding to the S100B binding site on ATAD3A (residues 290 to 310 [ac-RITVLEALRHPIQVSRRLLSR-am]) was purchased from Bio-Synthesis Inc. (Lewisville, TX), as were peptides of 16 amino acids (aa) (residues 290 to 305) and 11 amino acids (residues 290 to 300) in length. For all peptides, C termini were amidated (-am) and N termini were acetylated (ac-) to eliminate terminal charge effects. Peptides were delivered in lyophilized form. The peptide at residues 290 to 310 (P290-310) and P290-315 were resuspended in a chelexed solution consisting of 20 mM deuterated Tris, 25 mM NaCl, 0.35 mM NaN3, and ∼90% D2O (pH 6.5). P290-300 was resuspended in dimethyl sulfoxide (DMSO). The final peptide concentration in each solution was verified by using quantitative amino acid analysis.
Far-Western analysis.
Proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were first blocked for 2 h at 4°C in a solution containing 25 mM HEPES-KOH (pH 7.7), 25 mM NaCl, 3% bovine serum albumin (BSA), 20 mM DTT, and 0.05% NP-40 and incubated overnight with reticulocyte lysate (40 μl) containing 35S-labeled S100B (35S-S100B) in 10 ml of hybridizing buffer (20 mM HEPES-KOH [pH 7.7], 75 mM KCl, 0.1% BSA, 20 mM DTT, 0.05% NP-40, 10 μM ZnSO4, 0.3 mM CaCl2). After extensive washing, bound proteins were revealed by autoradiography.
Cell fractionation.
Cells grown in 100-mm plates were resuspended in 1 ml of buffer containing 0.28 M sucrose, 1 mM EDTA, and 10 mM Tris-HCl (pH 7.4). The cell suspension was homogenized with 20 passages through a 25-gauge needle. The homogenate was then centrifuged at 1,200 × g for 10 min. The supernatant was subsequently centrifuged at 11,000 × g for 15 min. The supernatant is called the cytosolic fraction. The pellet contains mitochondria and mitochondrion-associated membranes.
S100B pulldown assay.
U373 cells were lysed on ice in buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.3% Triton X-100) supplemented with protease inhibitor cocktails. Lysates were centrifuged for 10 min at 10,000 × g to remove insoluble material. Supernatants were supplemented with either 5 mM EDTA or EGTA or with 0.3 mM CaCl2 and 20 μM ZnSO4 and incubated for 30 min, with rotation at 4°C, with S100B-Sepharose beads (CnBr linkage) previously equilibrated either with 5 mM EDTA and EGTA or with 0.3 mM CaCl2 and 20 μM ZnSO4. The S100B beads were washed three times in incubation buffer and transferred into a new Eppendorf tube, and the beads were boiled in 1× Laemmli-SDS sample buffer with 20 mM DTT and 5 mM EDTA. Proteins were separated by SDS-PAGE and analyzed by Western blotting. In competition assays, competitors (S100 proteins or the chemically synthesized ATAD3A peptides) were added to cell extracts prior to incubation with S100B-Sepharose beads.
Immunoprecipitation.
Cells were lysed on ice in lysis buffer (40 mM Tris [pH 7.5], 150 mM NaCl, 0.3% Triton X-100) supplemented with protease inhibitor cocktails. Lysates were passed through a 26-gauge needle (15 times) and centrifuged to remove insoluble material. Supernatants were supplemented either with 5 mM EGTA or with 0.3 mM CaCl2 and 10 μM ZnSO4 and incubated with 5 μg anti-S100B monoclonal antibody (S16) together with protein G-Sepharose (Pharmacia) for 30 min with rotation at 4°C. The immunoprecipitates were washed three times with incubation buffer and transferred into a new Eppendorf tube, and the beads were boiled in 1× Laemmli buffer with 20 mM DTT. Proteins were separated by SDS-PAGE and analyzed by Western blotting.
Transfections.
Invitrogen eukaryotic vectors were used for the ectopic expression of ATAD3A and ATAD3A fragment mutants in U373 cells. All coding sequences were cloned into plasmid pcDNA3.1(−)/myc-His, which encodes a C-terminal Myc-His double tag. All DNA constructions used in this work were sequenced and proved to be identical to theoretical designed sequences. The delivery of plasmids into the cytoplasm of the U373B cells was performed by using the liposomal formulation Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions, in Opti-MEM culture medium (Gibco). For immunocytochemistry experiments, approximately 40,000 cells were seeded onto poly-l-lysine (Sigma)-coated coverslips. After 20 h, each coverslip was transfected with 0.5 μg of plasmid. For immunoprecipitation experiments, each petri dish with a confluence of approximately 70% was transfected with 7 μg of plasmid. After 2 h of incubation at 37°C in 5% CO2, the transfection was stopped by changing the medium to DMEM supplemented with 10% fetal bovine serum. The cells were cultured for 19 to 21 h prior to fixation or lysis.
Small interfering RNA (siRNA) transfection of OPC was performed by using the Amaxa rat oligodendrocyte Nucleofector kit according to the manufacturer's instructions (O-17 program). A total of 200 pM of ATAD3A siRNA [5′-GAAGUUGCUCCAUUUGUCC(dTdT)-3′] or control siRNA was used. After transfection, cells were replated onto 100-mm culture dishes. After 12 h, fresh culture medium was added. After 48 h, cells were seeded onto 60-mm plastic dishes or plated onto poly-l-lysine (Sigma)-coated glass coverslips.
Immunocytochemistry.
U373 cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton X-100 for 5 min, washed in Tris-buffered saline (TBS), and blocked in TBS containing 5% normal goat serum (NGS) for 30 min. After incubation with primary antibodies in NGS-TBS overnight at 4°C, cells were washed with TBS and stained with secondary antibodies. For OPC immunostaining, OPC were incubated with O4 hybridoma supernatant (1:5) in culture medium for 30 min at 37°C before the fixation step. After 3 washes with culture medium, cells were fixed with 1% PFA for 10 min and permeabilized with acetone (−20°C) for 10 min. Immunohistochemical staining was performed as described above. Images were obtained with a Zeiss (Axiovert 200 M) microscope or with a Leica (TCS SP2) confocal microscope.
Mass spectroscopy analysis.
Protein bands were manually excised from each of the two lanes of the gel and automatically prepared (EVO150; Tecan). Samples were washed several times by incubation in 25 mM NH4HCO3 for 15 min and then incubation in 50% (vol/vol) acetonitrile containing 25 mM NH4HCO3 for 15 min. Gel pieces were then dehydrated with 100% acetonitrile and then incubated with 7% H2O2 for 15 min before being washed again with the destaining solutions described above. A total of 0.15 μg of modified trypsin (sequencing grade; Promega) in 25 mM NH4HCO3 was added to the dehydrated gel spots for overnight incubation at 37°C. Peptides were then extracted from gel pieces in three 15-min sequential extraction steps with 30 μl of 50% acetonitrile, 30 μl of 5% formic acid, and, finally, 30 μl of 100% acetonitrile. The pooled supernatants were then dried under a vacuum. For nano-liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis, the dried extracted peptides were resuspended in water containing 2.5% acetonitrile and 2.5% trifluoroacetic acid. A nano-LC-MS/MS analysis was then performed (Ultimate 3000 [Dionex] and LTQ-Orbitrap [Thermo Fischer Scientific]). The method consisted of a 40-min gradient at a flow rate of 300 nl/min using a gradient from two solvents: gradient A (5% acetonitrile and 0.1% formic acid in water) and gradient B (80% acetonitrile and 0.08% formic acid in water). The system includes a 300-μm by 5-mm PepMap C18 precolumn and a 75-μm by 150-mm C18 column (Gemini C18 phase). MS and MS/MS data were acquired by using Xcalibur (Thermo Fischer Scientific) and processed automatically with Mascot Daemon software (Matrix Science). Consecutive searches against the SwissProt/Trembl decoy database were performed for each sample by using an intranet version of Mascot 2.0. For comparative analyses of proteins bound to S100B in the presence of the 290-305 and 293V/S-297L/S-290-305 ATAD3A peptides, the resulting data file from Mascot was filtered by using IrMa, a homemade tool used to filter and export the results to an Excel sheet, for example. The two Excel result sheets were compared by using an Excel macro which aligns data according to number or text. In this case, the alignment was done according to the protein name. To have a rough idea of the relative quantity of each protein on each fraction, we compared the spectral counts (numbers of MS2 spectra) for each protein aligned and determined the difference.
ITC.
The Ca2+-dependent interaction of S100A1 and S100B with ATAD3A was monitored by using a VP-ITC titration microcalorimeter (MicroCal Inc., Northampton, MA). Experiments were performed at 37°C. For all experiments, sample and titrant solutions contained 10 mM CaCl2, 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], 15 mM NaCl, 100 mM KCl, and 5% DMSO (pH 7.2). The titrant contained 400 μM peptide, and the ITC sample chamber (2-ml volume) contained 50 μM S100B. When zinc was present, a concentration of 60 μM ZnAc was present in sample and titrant solutions. For the titration of ATAD3A into S100A1, 1.5 mM ATAD3A was titrated into 2 ml of 100 μM S100A1. Protein concentrations refer to the subunit concentration and not the dimer concentration. Sample and protein solutions were degassed and preheated at 37°C prior to the running of the experiment.
S100B NMR sample preparation.
NMR buffer contained 20 mM Tris-d11, 10 mM CaCl2, 5 mM DTT, 25 mM NaCl, 0.35 mM NaN3, and 10% D2O (pH 6.5). The NMR sample was prepared by preheating a 500-μl sample of S100B (500 μM subunit concentration) and a separate stock of ATAD3A titrant at 37°C. The ATAD3A titrant was added incrementally with stirring to give a final sample that contained 470 μM S100B and 1,030 μM ATAD3A. A very small amount of precipitation was formed upon the addition of the peptide, which was removed by centrifugation. The resulting sample was stable for >2 weeks at room temperature, as determined by the lack of changes in the NMR spectra over this time period.
NMR spectroscopy.
All NMR experiments were performed at 37°C. Data from the two-dimensional (2D) heteronuclear single-quantum coherence (HSQC), three-dimensional (3D) 15N,15N-edited 3D heteronuclear multiple quantum coherence (HMQC)-nuclear overhauser effect spectroscopy (NOESY)-HSQC, and 15N-edited 3D NOESY experiments were collected with a Bruker DMX 600 NMR spectrometer (600.13 MHz for protons) using uniformly 15N-labeled S100B. In addition, data from carbon-decoupled 2D HSQC and 3D HNCA, HNCACB, and CBCA(CO)NH experiments were collected with a Bruker DMX 800 NMR spectrometer (800.27 MHz for protons) using uniformly 13C,15N-labeled S100B. Both magnets are equipped with four frequency channels and a triple-resonance z-axis-gradient 5-mm cryogenic probe head. Assignments for 1HN, 15N, 13Cα, and 13Cβ resonances were assigned sequence specifically by using standard multidimensional NMR spectroscopy techniques (1). Proton chemical shifts are reported with respect to the H2O signal, which is taken as 4.658 ppm downfield from the external trimethylsilyl proprionate (TSP) (0.00 ppm) at 37°C. Data were processed on a Linux Fedora 3.0 apparatus with NMRPIPE (8).
RESULTS
Identification of mitochondrial ATAD3A as a major calcium-dependent S100B binding protein.
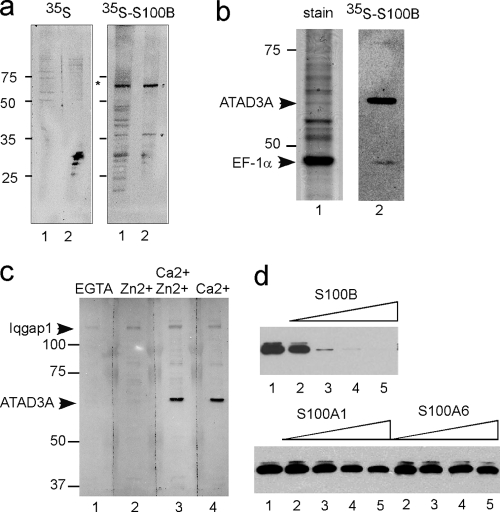
In oligodendrocyte progenitor cells (OPCs), the calcium and zinc binding S100B accumulates during differentiation (11). To identify high-affinity and Ca2+-dependent S100B target proteins in rat OPC, we used far-Western analysis. Total OPC proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and incubated with 35S-labeled S100B translated in rabbit reticulocytes in calcium buffer (Fig. 1a, lane 1). The 35S-S100B concentration used was in the 10 nM range, as determined by immunoassay titration, so only high-affinity protein targets were discovered under such conditions. The most intensively labeled protein that bound 35S-S100B had a molecular mass of 65 kDa and was enriched in mitochondrial fractions (Fig. 1a, right, lane 2). For identification, the human 65-kDa S100B binding protein was then partially purified from total astrocytoma U373 cell extracts on S100B-Sepharose beads. Proteins that bound to the S100B-Sepharose beads in the presence of Ca2+ and Zn2+ were separated by SDS-PAGE, and gels were either stained (Fig. 1b, lane 1) or processed for far-Western analysis (Fig. 1b, lane 2). The 65-kDa protein represented a minor protein associated with the S100B beads in the presence of Ca2+ and Zn2+, but it was the most intensively labeled protein with 35S-S100B. Matrix-assisted laser desorption ionization (MALDI) peptide mass fingerprints and tandem mass spectrometry identified the 65-kDa protein as a human mitochondrial protein termed ATAD3A. The major protein recovered within the S100B beads migrated with a molecular mass of 48 kDa and was identified to be the elongation factor EF-1α. EF-1α was also found to be labeled with 35S-S100B via far-Western analysis albeit with a much lower intensity than that of ATAD3A. The strict Ca2+ dependence of the interaction between S100B and ATAD3A was confirmed by far-Western analysis (Fig. 1c). The 180-kDa protein that binds 35S-S100B in the presence of Zn2+ alone corresponded to IQGAP1, a previously recognized Zn2+-dependent S100B binding protein (36).
FIG. 1.
Identification of ATAD3A as a high-affinity S100B binding protein. (a) Far-Western analysis identified a mitochondrion-associated 65-kDa protein as a high-affinity S100B binding protein in rat OPC. Proteins from total cell extracts (lanes 1) and mitochondrial fractions (lanes 2) were separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes. Transfer membranes were hybridized with free [35S]methionine (left) or [35S]methionine-labeled S100B (right) in buffer containing 0.3 mM CaCl2 and 10 μM ZnSO4. After extensive washing in binding buffer, membranes were subjected to autoradiography overnight. (b) Proteins from U373 cell extracts that bound to S100B-Sepharose beads in the presence of Ca2+ and Zn2+ were separated by 10% SDS-PAGE, and the gel was stained with Coomassie blue (lane 1) or transferred onto nitrocellulose membranes and analyzed by far-Western analysis using [35S]methionine-labeled S100B as described above (lane 2). The two proteins that bound 35S-S100B were identified by mass spectrometry as ATAD3A and EF-1α. (c) Far-Western analysis of U373A proteins bound to S100B-Sepharose with [35S]methionine-labeled S100B. Transfer membranes were hybridized with 35S-S100B in buffer containing EGTA (lane 1), 10 μM ZnSO4 (lane 2), 0.3 mM CaCl2 and 10 μM ZnSO4 (lane 3), or 0.3 mM CaCl2 (lane 4). Positions of IQGAP1 and ATAD3A are indicated. (d) Other S100 proteins do not compete with S100B for interactions with ATAD3A. The U373 cell extract was incubated with S100B-Sepharose beads in the absence (lanes 1) and in the presence of increasing concentrations of soluble S100B, S100A1, or S100A6, as indicated (lane 2, 1 μM; lane 3, 5 μM; lane 4, 10 μM; lane 5, 20 μM). ATAD3A bound to S100B-Sepharose beads was analyzed by 8% SDS-PAGE and Western blotting using an ATAD3A N-ter antibody.
To determine whether the S100B-ATAD3A interaction was specific, we next compared the interactions of ATAD3A with other EF-hand calcium binding proteins of the S100 family using a competitive binding assay. S100A1 and S100A6 were chosen because both proteins are most closely related to S100B in sequence comparisons, and both of these S100 proteins are capable of forming heterodimers with S100B (4, 5, 9). Competition binding studies of ATAD3A present in U373 cell extracts on S100B-Sepaharose beads revealed that purified S100B efficiently antagonized ATAD3A binding to S100B-Sepharose (Fig. 1d). S100A1 and S100A6 could also antagonize binding but only at high concentrations (Fig. 1d). We also determined that the binding of ATAD3A to S100B was not antagonized by S100A11 or by the ubiquitous EF-hand calcium binding calmodulin (not shown).
Characterization of ATAD3A and S100B proteins in oligodendrocyte progenitor cells.
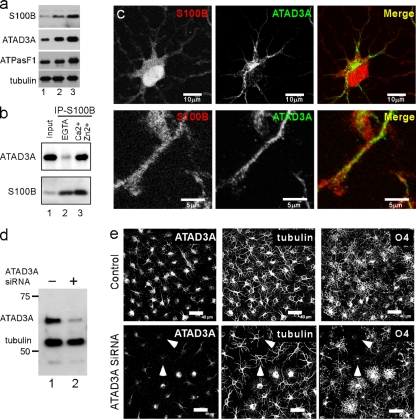
For an understanding of the meaning of the interaction between S100B and ATAD3A during oligodendrogenesis, it was necessary to refine the characterization of the two proteins in OPC. As previously shown with mouse OPC (11), S100B expression is upregulated during rat OPC differentiation (Fig. 2a). Consistent with genomic analyses (40; http://tinyurl.com/yk7hrp7), the ATAD3A protein was also upregulated in rat OPC during differentiation by comparisons of cytosolic marker proteins such as tubulin. However, the level of the mitochondrion-specific marker ATP synthase F1 subunit also significantly increased (Fig. 2a), so it is thus likely that OPC differentiation requires de novo mitochondrial protein synthesis in general. Using OPC extract prepared in detergent-containing buffer, we could confirm, by coimmunoprecipitation, the tight interaction between S100B and ATAD3A in the presence of Ca2+ and Zn2+ (Fig. 2b). However, a comparison of the subcellular localizations of the two proteins by indirect immunofluorescence confocal microscope analyses showed no overlap of S100B and ATAD3A immunoreactivities (Fig. 2c). Hence, if S100B does interact with ATAD3A in a cellular context, such an interaction should occur in the cytosol prior to ATAD3A mitochondrial targeting.
FIG. 2.
Characterization of ATAD3A and S100B in oligodendrocyte progenitor cells. (a) Western blot analysis of S100B, ATAD3A, ATP synthase F1 subunit, and tubulin in total extracts (20 μg) of exponentially growing OPC (lane 1) and OPC left for 24 h (lane 2) and 72 h (lane 3) in differentiation medium. (b) Rat OPC lysed in a buffer containing 40 mM Tris, 150 mM NaCl, and 0.3% Triton X-100 (pH 7.5) were incubated with S16 monoclonal S100B antibody in the presence of 5 mM EGTA (lane 2) or 0.3 mM CaCl2 and 10 μM ZnSO4 (lane 3). Total cell extract (lane 1) and immunoprecipitates (lanes 2 and 3) were analyzed by Western blotting with ATAD3A N-ter and S100B antibodies. IP, immunoprecipitation. (c) Rat OPC were double immunostained with anti-S100B S16 monoclonal antibody (red) and anti-ATAD3A N-ter antibody (green). Low-resolution (top [bar, 10 μm]) and high-resolution (bottom [bar, 5 μm]) confocal analysis of double-immunostained cells could not detect overlap between the two antibodies. (d) Exponentially growing OPC were mock transfected (SiRNA−) or transfected with ATAD3A siRNA (SiRNA+). After 4 days, cells were lysed in SDS sample buffer, and total proteins were separated by SDS-PAGE and analyzed by Western blotting using mixed anti-ATAD3A N-terminal (ATAD3A) and -β-tubulin (tubulin) antibodies. (e) OPC mock transfected (control) or transfected with ATAD3A siRNA were grown for 72 h in differentiation culture medium and triple immunostained with anti-ATAD3A N-terminal (ATAD3A), anti-β-tubulin monoclonal IgG (tubulin), and anti-O4 monoclonal IgM (O4) antibodies. Bar, 40 μm. White arrowheads point to cells that showed maximal ATAD3A downregulation and that are characterized by limited morphological transformation and an absence of O4 immunoreactivity.
In mouse OPC, S100B contributes to cell growth and differentiation (11). To evaluate the possible implication of ATAD3A during oligodendrogenesis, control siRNA and siRNA directed against rat ATAD3A production were transfected into OPC by electroporation using the Amaxa rat oligodendrocyte Nucleofector kit. ATAD3A downregulation by ATAD3A siRNA was analyzed by Western blotting (Fig. 2d) and indirect immunofluorescence (Fig. 2e). Forty-eight hours posttransfection, cells were shifted to defined culture medium without growth factors and supplemented with thyroid hormone. After 72 h, cells were fixed and triply immunostained with ATAD3A, β-tubulin, and O4 antibodies (Fig. 2e). In control cultures, cells adopted typical stellate-shaped processes and strong immunostaining with O4 antibodies. The average of two experiments revealed that 90% ± 5% cells stained positively for O4. In cultures treated with ATAD3A siRNA, ATAD3A was downregulated to different extents in cells with robust downregulation in 10% to 20% of cells. Cells that showed a robust downregulation of the ATAD3A protein exhibited a much-less-differentiated morphology systematically and were not able to be detected with an O4 antibody (Fig. 2e). The percentage of cells that did express O4 antigen in cultures treated with ATAD3A siRNA decreased to 70% ± 5%. The ATAD3A null phenotype is consistent with a more general function of the protein in the regulation of cell growth (20a).
Mapping of the interaction domains in the S100B-ATAD3A complex.
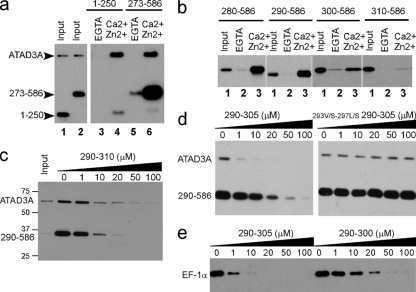
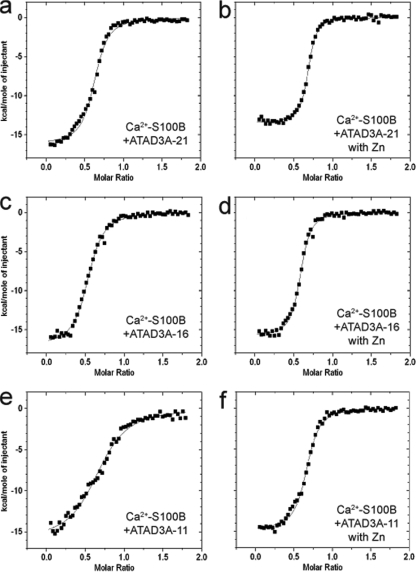
As a first step to extend the study of the S100B-ATAD3A interaction, we characterized the molecular basis of the interaction between the two proteins. In this regard, S100B-Sepharose pulldown assays were used to determine whether the N- and/or C-terminal half of ATAD3A contributed to complex formation with S100B. Because recombinant full-length ATAD3A and its C-terminal half spontaneously aggregated when expressed in bacteria, we used endogenous full-length ATAD3A and deletion mutants expressed in mammalian cells to define the S100B binding site. In one ATAD3A construct, the first 250 residues of ATAD3A were fused with Myc (ATAD3A N-ter half mutant 1-250-Myc), and in the other construct, residues at the C terminus of ATAD3A were combined with Myc (C-ter half mutant 273-586-Myc), and each construct was then tested for its ability to bind S100B-Sepharose in the presence or absence of Ca2+ and Zn2+. Each of these ATAD3A protein constructs was expressed in U373 cells, and binding interactions with S100B beads were compared to that of the endogenous full-length wild-type ATAD3A protein (Fig. 3). As shown in Fig. 3a, only C-ter half-mutant 273-586-Myc showed appreciable binding to S100B. Further deletion of the C-terminal-half mutant revealed that the high-affinity binding domain is localized between residues 290 and 310 of ATAD3A (Fig. 3b). To confirm the results from the mapping of the high-affinity S100B binding domain on ATAD3A, we next prepared a synthetic peptide corresponding to residues 290 to 310 and tested this peptide in competitive binding experiments. Total extracts of U373 cells transfected with the ATAD3A fragment at residues 290 to 586 were incubated with S100B-Sepharose beads in the presence of increasing concentrations of the ATAD3A290-310 peptide (P290-310), and it was found that P290-310 abolished binding to S100B beads for both the endogenous ATAD3A protein as well as the ATAD3A fragment at residues 290 to 586 (Fig. 3c). Next, the affinity of the ATAD3A290-310 peptide for S100B was determined directly in the presence of calcium, or calcium plus zinc, by using ITC (Fig. 4a and b). The dissociation constant (KD) for the S100B-P290-310 complex was found to be 0.6 ± 0.2 μM in the presence of Ca2+ and decreased to 0.2 ± 0.02 μM in the presence of Ca2+ and Zn2+ (Table 1). No binding of P290-310 was observed in the absence of calcium (data not shown). The increased affinity in the presence of Zn2+ is consistent with a positive regulatory function of Zn2+ on the Ca2+-dependent interaction of S100 with target proteins (19, 38, 39) The ATAD3A290-310 peptide represents one of the highest-affinity small-peptide targets for S100B (54) (Table 1).
FIG. 3.
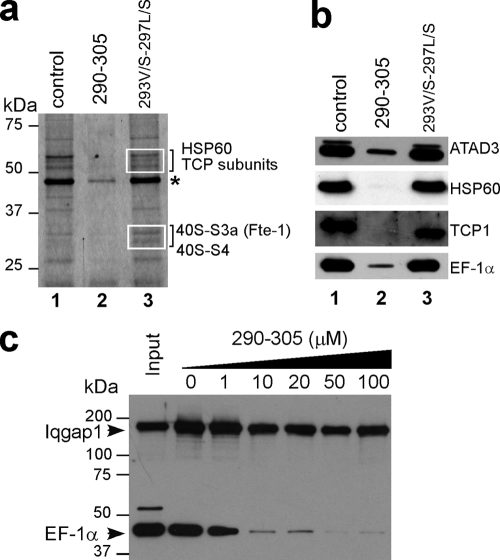
Characterization of the S100B binding domain of ATAD3A. (a and b) Pulldown mapping of the S100B binding site on ATAD3A. (a) ATAD3A fragments comprising residues 1 to 250 and 273 to 586 are expressed in U373 cells. The respective cell lysates (lanes 1 and2) were tested by S100B pulldown with EGTA (lanes 3 and 5)- or Ca2+ and Zn2+ (lanes 4 and 6)-containing buffer. (b) The fragments comprising residues 280 to 586, 290 to 586, 300 to 586, and 310 to 586 of ATAD3A were tested according to the same process described above (a). Lane 1, input; lane 2, EGTA; lane 3, Ca2+ and Zn2+. Endogenous ATAD3A (a) and its fragments (a and b) were revealed by Western blotting using a mixture of ATAD3A N-ter and C-ter antibodies. (c to e) Characterization of the minimal S100B binding domain on ATAD3A. (c) ATAD3A290-310 functions as a competitor for the Ca2+-dependent interaction of ATAD3A and the fragment at residues 290 to 586 with S100B-Sepharose beads. (d) ATAD3A290-305 but not its mutant counterpart 293V/S-297L/S-290-305 functions as a competitor for the Ca2+-dependent interaction of ATAD3A and the fragment at residues 290 to 586 with S100B-Sepharose beads. (e) Comparison of ATAD3A peptide fragments at residues 290 to 305 and 290 to 300 as competitors for the Ca2+-dependent interaction of EF-1α with S100B-Sepharose beads.U373 cell lysates were tested by S100B pulldown with Ca2+- and Zn2+-containing buffer in the presence of increasing concentrations of peptide competitor P290-305, as indicated. Endogenous ATAD3A and its fragment at residues 290 to 586 (c and d) or EF-1α (e) were revealed by Western blotting.
FIG. 4.
ITC titration of ATAD3A constructs into Ca2+-S100B in the absence and presence of zinc. Data represent the titration of 400 μM ATAD3A-21 (a and b), ATAD3A-16 (c and d), and ATAD3A-11 (e and f) into 50 μM Ca2+-S100B in the absence (a, c, and e) and presence (b, d, and f) of zinc, as monitored by ITC. All experiments were performed at 37°C with sample and titrant solutions containing 10 mM CaCl2, 10 mM TES, 15 mM NaCl, 100 mM KCl, and 5% DMSO (pH 7.2). For experiments containing zinc, a concentration of 60 μM ZnAc was used for the sample and titrant.
TABLE 1.
Dissociation constants, enthalpy values, and entropy terms measured for the titration of ATAD3A-derived peptides into S100B and S100A1 using ITCa
| Protein | ATAD3A peptide residues (aa length) | Avg KD (nM) ± SD | Avg ΔHapp (kcal/mol) ± SD | Avg TΔS (kcal/mol) ± SD |
|---|---|---|---|---|
| S100B | 290-310 (21) | 620 ± 170 | −12.7 ± 0.5 | −3.9 ± 0.3 |
| S100B + Zn | 290-310 (21) | 200 ± 20 | −13.6 ± 0.1 | −4.1 ± 0.2 |
| S100B | 290-305 (16) | 630 ± 190 | −15.5 ± 1.1 | −6.6 ± 1.3 |
| S100B + Zn | 290-305 (16) | 390 ± 40 | −15.3 ± 0.5 | −6.2 ± 0.5 |
| S100B | 290-300 (11) | 3,360 ± 1,400 | −16.5 ± 0.6 | −8.7 ± 0.8 |
| S100B + Zn | 290-300 (11) | 720 ± 190 | −13.5 ± 1.7 | −4.8 ± 1.7 |
The solution included 10 mM TES, 15 mM NaCl, 100 mM KCl, 10 mM CaCl2, and 5% DMSO (pH 7.2) at 37°C. Where zinc is present, a concentration of 60 μM ZnAc was used. KD, enthalphy (ΔHapp), and entropy term (TΔS) parameters represent the average values of multiple experiments, with the standard deviations reported as the uncertainty. Ca2+-S100A1 binds ATAD3A-21 with a KD of 1.2 ± 0.7 μM and has a second weak-binding-site KD of >50 μM.
Further deletion within the C terminus of the ATAD3A peptide identified the minimal S100B binding domain comprised of the sequence 290-RITVLEALRHPIQVSR-305. The ATAD3A290-305 peptide (P290-305) affinity for S100B was similar to that of P290-310 (Fig. 4c and d and Table 1); it was still able to compete with ATAD3A for binding to S100B beads (Fig. 3d). The mutagenesis of Val293 and Leu297 into hydrophilic Ser residues on P290-305 totally abrogated the capacity of the peptide to compete with ATAD3A for binding to S100B beads (Fig. 3d). This indicates that a sequence in ATAD3A, termed the VLEAL motif, is directly contributing to the interaction with S100B and that hydrophobic interactions are likely prevalent for that interaction. An “IQ motif” (residues 300 to 305) is also present in P290-305, which could contribute to the interaction with S100B (36, 42) (Table 2). To test this, we compared the binding of P290-305 and P290-300, a construct that lacks the “IQ” sequence, to S100B by ITC (Fig. 4 and Table 1). The results showed that the removal of the IQ motif significantly decreased the affinity of the 11-mer ATAD3A290-300 peptide interaction with S100B (KD = 3.4 ± 1.4 μM). Likewise, the removal of the IQ motif also decreased the efficiency of the 11-mer peptide to compete with EF-1α binding to S100B-Sepharose beads (Fig. 3e). Altogether, these results suggest that the IQ motif, although not dispensable, significantly contributes to the interaction between ATAD3A and S100B. Likewise, a hydrophobic interaction involving residues 293 and 297 of ATAD3A is also important for complex formation with calcium-loaded S100B.
TABLE 2.
Sequence alignment of S100B binding domainsa
| Binding domain | Sequence |
|---|---|
| ATAD3A | 290 RITVLEALRHPIQVSR 305 |
| p53 | 341 FRELNEALELKDAQAG 355 |
| Consensus | OXEAL*X1-3OQ |
| EF-1α | 213 GPTLLEALDQIQEPKR 228 |
| IQGAP1 | 542 IGLINEALDEGDAQKT 557 |
Sequence alignment of the minimal high-affinity S100B binding domains on ATAD3A (this study) and p53 (13) reveals a consensus sequence (O is hydrophobic, * is hydrophilic, and X is variable). This consensus sequence can be predicted for two recognized major S100B target proteins, EF-1α (this study) and IQGAP1 (36).
The interaction domain for S100B on ATAD3A defines a consensus Ca2+-dependent S100B binding sequence.
The S100B binding domain that we mapped on ATAD3A is very similar to the high-affinity and S100B-specific binding domain on the p53 protein (Table 2) (12). Such a domain can also be predicted on EF-1α and IQGAP1, two proteins that physically interact with S100B (Table 2). To investigate whether the S100B binding domain on ATAD3A may represent a consensus Ca2+-dependent S100B binding sequence, we next compared the effects of the 290-305 and 293V/S-297L/S-290-305 peptides on the binding of proteins present in U373 cell extracts to S100B-Sepharose beads (Fig. 5a). Gel electrophoresis and silver-staining analyses showed that the ATAD3A290-305 peptide (lane 2), but not its mutant counterpart (lane 3), antagonized the Ca2+-dependent binding of most proteins to S100B beads. Differential quantitative nano-LC-MS/MS analysis of proteins recovered in Fig. 5a, lanes 2 and 3. revealed that the major proteins antagonized by the ATAD3A290-305 peptide are linked with chaperone activities (see Materials and Methods). These include the subunits of the multichaperone complexes TCP and HSP60. Western blot analysis confirmed the sequencing data (Fig. 5b). Several other major proteins that we found to be associated with S100B beads were also sequenced and corresponded to proteins associated with translating ribosomes. These include the translational regulator GCN1 (molecular weight [MW], 292) and the 40S ribosomal proteins S3a (MW, 28.9) and S4 (MW, 29.3). The ATAD3A290-305 peptide was also able to antagonize the binding of other S100B target proteins such as tubulin and vimentin (not shown). Although the ATAD3A290-305 peptide does antagonize the binding of most proteins to S100B beads (Fig. 5a and b), it was not able to inhibit the Ca2+-independent interaction of the IQGAP1 protein with S100B (Fig. 5c). Altogether, these data suggest that the S100B binding domain on ATAD3A may represent a consensus Ca2+-dependent S100B binding sequence.
FIG. 5.
The ATAD3A peptide functions as a competitor for Ca2+-dependent target binding to S100B. (a) S100B-Sepharose beads were incubated with U373 cell extracts in Ca2+- and Zn2+-containing buffer in the absence (lane 1) or in the presence of 10 μM peptide competitor P290-305 (lane 2) or 293V/S-297L/S-290-305 peptide (lane 3). Bound proteins were separated by 10% SDS-PAGE, and the gel was silver stained. Proteins in the areas in squares were sequenced by nano-LC-MS/MS analysis as described in Materials and Methods. The major proteins whose binding is specifically antagonized by the P290-305 peptide are listed in the left margin. The asterisk indicates the position of the EF-1α protein. (b) Western blot analysis of the SDS-PAGE gel shown above (a) with corresponding antibodies. (c) S100B pulldown of EF-1α and IQGAP1 in the presence of increasing concentrations of peptide competitor P290-305 as indicated. Bound proteins were separated by 8% SDS-PAGE and analyzed by Western blotting using mixed anti-EF-1α and -IQGAP1 antibodies.
Structural organization of the S100B-ATAD3A complex resolved by NMR spectroscopy.
NMR spectroscopy has defined consensual mechanisms of the Ca2+-dependent interaction of S100B with target peptides (2, 6, 28, 45, 46). To model the S100B-ATAD3A peptide complex, we used NMR analyses (Fig. 6a to c). Like most S100 protein targets, S100B targets bind to the hydrophobic pocket exposed upon Ca2+ binding, consisting of residues along helix 4 and the hinge region (the loop connecting helix 2 and helix 3). To determine which residues of S100B are responsible for the tight interaction with ATAD3A, chemical shift perturbations were measured by NMR. Specifically, HSQC spectra were collected during titrations with the ATAD3A290-310 peptide (Fig. 6a). In such titrations, it was found that the peptide bound in the slow-exchange-time regimen on the chemical shift time scale, which is indicative of tight binding. As a result, data from a series of heteronuclear multidimensional NMR experiments [NOESY-HSQC, HMQC-NOESY-HSQC, HNCA, HNCACB, and CBCA(CO)NH] were collected, as necessary, to unambiguously complete the backbone 1H, 13C, and 15N resonance assignments of Ca2+-S100B bound to the ATAD3A peptide.
FIG. 6.
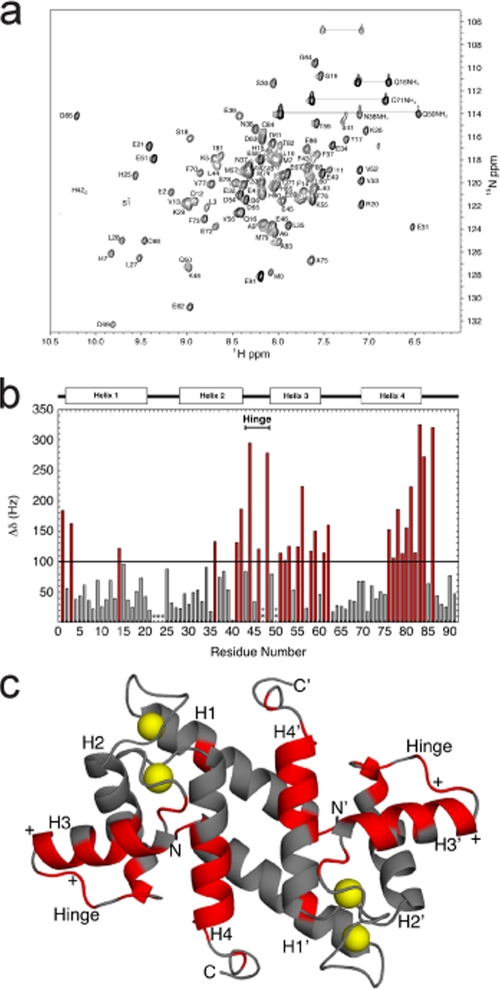
Interaction between calcium-bound S100B and the ATAD3A peptide (P290-305) examined by NMR spectroscopy. (a) Two-dimensional 1H-15N HSQC spectrum of 15N-labeled Ca2+-S100B bound to the ATAD3A peptide (unlabeled). The conditions of the NMR sample are described in Materials and Methods. (b) Histogram of chemical shift perturbations (Δδ in Hz for 1H + 15N) as a result of titrating the ATAD3A peptide into calcium-bound S100B. Residues that showed perturbations greater than 100 Hz are in red. (c) Ribbon diagram of the NMR structure of Ca2+-bound S100B (PDB accession number 2K7O), with those residues having chemical shift perturbations upon the addition of the ATAD3A peptide greater than 100 Hz in red. The yellow spheres represent calcium ions in the two EF hands (EF1 and EF2) in each subunit of dimeric calcium-bound S100B.
A comparison of the resonance assignments of Ca2+-S100B in the absence and in the presence of ATAD3A290-310 illustrated that there were a number of residues that displayed changes in the chemical shift upon the addition of the ATAD3A peptide (Fig. 6b and c). Affected residues were located throughout the protein, including at positions in helix 1 (S1, L3, and F14), helix 2 (I36, S41, and F43), the hinge region (L44, E46, and K48), helix 3 (E51, V52, V53, K55, V56, E58, and T59), helix 4 (F76, V77, S78, M79, V80, T81, and T82), and the C-terminal loop (A83, C84, and E86). Many of these residues also displayed changes in chemical shifts in S100B-p53 and S100B-TRTK12 complex formations indicative of the binding of ATAD3A290-310 to Ca2+-S100B in a similar fashion. In addition, previous studies demonstrated that helix 4 is extended upon target binding (28, 45), possibly explaining the large chemical shift changes for residues A83, C84, and E86. Most importantly, the seven amino acid side chains that form interactions with both p53 and TRTK-12 peptides (L44, V52, K55, V56, M79, V80, and A83) also displayed changes in the chemical shift upon binding ATAD3A. Taken together, these data all support the notion that the ATAD3A peptide binds to the same hydrophobic binding site on calcium-bound S100B as found previously for the binding of the p53 and TRTK-12 peptides. Likewise, these structural data combined with the high affinity of the ATAD3A peptide for Ca2+-S100B confirmed that the S100B binding domain on ATAD3A represents a canonical Ca2+-dependent S100B binding sequence.
S100B prevents aggregation of ATAD3A mutants.
To understand the regulatory function of S100B resulting from its interaction with ATAD3A, we undertook studies to characterize the functional role of the S100B binding domain of ATAD3A (aa 290 to 310).
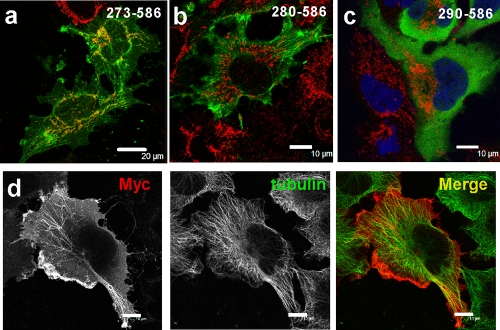
Using deletion mutants, we first found that the S100B binding site on ATAD3A overlaps with an aggregation-prone sequence and is contiguous to a mitochondrion-interacting region (Fig. 7a to c). For example, when expressed in U373 cells, the 273-586 ATAD3A-Myc mutant localized with the mitochondrial marker and also a decorated fibrous cytosolic structure (Fig. 7a). The deletion of 7 amino acids caused a relocalization of the 280-586 ATAD3A-Myc mutant at membrane ruffles with an enhanced immunostaining of cytosolic fibrous structures (Fig. 7b and d). The fibrous structures formed by the ATAD3A280-586 mutant protein often paralleled the mitochondrial network and did not colocalize with any known cytoskeletal proteins, including tubulin (Fig. 7d), actin, or intermediate filament proteins (data not shown). One possibility for such a unique staining pattern is that it could represent ATAD3A mutant aggregates that may form along translating ribosome tracks. The deletion of 10 additional amino acid residues at the N terminus of this construct resulted in a relocalization of the truncated 290-586 ATAD3A-Myc protein diffusely within the cytoplasm (Fig. 7c). These results indicate that the amino acid sequence at residues 273 to 290 comprises signals that function to regulate the mitochondrial localization and the cytoplasmic processing of ATAD3A mutants.
FIG. 7.
The S100B binding domain on ATAD3A promotes the aggregation of C-terminal ATAD3A mutants. (a to c) Indirect immunofluorescence analysis of U373 cells transfected with Myc-tagged ATDA3A mutants comprising fragments at residues 273 to 586 (a), 280 to 586 (b), and 290 to 586 (c) with anti-Myc (green) and anti-ATP synthase F1 (red). Bars, 20 μm (a) and 10 μm (b and c). (d) The 280-586 ATAD3A mutant aggregates under a fibrous structure in U373 cells. Transfected U373 cells with the 280-586 ATAD3A-Myc mutant were fixed with PFA and double immunostained with anti-Myc (red) and anti-β-tubulin (green) antibodies. Bar, 10 μm.
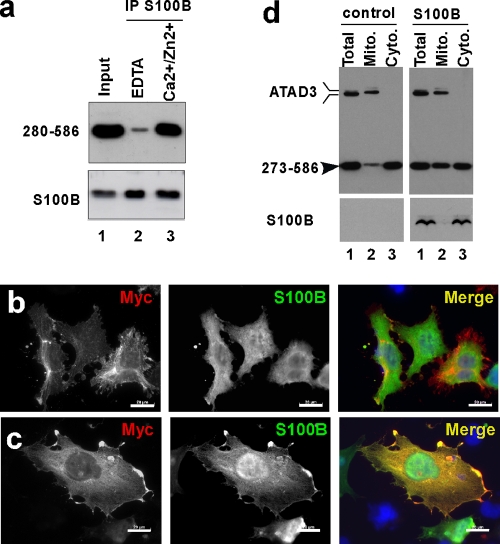
To investigate whether S100B regulates the processing of ATAD3A mutants, we next analyzed the interactions between S100B and the truncated 280-586 ATAD3A-Myc construct. When coexpressed in U373 cells, S100B and the 280-586 ATAD3A-Myc mutant coimmunoprecipitated in Ca2+/Zn2+-containing buffer, indicating that S100B can interact with 280-586 ATAD3A-Myc mutant aggregates (Fig. 8a). Indirect immunofluorescence analysis was used to evaluate the interaction between the two proteins in a cellular context (Fig. 8b and c). Due to the strict Ca2+-dependent interaction between the ATAD3A construct and S100B proteins and because of the low affinity of S100B for Ca2+ observed in vitro, we used mild membrane permeabilization methods to ensure a sufficient elevation in the free-calcium level in the cells and to shift the apo-S100B protein into its Ca2+-bound conformation. Double-transfected cells were incubated in culture medium containing 0.01% Triton X-100 in the presence of either 2 mM EDTA or 2 mM Ca2+ for 2 min prior to fixation with 4% PFA in 20 mM HEPES (pH 7.5) supplemented with 2 mM EDTA or 2 mM Ca2+. In EDTA-containing media, the 280-586 ATAD3A-Myc protein decorates the fibrous anchor structures, and S100B localized diffusely within the cytoplasm and the nuclei (Fig. 8b), whereas in Ca2+-containing media, the filamentous 280-586 ATAD3A-Myc aggregates disappeared, and the 280-586 ATAD3A-Myc protein colocalized with S100B in the cytoplasm and at membrane ruffles (Fig. 8c). The cytoplasmic relocalization of the 280-586 ATAD3A-Myc protein in response to the elevation of Ca2+ levels is dependent strictly on the presence of S100B, as demonstrated with control experiments performed without S100B being present (not shown). In summary, these results demonstrated that the S100B binding domain overlaps with the aggregation-prone sequence on the 280-586 ATAD3A-Myc mutant and that S100B can dissociate the 280-586 ATAD3A-Myc aggregates in a Ca2+-dependent manner as necessary for the relocalization of ATAD3A to the cytoplasm.
FIG. 8.
S100B dissociates ATAD3A mutant aggregates. (a) The 280-586 ATAD3A mutant coimmunoprecipitates with S100B. U373 cells were double transfected with the 280-586 ATAD3A-Myc mutant and S100B. Cells were lysed in buffer containing 40 mM Tris, 150 mM NaCl, and 0.3% Triton X-100 (pH 7.5) and incubated with S16 monoclonal S100B antibody in the presence of 5 mM EDTA (lane 2) or 0.3 mM CaCl2 and 10 μM ZnSO4 (lane 3). Total cell extract (lane 1) and immunoprecipitates (lanes 2 and 3) were analyzed by Western blotting with polyclonal ATAD3A N-ter and S100B antibodies. (b and c) S100B dissociates the 280-586 ATAD3A mutant aggregates in the presence of Ca2+. U375 cells doubly transfected with the 280-586 ATAD3A-Myc mutant and S100B were incubated in culture medium containing 0.01% Triton X-100 in the presence of 2 mM EDTA (b) or 2 mM Ca2+ (c) for 2 min prior to fixation with 4% PFA in 20 mM HEPES (pH 7.5). Cells were double immunostained with anti-Myc (red) and anti-S100B (green) antibodies. Bar, 20 μm. (d) S100B enhances the association of the 273-586 ATAD3A mutant with the mitochondrial fraction. U375 cells were double transfected with the 273-586 ATAD3A-Myc mutant and the control plasmid (control) or S100B plasmid (S100B). After 24 h, total cell extracts (lane 1), the mitochondrial fractions (lane 2), and the cytoplasmic fractions (lane 3) were analyzed by Western blotting with a mixture of ATAD3 C-ter and N-ter antibodies and anti-S100B S16 antibody.
We finally analyzed the effect of the ectopic expression of S100B on the mitochondrial localization of the 273-586 ATAD3A-Myc mutant (Fig. 8d). U373 cells were doubly transfected with the 273-586 ATAD3A-Myc mutant plasmid and the control or S100B plasmid. After 24 h, the amount of the 273-586 ATAD3A-Myc protein recovered in the mitochondrial and cytosolic fractions was analyzed by Western blotting. Two independent experiments revealed a significant increase in the amount of the 273-586 ATAD3A-Myc protein associated with the mitochondrial fraction in cells cotransfected with the S100B plasmid (Fig. 8d). Indirect immunofluorescence analysis showed no colocalization of S100B with the 273-586 ATAD3A-Myc protein bound to mitochondria (not shown). Altogether, these observations suggest that S100B could assist the 273-586 ATAD3A-Myc protein during its cytoplasmic processing, preventing aggregation and favoring its mitochondrial localization.
DISCUSSION
In OPC, S100B contributes to OPC differentiation in response to demyelinating insult (11). To understand the contribution of S100B to OPC differentiation, we searched for high-affinity S100B target proteins. S100B belongs to the S100 family of EF-hand calcium binding proteins. The conserved structure among the S100 proteins implies that these proteins will be able to interact with the same targets. Indeed, most cellular proteins that bind to S100B-Separose beads are also generally recovered associated with other S100 species in pulldown assays (19, 36). Hence, the criterion that should be considered to determine the specificity of the interaction of the individual S100 protein species with their respective targets is the affinity of the interaction. With this in mind, we used a far-Western analysis to identify high-affinity S100B target proteins in OPC. The results here identified the mitochondrial ATAD3A protein as the major protein in OPC that interacted with [35S]S100B at sub-μM concentrations. The specificity of the interaction between ATAD3A and S100B was next confirmed by competition assays with other related S100 species, including the closest homolog of S100B, the S100A1 protein (Fig. 1d).
To investigate possible interactions between the two proteins at the mitochondrial level, we used indirect immunofluorescence and confocal microscope analysis (Fig. 2c). The results showed no overlap between S100B and ATAD3A immunoreactivities. Hence, one has to consider that if S100B does interact with ATAD3A in a cellular context, such an interaction should occur prior to mitochondrial targeting. To understand the functional meaning of the interaction between S100B and ATAD3A, we next identified the interaction domain on ATAD3A for S100B. In these experiments, we identified a consensus S100B binding motif on ATAD3A comprised of residues 290 to 310 (P290-310). NMR spectroscopy analysis of the S100B-P290-310 peptide complex demonstrated that the ATAD3A290-310 peptide is binding to the same site on S100B as that seen with other S100B binding peptides (2, 6, 45). However, the affinity of the P290-310 peptide for S100B (KD of ∼0.2 μM) is higher than those reported previously for many other S100B binding peptides (54). This may be due to the fact that the ATAD3A290-310 peptide has an optimal hydrophobic binding site for interactions with S100B compared to some of the other S100B-peptide complexes studied thus far. Specifically, punctual and deletion mutagenesis identified the presence of two putative S100B binding motifs within the P290-310 peptide sequence (Fig. 3 and Table 2). The first motif, OXEAL*, is also found in other recognized calcium-dependent S100B target proteins, including the p53 tetramerization domain, IQGAP1, and EF-1α (Table 2). In ATAD3A, the 293VXEAL*298 motif is essential for the interaction with S100B, since the mutation of the hydrophobic 293V and L297 residues totally abrogates binding (Fig. 3d). The second motif, OQ, is similar to the IQ motif present in IQGAP1 known to bind S100B in a Ca2+-independent but Zn2+-dependent manner (36, 42). In ATAD3A, the IQ motif is not dispensable for Ca2+-dependent S100B binding to the P290-305 peptide but strengthens the interaction (Table 1 and Fig. 4). In IQGAP1, the presence of four IQ motifs (752LQARCR757, 782IQSQWR787, 812IQSLAR817, and 842IQAFIR847) may explain why the S100B-IQGAP1 interaction could not be fully antagonized by the P290-305 peptide (Fig. 5c).
We next analyzed the properties of the S100B binding domain on ATAD3A (aa 290 to 310) and found that it is contiguous to a domain on ATAD3A that is important for cytoplasmic ATAD3A processing and mitochondrial localization. With the 280-586 ATAD3A-Myc mutant, we showed that S100B binding dissociated ATAD3A mutant aggregates (Fig. 8a to c). With the 273-586 ATAD3A-Myc mutant, S100B increased the amount of the 273-586 ATAD3A-Myc mutant protein associated with the mitochondrial fraction (Fig. 8d). Altogether, these observations suggest that S100B protects the 273-586 ATAD3A mutant from aggregation to favor its mitochondrial targeting. In summary, we propose that S100B could function in assisting ATAD3A during its cytoplasmic processing prior to mitochondrial targeting. S100B could cooperate with other components of the cellular translational and folding machineries. Several arguments support this hypothesis. First, ATAD3A belongs to a family of ATPases characterized by a C terminus that harbors a conserved AAA ATPase domain allowing the formation of ring hexamers (17, 22). Computer-assisted analysis also revealed the presence of two 3,4-heptad repeat coiled-coil domains (CC1, amino acids 85 to 115; CC2, amino acids 180 to 220) within its cytoplasmic N terminus with high oligomerization probability. It is likely that proteins with multiple aggregation-prone sequences like ATAD3A require assisting cellular folding during synthesis (57). Second, proteomic analysis revealed that many proteins that bound to S100B-Sepharose beads are associated with the protein translational machinery, including translational regulators (EF-1α and GCN1), ribosomal proteins (S3a and S4), and multichaperone complex proteins (TCP-1 subunits and HSP-60) (Fig. 5a and b). EF-1α, which quantitatively represents the major S100B binding proteins here (Fig. 1b and 5a), plays a principal role in translation and catalyzes the GTP-dependent binding of aminoacyl-tRNA. EF-1α has also been demonstrated to interact with newly synthesized polypeptides with chaperone-like activity (26). Third, the S100B protein content in OPC correlates with increased mitochondrial protein synthesis demand (Fig. 2a). The coordinated mRNA localization and translation of nuclear mitochondrial proteins in the close vicinity of the mitochondrial outer membrane surface are essential for mitochondrial biogenesis (29, 31). However, actually little is known regarding the spatiotemporal regulation of nuclear mitochondrial protein synthesis and import. Fractionation experiments using OPC have shown that S100B can be recovered in a Ca2+-dependent manner with mitochondrial preparations that also contain ribosome-associated proteins, including EF-1α (Fig. 9). We believe that further studies to reconstitute the S100B-sensitive translational interactome at the mitochondrial surface should shed light on the molecular regulation of ATAD3A by S100B and may have implications for mitochondrial biogenesis in general. At this stage, we stress the fact that the apparent increase in the interaction of the 273-586 ATAD3A mutant with mitochondria in the presence of S100B does not required cell stimulation with Ca2+-mobilizing drugs. This is particularly significant if we consider that the high-affinity EF-hand calcium binding site on S100B for calcium (10 to 20 μM) is compatible only with microdomains of high Ca2+ concentrations found in the mitochondrial environment (7). Microdomains of high Ca2+ concentrations (10 to 100 μM) are due to a close spatial coupling between the endoplasmic reticulum and mitochondria (7, 44). It is thus likely that within the proximal mitochondrial environment, the high levels of Ca2+ may yield specific, rapid, and spatially limited changes in the activity of S100B to assist both protein synthesis and mitochondrial import processes. Another mechanism for the calcium-dependent S100B interaction inside cells is that the S100B-ATAD3A interaction could significantly increase the calcium affinity by decreasing the off rate of calcium, which was observed previously for S100B and for other S100-target protein interactions that have been observed at calcium concentrations as low as 100 nM (35, 55). Nonetheless, the proposed new function for S100B in assisting in the cytoplasmic folding of the target protein with the consensus S100B binding motif for proper subcellular localization should be validated with other S100B-specific target proteins.
FIG. 9.
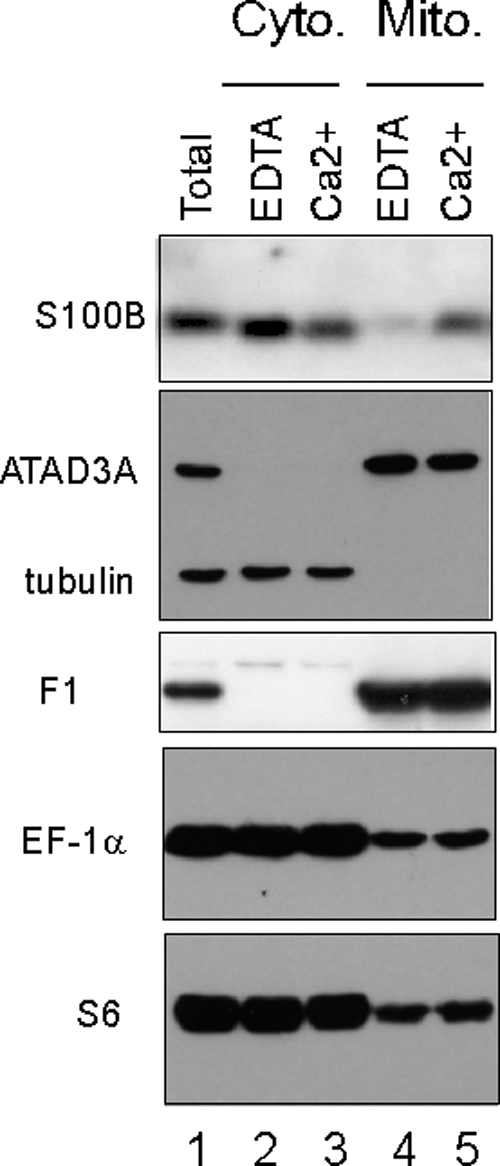
S100B and ribosomally associated EF-1α and S6 proteins associate with the mitochondrial fraction purified from rat OPC. Cytosolic (lanes 2 and 3) and mitochondrial (lanes 4 and 5) fractions were prepared from cultures of rat OPC in buffer supplemented with 5 mM EDTA (lanes 2 and 4) or 0.3 mM CaCl2 and 10 μM ZnSO4 (lanes 3 and 5), as described in Materials and Methods. Proteins were analyzed by Western blot analysis with anti-S100B antibodies, a mix of anti-ATAD3A and -β-tubulin antibodies, anti-ATP synthase F1, anti-EF-1α, and anti-S6, as indicated. Lane 1, total cell extract. Note that S100B associates with the mitochondrial fraction in a calcium-dependent manner.
A consensus S100B binding motif is present within the high-affinity S100B binding tetramerization domain (YF-O2 domain) of the p53 protein (Table 2) (12). On p53, the YF-O2 domain regulates oligomerization and nuclear translocation. In vitro, the binding of S100B to p53 prevents temperature-dependent p53 oligomerization at 37.5°C (3, 18). In vivo, the stable ectopic expression of S100B in rat embryo fibroblasts expressing temperature-sensitive p53val135 rescues wild-type p53 nuclear translocation and activities at a nonpermissive temperature (37.5°C) (48, 49). We propose that the interaction of S100B with nascent cytoplasmic p53val135 through its YF-O2 domain might contribute to prevent the p53val135 protein from thermal denaturation to favor its nuclear translocation and to rescue its nuclear wild-type activities. This hypothesis was discussed previously in more detail (48, 49). The p53 protein is also able to rapidly translocate to mitochondria under stress conditions to induce mitochondrial outer membrane permeabilization and cytochrome c release (15, 37). Further studies should investigate whether S100B can also contribute to p53 mitochondrial translocation. S100B overexpression can also lead to p53 inactivation (32). The dual effect of S100B on p53 functions could be explained by the presence of several S100B binding domains on the p53 sequence (18, 52). In contrast to the high-affinity tetramerization YF-O2 domain, the additional S100B binding sites on p53 can bind other S100 species (18, 52, 53), and their occupation can lead to p53 inactivation. The development of inhibitors of the S100B-p53 interaction to treat cancer is a subject of active research (34, 58). We suggest that these studies should take into account the multiple S100B binding sites on p53 and their possible dual activities in wild-type p53 processing and functions.
Acknowledgments
This work was supported by the Institut National du Cancer (INCA PL 114 to J.B.) and by the Association contre le Cancer (ARC no. 4829 to J.B.). In addition, grants from the National Institutes of Health (GM58888 and CA107745 to D.J.W.) supported this work.
We thank N. Bertacchi for excellent technical assistance.
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Baldisseri, D. M., R. R. Rustandi, Z. Zhang, C. Tang, C. L. Bair, A. Landar, A. Landar, D. B. Zimmer, and D. J. Weber. 1999. 1H, 13C and 15N NMR sequence-specific resonance assignments for rat apo-S100A1(alpha alpha). J. Biomol. NMR 14:91-92. [DOI] [PubMed] [Google Scholar]
- 2.Barber, K. R., K. A. McClintock, G. A. Jamieson, Jr., R. V. Dimlich, and G. S. Shaw. 1999. Specificity and Zn2+ enhancement of the S100B binding epitope TRTK-12. J. Biol. Chem. 274:1502-1508. [DOI] [PubMed] [Google Scholar]
- 3.Baudier, J., C. Delphin, D. Grunwald, S. Khochbin, and J. J. Lawrence. 1992. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc. Natl. Acad. Sci. U. S. A. 89:11627-11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudier, J., and D. Gerard. 1986. Ions binding to S100 proteins. II. Conformational studies and calcium-induced conformational changes in S100 alpha alpha protein: the effect of acidic pH and calcium incubation on subunit exchange in S100a (alpha beta) protein. J. Biol. Chem. 261:8204-8212. [PubMed] [Google Scholar]
- 5.Baudier, J., N. Glasser, and D. Gerard. 1986. Ions binding to S100 proteins. I. Calcium- and zinc-binding properties of bovine brain S100 alpha alpha, S100a (alpha beta), and S100b (beta beta) protein: Zn2+ regulates Ca2+ binding on S100b protein. J. Biol. Chem. 261:8192-8203. [PubMed] [Google Scholar]
- 6.Bhattacharya, S., E. Large, C. W. Heizmann, B. Hemmings, and W. J. Chazin. 2003. Structure of the Ca2+/S100B/NDR kinase peptide complex: insights into S100 target specificity and activation of the kinase. Biochemistry 42:14416-14426. [DOI] [PubMed] [Google Scholar]
- 7.Csordas, G., A. P. Thomas, and G. Hajnoczky. 1999. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J. 18:96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 9.Deloulme, J. C., N. Assard, G. O. Mbele, C. Mangin, R. Kuwano, and J. Baudier. 2000. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J. Biol. Chem. 275:35302-35310. [DOI] [PubMed] [Google Scholar]
- 10.Deloulme, J. C., T. Janet, D. Au, D. R. Storm, M. Sensenbrenner, and J. Baudier. 1990. Neuromodulin (GAP43): a neuronal protein kinase C substrate is also present in 0-2A glial cell lineage. Characterization of neuromodulin in secondary cultures of oligodendrocytes and comparison with the neuronal antigen. J. Cell Biol. 111:1559-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deloulme, J. C., E. Raponi, B. J. Gentil, N. Bertacchi, A. Marks, G. Labourdette, and J. Baudier. 2004. Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol. Cell. Neurosci. 27:453-465. [DOI] [PubMed] [Google Scholar]
- 12.Delphin, C., M. Ronjat, J. C. Deloulme, G. Garin, L. Debussche, Y. Higashimoto, K. Sakaguchi, and J. Baudier. 1999. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J. Biol. Chem. 274:10539-10544. [DOI] [PubMed] [Google Scholar]
- 13.Donato, R. 2003. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech. 60:540-551. [DOI] [PubMed] [Google Scholar]
- 14.Du, X. J., T. J. Cole, N. Tenis, X. M. Gao, F. Kontgen, B. E. Kemp, and J. Heierhorst. 2002. Impaired cardiac contractility response to hemodynamic stress in S100A1-deficient mice. Mol. Cell. Biol. 22:2821-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumont, P., J. I. Leu, A. C. Della Pietra III, D. L. George, and M. Murphy. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33:357-365. [DOI] [PubMed] [Google Scholar]
- 16.Eckert, R. L., A. M. Broome, M. Ruse, N. Robinson, D. Ryan, and K. Lee. 2004. S100 proteins in the epidermis. J. Invest. Dermatol. 123:23-33. [DOI] [PubMed] [Google Scholar]
- 17.Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93-114. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Fernandez, M. R., D. B. Veprintsev, and A. R. Fersht. 2005. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl. Acad. Sci. U. S. A. 102:4735-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentil, B. J., C. Delphin, G. O. Mbele, J. C. Deloulme, M. Ferro, J. Garin, and J. Baudier. 2001. The giant protein AHNAK is a specific target for the calcium- and zinc-binding S100B protein: potential implications for Ca2+ homeostasis regulation by S100B. J. Biol. Chem. 276:23253-23261. [DOI] [PubMed] [Google Scholar]
- 20.Geuijen, C. A., N. Bijl, R. C. Smit, F. Cox, M. Throsby, T. J. Visser, M. A. Jongeneelen, A. B. Bakker, A. M. Kruisbeek, J. Goudsmit, and J. de Kruif. 2005. A proteomic approach to tumour target identification using phage display, affinity purification and mass spectrometry. Eur. J. Cancer 41:178-187. [DOI] [PubMed] [Google Scholar]
- 20a.Gilquin, B., E. Taillebourg, N. Cherradi, A. Hubstenberger, O. Gay, N. Merle, N. Assard, M. O. Fauvarque, S. Tomohiro, O. Kuge, and J. Baudier. 2010. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell. Biol. 30:1984-1996. [DOI] [PMC free article] [PubMed]
- 21.Gires, O., M. Munz, M. Schaffrik, C. Kieu, J. Rauch, M. Ahlemann, D. Eberle, B. Mack, B. Wollenberg, S. Lang, T. Hofmann, W. Hammerschmidt, and R. Zeidler. 2004. Profile identification of disease-associated humoral antigens using AMIDA, a novel proteomics-based technology. Cell. Mol. Life Sci. 61:1198-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 23.He, J., C. C. Mao, A. Reyes, H. Sembongi, M. Di Re, C. Granycome, A. B. Clippingdale, I. M. Fearnley, M. Harbour, A. J. Robinson, S. Reichelt, J. N. Spelbrink, J. E. Walker, and I. J. Holt. 2007. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 176:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heizmann, C. W., G. Fritz, and B. W. Schafer. 2002. S100 proteins: structure, functions and pathology. Front. Biosci. 7:d1356-d1368. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann, M., N. Bellance, R. Rossignol, W. J. Koopman, P. H. Willems, E. Mayatepek, O. Bossinger, and F. Distelmaier. 2009. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One 4:e7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hotokezaka, Y., U. Tobben, H. Hotokezaka, K. Van Leyen, B. Beatrix, D. H. Smith, T. Nakamura, and M. Wiedmann. 2002. Interaction of the eukaryotic elongation factor 1A with newly synthesized polypeptides. J. Biol. Chem. 277:18545-18551. [DOI] [PubMed] [Google Scholar]
- 27.Hubstenberger, A., G. Labourdette, J. Baudier, and D. Rousseau. 2008. ATAD 3A and ATAD 3B are distal 1p-located genes differentially expressed in human glioma cell lines and present in vitro anti-oncogenic and chemoresistant properties. Exp. Cell Res. 314:2870-2883. [DOI] [PubMed] [Google Scholar]
- 28.Inman, K. G., R. Yang, R. R. Rustandi, K. E. Miller, D. M. Baldisseri, and D. J. Weber. 2002. Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J. Mol. Biol. 324:1003-1014. [DOI] [PubMed] [Google Scholar]
- 29.Kaltimbacher, V., C. Bonnet, G. Lecoeuvre, V. Forster, J. A. Sahel, and M. Corral-Debrinski. 2006. mRNA localization to the mitochondrial surface allows the efficient translocation inside the organelle of a nuclear recoded ATP6 protein. RNA 12:1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kube, E., T. Becker, K. Weber, and V. Gerke. 1992. Protein-protein interaction studied by site-directed mutagenesis. Characterization of the annexin II-binding site on p11, a member of the S100 protein family. J. Biol. Chem. 267:14175-14182. [PubMed] [Google Scholar]
- 31.Lelandais, G., Y. Saint-Georges, C. Geneix, L. Al-Shikhley, G. Dujardin, and C. Jacq. 2009. Spatio-temporal dynamics of yeast mitochondrial biogenesis: transcriptional and post-transcriptional mRNA oscillatory modules. PLoS Comput. Biol. 5:e1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., M. Blake, C. Tang, D. Zimmer, R. R. Rustandi, D. J. Weber, and F. Carrier. 2001. Inhibition of p53 transcriptional activity by the S100B calcium-binding protein. J. Biol. Chem. 276:35037-35041. [DOI] [PubMed] [Google Scholar]
- 33.Marenholz, I., C. W. Heizmann, and G. Fritz. 2004. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 322:1111-1122. [DOI] [PubMed] [Google Scholar]
- 34.Markowitz, J., A. D. MacKerell, Jr., and D. J. Weber. 2007. A search for inhibitors of S100B, a member of the S100 family of calcium-binding proteins. Mini Rev. Med. Chem. 7:609-616. [DOI] [PubMed] [Google Scholar]
- 35.Markowitz, J., R. R. Rustandi, K. M. Varney, P. T. Wilder, R. Udan, S. L. Wu, W. D. Horrocks, and D. J. Weber. 2005. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the C-terminal negative regulatory domain of p53. Biochemistry 44:7305-7314. [DOI] [PubMed] [Google Scholar]
- 36.Mbele, G. O., J. C. Deloulme, B. J. Gentil, C. Delphin, M. Ferro, J. Garin, M. Takahashi, and J. Baudier. 2002. The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. J. Biol. Chem. 277:49998-50007. [DOI] [PubMed] [Google Scholar]
- 37.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 38.Moroz, O. V., E. V. Blagova, A. J. Wilkinson, K. S. Wilson, and I. B. Bronstein. 2009. The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. J. Mol. Biol. 391:536-551. [DOI] [PubMed] [Google Scholar]
- 39.Moroz, O. V., W. Burkitt, H. Wittkowski, W. He, A. Ianoul, V. Novitskaya, J. Xie, O. Polyakova, I. K. Lednev, A. Shekhtman, P. J. Derrick, P. Bjoerk, D. Foell, and I. B. Bronstein. 2009. Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem. 10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, J. A., D. Maric, P. Lau, J. L. Barker, and L. D. Hudson. 2006. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J. Neurosci. 26:9881-9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada, M., T. Hatakeyama, H. Itoh, N. Tokuta, H. Tokumitsu, and R. Kobayashi. 2004. S100A1 is a novel molecular chaperone and a member of the Hsp70/Hsp90 multichaperone complex. J. Biol. Chem. 279:4221-4233. [DOI] [PubMed] [Google Scholar]
- 42.Pathmanathan, S., S. F. Elliott, S. McSwiggen, B. Greer, P. Harriott, G. B. Irvine, and D. J. Timson. 2008. IQ motif selectivity in human IQGAP1: binding of myosin essential light chain and S100B. Mol. Cell. Biochem. 318:43-51. [DOI] [PubMed] [Google Scholar]
- 43.Raponi, E., F. Agenes, C. Delphin, N. Assard, J. Baudier, C. Legraverend, and J. C. Deloulme. 2007. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia 55:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rizzuto, R., P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft, and T. Pozzan. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763-1766. [DOI] [PubMed] [Google Scholar]
- 45.Rustandi, R. R., D. M. Baldisseri, and D. J. Weber. 2000. Structure of the negative regulatory domain of p53 bound to S100B(betabeta). Nat. Struct. Biol. 7:570-574. [DOI] [PubMed] [Google Scholar]
- 46.Rustandi, R. R., A. C. Drohat, D. M. Baldisseri, P. T. Wilder, and D. J. Weber. 1998. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry 37:1951-1960. [DOI] [PubMed] [Google Scholar]
- 47.Santamaria-Kisiel, L., A. C. Rintala-Dempsey, and G. S. Shaw. 2006. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 396:201-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scotto, C., J. C. Deloulme, D. Rousseau, E. Chambaz, and J. Baudier. 1998. Calcium and S100B regulation of p53-dependent cell growth arrest and apoptosis. Mol. Cell. Biol. 18:4272-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scotto, C., C. Delphin, J. C. Deloulme, and J. Baudier. 1999. Concerted regulation of wild-type p53 nuclear accumulation and activation by S100B and calcium-dependent protein kinase C. Mol. Cell. Biol. 19:7168-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimamoto, S., M. Takata, M. Tokuda, F. Oohira, H. Tokumitsu, and R. Kobayashi. 2008. Interactions of S100A2 and S100A6 with the tetratricopeptide repeat proteins, Hsp90/Hsp70-organizing protein and kinesin light chain. J. Biol. Chem. 283:28246-28258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spiechowicz, M., A. Zylicz, P. Bieganowski, J. Kuznicki, and A. Filipek. 2007. Hsp70 is a new target of Sgt1—an interaction modulated by S100A6. Biochem. Biophys. Res. Commun. 357:1148-1153. [DOI] [PubMed] [Google Scholar]
- 52.van Dieck, J., M. R. Fernandez-Fernandez, D. B. Veprintsev, and A. R. Fersht. 2009. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J. Biol. Chem. 284:13804-13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dieck, J., D. P. Teufel, A. M. Jaulent, M. R. Fernandez-Fernandez, T. J. Rutherford, A. Wyslouch-Cieszynska, and A. R. Fersht. 2009. Posttranslational modifications affect the interaction of S100 proteins with tumor suppressor p53. J. Mol. Biol. 394:922-930. [DOI] [PubMed] [Google Scholar]
- 54.Wilder, P. T., J. Lin, C. L. Bair, T. H. Charpentier, D. Yang, M. Liriano, K. M. Varney, A. Lee, A. B. Oppenheim, S. Adhya, F. Carrier, and D. J. Weber. 2006. Recognition of the tumor suppressor protein p53 and other protein targets by the calcium-binding protein S100B. Biochim. Biophys. Acta 1763:1284-1297. [DOI] [PubMed] [Google Scholar]
- 55.Wright, N. T., B. L. Prosser, K. M. Varney, D. B. Zimmer, M. F. Schneider, and D. J. Weber. 2008. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 283:26676-26683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong, Z., D. O'Hanlon, L. E. Becker, J. Roder, J. F. MacDonald, and A. Marks. 2000. Enhanced calcium transients in glial cells in neonatal cerebellar cultures derived from S100B null mice. Exp. Cell Res. 257:281-289. [DOI] [PubMed] [Google Scholar]
- 57.Yam, A. Y., Y. Xia, H. T. Lin, A. Burlingame, M. Gerstein, and J. Frydman. 2008. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, Z., and Y. Li. 2009. Molecular dynamics simulation of S100B protein to explore ligand blockage of the interaction with p53 protein. J. Comput. Aided Mol. Des. 23:705-714. [DOI] [PubMed] [Google Scholar]