Abstract
RASSF2 is a novel proapoptotic effector of K-Ras. Inhibition of RASSF2 expression enhances the transforming effects of K-Ras, and epigenetic inactivation of RASSF2 is frequently detected in mutant Ras-containing primary tumors. Thus, RASSF2 is implicated as a tumor suppressor whose inactivation facilitates transformation by disconnecting apoptotic responses from Ras. The mechanism of action of RASSF2 is not known. Here we show that RASSF2 forms a direct and endogenous complex with the prostate apoptosis response protein 4 (PAR-4) tumor suppressor. This interaction is regulated by K-Ras and is essential for the full apoptotic effects of PAR-4. RASSF2 is primarily a nuclear protein, and shuttling of PAR-4 from the cytoplasm to the nucleus is essential for its function. We show that RASSF2 modulates the nuclear translocation of PAR-4 in prostate tumor cells, providing a mechanism for its biological effects. Thus, we identify the first tumor suppressor signaling pathway emanating from RASSF2, we identify a novel mode of action of a RASSF protein, and we provide an explanation for the extraordinarily high frequency of RASSF2 inactivation we have observed in primary prostate tumors.
Ras oncoproteins regulate a broad range of signaling pathways involved in the control of cell growth and transformation (reviewed in reference 31). Activating mutations in ras genes, primarily K-Ras, are found in approximately 30% of primary human tumors (31). Moreover, hyperactivation of Ras signaling pathways even in the absence of ras mutations has been reported in many tumor types (13). Thus, abnormal activation of Ras signaling appears to be a frequent component of tumor development.
Although activated forms of Ras promote growth and transformation, they can also induce apoptotic cell death (10). This is particularly apparent with K-Ras. The main Ras effector proteins identified to date that are implicated in mediating apoptosis are the RASSF proteins (12, 44).
The best-characterized member of the RASSF family is RASSF1A. RASSF1A is thought to act as a scaffold protein that may link Ras to multiple tumor suppressor pathways. In particular, RASSF1A has been shown to bind and activate the proapoptotic effectors MstI, a proapoptotic Ste20-related kinase (28), and MOAP-1, resulting in Bax activation (45). Overexpression of RASSF1A promotes apoptosis, and knockdown of RASSF1A impairs the apoptotic activity of activated K-Ras (45). Moreover, deletion of RASSF1A in transgenic mice promotes a modest increase in tumorigenesis (43). Thus, RASSF1A has the potential to be a Ras effector/tumor suppressor.
Expression of the RASSF1A protein is frequently lost in primary tumors due to promoter methylation, an epigenetic mechanism of gene silencing that plays a major role in the development of many cancers. Inactivation of RASSF1A expression has been shown to correlate with activation of Ras in tumors, suggesting that loss of RASSF1A-mediated growth-inhibitory signals is essential to subvert Ras apoptotic pathways, facilitating Ras driven tumorigenesis in vivo.
RASSF2 is structurally related to RASSF1A and may also serve as a proapoptotic, K-Ras-specific effector. RASSF2 binds to K-Ras in a GTP-dependent manner via the effector domain (47) and can be detected in an endogenous complex with K-Ras (4). Like RASSF1A it is inactivated in a variety of tumors by promoter hypermethylation (7, 16, 24, 25, 27, 32, 36, 47, 48). Overexpression of RASSF2 promotes apoptosis and cell cycle arrest (47). It also inhibits the growth of tumor cells and impairs tumor xenograft formation in nude mice (7, 47). Knockdown of RASSF2 expression by small interfering RNA (siRNA) leads to enhanced growth in soft agar and an enhanced transformation due to activated Ras (1). Thus, like RASSF1A, RASSF2 exhibits the properties of a Ras effector/tumor suppressor (47). However, the mechanism by which RASSF2 promotes cell death and tumor inhibition is completely unknown. It seems likely that it may differ from the mechanisms employed by RASSF1A, a primarily cytoplasmic protein, as RASSF2 localizes mostly to the nucleus (7, 29). As with RASSF1A, RASSF2 has no apparent intrinsic enzyme activity or DNA binding properties, and thus it may interact with other proapoptotic effectors/tumor suppressors to mediate cell death. We have found that prostate apoptosis response protein 4 (PAR-4), a key tumor suppressor in prostate cancer (5), may be one such protein.
PAR-4 appears to act at multiple levels that include activating both the FAS- and tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-controlled proapoptotic pathways, as well as inhibiting the NF-κB antiapoptotic pathway (5). PAR-4 is of note as it appears to be selective for inducing apoptosis in cancer cells but not in normal or immortalized cells (15). However, not all cancer cells are sensitive to PAR-4-mediated apoptosis. Those cancer cells resistant to PAR-4-induced cell death are resistant to nuclear translocation of PAR-4, a process necessary for the inhibition of NF-κB activity by PAR-4 (15, 23). The domain of PAR-4 responsible for its apoptotic activity has been mapped to the central core region of the protein (15) and confers resistance to tumor formation in vivo (49).
In order to determine the mechanism of action of RASSF2, we performed a two-hybrid screen. This screen identified PAR-4 as a direct binding partner of RASSF2. Further experiments confirmed that the interaction could be detected between the endogenous proteins and that it could be enhanced in the presence of activated K-Ras. Downregulation of RASSF2 impaired the ability of PAR-4 to kill cells. Thus, we established a K-Ras-RASSF2-PAR-4 signaling pathway.
PAR-4 must be translocated to the nucleus to induce apoptosis (15, 23); however, the mechanism by which this is accomplished is not known. RASSF2 is primarily a nuclear protein that directly binds PAR-4. Here we demonstrate that RASSF2 plays an essential role in the nuclear localization of PAR-4 and that activated K-Ras promotes the nuclear localization of PAR-4 in a RASSF2-dependent manner. Moreover, loss of RASSF2 confers resistance to TRAIL-induced PAR-4 nuclear localization and cell death in prostate cancer cells. As PAR-4 is so important to the development of prostate cancer, we performed an extensive analysis of the frequency of epigenetic inactivation of RASSF2 in primary prostate cancer. We determined that RASSF2 is inactivated in prostate cancer at a higher frequency than in any other cancer type yet investigated. Thus, we have identified the first tumor suppressor signaling pathway emanating from RASSF2 and shown that RASSF2 can link Ras to the key prostate tumor suppressor PAR-4. This may explain the high levels of inactivation observed for RASSF2 in prostate tumors and identifies RASSF2 as a key target for epigenetic therapy in prostate cancer.
MATERIALS AND METHODS
Yeast two-hybrid screen.
Yeast two-hybrid screening was performed as described previously (17). Briefly, the bait vector pGBKT7-RASSF2 (full length; NP_055552) was transformed in yeast strain Saccharomyces cerevisiae AH109 (RASSF2-BD) using the LiAc/polyethylene glycol method and selected on Trp- plates. Strain AH109 includes four reporter genes, ADE2, HIS3, lacZ, and MEL1, whose expression is regulated by GAL4-responsive upstream activating sequences and promoter elements. The bait, RASSF2-BD, was then used to screen a pretransformed MATCHMAKER brain library (Clontech, Palo Alto, CA), cloned in pACT2, by mating with the yeast strain S. cerevisiae Y187. A total of 4.4 × 106 clones were screened, of which 26 were positive for α-galactosidase expression. Positive clones were sequenced and identified using NCBI BLASTN/BLASTX. Clones were rescued and retransformed into AH109 to confirm positive interactions using cotransformation assays with empty pGBKT7 and pGBKT7-RASSF2 vectors. For yeast cotransformation assays, AH109 yeast cells were transformed with the appropriate vectors using the LiAc/polyethylene glycol method and Yeastmaker yeast transformation system 2 (Clontech). Cells were plated onto selection medium with added 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside and incubated for 4 to 8 days.
Cell lines and culture conditions.
PC-3 and Du-145 prostate cancer cells and H441 lung cancer cells were maintained in RPMI 1640 (Mediatech Inc., Herndon, VA) supplemented with 10% fetal bovine serum (FBS; Mediatech Inc.) and 1% penicillin-streptomycin (Mediatech Inc.). COS-7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech Inc) supplemented with 10% FBS and 1% penicillin-streptomycin.
Plasmids and transient transfections.
Red fluorescent protein (RFP)-RASSF2 was constructed by cloning a BglII/EcoRI cDNA fragment (47) into pHCR1-Red vector (Clontech, Mountain View, CA). Green fluorescent protein (GFP)-PAR-4 (21), Flag-tagged RASSF2 (47), and pCGN-K-Ras12V (18) have previously been described. Exponentially growing cells were transfected with 2 μg DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions and harvested 48 h later.
Knockdown of RASSF2 by short hairpin RNA.
Cells were transfected with the RASSF2 shRNA constructs shRNA sequence 1, 5′-GTGACAGAGCCACAGACTAGTATGAACACCCAAGTGTTCACACCAGCCTATGGCTCTGTCAC-3′, and sequence 2, 5′-GAAGACCTACAACTTGTACTA-3′, or corresponding vector/scrambled control (Openbiosystems, Huntsville, AL) using Lipofectamine 2000 and selected with puromycin to obtain a stable pool of cells.
K-Ras was knocked down using a validated siRNA mixture (sc35731) from Santa Cruz Biotechnology (Santa Cruz, CA) as per the manufacturer's recommendations.
Immunofluorescence.
GFP and RFP fluorescence microscopy was performed on cells grown on glass-bottom microwell dishes (MatTek Corp., Ashland, MA), and images were captured with an Olympus 1 × 50-FLA inverted fluorescent microscope (Optical Elements Corp., Dulles, VA) with an attached Spot Junior digital camera.
Growth inhibition assays.
PC-3 shRASSF2 or control cells were transfected with 1 μg GFP-PAR-4 or GFP-vector using Lipofectamine 2000 (Invitrogen) and selected in G418 (Mediatech Inc.) for 2 weeks, after which cells were fixed and stained with crystal violet.
Subcellular fractionation and Western blot analysis.
Total cell lysates were prepared by lysing the cells in radioimmunoprecipitation assay buffer (Sigma, St. Louis, MO) supplemented with 100 μg/ml leupeptin, 100 μg/ml aprotinin, and 1 mM sodium orthovanadate. The lysates were passed through a 21-gauge needle and centrifuged to remove debris. Nuclear and cytoplasmic fractions were prepared using the NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology, Rockford, IL) as described by the manufacturer. Equal amounts of protein were separated on NuPage Novex polyacrylamide gels (Invitrogen) and incubated with antibodies against GFP (sc-9996), PAR-4 (sc-1666), TFIIH (sc-293) (all from Santa Cruz Biotechnology Inc., Santa Cruz, CA), p38 (9212; Cell Signaling Technology, Danvers, MA), and RASSF2 (47). The signal was detected by enhanced chemiluminescence.
Immunoprecipitation.
Endogenous coimmunoprecipitations of RASSF2 and PAR-4 were performed using our RASSF2 antibody (47) and a commercial PAR-4 antibody (Santa Cruz) with Sepharose bead-conjugated Trueblot secondary antibodies (eBioscience). 293T cells were cotransfected with 2 μg of each plasmid using Lipofectamine 2000 (Invitrogen). Coimmunoprecipitation (co-IP) of exogenously expressed proteins was performed by immunoprecipitating the 293T cell lysates with Flag-Sepharose beads (Sigma) followed by Western analysis with anti-GFP (Santa Cruz).
Human prostate tissue samples.
Normal prostate epithelial tissues (n = 32) without histological evidence of prostate cancer were obtained from prostatectomy specimens from patients treated with cystectomy for invasive bladder cancer. The entire prostate was investigated in whole-mount sections to exclude patients with incidental prostate cancer. Benign prostatic hyperplasia tissues (BPH; n = 34) without histological evidence of prostate cancer were obtained from patients treated with transurethral resection of the prostate for BPH. Prostate cancer tissues (n = 82) with a minimum of 50% tumor cells were obtained from radical prostatectomy specimens from patients treated for prostate cancer. All primary prostate tissues were obtained from patients treated at the Department of Urology, Regensburg University Hospital. All of the tissues were snap-frozen and stored at −80°C. Five-micrometer-thick sections were cut for histological verification of the diagnosis by a pathologist (A. Hartmann). Tissue blocks were trimmed to maximize the yield of target cells. Relevant clinical data were collected from patients' clinical records (Table 1). The study was approved by the institutional review board at Regensburg University Hospital.
TABLE 1.
Clinical characteristics of patients and tissue samples
| Characteristic | Result for histological categorya |
||
|---|---|---|---|
| PCC (n = 82) | BPH (n = 34) | Normal (n = 32) | |
| Age (yrs) | |||
| Median | 66 | 65 | 65 |
| Range | 47-75 | 50-80 | 44-83 |
| Serum total PSA (ng/ml) | NA | ||
| Median | 10.24 | 3.7 | |
| Range | 0.01-256 | 0.15-21.88 | |
| No. (%) with PSA range of: | |||
| <4 | 8 (10) | 20/34 (59) | |
| 4.1-8 | 24 (29) | 7/34 (21) | |
| 8.1-12 | 17 (21) | 5/34 (15) | |
| >12 | 30 (37) | 2/34 (6) | |
| Unknown | 3 | 0 | |
| No. (%) at pathological stage: | NA | NA | |
| T2a | 4 (5) | ||
| T2b | 3 (4) | ||
| T2c | 22 (26) | ||
| T3a | 24 (29) | ||
| T3b | 29 (35) | ||
| Gleason sum | NA | NA | |
| Median | 7 | ||
| Range | 5-10 | ||
| No. (%) with sum of: | |||
| 5 | 4 (5) | ||
| 6 | 8 (10) | ||
| 7 | 43 (52) | ||
| 8 | 12 (15) | ||
| 9-10 | 15 (18) | ||
PSA, prostate-specific antigen; PCC, prostate cancer cells; NA, not applicable.
Bisulfite treatment of DNA and PCR amplification.
RASSF2 promoter methylation in prostate cancer cell lines was analyzed by PCR amplification of bisulfite-modified DNA as previously described (30). For primary samples, DNA was extracted from normal prostate epithelium tissues, BPH tissue, and prostate cancer tissues using Qiagen Genomic-tip 100/G columns (Qiagen) and bisulfite treated as previously described (33). Ten nanograms of bisulfite-treated DNA was analyzed in triplicate using the FastStart high-fidelity PCR system (Roche) and HeavyMethyl (HM) technology on a LightCycler 480 (Roche Diagnostics) (9). β-Actin (ACTB) analysis was performed prior to template dilution, following dilution and once more at the end of the study. Calibration curves for each assay were prepared with CpGenome Universal methylated DNA (Roche Applied Science) using standard concentrations between 400 ng and 20 ng/reaction mixture. The RASSF2 and ACTB primer and probe sequences used are available upon request.
Data analysis.
The amount of methylated DNA at the RASSF2 locus (percentage of methylated reference [PMR]) was calculated by dividing the RASSF2/ACTB ratio of a sample by the RASSF2/ACTB ratio of CpGenome Universal methylated DNA (Roche Applied Science) and multiplying by 100 as described previously (14). Box plot analysis was performed using log10 transformation of the mean PMR for each specimen.
Immunohistochemistry.
An AccuMaxc prostate cancer tissue array that included normal prostate tissues (ISU Abxis Co., Ltd., Seoul, Korea) was processed for immunohistochemical staining as per the manufacturer's instructions. Sections were blocked in 10% heat-inactivated normal goat serum (NGS) in 0.05% Tris-Triton X-100 and incubated overnight with rabbit polyclonal RASSF2 (47) diluted 1:150 with 1% NGS in Tris-Triton X-100. The sections were then washed three times for 5 min in Tris-Triton X-100 and incubated in goat anti-rabbit IgG-Texas Red (Molecular Probes, Eugene, OR).
RESULTS
RASSF2 interacts directly with PAR-4.
In order to identify proteins that interact with RASSF2, we performed a two-hybrid screen using RASSF2 as bait. We identified a single clone encoding amino acids 68 to 340 of the proapoptotic protein PAR-4 (NP_002574; gene symbol PAWR [NM_002583]). Yeast cotransformation assays confirmed that PAR-4 interacted with RASSF2 and not the GAL4 DNA binding domain alone (data not shown). Using cotransformation assays we also tested the RASSF members RASSF1A and RASSF4/AD037 (both full length) for their ability to interact with PAR-4 (amino acids 68 to 340). Neither RASSF1A nor RASSF4/AD037 interacted with PAR-4 in yeast (data not shown). To confirm that RASSF2 and PAR-4 interact in mammalian cells, lysates from H441 human lung tumor cells (which express RASSF2 [47]) containing equal amounts of protein were immunoprecipitated with a rabbit polyclonal RASSF2 antibody (47) or mock immunoprecipitated and subjected to Western blotting using a PAR-4 antibody (Fig. 1). The presence of endogenous PAR-4 protein in the immunoprecipitates confirmed that this interaction was physiologically relevant (Fig. 1). To define the domains involved in the interaction between RASSF2 and PAR-4, we performed coimmunoprecipitation experiments with a series of GFP-tagged RASSF2 deletion mutants (8) and full-length PAR-4 and found that PAR-4 bound to all the deletion mutants (data not shown), suggesting that PAR-4 binds to RASSF2 at multiple sites. A similar observation has been made for the interaction of RASSF1A, another member of the RASSF family, and the human homolog of the Drosophila melanogaster protein Salvador, hWW45 (22).
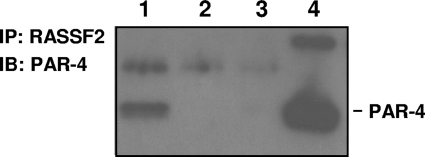
FIG. 1.
RASSF2 interacts with PAR-4 at physiologically relevant levels. Lysates with equal protein concentrations from H441 lung cancer cells were immunoprecipitated with an anti-RASSF2 antibody (lane 1) or control IgG (lane 2), fractionated on SDS gels, and immunoblotted (IB) with an anti-PAR-4 antibody. Lane 3 is a mock IP of a buffer with no lysate. The endogenous interaction between RASSF2 and PAR-4 was confirmed by the presence of PAR-4 in the proteins precipitated with the RASSF2 antibody (lane 1) and not with control IgG (lane 2). Lane 4 is a PAR-4 positive control lysate.
Activated K-Ras modulates the association of RASSF2 with PAR-4.
To determine the effect of activated K-Ras on the RASSF2-PAR-4 interaction, coimmunoprecipitation experiments were performed using Flag-tagged RASSF2 and GFP-tagged PAR-4 cotransfected into 293T cells. Lysates were immunoprecipitated (IP) with an anti-Flag antibody and subjected to Western blotting using an anti-GFP antibody. The results showed that RASSF2 and PAR-4 interact when exogenously expressed and that the interaction is enhanced in the presence of activated K-Ras (Fig. 2A). In some blots, we detected the presence of an additional, slightly smaller PAR-4 band in the IP. The relevance of this is not yet known. As further confirmation of the role of Ras in the RASSF2-PAR-4 interaction, we treated H441 cells, which contain an endogenous activated K-ras gene, with siRNA against K-ras or a scrambled control and then immunoprecipitated the cell lysates with PAR-4 before probing with RASSF2. Figure 2B shows that the K-Ras siRNA downregulated K-Ras while having no effect on the levels of RASSF2 or PAR-4 in the cells. However, the association of endogenous RASSF2 and endogenous PAR-4 was reduced when K-Ras was downregulated.
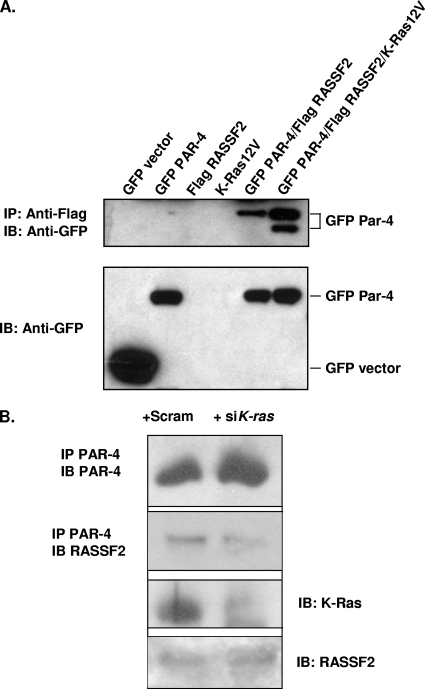
FIG. 2.
Activated K-Ras enhances the interaction between RASSF2 and PAR-4. (A) 293T cells were transfected with Flag-RASSF2 and GFP-PAR-4 or GFP-vector in the presence or absence of activated K-Ras, and lysates were prepared and immunoprecipitated with anti-Flag, fractionated on SDS gels, and immunoblotted (IB) with anti-GFP (top panel). Activated K-Ras enhanced the association between RASSF2 and PAR-4 (compare lanes 5 and 6). Aliquots of the input lysates were similarly blotted with anti-GFP to ensure equivalent GFP-PAR-4 expression levels (bottom panel). The lower band in lane 6, top panel, was consistently observed and may correspond to a cleaved form of PAR-4. (B) H441 cells were transfected with K-ras siRNA or scrambled control. The cells were then lysed and immunoprecipitated with anti-PAR-4 antibodies. The immunoprecipitate was then Western blotted for endogenous RASSF2. The top panel shows that the siRNA did not inhibit the expression of PAR-4. The second panel shows that the levels of RASSF2 coprecipitating with PAR-4 were reduced in the siRNA K-Ras-treated cells. The third panel shows that K-Ras protein was downregulated in the siRNA treated cells. The fourth panel shows that the total levels of RASSF2 in the pre-IP lysates were the same.
RASSF2 promotes nuclear localization of PAR-4.
We have previously shown that, unlike RASSF1A, RASSF2 localizes primarily to the nucleus (7, 47). PAR-4 requires nuclear localization for its apoptotic functions (15, 23), and it has been suggested that RASSF2 has the potential to serve as part of a nuclear transport system (29). Thus, we examined the effects of RASSF2 on the nuclear localization of PAR-4. COS-7 cells were transfected with RFP-RASSF2 and GFP-PAR-4 and then analyzed by fluorescence microscopy for subcellular localization of the proteins in live cells. Consistent with previous studies, when transfected alone RASSF2 was localized in the nucleus (7, 29, 47), whereas PAR-4 was located predominantly in the cytoplasm and on the cell membrane (Fig. 3A). However, in cells cotransfected with both proteins, GFP-PAR-4 colocalized with RASSF2 in the nucleus, as evidenced by the yellow color in Fig. 3A (bottom panel). Quantification of this effect showed a statistically significant shift of PAR-4 into the nucleus in the presence of RASSF2 (Fig. 3B). We then performed similar experiments examining the effects on endogenous PAR-4 in PC-3 cells. These experiments included a previously described deletion mutant of RASSF2 that lacks a key nuclear localization signal (NLS) (8). Wild-type RASSF2 was found to induce the nuclear localization of endogenous PAR-4, but the RASSF2 NLS mutant did not (Fig. 3C).
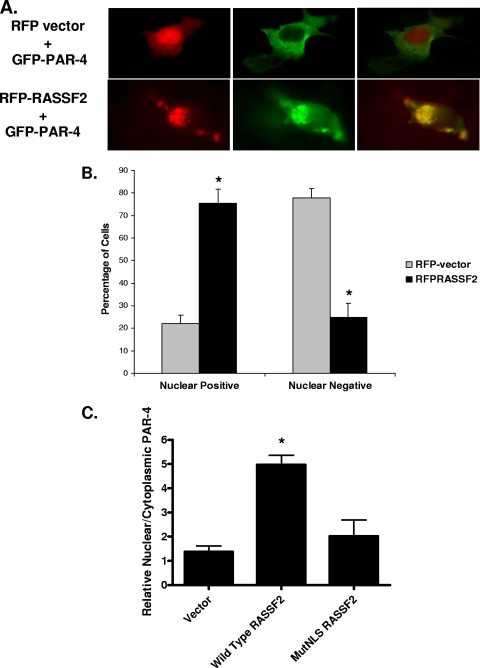
FIG. 3.
RASSF2 promotes the nuclear localization of PAR-4. (A) COS-7 cells were cotransfected with RFP vector or RFP-RASSF2 and GFP-PAR-4, and images were captured 24 h later using a fluorescence microscope as described in Materials and Methods. In the absence of RASSF2, PAR-4 localized to the plasma membrane and cytoplasm, but in the presence of RASSF2, PAR-4 colocalized with RASSF2 in the nucleus, as evidenced by the yellow color in the merged image. Magnification (all images), ×100. (B) Quantification of PAR-4 nuclear localization in the presence of RASSF2. Fifty randomly selected cells expressing both RFP-RASSF2 or RFP-vector and GFP-PAR-4 were scored for the presence of nuclear PAR-4. The bars show the means of triplicate experiments, and standard deviations are indicated. *, statistically significantly different (P < 0.01) from vector-transfected cells. (C) PC-3 human prostate tumor cells were transfected with RASSF2 or a deletion mutant of RASSF2 lacking at least one of the NLSs (MutNLS) and fractionated before Western blotting for endogenous PAR-4. The blots were quantified and used to generate a ratio of nuclear versus cytoplasmic PAR-4. The mutant of RASSF2 was impaired for inducing the nuclear localization of endogenous PAR-4. *, statistically different (P < 0.01) from vector-transfected cells.
Loss of RASSF2 expression enhances the tumorigenic phenotype of prostate cancer cells and confers resistance to PAR-4-mediated cell death.
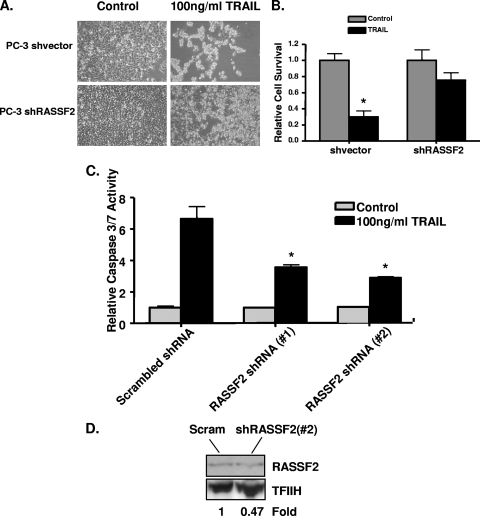
The prostate tumor cell line PC-3 is sensitive to PAR-4-induced apoptosis (40). In order to determine the biological significance of the RASSF2-PAR-4 interaction, we stably knocked down RASSF2 expression in PC-3 prostate cancer cells using an shRNA construct (Fig. 4A). Cells transfected with the empty vector served as a control. The cells were plated in soft agar, and those stably expressing RASSF2 shRNA demonstrated an enhanced ability to grow in soft agar compared to the control cells (Fig. 4B). These results are similar to those previously reported by Akino et al. with normal rat kidney cells and using siRNA to transiently downregulate RASSF2 (1). We obtained similar results with the prostate tumor cell line Du145 (data not shown). PC-3 cells were then transfected with either GFP-vector or GFP-PAR-4 and selected in G418, and 14 days later surviving colonies were stained with crystal violet. As expected, PAR-4 inhibited cell growth in the cells stably transfected with the empty shRNA vector; however, cells stably knocked down for RASSF2 were able to partially overcome this PAR-4 growth/survival-inhibitory effect to produce viable colonies (Fig. 4C). The PAR-4 growth-inhibitory effect observed in the vector control cells was apoptotic in nature, as evidenced by an increase in caspase 3/7 activity (Fig. 4D). No increase in caspase activity was observed in the cells knocked down for RASSF2 in response to PAR-4 (Fig. 4D). These results suggest that RASSF2 is required for PAR-4-mediated apoptotic cell death.
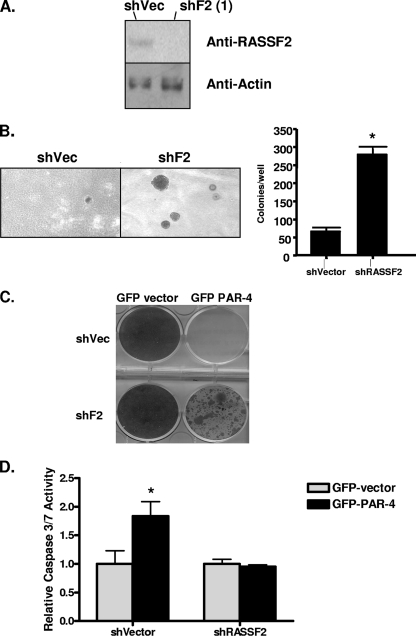
FIG. 4.
Loss of RASSF2 enhances tumorigenicity of prostate cancer cells and confers resistance to PAR-4-mediated cell death. (A) Western blot analysis of RASSF2 expression in PC-3 prostate cancer cells stably expressing a RASSF2 shRNA construct or corresponding vector control. Actin was used as a control for protein loading. (B) PC-3 cells stably expressing an shRNA to RASSF2 or vector control were plated in soft agar and scored for growth after 14 days. Quantification is shown in the adjacent panel. *, statistically different (P < 0.05) from cells transfected with the vector control. (C and D) The PC-3 prostate cancer cells stably expressing an shRNA to RASSF2 or a vector control were transfected with GFP-vector or GFP-PAR-4, and surviving colonies stained with crystal violet after 2 weeks of selection in G418 (C) or assayed for apoptosis via determination of caspase-3 and -7 activities (D). *, significantly different (P < 0.05) from cells transfected with GFP-vector.
Loss of RASSF2 protects prostate cancer cells from TRAIL-induced PAR-4 nuclear localization and apoptosis.
TRAIL-mediated apoptosis requires PAR-4 to fully manifest its apoptotic effects (3). To determine if RASSF2 is also required for TRAIL/PAR-4-mediated cell death, we treated PC-3 control cells and PC-3 cells knocked down for RASSF2 with 100 ng/ml recombinant TRAIL (rTRAIL) and measured cell survival. Whereas the vector control cells were sensitive to treatment with rTRAIL, the PC-3 shRASSF2 cells were resistant (Fig. 5A). Quantification of the number of surviving cells by trypan blue staining showed a 70% reduction in viable cells for the vector control cells (P < 0.05) but no statistically significant difference in cell number for the PC-3 shRASSF2 cells (Fig. 5B). TRAIL/PAR-4-mediated cell death is apoptotic in nature (3). Thus, we also measured the degree of TRAIL stimulated apoptosis in the RASSF2-positive and -negative matched pairs of cells. To determine the mechanism of cell death we repeated the experiments and assayed for caspase activation. We also included a second RASSF2 shRNA (Fig. 5D) to confirm the specificity of the effects. Figure 5C shows that the reduced cell death observed in the RASSF2 knockdown cells correlated with a reduction in the degree of apoptosis in both shRNA cell lines, as measured by caspase activation, induced by rTRAIL.
FIG. 5.
Loss of RASSF2 protects prostate cancer cells from TRAIL-induced apoptosis. (A and B) PC-3 cells stably transfected with a RASSF2 shRNA construct or a control vector were seeded at 1 × 105cells/well in six-well plates and treated with 100 ng/ml TRAIL, and cell death was estimated 72 h later by trypan blue exclusion. Bars show the means of triplicate experiments, and standard deviations are indicated. *, P < 0.05 compared to control cells. (C) Similar experiments were then performed to measure apoptosis. These included a second RASSF2 shRNA cell line (# 2). Cells were seeded at 3 × 103 cells/well in 96-well plates, and caspase-3 and -7 activities were measured after TRAIL treatment. Caspase activity is expressed relative to results in untreated cells. The bars show means of duplicate experiments. with standard deviations shown. *, significantly different (P < 0.05) from cells transfected with a scrambled shRNA. (D) Western blot analysis of RASSF2 expression in PC-3 prostate cancer cells stably expressing a RASSF2 shRNA (#2) construct (#2) or scrambled shRNA. TFIIH was used as a loading control.
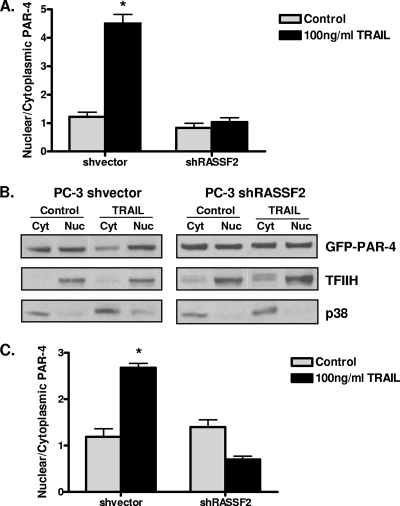
TRAIL causes nuclear translocation of PAR-4, which promotes apoptosis (42). We hypothesized that this effect may require RASSF2. PC-3 shRASSF2 or vector control cells were transfected with GFP-PAR-4 and treated with rTRAIL, and the amount of GFP-PAR-4 in the nuclear and cytoplasmic compartments was determined by Western blot analysis. Quantification of the distribution of PAR-4 is shown in Fig. 6A, with a representative Western blot shown in Fig. 6B. Treatment of the vector control cells with rTRAIL resulted in a 4- to 5-fold increase in the nuclear/cytoplasmic ratio of PAR-4, whereas no difference was observed in the cells knocked down for RASSF2.
FIG. 6.
Loss of RASSF2 impairs TRAIL-induced PAR-4 nuclear trafficking in prostate cancer cells. PC-3 cells stably expressing a RASSF2 shRNA construct or control vector were transfected with GFP-PAR-4 and treated with 100 ng/ml TRAIL for 1 h. (A) Cells were lysed, and nuclear (Nuc) and cytoplasmic (Cyt) fractions were prepared and analyzed by Western blotting for GFP-PAR-4. Densitometric quantitation of Western blot results is shown. Bars show means of triplicate experiments, with standard deviations indicated. *, statistically different (P < 0.01) from untreated cells. (B) Representative Western blot. TFIIH and p38 were used as markers for the nuclear and cytoplasmic fractions, respectively. Similar experiments were performed for the endogenous PAR-4 protein. (C) Densitometric quantitation of the Western blot results with endogenous PAR-4 in nuclear and cytoplasmic fractions in the RASSF2 (positive and negative) PC-3 cells. Bars show means of triplicate experiments, and standard deviations are indicated. *, P < 0.01 compared to untreated cells.
To ensure that these results were not merely an artifact of overexpressing exogenous PAR-4, we determined the levels of endogenous PAR-4 in the nuclear and cytoplasmic fractions in the PC-3 RASSF2-positive and -negative cells in the presence and absence of rTRAIL and found similar results. TRAIL caused an increase in the nuclear/cytoplasmic ratio of endogenous PAR-4 in the control cells, whereas no effect was observed in the cells knocked down for RASSF2 (Fig. 6C). These results show that nuclear transport of PAR-4 under the influence of TRAIL is dependent upon RASSF2.
Activated K-Ras promotes PAR-4 nuclear localization via RASSF2.
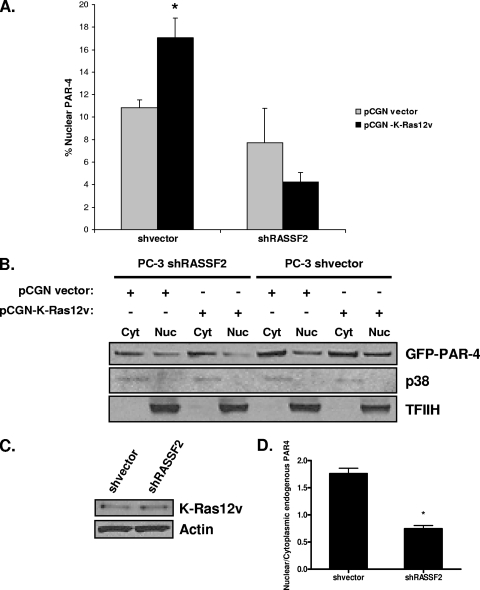
Activated K-Ras plays a role in activating PAR-4 (35), but the mechanism remains unclear. Here we have implicated RASSF2 as the K-Ras effector that modulates PAR-4 action. Thus, we sought to confirm that RASSF2 can serve as a connection between K-Ras and PAR-4. The PC-3 RASSF2-positive and -negative cells were transfected with PAR-4 in the presence or absence of activated K-Ras. The cells were then fractionated, and the effects of activated K-Ras on the nuclear localization of PAR-4 were analyzed. The percentage of nuclear PAR-4 in K-Ras-transfected cells was dramatically reduced in the RASSF2 knockdown cells. Quantification of this effect, derived from two separate experiments, is shown in Fig. 7A, and a representative assay is shown in Fig. 7B. Cell lysates were also examined by Western blotting to confirm that the RASSF2 positive and negative matched pairs expressed equal amounts of transfected K-Ras (Fig. 7C). We then performed similar experiments looking at the effects of K-Ras on the localization of endogenous PAR-4 in the positive and negative RASSF2 cells. Quantification of Western blots was used to generate a ratio of nuclear to cytoplasmic PAR-4 in the presence or absence of activated K-Ras in vector-transfected or RASSF2 shRNA-transfected cells (Fig. 7D). Thus, downregulation of RASSF2 inhibits the ability of K-Ras to promote endogenous PAR-4 nuclear localization.
FIG. 7.
Loss of RASSF2 reduces activated K-Ras-induced PAR-4 nuclear translocation. (A) PC-3 cells stably knocked down for RASSF2 and control cells were transfected with GFP-PAR-4 in the presence or absence of activated K-Ras. At 16 h posttransfection, cells were lysed and nuclear (Nuc) and cytoplasmic (Cyt) fractions were prepared and analyzed by Western blotting for GFP-PAR-4. The percentage of nuclear PAR-4 was quantitated by densitometry. Bars show means of duplicate experiments, and standard deviations are indicated. *, P < 0.05 compared to cells transfected with pCGN-vector. (B) Representative Western blots of the nuclear and cytoplasmic lysates. TFIIH and p38 were used as markers for the nuclear and cytoplasmic fractions, respectively. (C) Western blot analysis of the lysates using an anti-Ras antibody. Actin was used as a loading control. (D) The effects of activated K-Ras on endogenous PAR-4 localization were examined in the RASSF2-positive and -negative matched pair of PC-3 cells. Quantification of Western blot assay results was used to generate the ratio of nuclear versus cytoplasmic PAR-4 in Ras compared to vector-transfected cells for the matched pair. Data are means ± standard deviations of duplicate experiments. *, P < 0.01 compared to shVector-transfected cells.
The RASSF2 promoter is frequently methylated in prostate cancer.
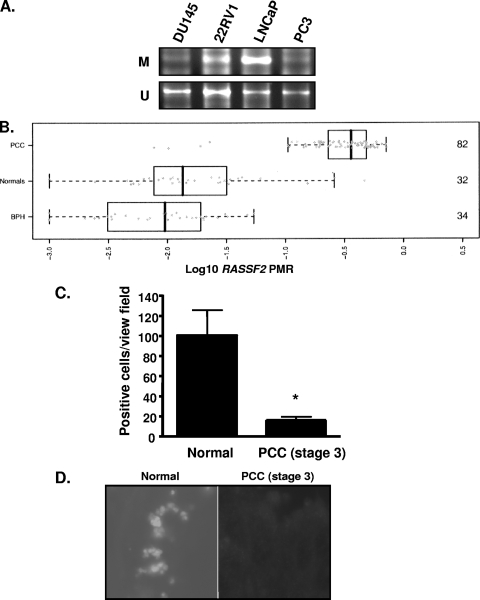
PAR-4 was first identified as a key component of apoptotic processes in prostate tumor cells (41). Here, we show that RASSF2 is essential for full function of PAR-4. Although RASSF2 is frequently epigenetically silenced in many human tumors by promoter methylation, its methylation status in prostate cancer has not been characterized. If PAR-4 is a vital tumor suppressor in the prostate, then perhaps it can be subverted indirectly by the inactivation of RASSF2. We thus determined the methylation status of the RASSF2 promoter in prostate cancer cell lines, benign prostate epithelium, and primary prostate tumors by real-time quantitative PCR of bisulfate-modified DNA. We found that RASSF2 was methylated in 50% of a small panel of prostate tumor cell lines (Fig. 8A) but in approximately 95% (78/82) of primary prostate cancers (Fig. 8B). This includes prostate cancers of all Gleason scores and all histopathological stages, indicating that RASSF2 methylation is an early event in prostate cancer development. In contrast, histologically confirmed benign prostate tissues were largely unmethylated at the RASSF2 promoter, with fewer than 5% of benign prostate tissues exhibiting a PMR value higher than 10% (Table 2). This is the highest level of RASSF2 inactivation yet reported in any tissue. To determine whether the very frequent aberrant promoter methylation of RASSF2 in primary prostate cancer correlates with frequent loss of RASSF2 protein expression in prostate cancer, we performed immunohistochemical analysis of a primary prostate cancer tissue array using our RASSF2 antibody. We found that reduction of the expression of RASSF2 protein in the prostate cancer samples compared to normal prostate was very pronounced (Fig. 8C and D). Thus, primary prostate tumors suffer frequent RASSF2 promoter methylation and loss of RASSF2 protein expression.
FIG. 8.
The RASSF2 promoter is methylated in prostate cancer. (A) RASSF2 promoter methylation in prostate cancer cell lines as determined by MSP. M, methylated; U, unmethylated. (B) RASSF2 promoter methylation expressed as the log10 transformation of the mean PMR in normal prostate (normals), benign prostatic hyperplasia tissue (BPH), and prostate cancer tissue (PCC). Numbers on the right represent the number of samples in each group. (C) RASSF2-positive cells in prostate cancer and normal prostate as determined by immunohistochemical staining. Data were obtained from an average of 10 scored samples. *, P < 0.05 compared to normal prostate. (D) Representative sections of normal prostate and prostate cancer stained for RASSF2 expression.
TABLE 2.
RASSF2 methylation status in primary prostate tissues
| Patient or sample characteristic |
RASSF2 methylation statusa for histological group |
|||||
|---|---|---|---|---|---|---|
| Prostate cancer |
BPH |
Normal |
||||
| M | U | M | U | M | U | |
| No. (%) in age range (yrs) | ||||||
| 40-49 | 1 (100) | 0 | NA | NA | 0 | 3 |
| 50-59 | 16 (100) | 0 | 0 | 5 | 1 (9) | 10 |
| 60-69 | 47 (94) | 3 | 0 | 11 | 1 (8) | 11 |
| >70 | 14 (93) | 1 | 0 | 18 | 1 (17) | 5 |
| No. (%) with serum total PSA (ng/ml) of: | NA | |||||
| <4 | 10 (91) | 1 | 0 | 20 | ||
| 4.1-8 | 22 (92) | 2 | 0 | 7 | ||
| 8.1-12 | 16 (100) | 0 | 0 | 5 | ||
| >12 | 30 (97) | 1 | 0 | 2 | ||
| No. (%) at pathological stage of: | NA | NA | ||||
| T2a | 4 (100) | 0 | ||||
| T2b | 3 (100) | 0 | ||||
| T2c | 22 (100) | 0 | ||||
| T3a | 25 (100) | 0 | ||||
| T3b | 24 (86) | 4 | ||||
| No. (%) with Gleason sum of: | NA | NA | ||||
| 5 | 4 (100) | 0 | ||||
| 6 | 8 (100) | 0 | ||||
| 7 | 40 (93) | 3 | ||||
| 8 | 11 (92) | 1 | ||||
| 9-10 | 15 (100) | 0 | ||||
M, methylated; U, unmethylated. The tissue was considered methylated at the RASSF2 promoter if the PMR value was >10%.
DISCUSSION
Although activated K-Ras is a powerful and notorious oncogene, it is also a surprisingly potent apoptotic agent (10). The mechanisms behind the apoptotic effects of K-Ras were largely unknown until the identification of RASSF family proteins as proapoptotic effectors of Ras (28, 46, 47). The best-studied RASSF family member is RASSF1A. RASSF1A can induce apoptosis via at least two different pathways: an MST1-mediated signaling pathway and a MOAP-1/Bax-mediated signaling pathway (45). In each case, RASSF1A appears to serve as a scaffolding molecule. The role of RASSF1A in supporting K-Ras-driven apoptosis has been confirmed by RASSF1A knockdown experiments that showed a reduced ability of K-Ras to induce apoptosis. The physiological role of RASSF family members in Ras-mediated apoptosis is supported by observations that tumors with high levels of active Ras tend to have lost the expression of RASSF family members due to promoter methylation (4, 24). Thus, we hypothesize that RASSF proteins are Ras effectors that must be inactivated to abrogate Ras apoptotic pathways in tumors. This implies that RASSF proteins may exhibit tumor suppressor functions, and this has been confirmed for RASSF1A by gene deletion experiments in vivo (43).
RASSF2 binds directly and specifically to K-Ras in a GTP-dependent manner via the effector domain (47). It also forms an endogenous complex with K-Ras in primary tissue (4). However, the signaling pathways modulated by RASSF2 to promote apoptosis are not known. They are likely to differ from those of RASSF1A, as the subcellular localization of RASSF2 is primarily nuclear (7) while RASSF1A appears to be primarily associated with the microtubule network (12). Moreover, there is significant divergence in their primary sequence (12).
RASSF2 was the third member of the RASSF family to be identified (47). It kills cells by apoptosis, promotes cell cycle arrest, and is downregulated in a variety of tumors (7, 16, 24, 25, 36, 47, 48), but its mechanism of action is unknown. We have now determined that RASSF2, but not RASSF1A, forms a direct and endogenous complex with a tumor suppressor called PAR-4 that plays a vital role in the development of prostate cancer. PAR-4 is a proapoptotic protein, initially identified as a key mediator of apoptosis in prostate cancer cells (41). PAR-4 is a particularly interesting protein, as it appears to function by activating apoptotic pathways, such as the FAS and TRAIL pathways, at the same time it inhibits proteins involved in survival pathways, such as Bcl-2 (39), PKCζ (11) (and thence AKT), and NF-κB (5).
Although the mode of action of PAR-4 is complex and still under investigation, it appears that nuclear translocation is essential for PAR-4 to induce apoptosis (15, 23). However, it is unclear as to how and why PAR-4 translocates to the nucleus. RASSF2 is a proapoptotic, primarily nuclear protein containing at least one NLS that binds importin-α (29). Thus, our finding that RASSF2 forms a direct, endogenous complex with PAR-4 suggested that RASSF2 might function, at least in part, by modulating translocation of PAR-4 from the cytoplasm to the nucleus. The observation that activated K-Ras appeared to enhance the association between PAR-4 and RASSF2 suggested that Ras might promote PAR-4 nuclear localization via RASSF2.
Further experiments confirmed that overexpression of RASSF2 promotes nuclear localization of PAR-4. To reduce the possibility of overexpression artifacts or cell-type-specific effects, we performed these experiments in PC-3 cells as well as COS-7 cells. Moreover, we confirmed that RASSF2 knockdown resulted in reduced levels of endogenous nuclear PAR-4 in PC-3 cells. We also confirmed that in the absence of RASSF2, activated K-Ras was significantly less able to promote PAR-4 nuclear localization. These results may explain mechanistically why knockdown of RASSF2 enhances Ras transforming activity (1) and why loss of RASSF2 expression confers partial resistance to PAR-4-mediated cell killing (Fig. 4). They may also explain our previous results showing that deletion of the NLS of RASSF2 severely impaired its ability to suppress the transformed phenotype (7). However, we acknowledge that Maruyama et al. have recently shown that a deletion mutant of RASSF2 that is defective for nuclear localization can maintain an apoptotic capacity (32). This suggests that, like RASSF1A, the role of RASSF2 is complex and probably involves multiple signaling pathways in addition to PAR-4, such as MST kinases (8).
Cotransfection of RASSF2 with K-Ras enhances the apoptotic effects of the Ras gene (47). A similar effect has previously been reported for PAR-4, as ectopic PAR-4 expression inhibits Ras-mediated cell growth (38) and induces apoptosis by inhibiting NF-κB activity and downregulation of its various proapoptotic targets (5, 20, 35). The connection we have identified between Ras, RASSF2, and PAR-4 may now explain this observation. It may also explain the previously reported effects of RASSF2 in inhibition of the NF-κB pathway (25).
Induced inactivation of RASSF2 enhances the transforming activity of K-Ras, and RASSF2 is frequently found to be downregulated by promoter methylation in activated Ras-containing primary tumors (1, 4). Thus, we propose that the RASSF2 pathway must be inactivated to facilitate transformation by K-Ras. Intriguingly, a similar observation has been made for PAR-4, as inactivation of PAR-4 is required for the full transforming effects of Ras to become manifest (38). Moreover, it has been shown that activation of Ras can actually promote the epigenetic inactivation of PAR-4 to facilitate Ras transformation. Perhaps Ras activation may have a similar effect on RASSF2 and even other members of the RASSF family.
PAR-4 expression is frequently downregulated in human tumors (2, 6, 26, 34), and Par-4 null mice develop spontaneous proliferative lesions predominantly in the endometrium and prostate (19). Here we show that RASSF2 is critical for normal PAR-4 function and for its response to K-Ras. Although prostate tumors rarely exhibit Ras mutations, aberrant activation of wild-type Ras due to defects in upstream regulators appears to be common and can play a key role in tumorigenesis (37). We now demonstrate for the first time that RASSF2 is frequently silenced by epigenetic mechanisms in primary prostate cancer (Fig. 8). The frequency of inactivation (∼95%) is the highest frequency yet recorded for this RASSF family member in primary human tumors. Further analysis confirmed that RASSF2 protein expression is frequently lost in primary prostate tumors. This suggests that disruption of PAR-4 function and its uncoupling from Ras by inactivation of RASSF2 plays a critical role in development of prostate cancer. The extremely high level of RASSF2 promoter methylation observed in prostate tumors has the potential to serve as a powerful diagnostic tool for the detection of prostate cancer. RASSF2 may also serve as an important target for epigenetic-based therapy for the prostate.
Acknowledgments
We thank Vivek Rangnekar (University of Kentucky, Lexington) for reagents and advice, Manuel Serrano and Pablo Fernandez-Marcos (Spanish National Cancer Research Center) for unpublished negative data, and Nina Niessl and Monika Kerscher for excellent technical assistance in processing the tissue specimens.
This work was funded in part by NIH grant 1P20 RR18733, NCI intramural funds (G.J.C.), and the Breast Cancer Campaign, Cancer Research UK (F.L.).
Footnotes
Published ahead of print on 5 April 2010.
REFERENCES
- 1.Akino, K., M. Toyota, H. Suzuki, H. Mita, Y. Sasaki, M. Ohe-Toyota, J. P. Issa, Y. Hinoda, K. Imai, and T. Tokino. 2005. The Ras effector RASSF2 is a novel tumor-suppressor gene in human colorectal cancer. Gastroenterology 129:156-169. [DOI] [PubMed] [Google Scholar]
- 2.Boehrer, S., K. U. Chow, E. Puccetti, M. Ruthardt, S. Godzisard, A. Krapohl, B. Schneider, D. Hoelzer, P. S. Mitrou, V. M. Rangnekar, and E. Weidmann. 2001. Deregulated expression of prostate apoptosis response gene-4 in less differentiated lymphocytes and inverse expressional patterns of par-4 and bcl-2 in acute lymphocytic leukemia. Hematol. J. 2:103-107. [DOI] [PubMed] [Google Scholar]
- 3.Boehrer, S., D. Nowak, E. Puccetti, M. Ruthardt, N. Sattler, B. Trepohl, B. Schneider, D. Hoelzer, P. S. Mitrou, and K. U. Chow. 2006. Prostate-apoptosis-response-gene-4 increases sensitivity to TRAIL-induced apoptosis. Leuk. Res. 30:597-605. [DOI] [PubMed] [Google Scholar]
- 4.Calvisi, D. F., S. Ladu, A. Gorden, M. Farina, E. A. Conner, J. S. Lee, V. M. Factor, and S. S. Thorgeirsson. 2006. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 130:1117-1128. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, M., S. G. Qiu, K. M. Vasudevan, and V. M. Rangnekar. 2001. Par-4 drives trafficking and activation of Fas and Fasl to induce prostate cancer cell apoptosis and tumor regression. Cancer Res. 61:7255-7263. [PubMed] [Google Scholar]
- 6.Cook, J., S. Krishnan, S. Ananth, S. F. Sells, Y. Shi, M. M. Walther, W. M. Linehan, V. P. Sukhatme, M. H. Weinstein, and V. M. Rangnekar. 1999. Decreased expression of the pro-apoptotic protein Par-4 in renal cell carcinoma. Oncogene 18:1205-1208. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, W. N., R. E. Dickinson, A. Dallol, E. V. Grigorieva, T. V. Pavlova, L. B. Hesson, I. Bieche, M. Broggini, E. R. Maher, E. R. Zabarovsky, G. J. Clark, and F. Latif. 2008. Epigenetic regulation of the ras effector/tumour suppressor RASSF2 in breast and lung cancer. Oncogene 27:1805-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper, W. N., L. B. Hesson, D. Matallanas, A. Dallol, A. von Kriegsheim, R. Ward, W. Kolch, and F. Latif. 2009. RASSF2 associates with and stabilizes the proapoptotic kinase MST2. Oncogene 28:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell, S. E., J. Distler, N. S. Goodman, S. H. Mooney, A. Kluth, A. Olek, I. Schwope, R. Tetzner, H. Ziebarth, and K. Berlin. 2004. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 32:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, A. D., and C. J. Der. 2003. The dark side of Ras: regulation of apoptosis. Oncogene 22:8999-9006. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Meco, M. T., M. M. Municio, S. Frutos, P. Sanchez, J. Lozano, L. Sanz, and J. Moscat. 1996. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell 86:777-786. [DOI] [PubMed] [Google Scholar]
- 12.Donninger, H., M. D. Vos, and G. J. Clark. 2007. The RASSF1A tumor suppressor. J. Cell Sci. 120:3163-3172. [DOI] [PubMed] [Google Scholar]
- 13.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Eads, C. A., R. V. Lord, K. Wickramasinghe, T. I. Long, S. K. Kurumboor, L. Bernstein, J. H. Peters, S. R. DeMeester, T. R. DeMeester, K. A. Skinner, and P. W. Laird. 2001. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 61:3410-3418. [PubMed] [Google Scholar]
- 15.El-Guendy, N., Y. Zhao, S. Gurumurthy, R. Burikhanov, and V. M. Rangnekar. 2003. Identification of a unique core domain of par-4 sufficient for selective apoptosis induction in cancer cells. Mol. Cell. Biol. 23:5516-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endoh, M., G. Tamura, T. Honda, N. Homma, M. Terashima, S. Nishizuka, and T. Motoyama. 2005. RASSF2, a potential tumour suppressor, is silenced by CpG island hypermethylation in gastric cancer. Br. J. Cancer 93:1395-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenton, S. L., A. Dallol, A. Agathanggelou, L. Hesson, J. Ahmed-Choudhury, S. Baksh, C. Sardet, R. Dammann, J. D. Minna, J. Downward, E. R. Maher, and F. Latif. 2004. Identification of the E1A-regulated transcription factor p120 E4F as an interacting partner of the RASSF1A candidate tumor suppressor gene. Cancer Res. 64:102-107. [DOI] [PubMed] [Google Scholar]
- 18.Fiordalisi, J. J., R. L. Johnson II, A. S. Ulku, C. J. Der, and A. D. Cox. 2001. Mammalian expression vectors for Ras family proteins: generation and use of expression constructs to analyze Ras family function. Methods Enzymol. 332:3-36. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Cao, I., A. Duran, M. Collado, M. J. Carrascosa, J. Martin-Caballero, J. M. Flores, M. T. Diaz-Meco, J. Moscat, and M. Serrano. 2005. Tumour-suppression activity of the proapoptotic regulator Par4. EMBO Rep. 6:577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Cao, I., M. J. Lafuente, L. M. Criado, M. T. Diaz-Meco, M. Serrano, and J. Moscat. 2003. Genetic inactivation of Par4 results in hyperactivation of NF-κB and impairment of JNK and p38. EMBO Rep. 4:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goswami, A., R. Burikhanov, A. de Thonel, N. Fujita, M. Goswami, Y. Zhao, J. E. Eriksson, T. Tsuruo, and V. M. Rangnekar. 2005. Binding and phosphorylation of par-4 by akt is essential for cancer cell survival. Mol. Cell 20:33-44. [DOI] [PubMed] [Google Scholar]
- 22.Guo, C., S. Tommasi, L. Liu, J. K. Yee, R. Dammann, and G. P. Pfeifer. 2007. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 17:700-705. [DOI] [PubMed] [Google Scholar]
- 23.Gurumurthy, S., A. Goswami, K. M. Vasudevan, and V. M. Rangnekar. 2005. Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol. Cell. Biol. 25:1146-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hesson, L. B., R. Wilson, D. Morton, C. Adams, M. Walker, E. R. Maher, and F. Latif. 2005. CpG island promoter hypermethylation of a novel Ras-effector gene RASSF2A is an early event in colon carcinogenesis and correlates inversely with K-ras mutations. Oncogene 24:3987-3994. [DOI] [PubMed] [Google Scholar]
- 25.Imai, T., M. Toyota, H. Suzuki, K. Akino, K. Ogi, Y. Sogabe, L. Kashima, R. Maruyama, M. Nojima, H. Mita, Y. Sasaki, F. Itoh, K. Imai, Y. Shinomura, H. Hiratsuka, and T. Tokino. 2008. Epigenetic inactivation of RASSF2 in oral squamous cell carcinoma. Cancer Sci. 99:958-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi, J., P. J. Fernandez-Marcos, A. Galvez, R. Amanchy, J. F. Linares, A. Duran, P. Pathrose, M. Leitges, M. Canamero, M. Collado, C. Salas, M. Serrano, J. Moscat, and M. T. Diaz-Meco. 2008. Par-4 inhibits Akt and suppresses Ras-induced lung tumorigenesis. EMBO J. 27:2181-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaira, K., N. Sunaga, Y. Tomizawa, N. Yanagitani, T. Ishizuka, R. Saito, T. Nakajima, and M. Mori. 2007. Epigenetic inactivation of the RAS-effector gene RASSF2 in lung cancers. Int. J. Oncol. 31:169-173. [PubMed] [Google Scholar]
- 28.Khokhlatchev, A., S. Rabizadeh, R. Xavier, M. Nedwidek, T. Chen, X. F. Zhang, B. Seed, and J. Avruch. 2002. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12:253-265. [DOI] [PubMed] [Google Scholar]
- 29.Kumari, G., P. K. Singhal, M. R. Rao, and S. Mahalingam. 2007. Nuclear transport of Ras-associated tumor suppressor proteins: different transport receptor binding specificities for arginine-rich nuclear targeting signals. J. Mol. Biol. 367:1294-1311. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J. W., J. K. Nagpal, C. Jeronimo, J. E. Lee, R. Henrique, M. S. Kim, K. L. Ostrow, K. Yamashita, V. van Criekinge, G. Wu, C. S. Moon, B. Trink, and D. Sidransky. 2008. Hypermethylation of MCAM gene is associated with advanced tumor stage in prostate cancer. Prostate 68:418-426. [DOI] [PubMed] [Google Scholar]
- 31.Malumbres, M., and M. Barbacid. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3:459-465. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama, R., K. Akino, M. Toyota, H. Suzuki, T. Imai, M. Ohe-Toyota, E. Yamamoto, M. Nojima, T. Fujikane, Y. Sasaki, T. Yamashita, Y. Watanabe, H. Hiratsuka, K. Hirata, F. Itoh, K. Imai, Y. Shinomura, and T. Tokino. 2008. Cytoplasmic RASSF2A is a proapoptotic mediator whose expression is epigenetically silenced in gastric cancer. Carcinogenesis 29:1312-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Model, F., N. Osborn, D. Ahlquist, R. Gruetzmann, B. Molnar, F. Sipos, O. Galamb, C. Pilarsky, H. D. Saeger, Z. Tulassay, K. Hale, S. Mooney, J. Lograsso, P. Adorjan, R. Lesche, A. Dessauer, J. Kleiber, B. Porstmann, A. Sledziewski, and C. Lofton-Day. 2007. Identification and validation of colorectal neoplasia-specific methylation markers for accurate classification of disease. Mol. Cancer Res. 5:153-163. [DOI] [PubMed] [Google Scholar]
- 34.Moreno-Bueno, G., P. J. Fernandez-Marcos, M. Collado, M. J. Tendero, S. M. Rodriguez-Pinilla, I. Garcia-Cao, D. Hardisson, M. T. Diaz-Meco, J. Moscat, M. Serrano, and J. Palacios. 2007. Inactivation of the candidate tumor suppressor par-4 in endometrial cancer. Cancer Res. 67:1927-1934. [DOI] [PubMed] [Google Scholar]
- 35.Nalca, A., S. G. Qiu, N. El-Guendy, S. Krishnan, and V. M. Rangnekar. 1999. Oncogenic Ras sensitizes cells to apoptosis by Par-4. J. Biol. Chem. 274:29976-29983. [DOI] [PubMed] [Google Scholar]
- 36.Park, H. W., H. C. Kang, I. J. Kim, S. G. Jang, K. Kim, H. J. Yoon, S. Y. Jeong, and J. G. Park. 2007. Correlation between hypermethylation of the RASSF2A promoter and K-ras/BRAF mutations in microsatellite-stable colorectal cancers. Int. J. Cancer 120:7-12. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, H. B., T. J. Phesse, and A. R. Clarke. 2009. K-ras and Wnt signaling synergize to accelerate prostate tumorigenesis in the mouse. Cancer Res. 69:94-101. [DOI] [PubMed] [Google Scholar]
- 38.Pruitt, K., A. S. Ulku, K. Frantz, R. J. Rojas, V. M. Muniz-Medina, V. M. Rangnekar, C. J. Der, and J. M. Shields. 2005. Ras-mediated loss of the pro-apoptotic response protein Par-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J. Biol. Chem. 280:23363-23370. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, G., M. Ahmed, S. F. Sells, M. Mohiuddin, M. H. Weinstein, and V. M. Rangnekar. 1999. Mutually exclusive expression patterns of Bcl-2 and Par-4 in human prostate tumors consistent with down-regulation of Bcl-2 by Par-4. Oncogene 18:623-631. [DOI] [PubMed] [Google Scholar]
- 40.Sells, S. F., S. S. Han, S. Muthukkumar, N. Maddiwar, R. Johnstone, E. Boghaert, D. Gillis, G. Liu, P. Nair, S. Monnig, P. Collini, M. P. Mattson, V. P. Sukhatme, S. G. Zimmer, D. P. Wood, Jr., J. W. McRoberts, Y. Shi, and V. M. Rangnekar. 1997. Expression and function of the leucine zipper protein Par-4 in apoptosis. Mol. Cell. Biol. 17:3823-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sells, S. F., D. P. Wood, Jr., S. S. Joshi-Barve, S. Muthukumar, R. J. Jacob, S. A. Crist, S. Humphreys, and V. M. Rangnekar. 1994. Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Differ. 5:457-466. [PubMed] [Google Scholar]
- 42.Shareef, M. M., N. Cui, R. Burikhanov, S. Gupta, S. Satishkumar, S. Shajahan, M. Mohiuddin, V. M. Rangnekar, and M. M. Ahmed. 2007. Role of tumor necrosis factor-alpha and TRAIL in high-dose radiation-induced bystander signaling in lung adenocarcinoma. Cancer Res. 67:11811-11820. [DOI] [PubMed] [Google Scholar]
- 43.Tommasi, S., R. Dammann, Z. Zhang, Y. Wang, L. Liu, W. M. Tsark, S. P. Wilczynski, J. Li, M. You, and G. P. Pfeifer. 2005. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 65:92-98. [PubMed] [Google Scholar]
- 44.van der Weyden, L., and D. J. Adams. 2007. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim. Biophys. Acta 1776:58-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vos, M. D., A. Dallol, K. Eckfeld, N. P. Allen, H. Donninger, L. B. Hesson, D. Calvisi, F. Latif, and G. J. Clark. 2006. The RASSF1A tumor suppressor activates Bax via MOAP-1. J. Biol. Chem. 281:4557-4563. [DOI] [PubMed] [Google Scholar]
- 46.Vos, M. D., C. A. Ellis, A. Bell, M. J. Birrer, and G. J. Clark. 2000. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J. Biol. Chem. 275:35669-35672. [DOI] [PubMed] [Google Scholar]
- 47.Vos, M. D., C. A. Ellis, C. Elam, A. S. Ulku, B. J. Taylor, and G. J. Clark. 2003. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J. Biol. Chem. 278:28045-28051. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Z., D. Sun, N. Van do, A. Tang, L. Hu, and G. Huang. 2007. Inactivation of RASSF2A by promoter methylation correlates with lymph node metastasis in nasopharyngeal carcinoma. Int. J. Cancer. 120:32-38. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, Y., R. Burikhanov, S. Qiu, S. M. Lele, C. D. Jennings, S. Bondada, B. Spear, and V. M. Rangnekar. 2007. Cancer resistance in transgenic mice expressing the SAC module of Par-4. Cancer Res. 67:9276-9285. [DOI] [PubMed] [Google Scholar]