Abstract
The human pathogen Chlamydia trachomatis exists as multiple serovariants that have distinct organotropisms for different tissue sites. Culture and epidemiologic data have demonstrated that serovar G is more prevalent, while serovar E is less prevalent, for rectal isolates from men having sex with men (MSM). The relative prevalence of these serovars is the opposite for isolates from female cervical infections. In contrast, the prevalence of serovar J isolates is approximately the same at the different tissue sites, and these isolates are the only C-class strains that are routinely cultured from MSM populations. These correlations led us to hypothesize that polymorphisms in open reading frame (ORF) sequences correlate with the different tissue tropisms of these serovars. To explore this possibility, we sequenced and compared the genomes of clinical anorectal and cervical isolates belonging to serovars E, G, and J and compared these genomes with each other, as well as with a set of previously sequenced genomes. We then used PCR- and restriction digestion-based genotyping assays performed with a large collection of recent clinical isolates to show that polymorphisms in ORFs CT144, CT154, and CT326 were highly associated with rectal tropism in serovar G isolates and that polymorphisms in CT869 and CT870 were associated with tissue tropism across all serovars tested. The genome sequences collected were also used to identify regions of likely recombination in recent clinical strains. This work demonstrated that whole-genome sequencing along with comparative genomics is an effective approach for discovering variable loci in Chlamydia spp. that are associated with clinical presentation.
Chlamydia trachomatis is an obligate intracellular human pathogen that is the leading cause of preventable blindness worldwide and is the most common sexually transmitted infectious bacterium in humans. The study of the biology of chlamydiae is complicated by their obligate intracellular development and the lack of a routine system for directed mutagenesis. Chlamydial isolates are differentiated into serovars based on serospecificity for the chlamydial major outer membrane protein (MOMP) (7), which is encoded by ompA (37). The serovars fall into biological groups associated with trachoma (serovars A to C), sexually transmitted noninvasive disease (serovars D to K), and invasive lymphogranuloma (serovars L1 to L3) (35). Comparative genomic analysis of ocular and urogenital chlamydial species has proven to be an effective approach for discovering genetic loci that are associated with observed tissue tropism (9, 10).
Studies conducted in Seattle, WA, and Birmingham, AL, have shown that serovar G rectal isolates are prevalent in men having sex with men (MSM), while serovar E rectal isolates are less prevalent (1, 5, 17). This prevalence of serovar G and rectal tropism differ from what has been observed in studies of female cervical populations in the same geographical regions, where the prevalence of serovar E was significantly higher than the prevalence of serovar G (38). It is not clear whether the causes of these differences are behavioral resulting from network bottlenecks or whether there are genuine biological differences between rectotropic and cervicotropic strains.
The limited examples of horizontally acquired DNA in chlamydial species suggest that lateral gene transfer and recombination are rare in these organisms. However, sequencing efforts have identified clear examples of recombination at a limited number of chlamydial loci, including ompA (6, 19-21, 27, 29). Recent studies have shown that chlamydiae contain the necessary machinery for recombination (4, 37) and that lateral gene transfer can be selected for in cell cultures following coinfection with strains carrying dissimilar drug markers both within and among chlamydial species (4, 13, 14, 42). The mechanisms of recombination and the role of recombination in chlamydial fitness in vivo remain to be investigated.
The distributions of serovar G strains in the heterosexual and MSM populations led us to hypothesize that strains with rectal tropism have variable genes or loci compared to other urogenital isolates. To test this hypothesis, we sequenced eight chlamydial isolates representing serovars D, E, F, J, and G that were collected from the female cervix, male urethra, or male rectum. PCR and restriction fragment length polymorphism (RFLP) assays were then developed to determine if candidate open reading frames (ORFs) identified in the genome sequence analysis were associated with the observed tropism for the rectal site of infection. A set of candidate ORFs that were associated with rectal tropism in serovar G isolates were discovered, and polymorphisms in the pmp genes were correlated with rectal tropism across all serovars tested. Analysis of the genomes also demonstrated that recombination appears to be common in clinical isolates and occurs at locations across the chlamydial genome.
MATERIALS AND METHODS
Chlamydia strains and genomic DNA preparation.
C. trachomatis clinical isolates Ds/2923, E/11023, E/150, F/70, G/9301, G/9768, G/11222, G/11074, and J/6276 were propagated from frozen samples stored at the University of Washington Chlamydia Repository (39). Isolate collection, clonal isolation, serotyping, and elementary body (EB) purification were conducted as previously described (41, 43). Purified EBs were incubated for 60 min with 4 U/ml RQ1 DNase (Promega), which was followed by treatment with 2 mM EGTA (RQ1 Stop solution; Promega) to inactivate the enzyme. DNA was then extracted from purified, DNase-treated EBs using a Qiagen genomic tip kit (Qiagen, Valencia, CA) by following the manufacturer's instructions. The initial suspension buffer used for these purifications was supplemented with dithiothreitol (5 mM) to facilitate EB lysis.
Genome sequencing and sequence analysis.
Isolates Ds/2923, E/11023, E/150, G/9301, G/9768, and G/11222 were sequenced using classical Sanger sequencing methods at the Joint Genome Institute (Walnut Creek, CA). DNA from isolates J/6276, G/11074, and F/70 was processed for Illumina-based sequencing using commercial DNA preparation kits (Illumina Inc., San Diego, CA) by following the manufacturer's instructions. Illumina-derived genomes were first assembled using the reference-guided assembly program Maq (28). Regions in reference-guided assembled genomes where Maq could not resolve the sequence were then compared to contiguous sequences assembled using the VCAKE de novo assembly software (22), and a single contiguous draft sequence was produced.
Whole-genome phylogenetic analysis was performed using the alignment program MAFFT with the default settings (24, 25). The sequences compared included sequences generated in this study, as well as previously published genomes for serovar D, A, and L2 strains (strains D/UW3 [GenBank accession number AE001273] [37], A/HAR-13 [GenBank accession number CP000051] [10], and L2-434/Bu [GenBank accession number AM884176] [45]). Pairwise genome alignments were produced using MAFFT with the following settings: iterative refinement, 2; default gap opening penalty, 1.53; and default gap extension penalty, 0.123. These alignments were used to determine the total number of substitutions and insertions or deletions (indels) in genome sequences. Regions where there was high variability between selected sequences were analyzed manually using the MacVector sequence analysis software (MacVector, Cary, NC), and counts were adjusted accordingly. Isolate genome sequences were compared with the previously published C. trachomatis D/UW3 genome sequence (GenBank accession number AE001273) (37) using Diffseq from the Emboss Bioinformatics suite (33), and an in-house single-nucleotide polymorphism (SNP) parsing program (Diffsort; http://people.oregonstate.edu/∼rockeyd/Diffsort) was used to determine the locations and translational effects of polymorphisms that were identified. Any gene variation not resolved by Diffsort was manually analyzed using the MacVector sequence analysis software.
Genome-wide recombination analysis.
DNA sequences were computationally extracted from selected isolates using sliding windows (1,000 nucleotide windows, 800-nucleotide slides) and were used as bioinformatic probes with a database consisting of template genome sequences (D/UW3, J/6276, G/9768, E/11023, and/or F/70 sequences). A comparison of the BLAST raw scores for each window was performed based on whether the window was more similar to a clade containing serovar J, G, or D or a clade containing serovar E or F or whether the probe sequence matched all template genomes equally. The following rules were used to assign a window to a genome sequence or clade. Queries that matched all serovars equally were plotted along the “All” line. Queries that did not match all genomes equally but in which one matched template was either a serovar E template or a serovar F template were grouped with the “E or F” clade. Queries that matched serovar J, G, or D more closely than either serovar E or serovar F were grouped with the “J, G, or D” clade. A single query did not match any of the template genomes and was categorized as “No Hit” in this analysis. Whole-genome results from this parsing were then graphed using the complete D/UW3 genome as a reference, beginning with ORF CT001 (37).
Regions where there was apparent recombination in strain Ds/2923 were then characterized using the ClustalW program, and identified loci were aligned with corresponding sequences from strains D/UW3 and E/11023. The resulting alignments were used to determine the number of informative sites shared by each strain. For this purpose, an informative site was any position in the sequences examined where there was a polymorphism in the template genomes analyzed. Insertions and deletions of any size were counted as one informative site. An identical approach was used for analysis of recombination in strain F/70, using strains D/UW3, J/6276, G/9768, and E/11023 as templates.
Preparation of clinical isolate DNA and PCR-based genotyping.
Clinical isolate genome sequence variation data were used to design PCR and RFLP assays for genotypic variation in a population of clinical isolates stored in the repository, using oligonucleotide primers and restriction endonucleases listed in Table S1 in the supplemental material. The isolates used in this study included chlamydiae collected in King County, WA (55 isolates), Lima, Peru (2 isolates) (34), and Birmingham, AL (6 isolates). The genes analyzed were selected based on variation between serovar G cervical and rectal isolates or variation between serovar G, E, and D isolates. This approach was also used to confirm that each ompA genotype was consistent with the MOMP phenotype identified by immunofluorescence (data not shown). McCoy cells in six-well trays were infected with cloned clinical isolates, and chlamydiae were grown for 48 h. Genomic DNA was extracted using a Qiagen DNeasy blood and tissue kit by following the manufacturer's instructions, using an initial suspension buffer supplemented with 5 mM dithiothreitol. Sequences in the polymorphic regions selected for SNP analysis were confirmed by traditional Sanger sequencing, using primers shown in Table S1 in the supplemental material. The Fisher exact test was used to determine any statistical association of identified SNPs with observed phenotypes. The alternate hypothesis in these statistical analyses was that there was no correlation between genotype and phenotype at each of the loci tested. Statistical significance was expressed using a P value of <0.01 or <0.001, and the significance data supported the hypothesis that differences at the loci tested correlated with tissue tropism for either the rectal or cervical site of infection.
Nucleotide sequence accession numbers.
The C. trachomatis clinical isolate genome sequences sequenced at the Joint Genome Institute have been deposited in the DDBJ/EMBL/GenBank database under the following accession numbers: D(s)/2923, ACFJ01000001; E/11023, CP001890; E/150, CP001886; G/9301, CP001930; G/9768, CP001887; and G/11222, CP001888. The strains sequenced using Illumina sequencing as part of the Whole Genome Shotgun Project have been deposited in the DDBJ/EMBL/GenBank database under the following accession numbers: J/6276, ABYD01000001; F/70, ABYF01000001; and G/11074, CP001889.
RESULTS
Comparative genome analysis of sequenced chlamydial isolates.
Pairwise alignment analysis using MAFFT (24, 25) of the genomes of 12 recent clinical isolates demonstrated that there were different levels of heterogeneity among strains (Table 1). The number of nucleotide substitutions between urogenital strains belonging to different serovars (not including L2-434/Bu) ranged from 6,494 (for G/11222 and E/11023) to 1,638 (for D/UW3 and G/11222), and the number of nucleotide differences between strains belonging to the same serovar ranged from 1,287 (for G/11074 and G/11222) to 3 (for G/9301 and G/9768). The serovar G strains showing the highest level of similarity were cultured from the male urethra and male rectum of different patients, and their collection dates were separated by more than 1 year. The sequence of a cervical nonfusogenic isolate, Ds/2923, was more similar to the sequences of serovar E and F isolates than to the published serovar D sequence (Table 1). A comparison of strains E/11023 and Ds/2923 identified 1,211 substitutions, while a comparison of strains D/UW3 and Ds/2923 identified 5,764 substitutions. The highest number of differences between genomes (8,811 nucleotides, 326 indels) was the number of differences between the genomes of the publicly available sequenced ocular strain A/HAR-13 (10) and the publicly available sequenced lymphogranuloma strain L2-434/Bu. This number represented a maximum level of variability of 0.87%.
TABLE 1.
Pairwise analysis of the numbers of substitutions and insertion-deletion events identified in genome sequences for a collection of C. trachomatis isolates
| Isolate | Genome size (bp) | No. of substitutions or no. of indel eventsa |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D/UW3 | A/HAR13 | L2-434/Bu | Ds/2923 | E/11023 | E/150 | G/9301 | G/9768 | G/11222 | G/11074 | J/6276 | F/70 | ||
| D/UW3 | 1,042,519 | 157 | 297 | 187 | 210 | 210 | 93 | 93 | 78 | 91 | 102 | 244 | |
| A/HAR13 | 1,044,459 | 3,696 | 326 | 251 | 254 | 254 | 151 | 151 | 169 | 152 | 172 | 261 | |
| L2-434/Bu | 1,038,842 | 8,163 | 8,811 | 319 | 309 | 314 | 310 | 310 | 309 | 308 | 312 | 318 | |
| Ds/2923 | 1,042,757 | 5,764 | 7,131 | 7,687 | 75 | 78 | 210 | 210 | 197 | 207 | 203 | 150 | |
| E/11023 | 1,043,025 | 6,135 | 7,143 | 7,737 | 1,211 | 54 | 231 | 231 | 220 | 228 | 235 | 118 | |
| E/150 | 1,042,996 | 6,138 | 7,027 | 7,766 | 1,325 | 1,130 | 205 | 205 | 197 | 203 | 225 | 135 | |
| G/9301 | 1,042,811 | 1,892 | 3,523 | 8,162 | 6,205 | 6,485 | 5,869 | 1 | 69 | 5 | 92 | 250 | |
| G/9768 | 1,042,810 | 1,893 | 3,524 | 8,163 | 6,206 | 6,486 | 5,776 | 3 | 69 | 6 | 92 | 250 | |
| G/11222 | 1,042,354 | 1,638 | 3,778 | 8,131 | 6,063 | 6,494 | 5,907 | 1,286 | 1,287 | 70 | 88 | 249 | |
| G/11074 | 1,042,875 | 1,893 | 3,529 | 8,153 | 6,207 | 6,490 | 5,874 | 41 | 42 | 1,287 | 90 | 247 | |
| J/6276 | 1,043,181 | 1,899 | 3,540 | 8,264 | 5,878 | 6,467 | 6,292 | 1,918 | 1,919 | 1,770 | 1,920 | 252 | |
| F/70 | 1,048,006 | 6,287 | 6,913 | 7,738 | 2,239 | 1,776 | 2,407 | 6,305 | 6,306 | 6,236 | 6,310 | 6,441 | |
The numbers of substitutions are indicated on the lower left, and the numbers of indels are indicated on the upper right.
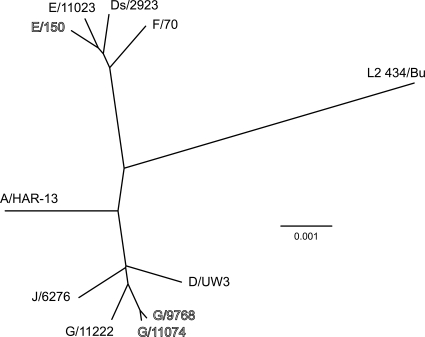
The whole-genome phylogenetic tree shown in Fig. 1 indicated that our sequenced urogenital serovars fell into at least two clades, one group containing serovars D, G, and J and a second group containing serovars E and F. These two groups were distinct from ocular strain A/HAR-13 and lymphogranuloma strain L2-434/Bu. In this analysis, the genome of strain Ds/2923, which was originally serotyped based on reactivity with serovar D-specific monoclonal antibodies, grouped in the clade with the serovar E and F strains. These data are parallel to the data shown in Table 1 and confirm that the genome of Ds/2923 is more similar to serovar E or F genomes than to the published serovar D genome.
FIG. 1.
Phylogenetic analysis of sequenced chlamydial isolates based on whole-genome alignment. The tree was constructed using a modified UPGMA (unweighted-pair group method using average linkages) algorithm as part of the MAFFT alignment program. Strains collected from the male rectum are indicated by outlined type, and cervical isolates are indicated by standard type. The branch lengths are proportional to the genetic distances between isolates.
Mapping of SNPs and greater changes in sequenced genomes of clinical isolates.
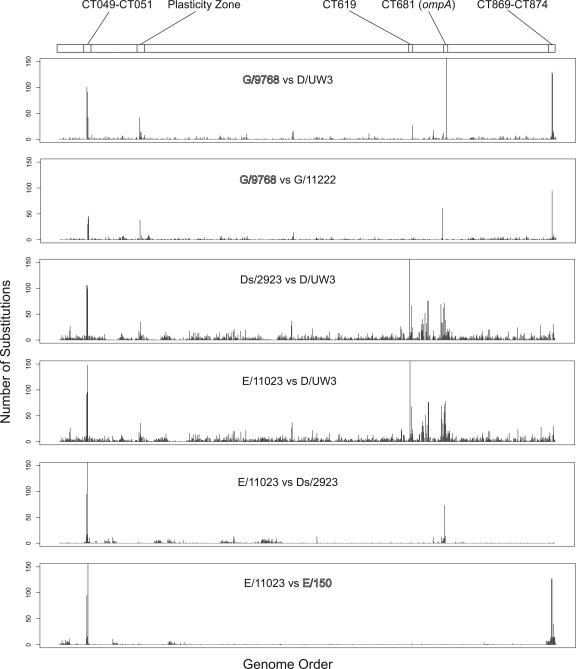
Diffsort was used to determine the number of substitutions per ORF in comparisons of selected clinical isolates. This study was undertaken to determine if variation is localized to specific regions in the chlamydial genome and to identify possible loci containing variable regions in different serovars or within serovars (Fig. 2). These analyses identified specific regions throughout the genome that exhibited higher levels of variation than the overall genome. These regions included possible recombination targets, including ORFs CT049 to CT051 (20), the plasticity zone ORFs CT144 to CT176, ompA, and the pmp genes (5, 9-11, 16, 32, 45, 46).
FIG. 2.
Numbers of substitutions per open reading frame for comparisons of strains. Each graph represents a Diffseq-generated whole-genome alignment for the strains indicated. The genes, beginning with CT001, are indicated on the x axis; the sequenced strain D/UW3 gene designations were used as a reference. The numbers of substitutions per ORF are indicated on the y axis. Strains collected from the male rectum are indicated by outlined type, and cervical isolates are indicated by standard type.
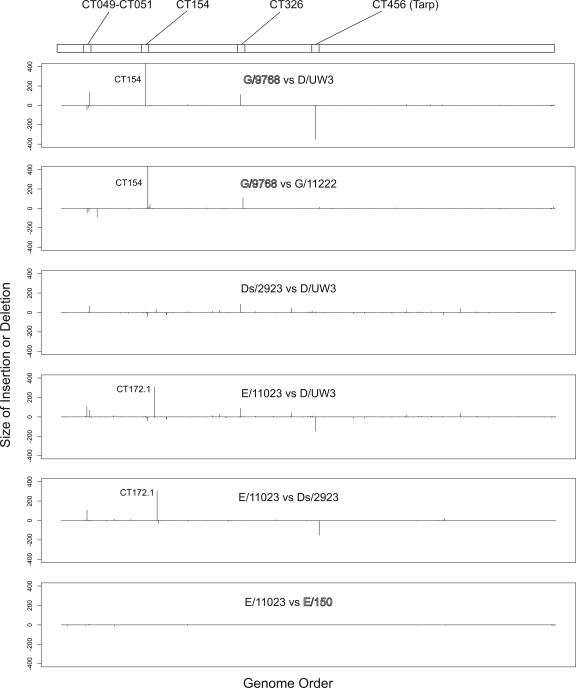
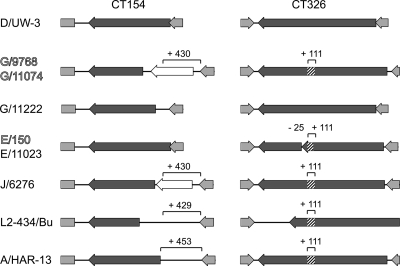
Similar pairwise analysis of clinical isolates identified a variety of insertions or deletions (indels) in the different strains (Fig. 3). The serovar G rectal strains had a 430-nucleotide insertion in the CT154 gene, which led to an N-terminal truncation of CT154 and a putative new ORF (CT154.1). A second change found in the serovar G rectal strains was an in-frame 111-nucleotide insertion in gene CT326. These polymorphisms were limited to the sequenced serovar G rectal isolates; the cervical serovar G isolate G/11222 contained neither of these insertions. Both rectal and cervical serovar E isolates lacked the insertion in CT154 but had the 111-nucleotide insertion in CT326. The structure of CT326 was complicated by a 25-nucleotide deletion in serovar E strains compared to D/UW3. This deletion led to a truncated N-terminal CT326 ORF and a C-terminal CT326 ORF (Fig. 4). Strain J/6726 and previously sequenced strains L2-434/Bu and A/HAR-13 also had the 111-nucleotide insertion in CT326, as well as similar but unique insertions in the CT154 region (Fig. 4). Consistent with the findings of Carlson et al. (10), insertions and deletions were located in ORF CT456 (encoding Tarp) in several of the sequenced isolates.
FIG. 3.
Locations of insertions or deletions in strains. The comparisons are identical to and in the same order as those shown in Fig. 2. The genes, beginning with CT001, are indicated on the x axis; the sequenced strain D/UW3 gene designations were used as a reference. The y axis indicates the lengths of the insertions (above the x axis) and deletions (below the x axis). For each graph the strain listed first has an insertion or a deletion at a given site compared to the strain listed second. For example, G/9768 has a 437-bp insertion in CT154 compared to D/UW3. Strains collected from the male rectum are indicated by outlined type, and cervical isolates are indicated by standard type.
FIG. 4.
Schematic diagram of open reading frames CT154 and CT326 in several sequenced C. trachomatis strains. The isolate designations are indicated on the left. Strains isolated from the male rectal site of infection are indicated by outlined type, and cervical isolates are indicated by standard type. The ORF examined is indicated by a dark gray arrow, and lighter gray arrows and boxes indicate flanking ORFs. The boxes with diagonal stripes indicate the locations of insertions in the genes. Putative new ORFs resulting from an insertion are indicated by open arrows. The numbers above the brackets indicate the lengths (in base pairs) of insertions based on a comparison with the D/UW3 sequence.
Evidence for recombination in genomes of sequenced clinical isolates.
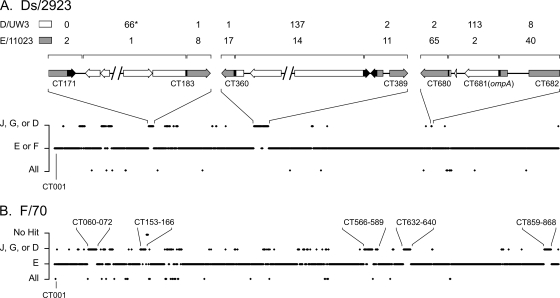
The apparent similarity of the isolate Ds/2923 genome to genomes of serovar E and F strains led us to hypothesize that regions other than ompA might show evidence of recombination in this strain. A BLAST-based similarity approach using sliding windows consisting of 1,000 nucleotides across the entire genome was used to uncover additional regions of recombination. These analyses confirmed that the majority of the Ds/2923 genome is similar to the genomes of serovar E and F isolates (Fig. 5A), while ompA and nearby sequences are most similar to D/UW3 sequences. Fine mapping of these regions in Ds/2923 demonstrated that the apparent upstream crossover point adjacent to ompA is in the rs2 gene (CT680), at a position previously described as a hot spot for recombination in chlamydial genomes (20). The downstream crossover point for this recombination event is located within ompA and results in a hybrid MOMP protein with variable domains from different serovars (48). These studies also uncovered several additional regions exhibiting higher levels of similarity to the genome of D/UW3 than to the genomes of serovar E or F strains. The clearest examples of this are in ORFs CT171 to CT183 and ORFs CT360 to CT388 (Fig. 5A). The differences between genomes in these regions included SNPs, indels as large as 308 nucleotides (CT171), and ORF fusions. These data support the conclusion that these regions were involved in recombination between chromosomes, leading to the mosaic Ds/2923 genome.
FIG. 5.
BLAST-based evidence for recombination in clinical C. trachomatis genome sequences. Complete genome sequences of strain Ds/2923 (A) and strain F/70 (B) were used as probes with a genome database containing sequences of D/UW3, G/9768, J/6276, E/11023, and (in panel A) F/70. The complete genome sequence of each probe was evaluated using BLAST analysis of a 1-kbp sliding window, and each window was given a score based on whether it was more similar to a clade containing serovar J, G, or D, it was more similar to a clade containing serovar E or F, or it was equally similar to all the genomes. The results for each window are plotted on the x axis beginning with ORF CT001 as defined for the D/UW3 genome (37). All ORF designations are also based on the D/UW3 genome sequence. Panel A includes ORF maps showing expanded views of three regions showing apparent recombination. In the ORF maps, a gray box or arrow indicates that the sequence is more similar to E/11023, an open box or arrow indicates that the sequence is more similar to D/UW3, and a black box or arrow indicates a region where apparent recombination events occurred. The brackets above the gene diagrams indicate the regions for which informative sites were examined and counted. The numbers above the brackets indicate the numbers of informative sites shared by the Ds/2923 and D/UW3 sequences (top line) or the Ds/2923 and E/11023 sequences (bottom line). The asterisk indicates that 1 of the 66 shared informative sites is a 308-nucleotide deletion that D/UW3 and Ds/2923 share compared to the E/11023 sequence. The identified areas in panel B are selected regions of the F/70 genome that are more similar to genomes of strains in the serovar J-serovar G-serovar D clade than to the genomes of serovar E strains.
To determine if genomes of other sequenced isolates exhibited a mosaic structure similar to that of Ds/2923, a BLAST similarity analysis was performed with isolate F/70 (Fig. 5B). For this experiment, the genome of F/70 was removed as a template in the serovar E-serovar F clade used for analysis of Ds/2923 and instead used as a probe of the remaining genomes. This analysis uncovered a set of regions where there was apparent recombination that were different than the regions observed in Ds/2923. One of the loci (ORFs CT153 to CT166) included the plasticity zone. This region also contained a sequence homologous to the TC0438 (tox) sequence found in Chlamydia muridarum (39). Another possible site of exchange in F/70 included ORFs CT859 through CT868. This region is upstream of the pmp genes (CT869 to CT872 and CT874) and has previously been hypothesized to be a locus where there was lateral gene transfer in C. trachomatis (19, 21).
Correlation of identified SNPs with tropism for the rectal or cervical site of infection.
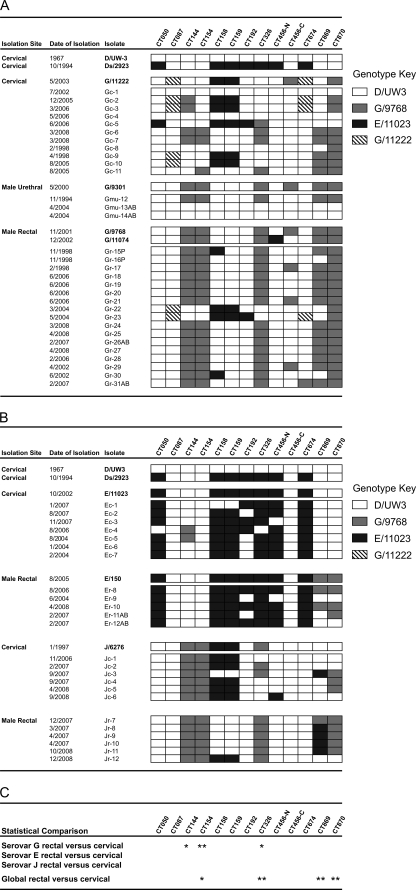
To determine if polymorphisms found in the isolates sequenced were associated with tropism for the rectal site of infection, loci with various degrees of difference were used for PCR or RFLP analysis. These loci included genomic regions with nucleotide substitutions (CT144, CT869, and CT870), regions with indels that affected the predicted protein length (CT154 and CT326) (Fig. 4), and regions with single-nucleotide polymorphisms that affect open reading frame length (CT158 and CT159). Analyses were conducted with a collection of clinical isolates representing serovars E, G, and J that were obtained from the cervix, male rectum, or male urethra. While in most cases there was no apparent or statistical difference between an SNP and tissue tropism, there were ORFs in which correlations were observed. Insertions in CT154 and CT326 (P < 0.001 and P < 0.01, respectively, Fisher's exact test) were associated with rectal tropism within serovar G (Fig. 4 and 6). Statistical analysis of these regions in serovar E and J isolates indicated that there was no association with genotype and tropism, but it is possible that some of these polymorphisms might be found to be significant (e.g., combinations of CT144, CT154, CT158, CT159, and CT326 in serovar J or pmp genes in serovars E and J) if higher numbers of isolates were investigated. Because a high proportion of our samples were collected in the Seattle area, it was possible that we examined a serovar G population that was restricted with respect to social network or geographic clustering. To address this concern, we included a limited number of rectal isolates belonging to serovar G collected in Alabama and Peru. Each of these isolates (n = 4) had the genotype associated with rectal infection for both ORF CT154 and ORF CT326. No significant association was found between polymorphisms in CT869 and CT870 and tropism for serovar G strains, but a comparison of rectal and cervical sites of infection across all isolates identified a significant association between selected polymorphisms in these two ORFs (P < 0.001) (Fig. 6C).
FIG. 6.
PCR- and RFLP-based genotype analysis of variable regions in clinical isolates. The strains, dates of isolation, and sites of isolation are indicated on the left. (A) Data for serovar G isolates. (B) Data for serovars E and J. (C) Results of statistical analyses with Fisher's exact test. One asterisk indicates that the P value is <0.01, and two asterisks indicate that the P value is <0.001. Isolates indicated by bold type are the isolates whose complete genome sequences were examined. Isolates whose designations end with P were collected from patients in Lima, Peru, and isolates whose designations end with AB were collected from patients in Birmingham, AL. All other isolates were collected from patients in King County, WA, area family planning clinics. The designations beginning with CT at the top indicate the gene targets for the PCR and RFLP analysis. CT456-N and CT456-C indicate variable regions that are present at the amino and carboxy termini of CT456. The different types of boxes indicate the genotypes detected, as follows: open boxes, D/UW3 genotype; gray boxes, rectal serovar G genotype; black boxes, serovar E genotype; diagonally striped boxes, cervical serovar G genotype.
While these analyses demonstrated that certain SNP patterns were associated with tropism for either the cervical or rectal site of infection, overall the chromosomes were mosaics of a variety of different SNP patterns. The mosaicism observed is most apparent in the serovar G cervical isolates shown in Fig. 6, but it is also evident in the serovar G rectal isolates (strains Gr-15P, Gr-16P, Gr-30, and Gr-30AB). This mosaicism also supports the conclusion that the analyses of MSM populations did not simply identify clonal expansion in a geographically restricted area or a closed social network.
DISCUSSION
The study of chlamydiae has benefitted greatly from advances in genome sequencing technology. These bacteria have small (∼1-Mb), largely syntenic, AT-rich genomes with very few repeat regions and almost no genomic islands. We (39) and other workers (10, 23) have explored the possibility that genome sequence analysis can be used to characterize functional roles of individual genes in the chlamydiae, particularly since there are no practical genetic tools this system.
In the present study, we used genome sequencing and PCR-based analyses of polymorphisms to examine chlamydial recombination in clinical isolates and to develop a technology for correlating the chlamydial genotype with the clinical phenotype. The genomes sequenced were collected at Seattle/King County sexual health clinics, and the isolation dates ranged from October 1994 to August 2005. Initial genome sequence data demonstrated that the maximum sequence divergence between clinical strains was 0.87% and that there was a minimum difference of three substitutions and a single nucleotide indel. The two strains showing the highest degree of similarity were serovar G isolates collected from the rectal (G/9768) and male urethral (G/9301) sites of infection, and more than a year separated the collection dates of these strains (November 2001 and May 2000, respectively). The largest insertion in any clinical strain sequenced was 4,668 bp long, and it was found in the previously sequenced strain F/70 and was shown to be variable in serovar J strains (9, 39). The overall level of variation observed among C. trachomatis strains is similar to the level of variation observed for members of the same genomic group of the obligate intracellular bacterium Coxiella burnetii (2), as well as for Rickettsia rickettsii isolates (15).
There are technical issues associated with generation of precise SNP counts for different strains. For example, we found 3,696 substitutions when the published sequences of strains D/UW3 and A/HAR13 were compared; this number is slightly higher than the number determined by Carlson et al. (10). The differences can be attributed to the specific aspects of the programs used to count SNPs. One example of this is the settings used in the MAFFT software to generate alignments for determining numbers of substitution and indel events. The default MAFFT settings, which were used in our study, result in a higher penalty for inserting a gap than for extending a gap. Adjusting the settings for a lower gap insertion penalty and a higher extension penalty resulted in slightly fewer substitutions at the cost of increasing the number of indel events. Such differences in the analysis programs lead to slight differences in counts, but the overall relationships among genomes are conserved.
Phylogeny-based characterization of the genomes revealed two clades for our sequences; one clade contained serovars E and F, and the second clade contained serovars D, G, and J. These relationships are in agreement with previously described relationships determined by phylogenetic analysis either by using a set of housekeeping genes (31) or by performing comparative studies of chlamydial phylogeny (9, 10, 32).
A BLAST-based analysis was used to examine if recombination is common in chlamydiae and could be reflected in the genomes collected. Strain Ds/2923 is a cervical isolate that was our original example of an IncA-negative, nonfusogenic strain (40), and its genome showed the best evidence of recombination in clinical strains. This strain, similar to all other IncA-negative strains that have been characterized, has a lesion in IncA that is correlated with the inactivated incA open reading frame, and inclusions formed by such strains do not fuse with inclusions formed by either IncA-negative or IncA-positive C. trachomatis (40). Although serovar-specific monoclonal antibodies identified Ds/2923 as a serovar D strain, our analyses demonstrated that most of its chromosome is more similar to the chromosomes of serovar E or F strains. A set of ORFs that may have been recombination targets in this strain includes ompA, the gene encoding MOMP, the major serovariant antigen in the chlamydiae. Possible recombination events involving ompA have been found in other studies. Early in the analysis of chlamydial gene sequences, hybrid ompA coding sequences were identified by several investigators (6, 27, 29, 48), and recent work by Gomes et al. (20) demonstrated that a recombination hot spot is just upstream of the ompA coding sequence. The Ds/2923 chromosome had an apparent recombination event exactly at the position identified by Gomes and colleagues. The downstream recombination event occurred between variable domains 3 and 4, which is also consistent with the results of other workers who have identified ompA sequences encoding mosaic MOMPs with variable domains in members of different prototype serovars (27, 48).
Studies of possible recombination sites in Ds/2923 were expanded by performing a BLAST-based similarity analysis of sequenced genomes. This analysis identified additional regions in the Ds/2923 chromosome that were targets for recombination between strains. These candidate recombination loci involve several sites, including the plasticity zone (3, 9, 10, 16, 32, 45, 46) and ORFs CT360 to CT389, a region encoding a set of metabolic pathways and hypothetical genes. Both of these genomic regions in Ds/2923 are more similar to the prototypic serovar D sequence than to our serovar E or F sequences. ORFs CT360 to CT389 include aaxABC (CT372 to CT374), which encode proteins that participate in an arginine-agmatine exchange system (36). Recently, Giles et al. used an Escherichia coli system to show that polymorphisms in aaxB led to inactivation of this gene in strain D/UW3 and serovar L2 and that function might be restored in D/UW3 by an R115G replacement (18). Genomic analysis demonstrated that isolates E/150 and E/11023 had this R115G replacement. Therefore, it is possible that there is phenotypic discrepancy at this locus, with serovar E and F strains being “wild type” and D/UW3 and the mosaic strain Ds/2923 being deficient in this exchange pathway.
A subsequent analysis of the genome of strain F/70 identified similar examples of likely recombination events, but at different loci. The data obtained are consistent with the results of in vitro analyses by DeMars et al. (13, 14) and our laboratory (42) and support the hypothesis that recombination is very common and involves many different locations across the chlamydial chromosome.
Our second hypothesis addressed the concept that genome sequencing of clinical strains could identify and help characterize genes and gene products important in the biology of the pathogen in vivo. Pioneering work in this area was conducted by Caldwell et al., who used sequence analysis of a limited number of ORFs to correlate the presence of trp synthesis genes with ocular or genital tropism in C. trachomatis (8). We chose a different clinical phenotype for analysis, tropism for the rectal site of infection, as a trait for study. Our analyses identified four loci that were statistically associated with a particular tropism, only one of which, the CT869-CT870 locus, was associated with rectal tropism across all serovars (P < 0.001). These ORFs encode two Pmp proteins, which are members of a family of chlamydial autotransporters important in different aspects of chlamydial biology (12, 26, 44, 47). The nucleotide variation found in these genes leads to amino acid changes that are not randomly distributed across the coding sequence, indicating that the variable regions may be parts of domains important in chlamydial biology. The variation and molecular evolution of the pmp genes is an active area of research (30), but the possible function of the different Pmp variants in attachment or intracellular development remains to be investigated.
Alterations in three other ORFs were statistically correlated with tropism only in serovar G. CT144 encodes a hypothetical 285-amino-acid protein that varies at 14 amino acids in the strains tested. Eleven of the 14 amino acid changes in CT144 were clustered in a 26-amino-acid region of the gene product (data not shown). Further study of CT144 might determine if this variable domain plays a role in tissue tropism or pathogenesis. ORF CT154.1 is the result of a 430-nucleotide insertion that was also found in the genomes of serovar A and L2 isolates (Fig. 4) and encodes a protein with no predicted function. Finally, the presence of a 111-nucleotide in-frame insertion in uncharacterized hypothetical gene CT326 was statistically correlated with tropism for the rectal culture site. Our association of these ORFs with tissue tropism is complicated by the fact that none of these ORFs were statistically associated with rectal tropism in serovar E or serovar J strains. It is possible that production of the different proteins leads to phenotypic differences in the context of serovar G, facilitating the apparent tissue tropism. Alternatively, it is possible that these SNP differences are in or linked to regions of the genome that encode unidentified proteins that collectively are important for this tropism. Possible functions of candidate proteins identified in this study are currently being explored in our laboratory.
Supplementary Material
Acknowledgments
Connie Celum and Will M. Geisler are gratefully acknowledged for supplying chlamydia isolates collected from individuals in Lima, Peru, and Birmingham, AL. Paul Richardson and Alla Lapidus are acknowledged for the genome sequencing and assembly conducted at the Joint Genome Institute. We thank Kelsi Sandoz for advice and consultation regarding PCR and RFLP analysis. Sara Weeks is acknowledged for technical assistance and editing of the manuscript.
This research was supported by grants A148769 and A1031448 from the National Institutes of Health.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 22 March 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Barnes, R. C., A. M. Rompalo, and W. E. Stamm. 1987. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J. Infect. Dis. 156:953-958. [DOI] [PubMed] [Google Scholar]
- 2.Beare, P. A., J. E. Samuel, D. Howe, K. Virtaneva, S. F. Porcella, and R. A. Heinzen. 2006. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J. Bacteriol. 188:2309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binet, R., and A. T. Maurelli. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl. Acad. Sci. U. S. A. 106:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, J. F., L. A. Koutsky, R. J. Suchland, and W. E. Stamm. 1999. Clinical features of Chlamydia trachomatis rectal infection by serovar among homosexually active men. Sex. Transm. Dis. 26:392-398. [DOI] [PubMed] [Google Scholar]
- 6.Brunham, R., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Invest. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell, H. D., and J. Schachter. 1982. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect. Immun. 35:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson, J. H., S. Hughes, D. Hogan, G. Cieplak, D. E. Sturdevant, G. McClarty, H. D. Caldwell, and R. J. Belland. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72:7063-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson, J. H., S. F. Porcella, G. McClarty, and H. D. Caldwell. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, J. H., H. Wood, C. Roshick, H. D. Caldwell, and G. McClarty. 2006. In vivo and in vitro studies of Chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59:1678-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crane, D. D., J. H. Carlson, E. R. Fischer, P. Bavoil, R. C. Hsia, C. Tan, C. C. Kuo, and H. D. Caldwell. 2006. Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. Proc. Natl. Acad. Sci. U. S. A. 103:1894-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMars, R., and J. Weinfurter. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J. Bacteriol. 190:1605-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMars, R., J. Weinfurter, E. Guex, J. Lin, and Y. Potucek. 2007. Lateral gene transfer in vitro in the intracellular pathogen Chlamydia trachomatis. J. Bacteriol. 189:991-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison, D. W., T. R. Clark, D. E. Sturdevant, K. Virtaneva, S. F. Porcella, and T. Hackstadt. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 76:542-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 17.Geisler, W. M., W. L. Whittington, R. J. Suchland, and W. E. Stamm. 2002. Epidemiology of anorectal chlamydial and gonococcal infections among men having sex with men in Seattle: utilizing serovar and auxotype strain typing. Sex. Transm. Dis. 29:189-195. [DOI] [PubMed] [Google Scholar]
- 18.Giles, T. N., D. J. Fisher, and D. E. Graham. 2009. Independent inactivation of arginine decarboxylase genes by nonsense and missense mutations led to pseudogene formation in Chlamydia trachomatis serovar L2 and D strains. BMC Evol. Biol. 9:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes, J. P., W. J. Bruno, A. Nunes, N. Santos, C. Florindo, M. J. Borrego, and D. Dean. 2007. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 17:50-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes, J. P., A. Nunes, W. J. Bruno, M. J. Borrego, C. Florindo, and D. Dean. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeck, W. R., J. A. Reinhardt, D. A. Baltrus, M. T. Hickenbotham, V. Magrini, E. R. Mardis, J. L. Dangl, and C. D. Jones. 2007. Extending assembly of short DNA sequences to handle error. Bioinformatics 23:2942-2944. [DOI] [PubMed] [Google Scholar]
- 23.Kari, L., W. M. Whitmire, J. H. Carlson, D. D. Crane, N. Reveneau, D. E. Nelson, D. C. Mabey, R. L. Bailey, M. J. Holland, G. McClarty, and H. D. Caldwell. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J. Infect. Dis. 197:449-456. [DOI] [PubMed] [Google Scholar]
- 24.Katoh, K., G. Asimenos, and H. Toh. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39-64. [DOI] [PubMed] [Google Scholar]
- 25.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiselev, A. O., W. E. Stamm, J. R. Yates, and M. F. Lampe. 2007. Expression, processing, and localization of PmpD of Chlamydia trachomatis serovar L2 during the chlamydial developmental cycle. PLoS One 2:e568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lampe, M. F., R. J. Suchland, and W. E. Stamm. 1993. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect. Immun. 61:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., J. Ruan, and R. Durbin. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millman, K. L., S. Tavare, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunes, A., P. Nogueira, M. Borrego, and J. Gomes. 2008. Chlamydia trachomatis diversity viewed as a tissue-specific coevolutionary arms race. Genome Biology. 9:R153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pannekoek, Y., G. Morelli, B. Kusecek, S. A. MorrŽ, J. M. Ossewaarde, A. A. Langerak, and A. van der Ende. 2008. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC. Microbiol. 8:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez, J., J. R. Lama, L. Kusunoki, H. Manrique, P. Goicochea, A. Lucchetti, M. Rouillon, M. Pun, L. Suarez, S. Montano, J. L. Sanchez, S. Tabet, J. P. Hughes, and C. Celum. 2007. HIV-1, sexually transmitted infections, and sexual behavior trends among men who have sex with men in Lima, Peru. J. Acquir. Immune Defic. Syndr. 44:578-585. [DOI] [PubMed] [Google Scholar]
- 35.Schachter, J. 1978. Chlamydial infections (first of three parts). N. Engl. J. Med. 298:428-435. [DOI] [PubMed] [Google Scholar]
- 36.Smith, C. B., and D. E. Graham. 2008. Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae. J. Bacteriol. 190:7431-7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 38.Suchland, R. J., L. O. Eckert, S. E. Hawes, and W. E. Stamm. 2003. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988-1996. Sex. Transm. Dis. 30:357-361. [DOI] [PubMed] [Google Scholar]
- 39.Suchland, R. J., B. Jeffrey, M. Xia, A. Bhatia, H. G. Chu, D. D. Rockey, and W. E. Stamm. 2008. Identification of concomitant infection with Chlamydia trachomatis IncA-negative mutant and wild-type strains: genomic, transcriptional and biological characterization. Infect. Immun. 76:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suchland, R. J., D. D. Rockey, S. K. Weeks, D. T. Alzhanov, and W. E. Stamm. 2005. Development of secondary inclusions in cells infected by Chlamydia trachomatis. Infect. Immun. 73:3954-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suchland, R. J., K. M. Sandoz, B. M. Jeffrey, W. E. Stamm, and D. D. Rockey. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob. Agents Chemother. 53:4604-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suchland, R. J., and W. E. Stamm. 1991. Simplified microtiter cell culture method for rapid immunotyping of Chlamydia trachomatis. J. Clin. Microbiol. 29:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, C., R. C. Hsia, H. Shou, C. L. Haggerty, R. B. Ness, C. A. Gaydos, D. Dean, A. M. Scurlock, D. P. Wilson, and P. M. Bavoil. 2009. Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect. Immun. 77:3218-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Connor, I. Goodhead, H. Norbertzcak, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeño-Tárraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wehrl, W., V. Brinkmann, P. R. Jungblut, T. F. Meyer, and A. J. Szczepek. 2004. From the inside out—processing of the chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51:319-334. [DOI] [PubMed] [Google Scholar]
- 48.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.