Abstract
Treponema pallidum subsp. pallidum is the causative agent of syphilis, a sexually transmitted disease characterized by widespread tissue dissemination and chronic infection. In this study, we analyzed the proteome of T. pallidum by the isoelectric focusing (IEF) and nonequilibrating pH gel electrophoresis (NEPHGE) forms of two-dimensional gel electrophoresis (2DGE), coupled with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis. We determined the identity of 148 T. pallidum protein spots, representing 88 T. pallidum polypeptides; 63 of these polypeptides had not been identified previously at the protein level. To examine which of these proteins are important in the antibody response to syphilis, we performed immunoblot analysis using infected rabbit sera or human sera from patients at different stages of syphilis infection. Twenty-nine previously described antigens (predominantly lipoproteins) were detected, as were a number of previously unidentified antigens. The reactivity patterns obtained with sera from infected rabbits and humans were similar; these patterns included a subset of antigens reactive with all serum samples tested, including CfpA, MglB-2, TmpA, TmpB, flagellins, and the 47-kDa, 17-kDa, and 15-kDa lipoproteins. A unique group of antigens specifically reactive with infected human serum was also identified and included the previously described antigen TpF1 and the hypothetical proteins TP0584, TP0608, and TP0965. This combined proteomic and serologic analysis further delineates the antigens potentially useful as vaccine candidates or diagnostic markers and may provide insight into the host-pathogen interactions that occur during T. pallidum infection.
Syphilis is a multistage progressive disease caused by the spirochete Treponema pallidum subsp. pallidum and is characterized by localized, disseminated, and chronic stages. Manifestations include the development of a localized lesion called a chancre during the primary stage and disseminated skin lesions and meningovascular syphilis during the secondary stage, followed by a period of latency lasting from months to decades. Chronic, debilitating symptoms develop during the tertiary stage, including granuloma-like lesions called gummas, neurosyphilis, and cardiovascular syphilis (38). Although syphilis can be successfully treated by antibiotics, it remains a significant public health problem, with an estimated 12 million new cases per year worldwide (41).
Continued improvement of diagnostic tests (particularly point-of-care tests) as well as the development of an effective vaccine for syphilis would aid greatly in the control of syphilis (4, 6). T. pallidum research, including the identification of antigens, has been hindered by the inability to culture the bacterium continuously in vitro, necessitating the propagation of organisms by experimental rabbit infection (28). In addition, the fragility and low protein content of the T. pallidum outer membrane have complicated the identification of surface proteins potentially useful in vaccines (5, 28).
The T. pallidum genome sequence (15) provides an additional tool for the analysis of potential antigens. The 1.14-Mb T. pallidum chromosome contains 1,039 open reading frames (ORFs) encoding predicted protein products, a smaller number than for any other spirochete genome sequenced to date (15). The average size of predicted proteins is 37,771 Da, ranging from 3,235 to 172,869 Da. Analysis of the translated genome of T. pallidum predicts an unusually basic proteome, with a mean pI of 8.1 and median pI of 8.5, with 66% of proteins having pIs of >7.0 (23). Small genome size and a predominance of basic proteins are more common in parasitic microorganisms, and the latter is thought to facilitate interaction of the organism with its host (20). Other pathogenic spirochetes also tend to have basic proteins; for example, the proteome of Borrelia burgdorferi has a mean pI of 8.36 and median pI of 9.03 (14, 29a), and 69% of Leptospira interrogans serovar Lai strain 56601 proteins have pIs greater than 7.0 (24, 33). A recent analysis of the T. pallidum genome indicates the presence of 46 putative lipoproteins, many fewer than the 127 predicted for B. burgdorferi (34).
The availability of the genome sequence made it possible to examine predicted T. pallidum ORFs for potential suitability as diagnostic or immunization tools. McKevitt et al. (22) and Brinkman et al. (3) created a protein expression library of 900 of the 1,039 T. pallidum proteins predicted from the genome sequence and examined the serologic reactivity of these proteins by enzyme-linked immunosorbent assays (ELISAs). They identified 106 antigens reactive with rabbit sera and 34 antigens reactive with sera from syphilis patients. This set of antigens was termed the T. pallidum immunoproteome. This approach permits identification of low-abundance T. pallidum antigens, since they may be expressed as recombinant proteins in much larger quantities. Conversely, proteins that are poorly expressed in Escherichia coli or do not fold correctly may not be detected, leading to false-negative results.
To provide a complementary set of data regarding the T. pallidum immunoproteome, we have performed proteomic analysis of T. pallidum proteins expressed during experimental rabbit infection. We used isoelectric focusing (IEF) and nonequilibrating pH gel electrophoresis (NEPHGE) forms of two-dimensional gel electrophoresis (2DGE) coupled with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis to identify T. pallidum polypeptides. Immunoblotting was subsequently used to identify antigens reactive with infected rabbit sera (IRS) and with human sera obtained at different stages of syphilis. This approach may permit identification of antigens that are not expressed well in E. coli and provides a more accurate picture of the level of protein expression in the intact organism. We have thereby characterized most of the major T. pallidum proteins expressed in infected tissue and identified a set of antigens reactive at all stages of infection, which could potentially be useful for the development of improved immunodiagnostic tests or for vaccines.
MATERIALS AND METHODS
Two-dimensional gel electrophoresis and immunoblotting.
T. pallidum subsp. pallidum (Nichols) was extracted from testicular tissue of infected rabbits and purified by Percoll density gradient centrifugation as described previously (17, 29). Organisms were resolved in the first dimension by either isoelectric focusing (IEF; pH 5 to 7) or nonequilibrium pH gel electrophoresis (NEPHGE; pH 3.5 to 10), as described by O'Farrell et al. (30, 31). For silver-stained gels and subsequent MALDI-TOF mass spectrometry (MS), 8 × 108 organisms were loaded per tube gel; for immunoblotting, 6 × 108 organisms were loaded per tube gel. Equilibrated tube gels were resolved by SDS-PAGE using 8 to 20% gradient gels in the second dimension, and the gels were stained using a Silver SNAP kit for mass spectrometry (Pierce) or transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) (37) at 150 V for 1.5 h at 4°C using a Trans-Blot cell (Bio-Rad). For immunoblot analysis, membranes were blocked overnight at 4°C in Tris-buffered saline solution containing 0.05% Tween 20 (TBST) and 1% bovine serum albumin (BSA) (blocking solution; Promega). Following incubation with primary antibody diluted in blocking solution (1:1,000 for rabbit serum and 1:500 for human serum) for 1 h at room temperature, the membranes were washed three times for 10 min each in TBST and then incubated with secondary antibody (goat anti-rabbit or goat anti-human IgG, allophycocyanin [AP] conjugate; Promega) at a concentration of 1:5,000, diluted in TBST, for 30 min at room temperature. Membranes were washed three times with TBST, followed by two washes with Tris-buffered saline (TBS) to remove inhibitory Tween 20. Membranes were developed for 3 min with Western Blue stabilized substrate for alkaline phosphatase (Promega), and development was stopped by washing with distilled water.
Sera used for immunoblots.
Rabbit sera were collected from three individual animals before infection (prebleed; noninfected rabbit sera [NRS]) or 84 days after intratesticular inoculation with T. pallidum (infected rabbit sera [IRS]). Sera were pooled from the three animals and used for immunoblotting as described above. Human serum samples had previously been collected in Texas from healthy human subjects and from patients diagnosed with primary, secondary, early latent, or late latent syphilis and are summarized in Table S1 in the supplemental material. Sera were pooled prior to immunoblotting experiments as noninfected human (10 sera), primary (3 sera), secondary (3 sera), early-latent-syphilis (8 sera), and late-latent-syphilis (13 sera) pools and used at a dilution of 1:500. Human syphilitic serum samples had rapid-plasma-reagin (RPR) titers ranging from 1:2 to 1:512. All human sera were collected under established guidelines with prior approval by the Committee for the Protection of Human Subjects, University of Texas Health Science Center at Houston.
2D spot preparation and MALDI-TOF MS analysis.
Protein spots of interest were excised manually (1.0 to 3.0 mm in diameter) from a set of four silver-stained gels with a OneTouch two-dimensional (2D) gel spot picker (The Gel Company, San Francisco, CA) and destained using the Silver SNAP kit for mass spectrometry according to the manufacturer's instructions. Excised spots were stored in wash buffer (25 mM ammonium bicarbonate, 50% acetonitrile; Sigma) at −20°C until in-gel trypsin digestion was performed. Destained 2D gel spots were treated with 0.2 M ammonium bicarbonate-50% acetonitrile for 15 min and dried completely in a CentriVap speed vacuum. Gel pieces were then rehydrated in 50 μl 50 mM ammonium bicarbonate containing 0.2 to 0.5 μg modified trypsin (Promega or Sigma) and digested for 20 h at 37°C. The supernatant was transferred to a clean microcentrifuge tube, and the gel fragments were extracted with 50 μl aqueous 50% acetonitrile-2% formic acid for 15 min and combined with the initial extract. The combined supernates were evaporated to 30 μl, acidified with trifluoroacetic acid to a pH of 3, and desalted using a C18 ZipTip (Millipore) as recommended by the manufacturer. Peptides were eluted from the ZipTip with 5 μl of an aqueous solution of 50% acetonitrile and 2% formic acid. Two microliters of the sample was spotted onto a 100-well stainless steel MALDI target plate and allowed to dry partially prior to the addition of 1 μl of a 1-mg/ml matrix solution (alpha-cyano-4-hydroxycinnamic acid), followed by complete drying. MALDI-TOF analyses were performed in reflector mode on an ABI/SCIEX 4700 proteomics analyzer TOF/TOF mass spectrometer, with the laser intensity adjusted manually to yield the best spectrum for each sample. The resulting spectra were calibrated manually utilizing the autodigestion products of trypsin as internal reference peaks with Data Explorer software, available from ABI. Proteins were identified using Protein Prospector (University of California, San Francisco, CA; http://prospector.ucsf.edu/) set to a mass accuracy of ±20 ppm and a missed cleavage allowance of 1. Mass fingerprints were compared to the predicted proteins in the NCBI database by use of a species-specific filter for T. pallidum or without the use of a species filter to identify nontreponemal contaminants.
RESULTS AND DISCUSSION
MALDI-TOF MS identification of T. pallidum proteins.
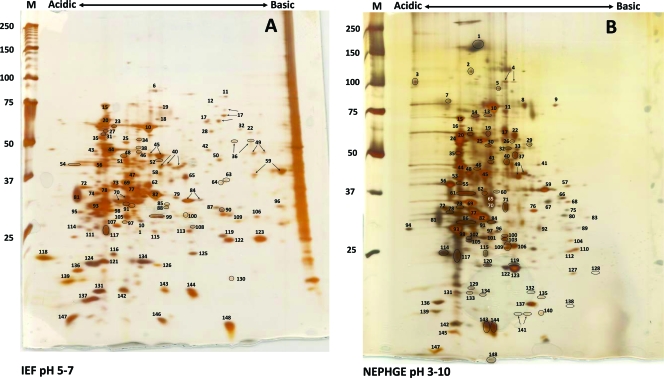
Rabbits have been used for many years to propagate T. pallidum in the absence of an effective in vitro culture system, and T. pallidum maintained in rabbits retains infectivity and virulence in humans (13, 21). We extracted T. pallidum subsp. pallidum Nichols from the testicular tissue of infected rabbits and purified the bacteria by Percoll gradient density centrifugation. Lysates of purified bacteria were separated by IEF and NEPHGE 2DGE; NEPHGE permits the separation of highly basic polypeptides (31). Proteins from silver-stained gels were analyzed by MALDI-TOF mass spectrometry. We identified the polypeptides present in 148 protein spots; of these, 144 corresponded to 88 different T. pallidum proteins, and 4 corresponded to rabbit proteins (Fig. 1 and Table 1). The 56 additional spots represented charge variants, minor size variants, or degradation products of T. pallidum proteins. The detection of only four rabbit products demonstrates the high degree of purification achievable by Percoll density gradient centrifugation (17).
FIG. 1.
Two-dimensional gel electrophoresis of T. pallidum proteins. T. pallidum lysates were separated by IEF at pH 5 to 7 (A) or NEPHGE at pH 3.5 to 10 (B) in the first dimension, followed by 8 to 20% SDS-PAGE in the second dimension. Gels were subsequently silver stained for protein visualization. Acidic and basic ends are denoted, and relative molecular mass markers (in kilodaltons) are indicated to the left of each gel. A T. pallidum lysate, resolved in the second dimension only, is shown at the right side of the IEF pH 5-to-7 gel (A). The identities of the numbered spots are presented in Table 1. Arrows indicate spots that were submitted separately for MALDI-TOF MS but returned the same identity. Circles demarcate some closely spaced spots to indicate more clearly which spots are labeled.
TABLE 1.
MALDI-TOF analysis of T. pallidum proteins separated by IEF and NEPHGE 2DGE and summary of reactivity of IRS and human syphilis patient sera against T. pallidum proteinsa
| Spot | TP ORF | Protein description | Predicted MW | Observed Mr | Predicted pI | No. of matching peptides | % coverage | Result for: |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IRS | Human sera at indicated phase |
|||||||||||
| Primary | Secondary | Early latent | Late latent | |||||||||
| 1 | TP1038 | Bacterioferritin (TpF1, antigen C1-5, 4D),oligomeric form | 206,808 | 166,126 | 5.3 | 6 | 50 | − | +++ | +++ | +++ | +++ |
| 2 | TP0408 | Chromosome segregation SMC protein homolog | 124,016 | 118,764 | 5.3 | 18 | 18 | − | − | − | − | − |
| 3 | TP0179 | Acidic hypothetical protein TP0179 | 66,536 | 101,782 | 4.3 | 14 | 22 | − | − | − | − | − |
| 4 | TP0524 | ATP-dependent protease LA (Lon-2) | 97,729 | 100,742 | 6.4 | 23 | 30 | − | − | − | − | − |
| 5 | TP0071 | ATP-dependent Clp protease subunit B (ClpB) | 98,982 | 96,316 | 6.0 | 26 | 36 | − | − | − | − | − |
| 6 | TP0767 | Translation elongation factor G (FusA-2) | 76,832 | 86,362 | 5.6 | 29 | 46 | + | − | − | − | − |
| 7 | TP0748 | CfpA degradation product | 78,540 | 83,771 | 5.9 | 10 | 18 | |||||
| 8 | Aconitase 2, probable rabbit contaminant | 85,762 | 80,021 | 5.9 | 10 | 18 | ||||||
| 9 | TP0748 | CfpA variant | 78,540 | 79,673 | 5.9 | 29 | 45 | |||||
| 10 | TP0748 | Cytoplasmic filament protein A (CfpA) | 78,540 | 78,809 | 5.9 | 46 | 61 | +++ | ++ | +++ | +++ | ++ |
| 11 | TP0886 | Polyribonucleotide nucleotidyltransferase (Pnp) | 76,333 | 74,602 | 6.2 | 26 | 41 | ++ | ++ | ++ | ++ | ++ |
| 12 | TP0216 | DnaK variant | 68,042 | 72,297 | 5.0 | 17 | 32 | |||||
| 13 | TP0748 | CfpA degradation product | 78,540 | 71,561 | 5.9 | 23 | 36 | |||||
| 14 | TP0984 | Heat shock protein 90 | 72,938 | 71,561 | 5.3 | 19 | 32 | − | − | − | − | − |
| 15 | TP0216 | Molecular chaperone DnaK | 68,042 | 68,945 | 5.0 | 39 | 64 | +++ | + | ++ | ++ | − |
| 16 | TP0216 | DnaK degradation product | 68,042 | 64,165 | 5.0 | 14 | 29 | |||||
| 17 | TP0122 | Phosphoenolpyruvate carboxykinase (PckA) | 68,126 | 65,606 | 6.2 | 24 | 40 | ++ | ++ | ++ | ++ | ++ |
| 18 | TP0104 | 5′-nucleotidase (UshA) | 62,557b | 63,286 | 5.9 | 24 | 39 | − | − | − | − | − |
| 19 | TP0426 | V-type ATPase, subunit A (AtpA-1) | 65,020 | 61,025 | 5.8 | 19 | 35 | − | + | ++ | + | − |
| 20 | TP0030 | Chaperonin GroEL | 57,981 | 61,025 | 5.0 | 14 | 30 | ++ | + | ++ | ++ | − |
| 21 | TP0748 | CfpA degradation product | 78,540 | 60,694 | 5.9 | 27 | 42 | |||||
| 22 | TP0056 | Oxaloacetate decarboxylase | 64,100 | 59,930 | 6.9 | 18 | 35 | − | + | + | − | − |
| 23 | TP0030 | GroEL degradation product | 57,981 | 58,627 | 5.0 | 35 | 57 | |||||
| 24 | TP0030 | GroEL degradation product | 57,981 | 57,030 | 5.0 | 18 | 41 | |||||
| 25 | TP0748 | CfpA degradation product | 78,540 | 56,715 | 5.9 | 29 | 37 | |||||
| 26 | TP0969 | Hypothetical protein TP0969 | 57,982b | 56,354 | 8.3 | 18 | 33 | − | − | − | − | − |
| 27 | TP0030 | GroEL degradation product | 57,981 | 56,136 | 5.0 | 36 | 60 | |||||
| 28 | TP0426 | AtpA-1 degradation product | 65,020 | 55,661 | 5.8 | 15 | 30 | |||||
| 29 | TP0584 | Hypothetical protein TP0584 | 53,514 | 55,043 | 9.1 | 9 | 20 | ++ | + | ++ | ++ | + |
| 30 | Protein disulfide isomerase family A, probable rabbit contaminant | 54,096 | 54,449 | 7.6 | 8 | 20 | ||||||
| 31 | TP0030 | GroEL degradation product | 57,981 | 53,852 | 5.0 | 35 | 55 | |||||
| 32 | TP0478 | Glucose-6-phosphate 1-dehydrogenase (Zwf) | 58,033 | 53,183 | 6.4 | 16 | 34 | − | − | − | − | − |
| 33 | TP0469 | Conserved hypothetical protein, containing TPR domains | 50,471b | 52,608 | 6.4 | 13 | 30 | − | − | − | − | − |
| 34 | TP0748 | CfpA degradation product | 78,540 | 51,079 | 5.9 | 31 | 41 | |||||
| 35 | TP0030 | GroEL degradation product | 57,927 | 50,830 | 5.0 | 23 | 41 | |||||
| 36 | TP0921 | NADH oxidase | 48,644 | 50,503 | 6.5 | 10 | 29 | − | + | + | − | − |
| 37 | TP0476 | Acetate kinase (Ack) | 49,196 | 49,698 | 6.6 | 17 | 36 | − | − | − | − | − |
| 38 | TP0765 | Cell division protein (FtsH) | 67,545 | 47,580 | 5.9 | 12 | 20 | − | − | − | − | − |
| 39 | TP0112 | Aminopeptidase C (PepC) | 51,243 | 47,052 | 5.9 | 9 | 28 | − | − | − | − | − |
| 40 | TP0921 | NADH oxidase | 48,644 | 46,750 | 6.5 | 17 | 32 | − | + | + | − | − |
| 41 | TP0108 | Diphosphate-fructose-6-phosphate 1-phosphotransferase | 50,162 | 46,301 | 8.3 | 11 | 23 | ++ | ++ | ++ | ++ | − |
| 42 | TP0505 | Hexokinase (Hxk) | 47,845 | 46,269 | 6.0 | 11 | 32 | − | + | + | + | + |
| 43 | TP0187 | Elongation factor Tu (EF-Tu), mature protein | 43,261 | 46,269 | 6.8 | 20 | 47 | − | − | − | − | − |
| 44 | TP0574 | Carboxypeptidase, 47 kDa | 45,100b | 45,635 | 5.4 | 19 | 47 | +++ | +++ | +++ | +++ | +++ |
| 45 | TP0187 | Elongation factor Tu (EF-Tu), mature protein | 43,261 | 44,925 | 5.6 | 23 | 67 | − | − | − | − | − |
| 46 | TP0390 | Cell division protein (FtsZ) | 43,839 | 44,762 | 5.4 | 18 | 59 | − | − | − | − | − |
| 47 | TP0574 | Carboxypeptidase degradation product | 45,100b | 44,267 | 5.4 | 25 | 51 | |||||
| 48 | Actin, probable rabbit contaminant | 41,738 | 43,205 | 5.3 | 14 | 40 | ||||||
| 49 | TP0538 | Phosphoglycerate kinase (Pgk) | 44,944 | 43,066 | 6.7 | 25 | 67 | − | − | − | − | − |
| 50 | TP0122 | PckA degradation product | 68,126 | 42,781 | 6.2 | 18 | 32 | |||||
| 51 | TP0748 | CfpA degradation product | 78,540 | 42,649 | 5.9 | 32 | 47 | |||||
| 52 | TP0659 | Flagellar hook-associated protein 3 (FlgL) | 45,569 | 42,550 | 5.6 | 10 | 30 | − | − | − | − | − |
| 53 | TP0400 | Flagellar motor protein (FliG) | 39,822 | 41,234 | 4.8 | 7 | 23 | + | + | ++ | + | + |
| 53 | TP0768 | Membrane protein (TmpA) | 34,833b | 41,234 | 5.2 | 15 | 59 | +++ | +++ | +++ | +++ | +++ |
| 54 | TP0748 | CfpA degradation product | 78,540 | 41,089 | 5.9 | 23 | 32 | |||||
| 55 | TP0216 | DnaK degradation product | 68,042 | 39,544 | 4.0 | 7 | 14 | |||||
| 56 | TP0684 | Methylgalactoside ABC transporter, periplasmic (MglB-2) | 41,013b | 39,544 | 5.2 | 11 | 40 | +++ | +++ | +++ | +++ | +++ |
| 57 | TP0747 | Basic hypothetical protein TP0747 | 37,780 | 38,976 | 9.2 | 16 | 54 | + | + | + | + | − |
| 58 | TP0448 | Uracil phosphoribosyltransferase, putative | 41,018 | 38,140 | 5.6 | 9 | 24 | − | − | − | − | − |
| 59 | TP0844 | Glyceraldehyde 3-phosphate dehydrogenase (Gap) | 38,084 | 37,680 | 8.4 | 20 | 53 | +++ | +++ | +++ | +++ | +++ |
| 60 | TP0655 | Spermidine/putrescine ABC transporter, periplasmic binding protein (PotD) | 37,195b | 37,079 | 6.4 | 7 | 17 | − | − | − | − | − |
| 61 | TP0574 | Carboxypeptidase degradation product | 45,100b | 36,855 | 5.4 | 21 | 48 | |||||
| 62 | TP0556 | Asparagine synthetase (AsnA) | 36,856 | 36,756 | 5.6 | 19 | 57 | − | − | − | − | − |
| 62 | TP0606 | 30S ribosomal protein S2 | 33,117 | 36,750 | 5.5 | 8 | 23 | − | − | − | − | − |
| 63 | TP0298 | Exported protein (Tpn38b) | 35,299b | 36,281 | 6.3 | 12 | 39 | − | − | − | − | − |
| 64 | TP0298 | Exported protein (Tpn38b) | 35,299b | 35,653 | 6.3 | 19 | 56 | − | − | − | − | − |
| 65 | TP0662 | Fructose-bisphosphate aldolase (CbbA) | 36,190 | 35,553 | 5.7 | 22 | 58 | − | − | − | − | − |
| 66 | TP0965 | Membrane fusion protein, putative | 31,675b | 35,553 | 9.4 | 10 | 33 | − | ++ | ++ | ++ | ++ |
| 67 | Malate dehydrogenase, probable rabbit contaminant | 35,597 | 34,836 | 8.8 | 10 | 37 | ||||||
| 68 | TP0249 | FlaA1 degradation product | 36,945b | 34,615 | 5.5 | 11 | 40 | |||||
| 69 | TP0249 | Flagellar filament outer layer protein (FlaA1) | 36,945b | 34,527 | 5.5 | 13 | 47 | +++ | +++ | +++ | +++ | +++ |
| 70 | TP0662 | CbbA degradation product | 36,190 | 34,308 | 5.7 | 18 | 52 | |||||
| 71 | TP0037 | d-Specific d-2-hydroxyacid dehydrogenase | 36,874 | 33,616 | 6.3 | 13 | 44 | − | − | − | − | − |
| 72 | TP0319 | Purine nucleoside receptor lipoprotein A (PnrA, TmpC) | 35,083b | 33,459 | 4.8 | 16 | 54 | + | ++ | ++ | + | − |
| 73 | TP0030 | GroEL degradation product | 57,981 | 33,280 | 5.0 | 15 | 33 | |||||
| 73 | TP0574 | Carboxypeptidase degradation product | 45,100b | 33,280 | 5.4 | 21 | 50 | |||||
| 74 | TP0792 | Flagellar filament 33-kDa core protein (FlaB2) degradation/charge variant | 31,348 | 33,032 | 7.0 | 7 | 35 | |||||
| 75 | TP0868 | FlaB1 degradation product | 31,175 | 33,022 | 6.9 | 9 | 39 | |||||
| 76 | TP0769 | Outer membrane protein (TmpB) | 34,353b | 32,854 | 8.8 | 12 | 40 | +++ | +++ | +++ | +++ | +++ |
| 77 | TP0868 | Flagellar filament 34.5-kDa core protein (FlaB1) | 31,175 | 32,661 | 6.9 | 10 | 45 | +++ | +++ | +++ | +++ | +++ |
| 78 | TP0684 | MglB-2 degradation product | 43,053 | 32,529 | 5.2 | 13 | 49 | |||||
| 79 | TP0792 | FlaB2 degradation product | 31,348 | 32,105 | 7.0 | 12 | 42 | |||||
| 80 | TP0792 | FlaB2 degradation product | 31,348 | 31,866 | 7.0 | 8 | 33 | |||||
| 81 | TP0971 | Lactoferrin and Zn2+ binding periplasmic lipoprotein (Tp34, TpD) | 20,639b | 31,625 | 4.7 | 9 | 59 | + | ++ | ++ | ++ | + |
| 82 | TP0792 | Flagellar filament 33-kDa core protein (FlaB2) | 31,348 | 31,422 | 7.0 | 10 | 48 | +++ | +++ | +++ | +++ | +++ |
| 83 | TP0870 | FlaB3 degradation product | 31,055 | 31,161 | 5.1 | 10 | 53 | |||||
| 84 | TP0163 | ABC transporter, periplasmic binding protein (TroA) | 31,182b | 31,036 | 6.2 | 15 | 59 | +++ | +++ | +++ | +++ | +++ |
| 85 | TP0094 | Phosphate acetyltransferase (Pta) | 36,538 | 30,466 | 7.0 | 8 | 28 | − | + | − | − | − |
| 86 | TP0030 | GroEL degradation product | 57,981 | 30,438 | 5.0 | 27 | 53 | |||||
| 87 | TP0870 | FlaB3 degradation product | 31,055 | 29,880 | 5.1 | 12 | 63 | |||||
| 88 | TP0574 | Carboxypeptidase degradation product | 45,100b | 29,822 | 5.4 | 14 | 29 | |||||
| 89 | TP0789 | Basic hypothetical protein TP0789 | 26,772b | 29,409 | 9.1 | 8 | 35 | − | − | − | − | − |
| 90 | TP0870 | FlaB3 degradation product | 31,055 | 29,382 | 5.1 | 7 | 32 | |||||
| 91 | TP0611 | ABC transporter, ATP-binding protein | 28,257 | 29,334 | 5.3 | 11 | 61 | − | − | − | − | − |
| 92 | TP0453 | Hypothetical protein TP0453, putative integral membrane protein | 28,453b | 29,260 | 9.3 | 8 | 33 | − | ++ | + | + | − |
| 92 | TP0605 | Elongation factor Ts | 31,783 | 29,260 | 8.1 | 11 | 41 | − | ++ | + | + | − |
| 93 | TP0870 | Flagellar filament 31-kDa core protein (FlaB3) | 31,055 | 29,260 | 5.1 | 20 | 62 | +++ | +++ | +++ | +++ | +++ |
| 94 | TP0870 | FlaB3 variant | 31,055 | 29,260 | 5.1 | 5 | 28 | |||||
| 95 | TP0870 | FlaB3 degradation product | 31,055 | 29,004 | 5.1 | 11 | 64 | |||||
| 96 | TP0862 | Peptidyl-prolyl cis-trans isomerase, FKBP-type, 22 kDa (FklB) | 25,818b | 28,633 | 8.3 | 8 | 30 | − | + | ++ | − | − |
| 97 | TP0821 | Lipoprotein (Tpn32) | 26,887b | 28,415 | 6.7 | 8 | 37 | − | − | − | − | − |
| 97 | TP0139 | TrkA domain protein, putative potassium transport protein | 25,324 | 28,415 | 5.8 | 9 | 41 | − | − | − | − | − |
| 98 | TP0664 | Flagellar filament outer layer protein homolog (FlaA2) | 24,983b | 28,307 | 5.7 | 14 | 64 | − | − | − | − | − |
| 99 | TP0030 | GroEL degradation product | 57,981 | 28,008 | 5.5 | 10 | 17 | |||||
| 99 | TP0574 | Carboxypeptidase degradation product | 45,100b | 28,008 | 5.4 | 23 | 46 | |||||
| 100 | TP0290 | Conserved hypothetical protein TP0290 | 30,939 | 27,984 | 6.2 | 12 | 34 | − | − | − | − | − |
| 100 | TP0168 | Phosphoglycerate mutase (Pgm) | 28,358 | 27,984 | 6.3 | 16 | 52 | − | − | − | − | − |
| 101 | TP0030 | GroEL degradation product | 57,981 | 27,630 | 5.0 | 17 | 29 | |||||
| 102 | TP0608 | Hypothetical protein TP0608 | 31,910 | 27,525 | 8.7 | 14 | 42 | ++ | ++ | ++ | + | + |
| 103 | TP0537 | Triosephosphate isomerase (Tpi) | 26,541 | 27,101 | 6.1 | 10 | 48 | − | − | − | − | − |
| 104 | TP0964 | ABC transporter, ATP-binding protein | 25,177 | 26,884 | 9.2 | 6 | 35 | − | − | − | − | − |
| 105 | TP0608 | Hypothetical protein TP0608 | 31,910 | 26,632 | 8.7 | 14 | 42 | ++ | ++ | ++ | + | + |
| 106 | TP0115 | Phosphomethypyrimidine kinase (ThiD) | 28,943 | 26,292 | 6.4 | 14 | 68 | − | − | + | − | − |
| 107 | TP0249 | FlaA1 degradation product | 36,945b | 26,202 | 5.5 | 11 | 28 | |||||
| 108 | TP0734 | Purine nucleoside phosphorylase (DeoD) | 25,318 | 25,916 | 6.0 | 8 | 45 | − | − | − | − | − |
| 109 | TP0663 | Tromp-2, FlaA homolog (28-kDa outer membrane protein) | 24,799b | 25,761 | 7.0 | 12 | 55 | − | − | − | − | − |
| 110 | TP0769 | TmpB degradation product | 36,961 | 25,598 | 8.8 | 11 | 29 | |||||
| 111 | TP0424 | V-type ATPase, subunit E, putative | 24,978 | 25,467 | 5.1 | 8 | 35 | − | − | − | − | − |
| 112 | TP0769 | TmpB degradation product | 36,961 | 25,306 | 8.8 | 9 | 25 | |||||
| 113 | TP0115 | Phosphomethypyrimidine kinase (ThiD) | 28,943 | 25,205 | 6.4 | 17 | 64 | − | − | − | − | − |
| 114 | TP0971 | Tp34 degradation product | 22,085 | 24,986 | 4.7 | 8 | 53 | |||||
| 115 | TP0554 | Phosphoglycolate phosphatase (Gph-2) | 24,577 | 21,751 | 5.9 | 7 | 41 | − | − | − | − | − |
| 116 | TP0037 | d-Specific d-2-hydroxyacid dehydrogenase degradation product | 36,874 | 21,371 | 6.3 | 9 | 35 | |||||
| 117 | TP0259 | Membrane lipoprotein TpE (LysM domain protein) | 22,865 | 21,332 | 4.9 | 10 | 37 | +++ | +++ | +++ | +++ | ++ |
| 118 | TP0349 | Peptidyl-prolyl cis-trans isomerase, FKBP-type (SlyD) | 18,429 | 20,852 | 4.9 | 5 | 34 | + | + | − | − | − |
| 119 | TP0509 | Alkyl hydroperoxide reductase (AhpC) | 20,709 | 20,601 | 8.7 | 9 | 34 | − | − | − | − | − |
| 120 | TP0568 | 4-hydroxy-2-oxoglutarate aldolase/2-dehydro-3-deoxyphosphogluconate aldolase (Eda) | 22,074 | 20,496 | 5.8 | 9 | 41 | − | − | − | − | − |
| 121 | TP0249 | FlaA1 degradation product | 36,945b | 20,345 | 5.5 | 9 | 22 | |||||
| 122 | TP0509 | Alkyl hydroperoxide reductase (AhpC) | 20,709 | 20,204 | 6.4 | 14 | 74 | − | − | − | − | − |
| 123 | TP0509 | Alkyl hydroperoxide reductase (AhpC) | 20,709 | 20,204 | 6.4 | 11 | 59 | + | + | + | + | + |
| 124 | TP0249 | FlaA1 degradation product | 36,945b | 19,993 | 5.5 | 17 | 51 | |||||
| 125 | TP0748 | CfpA degradation product | 78,540 | 19,909 | 5.9 | 15 | 21 | |||||
| 126 | TP0249 | FlaA1 degradation product | 36,945b | 19,851 | 5.5 | 11 | 36 | |||||
| 127 | TP0748 | CfpA degradation product | 78,540 | 19,593 | 5.9 | 10 | 18 | |||||
| 128 | TP0201 | Ribosomal protein L5 (RplE) | 20,806 | 19,593 | 9.7 | 15 | 60 | − | − | − | − | − |
| 129 | TP0259 | Hypothetical protein TP0259 degradation product | 22,865 | 18,199 | 4.9 | 7 | 20 | |||||
| 130 | TP0664 | FlaA2 degradation product | 26,822 | 18,062 | 5.7 | 14 | 52 | |||||
| 131 | TP0249 | FlaA1 degradation product | 36,945b | 17,812 | 5.5 | 10 | 31 | |||||
| 132 | TP0239 | Ribosomal protein L10 (RplJ) | 19,566 | 17,812 | 7.8 | 10 | 54 | − | − | − | − | − |
| 133 | TP0249 | FlaA1 degradation product | 36,945b | 17,757 | 5.5 | 11 | 39 | |||||
| 134 | TP1038 | Bacterioferritin (TpF1, antigen C1-5, 4D) | 17,234 | 17,558 | 5.3 | 15 | 92 | − | − | − | − | − |
| 135 | TP0437 | Basic hypothetical protein TP0437 | 19,893 | 17,620 | 9.5 | 10 | 59 | − | − | − | − | − |
| 136 | TP0365 | Chemotaxis protein (CheX) | 16,612 | 16,887 | 4.5 | 8 | 53 | + | + | + | − | − |
| 137 | TP0366 | Chemotaxis response regulator (CheY) | 15,736 | 16,698 | 7.8 | 11 | 70 | − | − | − | − | − |
| 138 | TP0060 | Ribosomal protein L9 (RplI) | 17,504 | 16,209 | 9.0 | 10 | 45 | − | − | − | − | − |
| 139 | TP0925 | Flavodoxin | 15,794 | 16,143 | 4.4 | 9 | 55 | + | + | + | − | − |
| 140 | TP0435 | Lipoprotein, 17 kDa (Tpp17) | 13,361b | 15,962 | 8.9 | 9 | 45 | +++ | +++ | +++ | +++ | +++ |
| 141 | TP0435 | Lipoprotein, 17 kDa (Tpp17) | 13,361b | 15,946 | 8.9 | 9 | 44 | +++ | +++ | +++ | +++ | +++ |
| 142 | TP0171 | Lipoprotein, 15 kDa (Tpp15) | 13,176b | 15,103 | 6.7 | 11 | 52 | +++ | +++ | +++ | +++ | +++ |
| 143 | TP0823 | Desulfoferrodoxin (Rbo) | 13,802 | 14,995 | 5.8 | 8 | 62 | − | − | − | − | − |
| 144 | TP0823 | Desulfoferrodoxin (Rbo) | 13,802 | 14,857 | 5.8 | 6 | 62 | − | − | − | − | − |
| 145 | TP0171 | Lipoprotein, 15 kDa (Tpp15) | 15,670 | 14,496 | 6.7 | 8 | 44 | +++ | +++ | +++ | +++ | +++ |
| 146 | TP0356 | RNA-binding protein, putative | 11,956 | 13,404 | 7.8 | 7 | 40 | − | − | − | − | − |
| 147 | TP0919 | Thioredoxin (Trx) | 11,391 | 12,911 | 4.5 | 5 | 45 | − | − | − | − | − |
| 148 | TP1013 | Chaperonin (GroES) | 9,441 | 12,342 | 6.1 | 10 | 85 | − | − | − | − | − |
Immunoreactivity was determined by 2DGE immunoblot analysis (Fig. 2 to 4) for a pool of three infected rabbit sera (Fig. 2) or pools of sera from patients with primary, secondary, early-latent, or late-latent syphilis (Fig. 3; see also Table S2 in the supplemental material). Reactivity was evaluated subjectively as nonreactive (−), weakly reactive (+), moderately reactive (++), or highly reactive (+++). Degradation products, size variants, and rabbit contaminants were excluded from this analysis.
Calculated after removal of the putative signal sequence.
Multiple distinct spots in the same gel were often identified as the same protein and apparently represent charge or molecular mass variants; examples of these variants are indicated by arrows in Fig. 1. This phenomenon was observed at a higher frequency with IEF than with NEPHGE 2DGE, most likely due to the higher resolution and lower compression level of the gradient in the IEF gels (for example, compare spots 143 and 144 on the IEF and NEPHGE gels in Fig. 1). Fifty-one of the spots identified represented apparent mass variants, based on deviation from the predicted molecular weight (MW). All of these spots corresponded to abundant proteins for which a major spot of the expected size and pI was identified; these included the cytoplasmic filament protein CfpA (9 mass variants), the 47-kDa carboxypeptidase (5 mass variants), the flagellar proteins FlaA1, FlaB1, FlaB2, and FlaB3, and the FlaA1 paralog FlaA2 (a total of 16 mass variants).
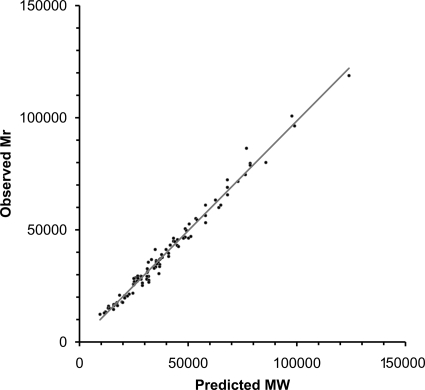
The observed Mr and predicted MW of intact polypeptides were compared to further verify the MS identifications (Table 1 and Fig. 2). In this analysis, the MW values were adjusted to take into account either experimentally verified or predicted cleavage of the polypeptides by either signal peptidase I or signal peptidase II; for the latter, the cleavage points of 46 predicted lipoproteins as determined by Setubal et al. (34) were utilized. No attempt was made to correct the predicted MWs for effects of lipidation or other potential modifications. Four polypeptides (the dodecameric form of TpF1, hypothetical protein TP0179, FtsH, and Tp34 [which migrates as a smear, as determined by SDS-PAGE]) were considered outliers, with ratios of observed Mr to predicted MW of 0.68, 1.57, 0.70, and 1.53, respectively. The TpF1 dodecamer likely migrates aberrantly because of its multimeric conformation. TP0179 was found to have an Mr much greater than that corresponding to its predicted size (101.9 kDa, compared to 66.5 kDa); the predicted gene may be truncated by a sequence error, because inclusion of the adjacent gene (TP0178) in the TP0179 reading frame results in a predicted molecular mass of ∼101 kDa. Tp34 migrates as a smear, as determined by SDS-PAGE, for unknown reasons. The reason for the discrepancy in the Mr of FtsH is not known; the spot identified may be a degradation product.
FIG. 2.
Correlation between predicted molecular weights of T. pallidum proteins and Mr values obtained in 2DGE patterns in this study. Molecular weights take into account removal of predicted signal peptides. Apparent degradation products were excluded.
Exclusion of the mass variants and the four outliers from the list of proteins identified resulted in a high degree of correlation between the observed Mr and predicted MW (Fig. 2); the mean ratio ± standard deviation (SD) for observed Mr/predicted MW was 1.01 ± 0.08 (R2 = 0.9849). In contrast, the mass variants had a poor correlation (ratio = 0.75 ± 0.22; R2 = 0.39) (data not shown). Therefore, the mass variants appear to represent degradation products. These may be naturally occurring breakdown products or may arise during the purification of T. pallidum from rabbit tissue.
Although the majority of spots identified corresponded to a single protein, there were a few spots where MALDI-TOF data indicated a mixture of two protein species. Examples are spot 53 (TmpA and FliG), spot 62 (AsnA and 30S ribosomal protein S2), spots 73 and 99 (GroEL and 47-kDa carboxypeptidase degradation products), spot 92 (hypothetical protein TP0453 and elongation factor Ts), spot 97 (hypothetical protein TP0139 and lipoprotein Tpn32), and spot 100 (Pgm and hypothetical protein TP0290). Thus, these spots appear to be composed of multiple protein species with similar molecular masses and pIs that were not resolved from one another under the electrophoretic conditions utilized.
A number of the more-abundant polypeptides that we describe here have previously been identified by other methods, such as N-terminal sequencing or immunoblotting with monoclonal antibodies (18); these proteins are CfpA, GroEL, DnaK, the 47-kDa carboxypeptidase, TmpA, TmpB, the 17-kDa and 15-kDa lipoproteins, the purine nucleoside receptor lipoprotein PnrA (TmpC) (8), the lactoferrin-binding periplasmic lipoprotein Tp34 (TpD) (7), and the flagellar proteins FlaA1, FlaB1, FlaB2, and FlaB3. As in previous studies, we found that the most-abundant proteins observed by silver staining were flagellins, CfpA, chaperonins, and several lipoproteins, including MglB-2, TmpA, TmpC, and the 47-kDa, 17-kDa, and 15-kDa proteins. High-level expression of lipoprotein genes is typical of Treponema species and other spirochetes.
In addition to confirming previous protein identities, we identified 63 proteins that had not been described previously by electrophoresis or immunoblotting (Fig. 1 and Table 1). These proteins can be categorized by their predicted functions: carbohydrate metabolism (13 proteins), cell division (2 proteins), lipoproteins or structural proteins (10 proteins), flagellum-associated proteins (8 proteins), nucleotide metabolism, degradation, or salvage proteins (4 proteins), molecular chaperones (4 proteins), chemotaxis proteins (2 proteins), energy metabolism enzymes (7 proteins), ABC transporters (7 proteins), proteases (2 proteins), proteins involved in translation (9 proteins), amino acid and cofactor biosynthesis proteins (3 proteins), iron storage proteins (1 protein), cellular detoxification proteins (1 protein), and hypothetical proteins with unknown function (12 proteins). We also determined that the hypothetical protein TP0259 is the lipoprotein TpE; this T. pallidum gene product had been described previously (19), but its sequence was not published.
The protein expression that we observed with 2DGE was consistent with T. pallidum mRNA level data reported previously (35). We identified the proteins corresponding to nearly all of the highly expressed mRNAs reported by Šmajs et al. (those with cDNA/DNA signal ratios of 4.0 or higher) by 2DGE and MALDI-TOF MS (Fig. 1 and Table 1). The majority of proteins with corresponding high transcript levels identified in that study that we did not detect were ribosomal proteins. We identified only four ribosomal proteins, ribosomal proteins S2 (TP0606; spot 62), L5 (TP0201; spot 128), L10 (TP0239; spot 132), and L9 (TP0060; spot 138), in contrast to the 11 ribosomal proteins reported to be transcribed at high levels (35).
Serologic reactivity of T. pallidum proteins.
The IEF and NEPHGE 2DGE patterns obtained were highly reproducible, enabling us to reliably correlate seroreactive proteins in Western blots with the corresponding silver-stained gels. We first examined the T. pallidum proteome for serological reactivity by immunoblotting with pooled sera from rabbits infected for 84 days. At this time postinfection, rabbits develop “chancre immunity,” i.e., resistance to reinfection from intradermal inoculation. In addition, the seroreactivity of human sera from patients at different stages of syphilitic infection to the 2DGE-separated proteins was determined. Our goal was to identify antigens that were consistently reactive at all stages of infection as well as those exhibiting differential reactivity at each stage of infection. A summary of the serologic reactivity against T. pallidum proteins can be found in Table 1. Degradation products were excluded from this analysis.
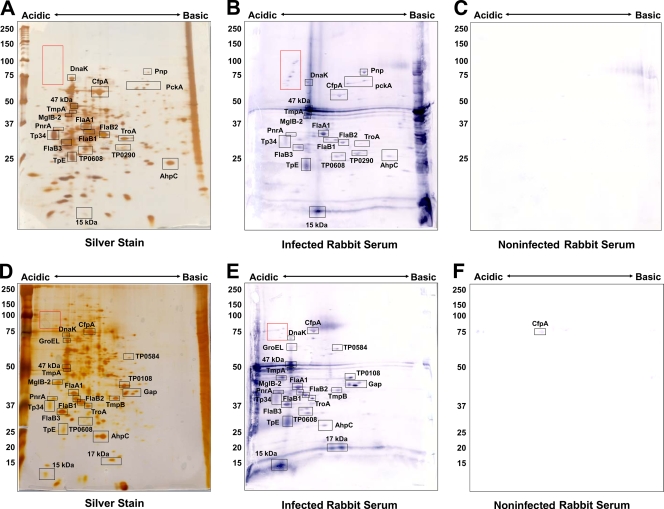
The infected-rabbit-serum (IRS) pool was reactive with a total of 33 T. pallidum proteins in the 2DGE immunoblots (Fig. 3B and E and Table 1). The majority of proteins reactive in 2DGE patterns were previously described antigens (26), including flagellar proteins and lipoproteins, including the ABC transport proteins MglB-2 and TroA (Table 1). This study also confirmed IRS serologic reactivity against phosphoenolpyruvate carboxykinase (PckA; TP0122; spot 17), translation elongation factor G (FusA-2; TP0767; spot 6), and chemotaxis protein X (CheX; TP0365; spot 136), which were identified as antigens by McKevitt et al. (22).
FIG. 3.
Immunoreactivity of T. pallidum proteins separated by 2DGE with rabbit sera. T. pallidum lysates were separated by IEF at pH 5 to 7 (A to C) or NEPHGE at pH 3.5 to 10 (D to F) in the first dimension, followed by 8 to 20% SDS-PAGE in the second dimension. Gels were subsequently silver stained (A, D) or immunoblotted with a 1:1,000 dilution of infected (B, E) or noninfected (C, F) rabbit sera. Black boxed areas indicate major polypeptides that were reactive with each serum pool. Red boxed areas indicate unidentified acidic proteins. Acidic and basic ends are denoted, and relative molecular mass markers (in kilodaltons) are indicated to the left of each gel.
Thirty-two of the 106 T. pallidum proteins found to be reactive with IRS by McKevitt et al. (22) were identified by MALDI-TOF MS. Surprisingly, only 16 of these 32 antigens were reactive with rabbit sera in the present study. Although the other 16 antigens reported by McKevitt et al. were detected by silver staining and MS, they were not reactive with IRS in our study. However, four of those proteins were reactive with human sera (see below), including bacterioferritin TpF1 (TP1038; spot 1), oxaloacetate decarboxylase (TP0056; spot 22), the integral membrane protein (TP0453; spot 92), and peptidyl-prolyl cis-trans isomerase FklB (TP0862; spot 96), indicating that a sufficient amount of these proteins was present for detection of serological reactivity by immunoblotting. Possible explanations for these results are that the immunoblot reactivity in our studies was less sensitive than the reactivity obtained with the ELISA format utilized by McKevitt et al. and that the human patient sera were more reactive to some antigens than were the IRS in our analysis. Many of the most-reactive antigens identified in the McKevitt et al. study were not detected by the 2DGE immunoblotting method, including rare lipoprotein A (RlpA; TP0993), glycerophosphodiester phosphodiesterase (GlpQ; TP0257), thioredoxin (TP0100), and the hypothetical proteins TP0957, TP0625, TP0956, TP0463, TP0567, TP0326, and TP0772. Those proteins were not identified by silver staining and subsequent MALDI-TOF MS, indicating that there may not have been sufficient protein present for detection of serological reactivity against those proteins. Overexpression of those proteins in the McKevitt et al. study may have provided adequate protein levels for rabbit serological reactivity to be observed (22). Alternately, some of these proteins may have been among the proteins that were not selected for MALDI-TOF analysis. For example, several faint spots between 22 and 38 kDa were visible by immunoblotting but were of insufficient quantities to be identified by mass spectrometry. All but two of the antigens not identified by IRS are within that size range.
A number of previously unreported antigens were detected by immunoblotting with IRS in this study, including diphosphate-fructose-6-phosphate 1-phosphotransferase (TP0108; spot 41), flavodoxin (TP0925; spot 139), the FKBP-type peptidyl-prolyl cis-trans isomerase SlyD (TP0349; spot 118), polyribonucleotide nucleotidyltransferase (Pnp; TP0886; spot 11), glyceraldehyde 3-phosphate dehydrogenase (Gap; TP0844; spot 59), and hypothetical protein TP0608 (spots 102 and 105) (Fig. 3). We also found one antigen, CfpA, to be weakly reactive with sera from uninfected animals (Fig. 3C and E).
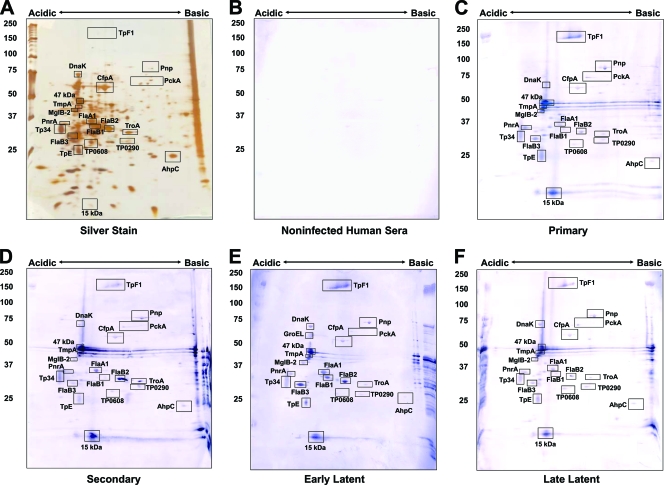
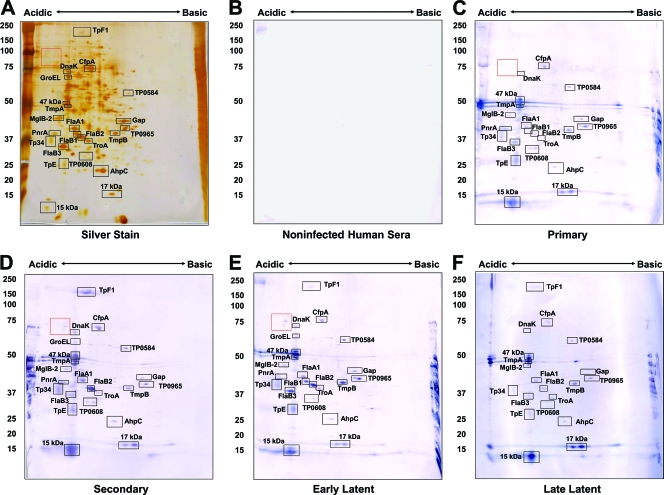
The immunoreactivity of the identified T. pallidum proteins with human sera collected from patients diagnosed with primary, secondary, early latent, or late latent syphilis was also examined. Sera from patients at each stage were pooled as described in Materials and Methods to provide an analysis of reactivity during the course of infection. As expected, the highest level of reactivity occurred with sera from secondary- and early-latent-syphilis patients (Fig. 4C and 5C and Table 1). All of the proteins that were strongly reactive with infected rabbit sera were also reactive with sera from syphilis patients. Many of these are lipoproteins, such as the purine nucleoside periplasmic binding protein PnrA (TmpC) (8), the lactoferrin-binding periplasmic lipoprotein Tp34 (TpD) (7), the 47-kDa carboxypeptidase (9), TmpA, TmpB, TpE, and the 17-kDa and 15-kDa lipoproteins (Fig. 3 to 5 and Table 1), and were reactive with sera from patients at all stages of syphilis. Lipoproteins thus appeared to elicit the strongest antibody response, even if they are expressed at low levels, as is the case for the 17-kDa lipoprotein (TP0435; spot 141). This protein, as well as the 47-kDa lipoprotein, has been demonstrated previously to be highly antigenic and is currently used in T. pallidum diagnostic tests (11, 40). The strong immunogenicity observed with these lipoproteins appears to be dependent on the lipid moiety, as its removal diminishes the production of inflammatory cytokines and activation of immune effector cells (1). The induction of antibody responses against the B. burgdorferi lipoprotein outer surface protein A is highly dependent upon lipidation (10). Therefore, the lipid portion of T. pallidum lipoproteins is likely acting as an intrinsic adjuvant to stimulate the antibody response against these proteins.
FIG. 4.
Immunoreactivity of T. pallidum proteins separated by IEF (pH 5 to 7) 2DGE with human sera. T. pallidum lysates were separated by IEF at pH 5 to 7 in the first dimension, followed by 8 to 20% SDS-PAGE in the second dimension. Gels were subsequently silver stained (A) or immunoblotted with a 1:500 dilution of pooled human sera from healthy blood donors (B), primary-syphilis patients (C), secondary-syphilis patients (D), early-latent-syphilis patients (E) or late-latent-syphilis patients (F). Black boxed areas indicate major polypeptides that were reactive with each serum pool. Acidic and basic ends are denoted, and relative molecular mass markers (in kilodaltons) are indicated to the left of each gel.
FIG. 5.
Immunoreactivity of T. pallidum proteins separated by NEPHGE (pH 3.5 to 10) 2DGE with human sera. T. pallidum lysates were separated by NEPHGE at pH 3.5 to 10 in the first dimension, followed by 8 to 20% SDS-PAGE in the second dimension. Gels were subsequently silver stained (A) or immunoblotted with a 1:500 dilution of pooled human sera from healthy blood donors (B), primary-syphilis patients (C), secondary-syphilis patients (D), early-latent-syphilis patients (E), or late-latent-syphilis patients (F). Black boxed areas indicate major polypeptides that were reactive with each serum pool. Acidic and basic ends are denoted, and relative molecular mass markers (in kilodaltons) are indicated to the left of each gel.
Tp34 (TpD; spot 81), also a lipoprotein, was reactive with all the serum pools, although at lower levels with human late-latent-syphilis sera and infected rabbit sera. Two lipidated periplasmic ABC transport proteins, MglB-2 and TroA, were also reactive at all stages of infection. In addition, a number of antigens that have not previously been reported were identified, including hypothetical protein TP0584 (spot 29), the V-type ATPase AtpA-1 (TP0425; spot 19), and hypothetical protein TP0608 (spots 102 and 105). These proteins were reactive with all sera tested (Fig. 3 to 5). Five antigens found to be reactive with infected human sera (but not with infected rabbit sera) included hexokinase (TP0505; spot 42), hypothetical protein TP0965 (spot 66), phosphate acetyltransferase (Pta; TP0094; spot 85), the integral membrane protein TP0453 (spot 92), and peptidyl-prolyl cis-trans isomerase FklB (TP0862; spot 96). These antigens were not identified as significantly reactive proteins in the Brinkman et al. study of the reactivity of recombinant T. pallidum proteins with human syphilitic sera (3).
We observed a number of acidic, high-molecular-weight spots that were strongly reactive with infected rabbit sera and sera from primary-, secondary-, and early-latent-syphilis patients (red boxes in Fig. 3 to 5). These polypeptides ranged in size from approximately 70 kDa to 120 kDa in molecular mass (Fig. 3 and 5). Due to their low abundance, we were able to detect only one of the spots by silver staining-MS (spot 7; CfpA variant). A search of the T. pallidum genome revealed 13 proteins of appropriate predicted molecular masses with predicted pIs of <6.0 that were not identified by silver staining-MS (see Table S2 in the supplemental material); these therefore represent candidate proteins for this group.
One antigen of interest that appears to be uniquely reactive in human infection is the oligomeric form of the bacterioferritin protein TpF1 (TP1038; spot 1) (Fig. 3 and 4). TpF1 functions as a dodecamer to bind iron (36) but has been observed to migrate at several molecular masses on SDS-PAGE gel, ranging from160 kDa to >400 kDa, in the oligomeric form (12, 25). Multiple identical subunits form a ring structure held together by disulfide bonds, creating a very stable oligomer (32). In its unreduced form, the basic TpF1 oligomer typically migrates at 190 kDa, but reduction by mercaptoethanol results in migration of an oligomer at 160 kDa and dissociated monomers at 19 kDa (32). We observed the 160-kDa form by 2DGE and Western blot analysis. We did not observe serologic reactivity against the monomeric form of TpF1 (spot 134), which is consistent with previous findings. In prior studies by Borenstein et al. (2), TpF1 was cloned and expressed as a recombinant protein from E. coli, and serologic reactivity from syphilis patients against a 190-kDa oligomeric form of the protein was observed. However, no reactivity against the dissociated 19-kDa monomeric form of the expressed protein was observed (2). Furthermore, immunization of rabbits with recombinant TpF1 provided partial protection against challenge with viable treponemes (2). It may also be of interest to determine the identity of the low-abundance polypeptides in the “red box” as proteins that might be of diagnostic or immunogenic value.
We observed stronger reactivity with pooled sera from primary-syphilis patients than expected. The serum pool used for Fig. 4C and 5C was comprised of three samples, i.e., the two tested for Fig. S1 in the supplemental material and an additional serum sample with an RPR titer of 1:64; insufficient serum from the latter sample was available for performance of a separate immunoblot analysis. Therefore, two of the sera from the primary-syphilis-patient pool were examined for seroreactivity with NEPHGE 2DGE immunoblots (see Fig. S1 in the supplemental material). Figure S1B in the supplemental material shows the reactivity of serum from a primary syphilis patient with an RPR titer of 1:16, and Fig. S1C in the supplemental material exhibits the reactivity of serum from a primary syphilis patient with an RPR titer of 1:64. As expected, the serum sample with an RPR titer of 1:16 was reactive to fewer proteins than the serum sample with an RPR titer of 1:64. For example, reactivity to TpF1, hypothetical protein TP0965, and AhpC was not detectable in the sample with an RPR titer of 1:16 (see Fig. S1B in the supplemental material). The high RPR titers of two of these samples correlate with the unexpected strong reactivity that we observed with pooled primary-syphilis sera.
The antibody reactivities obtained with human sera and IRS in the 2DGE immunoproteome analysis correlated well in general, with some differences (Table 1). Of the 87 T. pallidum polypeptides identified by 2DGE-MS, 40 were found to be reactive with sera from humans at some stage of infection, whereas only 32 were reactive with IRS; 31 of the proteins were reactive with both human sera and IRS. Nine of the human serum-reactive proteins were not detectably reactive with IRS, whereas only one of the IRS-reactive proteins was not reactive with the human sera tested. In most cases, these represented faint reactions indicative of low antibody titers. However, moderate immunoblot reactivity with some human serum pools against AtpA-1 (V-type ATPase, subunit a; TP0424), elongation factor Ts (TP0605), and FklB (peptidyl-prolyl cis-trans isomerase; TP0862) was observed (Table 1), whereas a reaction with IRS was not detected. The IRS used in this study were collected at 84 days postinfection and are considered to be highly reactive. Therefore, the apparent differential reactivity observed for these three proteins may reflect differences between the antibody responses of humans and rabbits to certain T. pallidum polypeptides, but additional studies using purified proteins would be needed to verify this finding. Other possible explanations for the differences observed in rabbit and human immunoreactivity are that (i) multiple time points were tested for humans, compared to the single time point tested for rabbits, (ii) fewer subjects were used in the rabbit serum pool than in the human serum pools, resulting in a smaller array of immunoreactive proteins, and (iii) laboratory rabbits are more inbred than the human population and thus may have reduced antibody repertoire diversity.
Comparison of the antigenicity results obtained in the present study with those obtained in prior recombinant protein immunoproteome analyses (3, 22) indicates that the two approaches provide overlapping but somewhat disparate results. In the current study, moderate to high levels of reactivity of human serum pools against CfpA (TP0748), the three flagellar filament core proteins FlaB1, FlaB2, and FlaB3 (TP0868, TP0792, and TP0870, respectively), flagellar motor protein FliG (TP0400), the V-type ATPase subunit AtpA-1 (TP0426), hypothetical protein TP0584, and elongation factor Ts (TP0606) were observed (Fig. 4 and 5 and Table 1; see also Fig. S1 in the supplemental material), whereas these proteins were nonreactive in both of the prior studies using expression of recombinant proteins in E. coli (3, 22). In addition, Tpp15 (TP0171), GroEL (TP0030), PckA (TP0122), hypothetical protein TP0453, and membrane fusion protein TP0965 were reactive in the current study and the prior IRS analysis (22) but not in the prior human serum analysis (3). The lack of reactivity of the flagellar core proteins and CfpA in the prior immunoproteome studies was particularly surprising, in that these proteins had been shown previously to be highly immunogenic and to induce antibody responses during infection (reviewed in references 27 and 29). The lack of reactivity in the immunoproteome studies (3, 22) may have been due to poor expression, rapid degradation, or improper folding with loss of antibody binding activity. For some of the relatively minor spots in the 2DGE pattern, it is possible that the antigenic reactivity detected in the immunoblots was due to comigrating proteins that were not detected in the MS analysis, yielding a “false-positive” result. There were also 5 proteins for which clones were not obtained in the previous recombinant protein studies but were found to be highly reactive by 2DGE immunoblot analysis; these proteins were phosphofructokinase (Pfk; TP0108), flagellar sheath protein (FlaA-1; TP0249), membrane lipoprotein TpE (TP0259), hypothetical protein TP0608, and polyribonucleotide nucleotidyltransferase (Pnp; TP0886) (Table 1).
Conversely, four proteins identified by 2DGE-MS were not found to be reactive with human sera by immunoblot analysis in our study but were reactive in the prior recombinant protein analyses (3, 22). These proteins were FlaA2/Tromp2 (TP0663), translation elongation factor G (FusA-2), hypothetical protein TP0789, and lipoprotein Tpn32 (TP0821). All of these are relatively minor spots in the 2D gels and may be present in too small of a quantity to yield a visible antibody reaction under the conditions used in the current study. An even greater discrepancy was observed with the IRS chemiluminescence enzyme immunoassay (EIA) recombinant protein results reported by McKevitt et al., in which 17 proteins reactive in this prior study were not detectably reactive by our 2DGE IRS immunoblot analysis. However, 11 and 13 of these proteins were not reactive with human syphilis sera in the current study (Fig. 4 and 5; see also Fig. S1 in the supplemental material) or in the Brinkman et al. analysis (3). Therefore, many of these disparities may have resulted from a low positive-value threshold or procedural differences, resulting in detection of weakly positive or potentially false-positive results.
We assessed whether polypeptides expressed at high levels were more likely to evoke a strong antibody response than proteins expressed at low levels. To provide a rough estimate of expression and relative antigenicity, we compared the apparent amounts of protein of 22 polypeptides (see Table S3 in the supplemental material) in the stained gels to the intensities of antibody staining using ImageQuantTL, version 7.0 (General Electric), software. Immunostaining intensity did not correlate with silver staining intensity, as exemplified by the values obtained with early-latent-syphilis human sera and with IRS (see Fig. S2 in the supplemental material). Several low-abundance proteins exhibited high immunoreactivity, whereas certain abundant proteins had low immunoreactivity. Of particular interest was the very strong reactivity observed for the 15-kDa lipoprotein, TpF1, and the unidentified polypeptides highlighted in the red box (Fig. 3 to 5; see also Fig. S1 in the supplemental material). TpF1 and the 15-kDa lipoprotein account for >25% of the total immunostaining intensity obtained with early-latent-syphilis human sera. However, when the silver staining intensities of all immunoreactive proteins were quantitated, these proteins were found to represent <0.6% of the total reactive protein. In IRS stained immunoblots, the proteins in the red box and the 15-kDa lipoprotein account for <1% of the total reactive protein, while accounting for 20% of the immunoreactivity. An important caveat to note is that silver staining is not very quantitative, especially for smaller proteins in higher-percentage polyacrylamide (16). However, staining intensity tends to be relatively reduced for highly expressed proteins rather than proteins expressed at low levels, suggesting that our quantitation might overrepresent any correlation between the quantity and immunogenicity of a polypeptide. Thus, the immunogenicity of a polypeptide does not appear to be closely related to its abundance.
In contrast, a relatively good correlation was obtained between the reactivity of individual proteins with human sera from patients at different stages or with IRS, as exemplified by the comparisons of early-latent-syphilis sera/primary-latent-syphilis sera and early-latent-syphilis sera/IRS shown in Fig. S3 in the supplemental material. This analysis further emphasized differences in the reactivities of sera from infected humans and rabbits (see Fig. S3A in the supplemental material). Most notably, TpF1 (spot 1) had ∼12-fold-higher staining intensity with human early-latent-syphilis sera than with IRS, and the unidentified “red box” antigens were essentially nonreactive with human sera but were highly reactive with IRS. In addition, FlaB2 (spot 82) reacted 3.4 times more intensely with human early-latent-syphilis sera than with IRS, and IRS were ∼3-fold more reactive with TpE (spot 117) and TmpA (spot 53) than early-latent-syphilis sera. Removal of these 5 “outliers” from the correlation shown in Fig. S3A in the supplemental material increased the R2 value from 0.364 to 0.897. These results indicate that the immune responses to some T. pallidum proteins may differ in humans and experimentally infected rabbits. Overall, the 22 polypeptides analyzed quantitatively had similar reactivities with human primary- and early-latent-syphilis serum samples (see Fig. S3B in the supplemental material).
In reality, it is likely that nearly all bacterial proteins induce an adaptive immune response during an infection, due to the foreign nature of these proteins and the exquisite sensitivity of the immune system. The degree of immunogenicity of T. pallidum proteins may therefore represent a continuum. Only those proteins with the highest-level responses are potentially useful for immunodiagnostics, while those that are surface exposed are most likely to be immunoprotective. The data presented in this study confirmed the identity of previously reported antigens. Many new immunoreactive T. pallidum proteins were also revealed by 2DGE and MS analysis, demonstrating the value of analyzing the immunoproteome by a variety of methods. These antigens may provide useful future directions for the development of vaccines and immunodiagnostics. Five antigens of particular interest are the bacterioferritin TpF1, the integral membrane protein TP0453, TP0965 (a putative membrane fusion protein), and the hypothetical proteins TP0584 and TP0608. All five of these antigens were reactive with sera from patients with primary syphilis, suggesting that these antigens might be useful in early diagnostic studies. TP0453 has been tested by an enzyme immunoassay in the serodiagnosis of syphilis, was found to be highly reactive with sera from primary-syphilis patients, and exhibited 100% specificity and sensitivity when reacted with sera from syphilis, relapsing-fever, Lyme disease, or leptospirosis patients (39). The outer membrane location of TP0453 (18) may also make this antigen useful for vaccine development. TP0965, TP0584, and TP0608 were reactive with sera from patients at all syphilis stages, indicating that these antigens may also be useful in the serodiagnosis of syphilis. The cellular location and protective activity of these antigens have yet to be determined. Two-dimensional gel electrophoresis coupled with MALDI-TOF MS and serological analysis is a valuable tool for the identification of new antigens and virulence factors and can be applied to a variety of microbiological systems. These tools are especially useful for organisms like T. pallidum that cannot be cultured in vitro.
Supplementary Material
Acknowledgments
We thank Jerrilyn Howell for her assistance with rabbit infections and immunization.
This work was supported by NIH grant R03 AI69107 and the Greer Professorship in Biomedical Sciences (S.J.N.).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 12 April 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akins, D. R., B. K. Purcell, M. M. Mitra, M. V. Norgard, and J. D. Radolf. 1993. Lipid modification of the 17-kilodalton membrane immunogen of Treponema pallidum determines macrophage activation as well as amphiphilicity. Infect. Immun. 61:1202-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borenstein, L. A., J. D. Radolf, T. E. Fehniger, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1988. Immunization of rabbits with recombinant Treponema pallidum surface antigen 4D alters the course of experimental syphilis. J. Immunol. 140:2415-2421. [PubMed] [Google Scholar]
- 3.Brinkman, M. B., M. McKevitt, M. McLoughlin, C. Perez, J. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 44:888-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2006. Together we can. The National Plan to Eliminate Syphilis from the United States. U.S. Department of Health and Human Services, Washington, DC.
- 5.Cox, D. L., P. Chang, A. W. McDowall, and J. D. Radolf. 1992. The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect. Immun. 60:1076-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen, P. A., and C. E. Cameron. 2006. Progress towards an effective syphilis vaccine: the past, present and future. Expert Rev. Vaccines 5:67-80. [DOI] [PubMed] [Google Scholar]
- 7.Deka, R. K., C. A. Brautigam, F. L. Tomson, S. B. Lumpkins, D. R. Tomchick, M. Machius, and M. V. Norgard. 2007. Crystal structure of the Tp34 (TP0971) lipoprotein of Treponema pallidum. J. Biol. Chem. 282:5944-5958. [DOI] [PubMed] [Google Scholar]
- 8.Deka, R. K., C. A. Brautigam, X. F. Yang, J. S. Blevins, M. Machius, D. R. Tomchick, and M. V. Norgard. 2006. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J. Biol. Chem. 281:8072-8081. [DOI] [PubMed] [Google Scholar]
- 9.Deka, R. K., M. Machius, M. V. Norgard, and D. R. Tomchick. 2002. Crystal structure of the 47-kDa lipoprotein of Treponema pallidum reveals a novel penicillin-binding protein. J. Biol. Chem. 277:41857-41864. [DOI] [PubMed] [Google Scholar]
- 10.Erdile, L. F., M. A. Brandt, D. J. Warakomski, G. J. Westrack, A. Sadziene, A. G. Barbour, and J. P. Mays. 1993. Role of attached lipid in immunogenicity of Borrelia burgdorferi OspA. Infect. Immun. 61:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fears, M. B., and V. Pope. 2001. Syphilis Fast latex agglutination test, a rapid confirmatory test. Clin. Diagn. Lab. Immunol. 8:841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehniger, T. E., A. M. Walfield, T. M. Cunningham, J. D. Radolf, J. N. Miller, and M. A. Lovett. 1984. Purification and characterization of a cloned protease-resistant Treponema pallidum-specific antigen. Infect. Immun. 46:598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, T. J., R. C. Johnson, and M. Smith. 1976. Accidental laboratory infection with Treponema pallidum. J. Am. Vener. Dis. Assoc. 3:76-78. [PubMed] [Google Scholar]
- 14.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 16.Guevara, J. J., D. A. Johnston, L. S. Ramagli, L. S. Martin, B. S. Capetillo, and L. V. Rodriguez. 1982. Quantitative aspects of silver deposition in proteins resolved in complex polyacrylamide gels. Electrophoresis 3:197-205. [Google Scholar]
- 17.Hanff, P. A., S. J. Norris, M. A. Lovett, and J. N. Miller. 1984. Purification of Treponema pallidum, Nichols strain, by Percoll density gradient centrifugation. Sex. Transm. Dis. 11:275-286. [DOI] [PubMed] [Google Scholar]
- 18.Hazlett, K. R., D. L. Cox, M. Decaffmeyer, M. P. Bennett, D. C. Desrosiers, C. J. La Vake, M. E. La Vake, K. W. Bourell, E. J. Robinson, R. Brasseur, and J. D. Radolf. 2005. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 187:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hindersson, P., D. Thomas, L. Stamm, C. Penn, S. Norris, and L. A. Joens. 1992. Interaction of spirochetes with the host. Res. Microbiol. 143:629-639. [DOI] [PubMed] [Google Scholar]
- 20.Knight, C. G., R. Kassen, H. Hebestreit, and P. B. Rainey. 2004. Global analysis of predicted proteomes: functional adaptation of physical properties. Proc. Natl. Acad. Sci. U. S. A. 101:8390-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson, H. J., E. W. Thomas, S. Olansky, B. I. Kaplan, L. DeMello, and J. C. Cutler. 1956. Inoculation syphilis in human volunteers. Medicine 35:33-82. [DOI] [PubMed] [Google Scholar]
- 22.McKevitt, M., M. B. Brinkman, M. McLoughlin, C. Perez, J. K. Howell, G. M. Weinstock, S. J. Norris, and T. Palzkill. 2005. Genome scale identification of Treponema pallidum antigens. Infect. Immun. 73:4445-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nally, J. E., J. P. Whitelegge, and J. A. Carroll. 2007. Proteomic strategies to elucidate pathogenic mechanisms of spirochetes. Proteomics Clin. Appl. 1:1185-1197. [DOI] [PubMed] [Google Scholar]
- 24.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-De-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. De Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noordhoek, G. T., A. Cockayne, L. M. Schouls, R. H. Meloen, E. Stolz, and J. D. A. van Embden. 1990. A new attempt to distinguish serologically the subspecies of Treponema pallidum causing syphilis and yaws. J. Clin. Microbiol. 28:1600-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norris, S. J., J. F. Alderete, N. H. Axelsen, M. J. Bailey, S. A. Baker-Zander, J. B. Baseman, P. J. Bassford, R. E. Baughn, A. Cockayne, P. A. Hanff, P. Hindersson, S. A. Larsen, M. A. Lovett, S. A. Lukehart, J. N. Miller, M. A. Moskiphidis, F. Miller, M. V. Norgard, C. W. Penn, L. V. Stamm, J. D. van Embden, and K. Wicher. 1987. Identity of Treponema pallidum susp. pallidum polypeptides: correlation of sodium dodecyl sulfate-polyacylamide gel electrophoresis results from different laboratories. Electrophoresis 8:77-92. [Google Scholar]
- 27.Norris, S. J., N. W. Charon, R. G. Cook, M. D. Fuentes, and R. J. Limberger. 1988. Antigenic relatedness and N-terminal sequence homology define two classes of periplasmic flagellar proteins of Treponema pallidum subsp. pallidum and Treponema phagedenis. J. Bacteriol. 170:4072-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norris, S. J., D. L. Cox, and G. M. Weinstock. 2001. Biology of Treponema pallidum: correlation of functional activities with genome sequence data. J. Mol. Microbiol. Biotechnol. 3:37-62. [PubMed] [Google Scholar]
- 29.Norris, S. J., and the Treponema pallidum Polypeptide Research Group. 1993. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunologic roles. Microbiol. Rev. 57:750-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Nowalk, A. J., C. Nolder, D. R. Clifton, and J. A. Carroll. 2006. Comparative proteome analysis of subcellular fractions from Borrelia burgdorferi by NEPHGE and IPG. Proteomics 6:2121-2134. [DOI] [PubMed] [Google Scholar]
- 30.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 31.O'Farrell, P. Z., H. M. Goodman, and P. H. O'Farrell. 1977. High-resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12:1133-1142. [DOI] [PubMed] [Google Scholar]
- 32.Radolf, J. D., L. A. Borenstein, J. Y. Kim, T. E. Fehniger, and M. A. Lovett. 1987. Role of disulfide bonds in the oligomeric structure and protease resistance of recombinant and native Treponema pallidum surface antigen 4D. J. Bacteriol. 169:1365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 34.Setubal, J. C., M. Reis, J. Matsunaga, and D. A. Haake. 2006. Lipoprotein computational prediction in spirochaetal genomes. Microbiology 152:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Šmajs, D., M. McKevitt, J. K. Howell, S. J. Norris, W. W. Cai, T. Palzkill, and G. M. Weinstock. 2005. Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J. Bacteriol. 187:1866-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thumiger, A., A. Polenghi, E. Papinutto, R. Battistutta, C. Montecucco, and G. Zanotti. 2006. Crystal structure of antigen TpF1 from Treponema pallidum. Proteins 62:827-830. [DOI] [PubMed] [Google Scholar]
- 37.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.United States Public Health Service. 1968. Syphilis: a synopsis. U.S. Government Printing Office, Washington, DC.
- 39.Van Voorhis, W. C., L. K. Barrett, S. A. Lukehart, B. Schmidt, M. Schriefer, and C. E. Cameron. 2003. Serodiagnosis of syphilis: antibodies to recombinant Tp0453, Tp92, and Gpd proteins are sensitive and specific indicators of infection by Treponema pallidum. J. Clin. Microbiol. 41:3668-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO/TDR. 2003. Laboratory-based evaluation of rapid syphilis diagnostics. WHO, Geneva, Switzerland.
- 41.World Health Organization. 2001. Global prevalence and incidence of selected curable sexually transmitted diseases: overview and estimates. WHO/HIV_AIDS/2001.02. World Health Organization, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.