Abstract
Borrelia burgdorferi, the causative agent of Lyme disease, has a complex genome consisting of a linear chromosome and up to 21 linear and circular plasmids. These plasmids encode numerous proteins critical to the spirochete's infectious cycle and many hypothetical proteins whose functions and requirements are unknown. The conserved linear plasmid lp54 encodes several proteins important for survival in the mouse-tick infectious cycle, but the majority of the proteins are of unknown function and lack homologs outside the borreliae. In this study we adapted the Cre-lox recombination system to create large deletions in the B. burgdorferi genome. Using Cre-lox, we systematically investigated the contribution of 14 adjacent genes on the left arm of lp54 to the overall infectivity of B. burgdorferi. The deletion of the region of lp54 encompassing bba07 to bba14 had no significant effect on the infectious cycle of B. burgdorferi. The deletion of bba01 to bba07 resulted in a slight growth defect but did not significantly affect the ability of B. burgdorferi to complete the infectious cycle. This study demonstrated the utility of the Cre-lox system to efficiently explore gene requirements in B. burgdorferi and surprisingly revealed that a large number of the highly conserved proteins encoded on lp54 are not required to complete the infectious cycle.
The genome of Borrelia burgdorferi strain B31 consists of a ∼900-kb linear chromosome and more than 20 linear and circular plasmids (11, 18). Within the chromosome are many genes that encode basic housekeeping proteins for DNA replication, transcription, translation, solute transport, and energy metabolism, while the majority of open reading frames (ORFs) on the plasmids encode proteins of unknown function (11, 18). Genetic studies of B. burgdorferi are inefficient and hampered by the loss of critical virulence plasmids during in vitro propagation, making functional characterizations of these plasmid ORFs difficult (20, 25, 30, 31, 38, 48, 59).
Among the plasmids shown to be critical for the overall fitness of Lyme disease spirochetes is the linear plasmid lp54 (3, 39, 60), which is present in all characterized B. burgdorferi isolates and is stably maintained during in vitro propagation (24, 35, 40, 54, 58). In contrast, other linear plasmids, such as lp25 and lp28-1, carry important virulence factors but are relatively unstable during in vitro cultivation (11, 19, 31, 38, 39, 59). Relatively few of the lp54-encoded proteins have been characterized in vitro or in vivo, and most lack homologs outside the borreliae (11, 18).
Although direct evidence is limited, analyses of environmentally regulated genes and proteins suggested that lp54 encodes proteins important to the spirochete in its natural infectious cycle. Many of the hypothetical ORFs on lp54 are regulated by temperature (33), pH (10), or both (41), which correlates with the environmental conditions that distinguish the tick vector and mammalian host. Array studies indicate that 32 of the 76 ORFs on lp54 are temperature regulated, a higher percentage of regulated genes than on the chromosome or any other plasmid in the genome (33). As most of these environmentally regulated ORFs are hypothetical and gene inactivation in infectious B. burgdorferi remains inefficient, the prospect of individually inactivating all 76 genes on lp54 is daunting.
In order to more efficiently manipulate the B. burgdorferi genome, we adapted the Cre-lox recombination system to delete large segments of lp54. This system utilizes the P1 bacteriophage Cre protein (51), which is a tyrosine site-specific DNA recombinase (1), and two 34-bp recognition sites called loxP (22, 23). The loxP sites are inserted into the genome in a directly repeated orientation flanking the targeted DNA. The sequential breaking and rejoining of the loxP sites by the Cre recombinase result in the functional deletion of one site and the intervening target sequence. The Cre-lox system requires no accessory proteins or host cofactors and proceeds efficiently with both supercoiled and linear DNA. After demonstrating that Cre-loxP functioned efficiently in B. burgdorferi, we undertook a systematic deletion mapping of the left arm of lp54, from bba01 to bba14. The resulting B. burgdorferi mutants were assessed for their in vitro growth phenotypes and in vivo competence in the mouse-tick infectious cycle.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. burgdorferi strains used in this study are described in Table 1. Spirochetes were grown in Barbour-Stoenner-Kelly II (BSKII) medium (2) supplemented with 6% rabbit serum (Pel Freez Biologicals, Rogers, AZ) at 35°C or in solid BSK medium incubated at 35°C under 2.5% CO2 (42). All DNA manipulations in Escherichia coli were performed with TOP10 cells (Invitrogen, Carlsbad, CA).
TABLE 1.
B. burgdorferi strains used in this study
| Strain | Description | Reference |
|---|---|---|
| B314 | Attenuated B. burgdorferi strain B31 clone lacking all linear plasmids, including lp54 | 46 |
| B31-A34 | Transformable, noninfectious B31 clone lacking lp5, lp25, lp28-1, lp28-4, lp56, lp36, cp-9, and cp32-6, used for testing the Cre-lox system | 26 |
| B31-A3 | Transformable, infectious B31 clone lacking cp9; background of lp54 mutants listed below | 15 |
| A3-A07 | Streptomycin resistance cassette and loxP insertion into bba07 | This study |
| A3-A01/A07 | Streptomycin resistance cassette and loxP insertion into bba07, kanamycin resistance cassette and loxP insertion into bba01 | This study |
| A3-A07/A14 | Streptomycin resistance cassette and loxP insertion into bba07, kanamycin resistance cassette and loxP insertion into bba14 | This study |
| A3ΔA1-7 | Deletion of bba01 to bba07 loci and retention of only the streptomycin resistance cassette | This study |
| A3ΔA7-14 | Deletion of bba07 to bba14 loci and retention of only the kanamycin resistance cassette | This study |
Construction of the GFP expression vector.
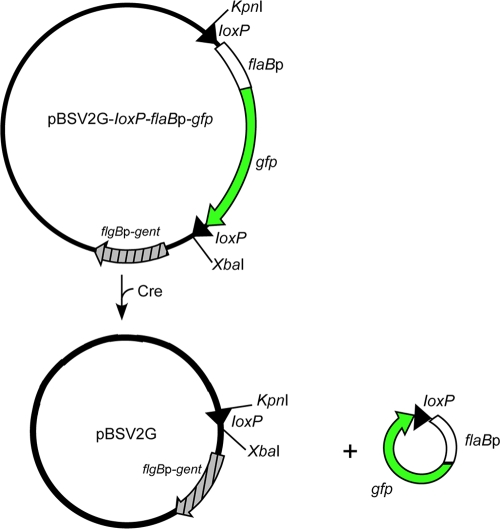
The green fluorescent protein (GFP) expression vector containing flanking loxP sites (Fig. 1) was constructed as follows. Primers 1 and 2 (Table 2), containing 5′ and 3′ loxP sites, were used to amplify flaBp-gfp from pBSVΦ-flaBp-gfp (9) with Expand high-fidelity polymerase (Roche, Indianapolis, IN), and the resulting PCR fragment was cloned into pCR2.1-TOPO (Invitrogen). loxP-flaBp-gfp was removed from the pCR2.1-TOPO vector by digestion with KpnI and PstI and ligated into pBSV2G (14), yielding pBSV2G-loxP-flaBp-gfp (Fig. 1 and Table 3).
FIG. 1.
Schematic diagram illustrating the loxP/Cre-mediated deletion of the gene encoding GFP. The introduction of Cre recombinase into B. burgdorferi containing flaBp-gfp flanked by loxP sites should result in recombination between loxP sites, the excision and loss of gfp from shuttle vector pBSV2G (15) due to the absence of replication factors in this region of the plasmid, and a loss of fluorescence by the cell.
TABLE 2.
Oligonucleotides used in this study
| Primer | Designation | Sequencea |
|---|---|---|
| 1 | FlaBp-loxP 5′ | ATAACTTCGTATAGCATACATTATACGAAGTTATtgtctgtcgcctcttgtggc |
| 2 | GFP-loxP 3′ PstI | ctgcagATAACTTCGTATAGCATACATTATACGAAGTTATttatttgtagagctcatcca |
| 3 | Cre 5′ NdeI | catatgtccaatttactgaccgtac |
| 4 | Cre 3′ XbaI | tctagactaatcgccatcttccagcagg |
| 5 | 229F | ggtaccgattaatccaagctcaataa |
| 6 | 1228R | ggctcaatattgcgtttaag |
| 7 | LoxflgF-1 | ggatccATAACTTCGTATAGCATACATTATACGAAGTTATtacccgagcttcaaggaaga |
| 8 | KanR-1 | ggatccttagaaaaactcatcgagct |
| 9 | 4297F | ggtaccggcttgcagctgccttaata |
| 10 | 5391R | taaaagctgtagtcaagccc |
| 11 | LoxflaBF | gagctcATAACTTCGTATAGCATACATTATACGAAGTTATctgtcgcctcttgtggcttc |
| 12 | aadaR | gagctcttatttgccgactaccttgg |
| 13 | 8398F | acgcgtcaactctggcattgacgctt |
| 14 | 9198R | aaaaaagtcccagttttggg |
| 15 | LoxflgF-2 | aagcttATAACTTCGTATAGCATACATTATACGAAGTTATtacccgagcttcaaggaaga |
| 16 | KanR-2 | aagcttttagaaaaactcatcgagct |
| 17 | pBSV2* 5′ | ttttattatagagttcatatacatgtttttctaaatctataa |
| 18 | pBSV2* 3′ | acatgtatatgagttctataataaaaaatattctagtgaggg |
| 19 | pncA+RBS EcoRI 5′ | ccggaattcttgtatcaagggatattgcc |
| 20 | pncA XbaI 3′ | gctctagattatatattaagcttactttggctg |
| 21 | Cre+RBS XbaI 5′ | gctctagataagaacaataaaaaggaaaatttatgtccaatttactgaccgtacacc |
| 22 | cre SphI 3′ | acatgcatgcctaatcgccatcttccagcagg |
| 23 | pncA prom 5′ EcoRI | cggaattcctatgctatccccttgttcaagtc |
| 24 | pncA prom 3′ EcoRI | cggaattccttttgtcaataattatatatttggg |
Restriction enzyme sites are indicated in boldface type, and the loxP sequences are in uppercase type.
TABLE 3.
Plasmid constructs used in this study
| Plasmid | Description | Reference(s) |
|---|---|---|
| pBSV2G-loxP-flaBp-gfp | GFP expression vector with loxP sites flanking flaBp-gfp | 10, 15; this study |
| pBSV25-flgBp-cre | Cre expression vector used to excise flaBp-gfp in pilot expt | 58; this study |
| pABA07 | Suicide vector used for inserting loxP-Strr into bba07 | This study |
| pABA01 | Suicide vector used for inserting loxP-Kanr into bba01 | This study |
| pABA14 | Suicide vector used for inserting loxP-Kanr into bba14 | This study |
| pBSV2* | Created by disruption of the EcoRI site found at bp 1852 on pBSV2 | 59; this study |
| pBSV2G-pncAp-pncA-cre | Cre expression vector used to excise targeted regions of lp54 | 15; this study |
Construction of pBSV25-flgB-cre.
The gene encoding Cre recombinase was kindly provided by Michael Culbertson (University of Wisconsin-Madison) and was amplified with Taq polymerase (New England Biolabs, Ipswich, MA) by using primers 3 and 4 (Table 2) and cloned into pBSV2. The cre gene was digested from pBSV2 with NdeI and XbaI and cloned into pBSV2ex (21) to create the cre expression construct pBSV2ex-flgBp-cre. The flgBp-cre fragment was removed from pBSV2ex by digestion with NotI and cloned into pBSV25 (52), yielding pBSV25-flgBp-cre (Table 3).
Suicide vector construction.
The pABA01 suicide vector, used to introduce a loxP site with a kanamycin resistance cassette (6) into bba01, was constructed as follows. The region at positions 229 to 1228 (GenBank) of lp54 was amplified with Expand high-fidelity polymerase (Roche) from B31-A3 by using primers 5 and 6 (Table 2) and cloned into pCR-XL-TOPO (Invitrogen). A KpnI site was engineered into the 5′ end of primer 5, allowing the removal of the BamHI site within pCR-XL-TOPO by digestion with KpnI and religation. The flgBp-driven kanamycin marker with a 5′ loxP site was amplified from shuttle vector pBSV2 (53) by using primers 7 and 8 (Table 2) and ligated into the cloned fragment of lp54 at positions 229 to 1228 by using a native BamHI site at position 761, yielding pABA01 (Table 3).
The pABA07 suicide vector, used to introduce a loxP site with the streptomycin resistance cassette aadA (17) into bba07, was constructed as follows. The region at positions 4297 to 5391 of lp54 was amplified with Expand high-fidelity polymerase (Roche) from B31 clone A3 by using primers 9 and 10 (Table 2) and cloned into pCR-XL-TOPO. A KpnI site was engineered into the 5′ end of primer 9, allowing the removal of the SacI site within pCR-XL-TOPO by digestion with KpnI and religation. The flaBp-driven spectinomycin-streptomycin resistance cassette (aadA), with a 5′ loxP site, was amplified by using primers 11 and 12 (Table 2). This fragment was inserted into the cloned region of lp54 at positions 4297 to 5391 by using a native SacI site at position 4754 to yield pABA07 (Table 3).
The pABA14 suicide vector, used to introduce a loxP site with a kanamycin resistance cassette into bba14, was constructed as follows. The region of lp54 at positions 8398 to 9198 was amplified with Expand high-fidelity polymerase (Roche) from B31 clone A3 by using primers 13 and 14 (Table 2) and cloned into pCR-XL-TOPO (Invitrogen). An MluI site was engineered into the 5′ end of primer 13 to allow the removal of the HindIII site in pCR-XL-TOPO by digestion with MluI and religation. The loxP-flgBp-kan cassette in pABA01 was amplified by using primers 15 and 16 (Table 2) and ligated into the cloned region of positions 8398 to 9198 of lp54 by using a native HindIII site at position 8915, yielding pABA14 (Table 3).
Construction of the pBSV2* shuttle vector.
Shuttle vector pBSV2* was constructed by altering the sequence of the second EcoRI site located outside the multiple-cloning site on pBSV2 (53) by PCR-based mutagenesis. Primers 17 and 18 (Table 2) include the targeted mutation GAGTTC for the EcoRI site found at bp 1852 on pBSV2 as well as 16 overlapping bases on the 3′ terminus of each primer to avoid primer self-extension. Reaction mixtures and parameters for the PCR followed the protocol described previously by Zheng et al., with the slight modifications of 20 cycles at 94°C for 1 min and an extension time of 16 min at 68°C (61). The PCR product was purified by use of a QIAquick PCR purification kit (Qiagen, Valencia, CA) and treated with DpnI (New England Biolabs) to digest the template DNA. The resulting modified pBSV2 PCR product was used to transform E. coli Top10F′ cells (Invitrogen) and selected on LB plates with kanamycin. Plasmid DNA was digested with EcoRI and sequenced to ensure that there were no mutations other than the disruption of the targeted EcoRI site. The resulting modified vector, termed pBSV2*, was found to be stable during the in vitro propagation of B. burgdorferi through 58 generations (the longest time period tested) in the absence of selection.
Construction of the pBSV2G-pncAp-pncA-cre vector.
A 679-bp DNA fragment containing the pncA open reading frame and its ribosome binding site with EcoRI and XbaI ends was PCR amplified from wild-type B. burgdorferi genomic DNA by using Taq polymerase (Invitrogen) and primers 19 and 20 (Table 2). A 1,066-bp DNA fragment containing the cre open reading frame with XbaI and SphI ends was PCR amplified from pBSV25-flgBp-cre by using Taq polymerase (Invitrogen) and primers 21 and 22 (Table 2). Primer 17 also included the 24-bp intergenic sequence between the guaA and guaB genes on the B. burgdorferi cp26 plasmid, which provided a ribosome binding site for the cre open reading frame. The pncA and cre PCR products were digested with EcoRI/XbaI and XbaI/SphI, respectively, and ligated into pBSV2*, which was digested with EcoRI/SphI. The structure and sequence of the pBSV2* pncA-cre plasmid were verified by restriction digestion and sequence analysis. The pBSV2* pncA-cre plasmid was digested with EcoRI, treated with Antarctic phosphatase (NEB), and gel purified by using the Qiagen Mini-Elute gel purification kit. A 906-bp DNA fragment containing the pncA promoter region with EcoRI ends was PCR amplified from B. burgdorferi genomic DNA by using Taq polymerase (Invitrogen) and primers 23 and 24 (Table 2). This DNA fragment was digested with EcoRI and cloned into the prepared pBSV2*pncA-cre plasmid. The structure and sequence of plasmid pBSV2*pncAp-pncA-cre were verified by restriction digestion and sequence analysis. The synthesis of Cre by both E. coli and B. burgdorferi harboring plasmid pBSV2* pncAp-pncA-cre was detected by immunoblot analysis of bacterial lysates using an anti-Cre polyclonal antibody (Covance, Madison, WI). Because a Cre expression vector carrying gentamicin resistance was required for this study, the pncAp-pncA-cre fragment was cut from the pBSV2* vector by using the NotI sites flanking the multiple-cloning site and ligated into NotI-digested pBSV2G (14), yielding pBSV2G-pncAp-pncA-cre (Table 3).
B. burgdorferi transformations.
Strain B31-A34 was transformed with 10 μg pBSV2G-loxP-flaBp-gfp (Table 3) under standard electroporation conditions (47), immediately resuspended in 5 ml BSKII liquid medium, and allowed to recover for 24 h at 35°C. Cells were then plated onto solid BSKII medium containing 40 μg/ml gentamicin and grown at 35°C. Positive transformants were confirmed by PCR for the presence of gfp and sequenced to ensure that the loxP sites were intact. Fluorescence microscopy confirmed the expression of GFP in all PCR-positive clones. A B31-A34 pBSV2G-loxP-flaBp-gfp clone was randomly selected for the introduction of the Cre expression vector pBSV25-flgBp-cre (Table 3). Transformations with 20 μg of pBSV25-flgBp-cre or the pBSV25 empty vector control were conducted as described above, and cells were plated in solid BSKII medium containing 200 μg/ml kanamycin. Positive transformants were investigated by PCR and sequencing for the presence of cre, gfp, and the gentamicin resistance cassette aacCI. Fluorescence microscopy was used to assess the presence or loss of GFP expression in the clones.
The loxP insertion mutants were created in the infectious clone B31-A3 (15) by using standard electroporation conditions (47). The A3-A07 loxP insertion mutant was obtained by electroporation with 20 μg of pABA07 DNA (Table 3), and transformants were selected in solid BSKII medium containing 50 μg/ml streptomycin and grown at 35°C. Positive transformants were confirmed by PCR for the presence of the streptomycin resistance cassette aadA and sequenced to confirm that the loxP site was intact. A single transformant containing all the plasmids present in parental strain B31-A3 was randomly chosen and designated A3-A07. This clone was electroporated with 20 μg of either pABA01 or pABA14 DNA (Table 3), and transformants were plated in the presence of 50 μg/ml streptomycin and 200 μg/ml kanamycin. Positive transformants were confirmed by PCR for the presence of the kanamycin resistance cassette and sequenced to confirm that both loxP sites were intact.
Twenty micrograms of methylated vector pBSV2G-pncAp-pncA-cre (Table 3) (13) was electroporated into the double-loxP insertion mutants, and bacteria were plated in the presence of 40 μg/ml gentamicin. Colonies positive by PCR for pncA-cre were grown in 5 ml BSKII medium, and total genomic DNA was isolated. PCR for bba01 to bba07 and bba07 to bba14 in the respective mutants and Southern blot analysis of genomic DNA confirmed the loss of targeted regions of lp54. The plasmid profiles of all transformants were identical to that of parental clone B31-A3 as determined by PCR using the Purser-Norris primer set (39).
Fluorescence microscopy.
One milliliter of a mid-log-phase B. burgdorferi culture was pelleted at 8,000 rpm for 10 min, washed once in phosphate-buffered saline (PBS), resuspended in GTE (50 mM glucose-20 mM Tris-HCl [pH 7.5]-10 mM EDTA) with a 1:100 dilution of FM4-64 membrane stain (Invitrogen), and incubated for 15 min. Cells were placed onto a glass slide, and images of GFP fluorescence and FM4-64 membrane staining were taken with a Nikon E800 microscope and a Photometrics CoolSnap HQ camera.
In vitro growth analysis.
Strains inoculated from frozen stocks were grown to mid-log phase and then diluted in triplicate to 105 spirochetes/ml (time zero) in 5 ml BSKII medium and incubated at 35°C. Triplicate cultures were counted at 24-h intervals in Petroff-Hauser chambers to assess the growth rate.
Mouse-tick infection studies.
Mouse studies were carried out in accordance with guidelines of the National Institutes of Health. All infection studies were done according to protocols approved by the Rocky Mountain Laboratories Animal Care and Use Committee. The Rocky Mountain Laboratories are accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). These studies were done by using 6- to 8-week-old female RML mice, an outbred strain of Swiss-Webster mice reared at the Rocky Mountain Laboratories breeding facility. Mice were retro-orbitally bled and needle inoculated with 5 × 103 B. burgdorferi spirochetes (4 × 103 spirochetes intraperitoneally and 1 × 103 spirochetes subcutaneously). At 3 weeks postinjection, mice were bled again to assess seroreactivity to B. burgdorferi proteins. Two ear punches (3 mm) were taken from seropositive mice, placed into BSKII medium, and incubated at 35°C to confirm infection by the reisolation of B. burgdorferi. Approximately 100 Ixodes scapularis larvae (Oklahoma State University) were fed to repletion on each infected mouse. Some fed larvae were ground and plated in solid BSKII medium at 10 days postfeeding to assess the acquisition of B. burgdorferi from infected mice. The remaining fed I. scapularis larvae were allowed to molt to nymphs and recover (approximately 10 weeks after larval feeding) before feeding on naïve mice to assess the persistence of B. burgdorferi mutant strains in ticks and transmission to mice.
RESULTS
Cre recombinase functions in B. burgdorferi.
To test the feasibility of using the Cre-lox system in B. burgdorferi, Cre recombinase was introduced into a B. burgdorferi strain containing a shuttle vector encoding GFP with flanking loxP sites. Cre-mediated recombination should result in the excision and loss of gfp from the shuttle vector (Fig. 1), which could easily be observed by a loss of fluorescence.
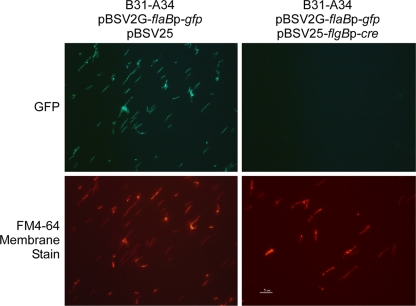
The GFP expression vector pBSV2G-loxP-flaBp-gfp was transformed into high-passage strain B31-A34 by electroporation as described above. Positive B. burgdorferi transformants containing the GFP shuttle vector were clearly visible by fluorescence microscopy (Fig. 2), and sequence analysis confirmed that intact loxP sites flanked flaBp-gfp in the transformants. The subsequent transformation of a GFP-containing clone with the Cre expression vector pBSV25-flgBp-cre yielded only four transformants. All pBSV2G-loxP-flaBp-gfp/pBSV25-flgBp-cre double transformants no longer fluoresced (Fig. 2), and sequencing of the rescued pBSV2G-loxP-flaBp-gfp shuttle vector confirmed that flaBp-gfp had been excised, leaving only a single loxP site. This pilot experiment demonstrated that Cre functioned efficiently in B. burgdorferi and could be used to engineer mutations in the spirochete. We noted, however, that the transformation of B. burgdorferi with pBSV25-flgBp-cre was relatively difficult compared to transformation with the empty pBSV25 shuttle vector. Control transformations with pBSV25 resulted in several hundred transformants, compared to only four with pBSV25-flgBp-cre. These results suggested that the strong expression of Cre by the flgB promoter might be deleterious, although Cre activity within wild-type B. burgdorferi should not produce any rearrangements or deletions, as there are no sequences resembling intact loxP sites present within the genome. However, subsequent experiments indicated that the low transformation frequency was not due to an overexpression of Cre, leading us to believe that the flgBp-cre insert sequence might be recognized by endogenous B. burgdorferi restriction enzymes and thus represent a barrier to transformation (27, 32).
FIG. 2.
Loss of fluorescence by B. burgdorferi carrying loxP-flanked GFP after introduction of Cre recombinase. Strain B31-A34 was first transformed with shuttle vector pBSV2G-loxP-flaBp-gfp (Fig. 1) and subsequently transformed with the compatible shuttle vector pBSV25 or pBSV25-flgBp-cre, which encodes Cre recombinase. (Top) Presence or absence of GFP fluorescence in these strains as visualized by fluorescence microscopy. (Bottom) Cells were counterstained with the membrane stain FM4-64 to visualize all spirochetes in the same field.
Cre-mediated deletions of lp54.
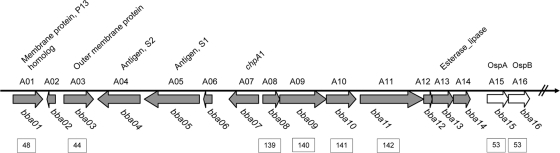
Following the demonstration of Cre-lox recombination in B. burgdorferi with GFP, we utilized this system to investigate the in vivo requirement for the region encompassing bba01 to bba14 of lp54 in the infectious clone B31-A3. Although several of the proteins encoded by genes in this region of lp54 were described in previous studies (10, 16, 36, 37), their requirement during the mouse-tick cycle has not been investigated. We engineered the deletions in two parts, bba01 to bba07 and bba07 to bba14, which encompass all genes on the left arm of lp54, between the telomere and the ospAB locus (bba15 and bba16) (Fig. 3).
FIG. 3.
Schematic diagram of the targeted region encompassing bba01 to bba14 of lp54, extending from the left telomere to the ospAB operon. Boxed numbers beneath the diagram indicate previously identified members of paralogous gene families (11, 18). Protein designations above the diagram and gene designations below are based on previous studies and annotations (10, 11, 16, 18, 36, 37).
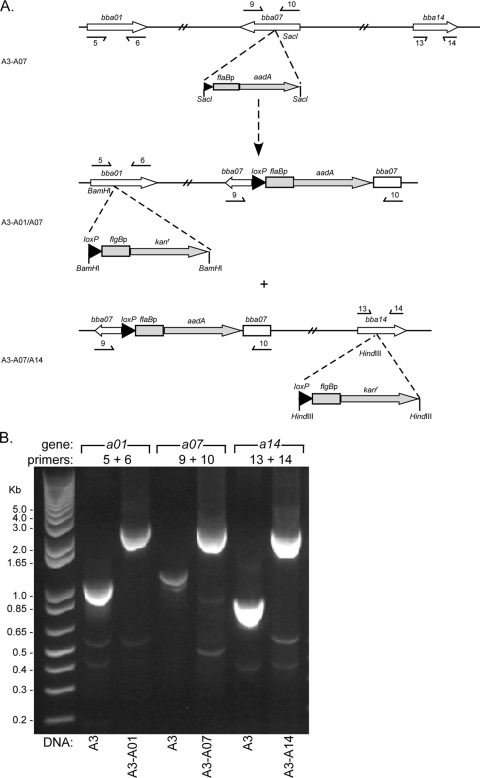
A loxP site carrying the streptomycin resistance cassette flaBp::aadA was introduced into the bba07 gene by allelic exchange using the pABA07 suicide vector in the B31-A3 background (Fig. 4). PCR of the bba07 locus with primers 9 and 10 (Table 2) showed the expected increase in size associated with a successful insertion of the aadA cassette in the gene (Fig. 4B), and sequencing confirmed that the loxP site was intact. Analysis of plasmid content by PCR confirmed that the clone retained all B31-A3 (wild-type)-associated plasmids.
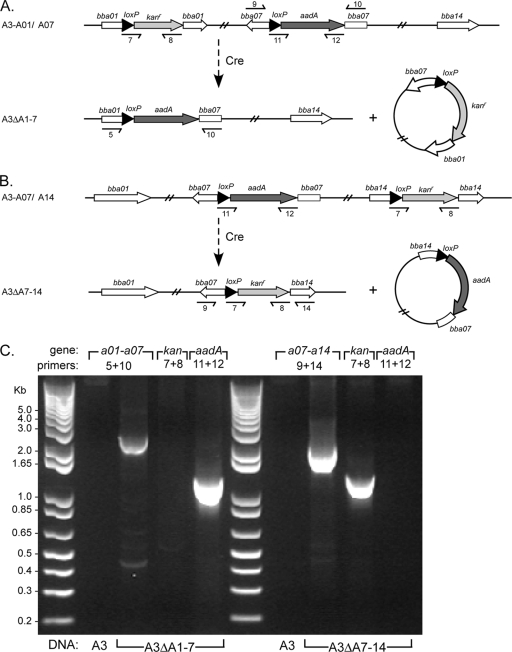
FIG. 4.
Insertion of loxP into targeted lp54 loci. (A) Schematic diagram showing how a loxP site (filled arrowheads) and an adjacent selectable marker conferring resistance to streptomycin or kanamycin (aadA and Kanr, respectively) were introduced by allelic exchange into bba07 and subsequently into either bba01 or bba14 on lp54. The relevant restriction enzyme sites used in cloning allelic exchange constructs are indicated. Small arrows beneath bba07, bba01, and bba14 indicate the positions of oligonucleotides (Table 2) used with PCR to confirm loxP insertions into these loci, as shown below (B). The designations of the resulting strains (Table 1) are shown on the left. (B) PCR amplification of targeted lp54 loci demonstrating an increase in fragment size after loxP insertion. The gene target and PCR primers are indicated above the lanes, and the source of template DNA is shown below the lanes. PCR primer positions and sequences are shown above (A) and in Table 2, respectively. The relative mobility of DNA size standards (kb) is shown on the left.
The subsequent insertion of a second loxP site with a kanamycin resistance marker by allelic exchange into the bba01 or bba14 gene using the suicide vector pABA01 or pABA14, respectively, resulted in the successful disruption of these targeted loci with loxP-kan (Fig. 4). PCR for bba01 or bba14 confirmed the size increase associated with the kanamycin cassette (Fig. 4b), and sequencing confirmed that the loxP sites were intact. The resulting double-loxP insertion mutants, A3-A01/A07 and A3-A07/A14, retained all plasmids associated with the parental clone B31-A3.
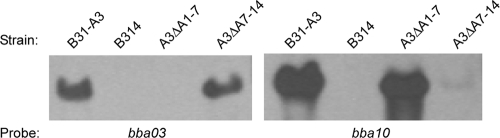
The lp54 loxP insertion mutants A3-A01/A07 and A3-A07/A14 were subsequently transformed with in vitro-methylated Cre expression vector pBSV2G-pncAp-pncA-cre to create deletions of these regions of lp54 (Fig. 5). Read-through from the pncA promoter was used instead of flgBp-cre to limit Cre production. Electroporation with the methylated Cre expression vector yielded three positive transformants in the A3-A01/A07 insertion mutant and two positive transformants in the A3-A07/A14 loxP insertion mutant. All pBSV2G-pncAp-pncA-cre transformants were positive by PCR for pncA-cre and negative by PCR for the targeted regions of lp54, indicating the successful excision of the loxP-flanked regions by the Cre recombinase (Fig. 5C). Southern blot analysis of the resulting lp54 deletion mutants, A3ΔA1-7 and A3ΔA7-14, confirmed the loss of the targeted regions of lp54 within the respective mutants (Fig. 6). Deletion mutants with plasmid profiles identical to that of wild-type clone B31-A3 were chosen for use in subsequent studies. Taken together, these results indicate that the Cre-lox system was used successfully to delete multiple loci on lp54 in infectious B. burgdorferi.
FIG. 5.
Deletion of loxP-flanked regions of lp54 after introduction of Cre recombinase. (A and B) Schematic diagrams showing the excision of the intervening DNA between loxP sites (filled arrowheads) present in the lp54 loci bba07 and bba01 (A) or bba07 and bba14 (B) as a result of Cre-mediated recombination. A recombined loxP site and the adjacent resistance cassette are present on a nonreplicating circular DNA fragment. Small arrows beneath bba07, bba01, and bba14 indicate the positions of oligonucleotides (Table 2) used with PCR to confirm the deletion of the intervening sequences in Cre transformants, as shown below (C). The designations of the resulting strains (Table 1) are shown on the left. (C) PCR amplification of targeted lp54 regions in the wild type and deletion mutants. Smaller products spanning the deleted regions were amplified from mutant strains, whereas the larger lp54 fragments were not efficiently amplified from wild-type A3. PCR amplification of the kan and aadA genes demonstrates the presence or absence of these antibiotic resistance cassettes as a consequence of the lp54 deletions. The gene target and PCR primers are indicated above the lanes, and the template DNA is indicated below the lanes. Primer positions are indicated above (A and B), and sequences are shown in Table 2. The relative mobility of DNA size standards (kb) is shown on the left.
FIG. 6.
Southern blots confirming the deletion of multigene segments of lp54 by loxP/Cre-mediated recombination. Genomic DNAs prepared from the wild type (B31-A3), a strain lacking lp54 (B314), and the lp54 deletion mutants (A3ΔA1-7 and A3ΔA7-14) were subjected to Southern blot analysis with probes specific to bba03 and bba10 to confirm the loss of targeted regions of lp54 containing these loci in the respective loxP/Cre deletion mutants. The source of DNA is identified above the lanes, and the probe is specified below.
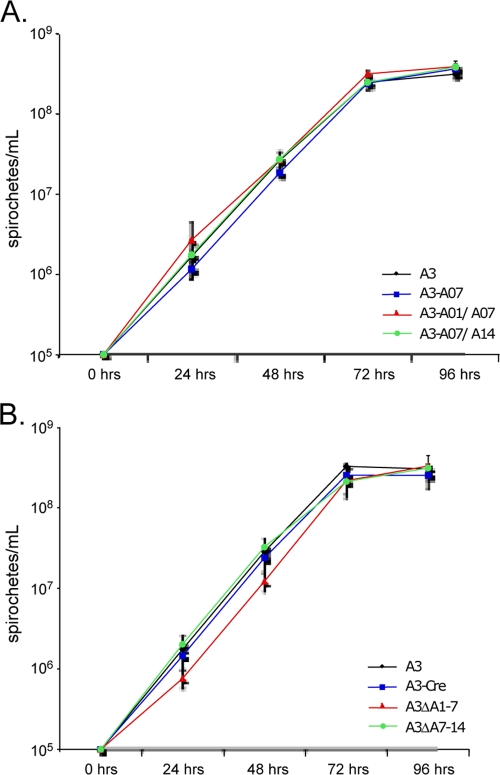
Growth phenotype of mutants lacking bba01 to bba07.
The requirement for the deleted regions of lp54 during in vitro growth was assessed by counting triplicate cultures of mutant and wild-type strains at 24-h intervals during growth in liquid media. None of the mutants displayed a significant growth phenotype when grown in liquid BSKII, and Cre production alone did not impair growth relative to that of wild-type B. burgdorferi (Fig. 7). Although there was no detectable effect on spirochete morphology or growth rate in liquid media, the A3ΔA1-7 mutant displayed a 2- to 3-day lag in colony formation compared to wild-type B31-A3 and other mutants, suggesting that genes within the region of bba01 to bba07 of lp54 may contribute to bacterial replication under specific environmental conditions.
FIG. 7.
In vitro growth curves. Wild-type B. burgdorferi (A3) and loxP insertion mutants (A3-A07, A3-A01/A07, and A3-A07/A14) (A) or lp54 deletion mutants (A3ΔA1-7 and A3ΔA7-14) and wild-type B. burgdorferi containing Cre on a shuttle vector (A3-Cre) (B) were grown at 35°C in BSKII medium from a starting concentration of 105 spirochetes/ml. Spirochetes were counted at 24-h intervals in Petroff-Hauser chambers, and the mean number of spirochetes per ml was determined from triplicate cultures of each strain, as shown.
Genes in targeted lp54 regions are not essential for mouse infectivity.
The contributions of genes encompassing bba01 to bba14 in establishing infection in a mammalian host were investigated by injecting mice with a standard inoculum of 5 × 103 organisms of either B31-A3, the lp54 loxP insertion mutants A3-A01/A07 and A3-A07/A14, and the lp54 deletion mutants A3ΔA1-7 and A3ΔA7-14. The 50% infectious dose (ID50) of wild-type clone B31-A3 is ∼103 organisms by this route (26, 55). At 3 weeks postinoculation, all mice were seropositive for B. burgdorferi antigens, and reisolates were obtained from all harvested tissues for all strains (Tables 4 and 5). These results indicate that the genes encompassing bba01 to bba14 are not critical to the spirochete's ability to infect and disseminate in a mammalian host.
TABLE 4.
Mouse infectivity of loxP insertion mutants by needle inoculation
| B. burgdorferi strain | No. of infected mice/no. of mice injected with 5 × 103 spirochetesa | No. of tissue reisolates/no. of injected mice (ear, bladder, joint)b |
|---|---|---|
| A3 | 3/3 | 3/3, 3/3, 3/3 |
| A3-A07 | 3/3 | 3/3, 3/3, 3/3 |
| A3-A01/A07 | 3/3 | 3/3, 3/3, 3/3 |
| A3-A07/A14 | 3/3 | 3/3, 3/3, 3/3 |
Number of infected mice/number of mice injected with 5 × 103 spirochetes (4 × 103 spirochetes intraperitoneally and 1 × 103 spirochetes subcutaneously). Seroconversion was determined by immunoblot analysis with B. burgdorferi lysate 3 weeks postinoculation.
Number of tissue reisolates/number of injected mice, for ear, bladder, and joint. Mice were euthanized and tissues were cultured at 4 weeks postinjection.
TABLE 5.
Mouse infectivity of lp54 deletion mutants by needle inoculation
| B. burgdorferi strain | No. of infected mice/no. of mice injected with 5 × 103 spirochetesa | No. of tissue reisolates/no. of injected mice (ear, bladder, joint)b | No. of spirochetes/larval tick (mouse 1; mouse 2)c |
|---|---|---|---|
| A3 | 3/4 | 3/4, 3/4, 3/4 | 2,500; 933 |
| A3ΔA1-7 | 4/4 | 4/4, 4/4, 4/4 | 27,332; 866 |
| A3ΔA7-14 | 4/4 | 4/4, 4/4, 4/4 | 6,333; 4 |
Number of infected mice/number of mice injected with 5 × 103 spirochetes (4 × 103 spirochetes intraperitoneally and 1 × 103 spirochetes subcutaneously). Seroconversion was determined by immunoblot analysis with B. burgdorferi lysate 3 weeks postinoculation.
Number of tissue reisolates/number of injected mice, for ear, bladder, and joint at 5 weeks postinoculation. All three tissues from infected mice were positive.
Larval ticks were fed to repletion on two seropositive mice for each strain. Ten days postfeeding, three fed larvae from each mouse were pooled, crushed, and plated to determine the number of viable spirochetes. The average number of spirochetes per tick was calculated from the total number of colonies obtained per pool of three larvae for each mouse. Mean numbers of spirochetes per tick for the A3ΔA1-7 and A3ΔA7-14 mutants were not significantly different from those of the wild type (A3) as determined by a two-tailed, unpaired t test (P = 0.3 and P = 0.7, respectively), performed by use of GraphPad Prism 5 software.
Tick acquisition, survival, and transmission of B. burgdorferi are not affected by lp54 deletions.
To complete the analyses of the infectious cycle of the lp54 deletion mutants, we investigated their acquisition by feeding ticks, replication and persistence within the tick midgut through the molting stage, and the ability to infect naïve hosts by tick bite. Approximately 100 naïve larval I. scapularis ticks were fed to repletion (3 to 5 days) on mice persistently infected with A3 (wild-type), A3ΔA1-7, or A3ΔA7-14 spirochetes. At 10 days postfeeding, ticks from each mouse were ground and plated in solid BSK medium to quantitatively assess spirochete numbers in the ticks. Spirochetes from mice infected with either mutant colonized larval ticks at densities similar to that of wild-type B. burgdorferi (Table 5), demonstrating no requirement for these lp54 genes for spirochete acquisition by feeding ticks.
We next investigated the contributions of genes in the deleted region of bba01 to bba14 of lp54 for spirochete survival through the tick molt and transmission to a new host. Cohorts of 20 I. scapularis nymphs infected with the wild type, A3ΔA1-7, or A3ΔA7-14 were fed to repletion on naïve mice. Ten days later, nymphs were ground and plated to determine the number of viable spirochetes in these ticks. As shown in Table 6, spirochete burdens were similar between the mutant- and wild-type-infected ticks, indicating that genes within the region of bba01 to bba14 are not required for spirochete survival or replication within the tick. At 3 weeks after tick feeding, mouse infection with the lp54 mutants was assessed. Two out of three mice fed on by A3ΔA1-7-infected nymphs did not become infected, whereas all three mice fed on by ticks carrying either the wild-type or the A3ΔA7-14 strain were infected. To extend this finding, we repeated the tick challenge of A3ΔA1-7-infected nymphs with four additional naïve mice, and three became infected, resulting in a total of four of seven mice acquiring A3ΔA1-7 infection following a tick bite (Table 6). Although there may be a slight attenuation in the A3ΔA1-7 mutant's ability to infect mice by tick bite, these differences were not statistically significant.
TABLE 6.
Mouse infectivity of lp54 deletion mutants by tick transmission
| B. burgdorferi strain | No. of infected mice/no. of mice fed on by 20 infected nymphsa | No. of tissue reisolates/no. of mice fed on by infected ticks (ear, bladder, joint)b | No. of spirochetes/nymphal tick (mouse 1; mouse 2; mouse 3)c |
|---|---|---|---|
| A3 | 3/3 | 3/3, 3/3, 3/3 | 100,000; 130,000; 140,000 |
| A3ΔA1-7 | 4/7 | 4/7, 4/7, 4/7 | 100,000; 140,000; 130,000 |
| A3ΔA7-14 | 3/3 | 3/3, 3/3, 3/3 | 140,000; 120,000; 140,000 |
Seroconversion was determined by immunoblot analysis with B. burgdorferi lysate 3 weeks postfeeding. The number of mice infected by either A3- or A3ΔA1-7-infected ticks was not significantly different using 20 ticks per mouse (P = 0.48, performed by Fisher's two-tailed exact-probability test).
Number of tissue reisolates/number of mice fed on by infected ticks, for ear, bladder, and joint. All three tissues from infected mice were positive.
Twenty infected nymphal ticks were fed to repletion on three naïve mice per strain. Ten days postfeeding, three fed nymphs from each mouse were pooled, crushed, and plated to determine the number of viable spirochetes. The average number of spirochetes per tick was calculated from the total number of colonies obtained per pool of three nymphs. Mean numbers of spirochetes per tick were not significantly different between strains as determined by one-way analysis of variance (ANOVA) (P = 0.75), performed by use of GraphPad Prism 5 software.
DISCUSSION
The genome of B. burgdorferi contains a number of plasmids, including cp26, lp25, lp28-1, lp36, and lp54, that encode factors essential for the spirochete's survival at different points during its infectious cycle. The persistence of B. burgdorferi in nature depends upon the completion of an entire infectious cycle through the tick vector and mammalian host; thus, the presence of critical gene functions on extrachromosomal elements in B. burgdorferi contradicts the standard definition of plasmids as nonessential replicons. The largest of the B. burgdorferi strain B31 plasmids, lp54, encodes proteins important for both mammalian and tick infection. These proteins include outer surface protein A (OspA) (BBA15), which is important for persistence within the tick (3, 34), and decorin binding protein A (DbpA) and DbpB (BBA24 and BBA25, respectively), which contribute to mammalian infection (5, 49, 50, 57). Other lp54-encoded proteins that may play a significant role in vivo include CspA or Crasp1 (BBA68), which was shown to bind complement regulatory proteins and confer serum resistance (28, 29), and OppAV (BBA34), which is a peptide binding component of oligopeptide permease that could contribute to nutrient acquisition (7).
Additional observations underline the importance of lp54 to the life cycle of B. burgdorferi. In a comparative genomic study, Qiu and colleagues reported previously that only two plasmids, cp26 and lp54, appear to be universally conserved among Lyme disease species (40); other studies reached the same conclusion (24, 35, 54). Unlike the other linear plasmids, lp54 is remarkably stable during the in vitro propagation of B. burgdorferi. The loss of this plasmid has been reported as an escape mechanism only when selective pressure was exerted against it (44, 45), suggesting the presence of genes encoding proteins or regulatory RNAs that are important to the spirochete's fitness in vitro. Interestingly, the loss of lp54 correlated with the inability of these strains to form colonies on solid medium (44, 46). Finally, microarray analyses in which environmental conditions were adjusted to mimic mammalian or tick conditions revealed that lp54 encodes the highest percentage of regulated proteins of any B. burgdorferi replicon, including the chromosome (8, 33, 41, 56). The conserved nature of lp54, its stability during in vitro propagation, and the indication that many of its genes are differentially expressed in either the tick or the mammal suggest that lp54 is likely to be essential for the spirochete's survival throughout the infectious cycle and also contributes to efficient growth in vitro.
Although data indicate that lp54 is important, the determination of precisely which genes are responsible is challenging because there are 76 predicted ORFs on lp54, the majority of which encode proteins of unknown function (11, 18). Therefore, we adapted the Cre-lox system to function in B. burgdorferi as an efficient screening tool with which to focus on loci relevant to fitness and survival in vivo. The use of Cre to engineer large deletions within the chromosome or a plasmid presents unique advantages over other available methods for genetic manipulation, such as allelic exchange or deletion walking by telomere insertion (4, 12, 43). The low transformation efficiency of infectious B. burgdorferi precludes the targeted inactivation of each individual gene as a practical means to investigate the roles of all lp54 gene products in vivo (15). Telomere insertion can be used to delete large segments from the ends of linear DNAs in B. burgdorferi (4, 12), whereas the Cre-lox system permits the introduction of internal deletions on both linear and circular replicons.
Based on a pilot study of loxP-flanked GFP as a reporter for Cre activity (Fig. 1 and 2), we modified the Cre-loxP system for use in the infectious clone B31-A3. First, we adjusted Cre expression by replacing the strong flgB promoter with that of pncA, which is expressed at a lower level (33, 38). Although we initially thought that Cre overexpression was toxic, resulting in poor transformation frequencies, read-through from the weaker pncA promoter did not increase the yield of transformants. In addition, Cre functioned efficiently upon the successful introduction into the B31-A3 background without observable adverse effects. Cre expression did not impede growth (Fig. 7B) or attenuate the spirochete's ability to complete the infectious cycle (Tables 5 and 6), and the shuttle vector encoding Cre was stably maintained by B. burgdorferi in infected mice and during in vitro growth in the absence of antibiotic selection (data not shown). Subsequently, we concluded that the cre gene sequence, and not the gene product, likely diminished the transformation efficiency, perhaps because it carries a site(s) recognized by an endogenous restriction enzyme of B. burgdorferi (27, 32). This limitation was addressed by the in vitro methylation of cre-containing plasmid DNA prior to the transformation of the loxP insertion mutants (13). Methylation was shown previously to increase the transformation efficiency in B. burgdorferi strain B31 by bypassing the restriction barriers encoded by lp56 (13). This proved to be an essential step in obtaining Cre transformants in the infectious clone B31-A3, as attempts to introduce the cre locus on unmethylated shuttle vector DNA into low-passage strains were unsuccessful.
With these modifications to the Cre-loxP system, we engineered two sets of deletions in infectious B. burgdorferi, each removing seven genes at a time from the left arm of lp54 (Fig. 3). The resulting deletion mutants, A3ΔA1-7 and A3ΔA7-14, maintained all the plasmids present in the parental wild-type clone and were assessed for both in vitro and in vivo phenotypes. All strains grew comparably in liquid medium (Fig. 7), but the A3ΔA1-7 strain took several days longer to form colonies in solid media than the wild-type and other mutant strains. This phenotype is consistent with the previously reported observation that B. burgdorferi escape mutants lacking lp54 had simultaneously lost the ability to form colonies in solid media (44, 46). Although we have not identified the gene product(s) responsible for this phenotype, the loxP insertions in bba01 and bba07 rule out those genes as candidates, since the respective mutants did not display any delay in colony formation. This finding suggests that at least one of the genes within the segment of bba02 to bba06 of lp54 contributes to growth under specific environmental conditions.
All B. burgdorferi mutant strains (both deletion and loxP insertion mutants) were able to infect mice at wild-type levels by needle inoculation (Tables 4 and 5). Furthermore, all mutant strains were acquired by naïve ticks that fed on infected mice, and these strains were maintained by ticks through the molting stage to the nymphal stage at wild-type levels (Table 5 and 6). The subsequent feeding of the infected nymphs on naïve mice demonstrated no defect in the A3ΔA7-14 mutant's ability to cause infection by tick bite. However, the A3ΔA1-7 mutant's ability to cause infection by tick bite may be slightly attenuated under our standard challenge of 20 ticks per mouse, as three of the seven mice did not become infected (Table 6). Although these findings are not statistically significant, they do suggest that a minor contribution might be made by one or more genes within the region of bba01 to bba07 at this stage of the infectious cycle.
Considering the highly conserved and regulated nature of lp54, we found it surprising that the first 14 genes of the plasmid could be deleted without significantly affecting the overall fitness of B. burgdorferi. Although little is known about the genes encompassing bba08 to bba14, previously characterized genes within the region of bba01 to bba07 suggested possible contributions by one or more of these genes during the infectious cycle. Although our data demonstrate that this region of lp54 does not encode any proteins or small regulatory RNAs that are required for mouse or tick infectivity, this study demonstrates the efficiency and usefulness of the Cre-lox system as a screening tool to scan the B. burgdorferi genome for important loci. The stability of lp54 coupled with its highly conserved and differentially expressed gene content argue that the continued investigation of lp54 with Cre-lox and other genetic tools is essential for a better understanding of the biology of the Lyme disease spirochete.
Acknowledgments
We thank Michael Culbertson, University of Wisconsin, for providing the cre gene. We thank John Leong for sharing unpublished data regarding in vitro methylation of E. coli DNA. We thank Jonah Cullen for technical assistance. We gratefully acknowledge the expert assistance of Anita Mora and Gary Hettrick in figure preparation. We thank Paul Beare and Chris Bosio for critical reading of the manuscript and helpful comments.
This research was supported by the Intramural Research Program of the NIH NIAID.
Editor: S. M. Payne
Footnotes
Published ahead of print on 15 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. Narayana, L. S. Pierson III, et al. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Battisti, J. M., J. L. Bono, P. A. Rosa, M. E. Schrumpf, T. G. Schwan, and P. F. Policastro. 2008. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect. Immun. 76:5228-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaurepaire, C., and G. Chaconas. 2005. Mapping of essential replication functions of the linear plasmid lp17 of Borrelia burgdorferi by targeted deletion walking. Mol. Microbiol. 57:132-142. [DOI] [PubMed] [Google Scholar]
- 5.Blevins, J. S., K. E. Hagman, and M. V. Norgard. 2008. Assessment of decorin-binding protein A to the infectivity of Borrelia burgdorferi in the murine models of needle and tick infection. BMC Microbiol. 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 8.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll, J., P. Stewart, P. Rosa, and C. Garon. 2003. An enhanced GFP reporter system to monitor gene expression in Borrelia burgdorferi. Microbiology 149:1819-1828. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 12.Chaconas, G., P. E. Stewart, K. Tilly, J. L. Bono, and P. Rosa. 2001. Telomere resolution in the Lyme disease spirochete. EMBO J. 20:3229-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Q., J. R. Fischer, V. M. Benoit, N. P. Dufour, P. Youderian, and J. M. Leong. 2008. In vitro CpG methylation increases the transformation efficiency of Borrelia burgdorferi strains harboring the endogenous linear plasmid lp56. J. Bacteriol. 190:7885-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias, A. F., J. L. Bono, J. J. Kupko, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 15.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, S., S. Das, T. Lam, R. A. Flavell, and E. Fikrig. 1995. A 55-kilodalton antigen encoded by a gene on a Borrelia burgdorferi 49-kilobase plasmid is recognized by antibodies in sera from patients with Lyme disease. Infect. Immun. 63:3459-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank, K. L., S. F. Bundle, M. E. Kresge, C. H. Eggers, and D. S. Samuels. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J. Bacteriol. 185:6723-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 19.Grimm, D., C. H. Eggers, M. J. Caimano, K. Tilly, P. E. Stewart, A. F. Elias, J. D. Radolf, and P. A. Rosa. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm, D., A. F. Elias, K. Tilly, and P. A. Rosa. 2003. Plasmid stability during in vitro propagation of Borrelia burgdorferi assessed at a clonal level. Infect. Immun. 71:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyard, C., J. Battisti, S. Raffel, M. Schrumpf, A. Whitney, J. G. Krum, S. Porcella, P. Rosa, F. DeLeo, and T. Schwan. 2006. Relapsing fever spirochetes produce a serine protease that provides resistance to oxidative stress and killing by neutrophils. Mol. Microbiol. 60:710-722. [DOI] [PubMed] [Google Scholar]
- 22.Hoess, R. H., and K. Abremski. 1984. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. U. S. A. 81:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoess, R. H., M. Ziese, and N. Sternberg. 1982. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. U. S. A. 79:3398-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer, R., O. Kalu, J. Purser, S. Norris, B. Stevenson, and I. Schwartz. 2003. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun. 71:3699-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jewett, M. W., R. Byram, A. Bestor, K. Tilly, K. Lawrence, M. N. Burtnick, F. Gherardini, and P. A. Rosa. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jewett, M. W., K. Lawrence, A. C. Bestor, K. Tilly, D. Grimm, P. Shaw, M. VanRaden, F. Gherardini, and P. A. Rosa. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64:1358-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawabata, H., S. J. Norris, and H. Watanabe. 2004. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72:7147-7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenedy, M. R., S. R. Vuppala, C. Siegel, P. Kraiczy, and D. R. Akins. 2009. CspA-mediated binding of human factor H inhibits complement deposition and confers serum resistance in Borrelia burgdorferi. Infect. Immun. 77:2773-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421-2429. [DOI] [PubMed] [Google Scholar]
- 30.Labandeira-Rey, M., J. Seshu, and J. T. Skare. 2003. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect. Immun. 71:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70:4798-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. G. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling temperature-induced changes in Borrelia burgdorferi gene expression using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Invest. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer, N., C. Fraser, and S. Casjens. 2000. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J. Bacteriol. 182:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinne, M., K. Denker, E. Nilsson, R. Benz, and S. Bergstrom. 2006. The BBA01 protein, a member of paralog family 48 from Borrelia burgdorferi, is potentially interchangeable with the channel-forming protein P13. J. Bacteriol. 188:4207-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinne, M., Y. Ostberg, P. Comstedt, and S. Bergström. 2004. Molecular analysis of the channel-forming protein P13 and its paralogue family 48 from different Lyme disease Borrelia species. Microbiology 150:549-559. [DOI] [PubMed] [Google Scholar]
- 38.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 39.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu, W.-G., S. E. Schutzer, J. F. Bruno, O. Attie, Y. Xu, J. J. Dunn, C. Fraser, S. R. Casjens, and B. Luft. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. U. S. A. 101:14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosa, P. A., K. Tilly, and P. E. Stewart. 2005. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3:129-143. [DOI] [PubMed] [Google Scholar]
- 44.Sădziene, A., P. A. Rosa, P. A. Thompson, D. M. Hogan, and A. G. Barbour. 1992. Antibody-resistant mutants of Borrelia burgdorferi: in vitro selection and characterization. J. Exp. Med. 176:799-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadziene, A., P. A. Thompson, and A. G. Barbour. 1993. In vitro inhibition of Borrelia burgdorferi growth by antibodies. J. Infect. Dis. 167:165-172. [DOI] [PubMed] [Google Scholar]
- 46.Sadziene, A., B. Wilske, M. S. Ferdows, and A. G. Barbour. 1993. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect. Immun. 61:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, Y., Q. Xu, K. McShan, and F. T. Liang. 2008. Both decorin-binding proteins A and B are critical for overall virulence of Borrelia burgdorferi. Infect. Immun. 76:1239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, Y., Q. Xu, S. V. Seemanapalli, K. McShan, and F. T. Liang. 2006. The dbpBA locus of Borrelia burgdorferi is not essential for infection of mice. Infect. Immun. 74:6509-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sternberg, N., B. Sauer, R. Hoess, and K. Abremski. 1986. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J. Mol. Biol. 187:197-212. [DOI] [PubMed] [Google Scholar]
- 52.Stewart, P. E., G. Chaconas, and P. Rosa. 2003. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J. Bacteriol. 185:3202-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 54.Terekhova, D., R. Iyer, G. P. Wormser, and I. Schwartz. 2006. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J. Bacteriol. 188:6124-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weening, E. H., N. Parveen, J. P. Trzeciakowski, J. M. Leong, M. Hook, and J. T. Skare. 2008. Borrelia burgdorferi lacking DbpBA exhibits an early survival defect during experimental infection. Infect. Immun. 76:5694-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y., and R. C. Johnson. 1995. Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J. Clin. Microbiol. 33:2679-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu, Y., C. Kodner, L. Coleman, and R. C. Johnson. 1996. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect. Immun. 64:3870-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, X. F., U. Pal, S. M. Alani, E. Fikrig, and M. V. Norgard. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]