Abstract
When confronted with metabolic stress, replicative Legionella pneumophila bacteria convert to resilient, infectious cells equipped for transmission. Differentiation is promoted by the LetA/LetS two-component system, which belongs to a family of signal-transducing proteins that employ a four-step phosphorelay to regulate gene expression. Histidine 307 of LetS was essential to switch on the transmission profile, but a threonine substitution at position 311 (T311M) suggested a rheostat-like function. The letS(T311M) bacteria resembled the wild type (WT) for some traits and letS null mutants for others, whereas they displayed intermediate levels of infectivity, cytotoxicity, and lysosome evasion. Although only 30 to 50% of letS(T311M) mutants became motile, flow cytometry determined that every cell eventually activated the flagellin promoter to WT levels, but expression was delayed. Likewise, letS(T311M) mutants exhibited delayed induction of RsmY and RsmZ, regulatory RNAs that relieve CsrA repression of transmission traits. Transcriptional profile analysis revealed that letS(T311M) mutants expressed the flagellar regulon and multiple other transmissive-phase loci at a higher cell density than the WT. Accordingly, we postulate that the letS(T311M) mutant may relay phosphate less efficiently than the WT LetS sensor protein, leading to sluggish gene expression and a variety of phenotypic profiles. Thus, as first described for BvgA/BvgS, rather than acting as on/off switches, this family of two-component systems exhibit rheostat activity that likely confers versatility as microbes adapt to fluctuating environments.
In aquatic reservoirs, the Gram-negative bacterium Legionella pneumophila resides within biofilm communities (26). When ingested by various species of amoebae or ciliated protozoa, the microbe avoids digestion and instead establishes a protective intracellular niche (26). Consequently, if humans inhale aerosols contaminated with L. pneumophila, the bacteria can parasitize alveolar macrophages and cause the acute pneumonia Legionnaires' disease (30, 40). Due to the disparate conditions under which L. pneumophila persists, the pathogen must employ strategies that enable swift adaptations to environmental fluctuations.
One mechanism by which L. pneumophila acclimates to its surroundings is by altering its cellular physiology, a process known as differentiation (45). When either protozoa or macrophages engulf transmissive-phase L. pneumophila, the microbes avoid lysosomal degradation and instead establish vacuoles isolated from the endosomal network, a process mediated by the Dot/Icm type IV secretion system (T4SS) (5, 54, 58, 69) and the shedding of vesicles rich in lipopolysaccharide (24). If conditions in the vacuole are favorable, L. pneumophila represses its transmissive traits and instead undergoes robust replication (25, 46). Once its nutrients are exhausted, bacterial replication halts, and the progeny induce traits that promote escape from their spent host, survival in the extracellular milieu, and the ability to infect subsequent phagocytic cells (27, 46, 57, 77).
From studies of synchronous broth cultures, many of the regulatory elements that govern the reciprocal phases displayed by L. pneumophila during its life cycle have been discerned. During the exponential (E) phase of growth, the posttranscriptional regulator CsrA and the sRNA chaperone Hfq suppress transmissive-phase traits and promote replication (25, 41, 46). However, once E-phase L. pneumophila experiences nutrient deprivation, cell division stops, and the enzymes RelA and SpoT produce the alarmone ppGpp (27, 77). Activation of the stringent response pathway leads to an accumulation of ppGpp in the bacterial cytosol (27, 77). As a result, transcription factors such as the alternative sigma factors RpoN, RpoS, and FliA likely recruit RNA polymerase to a new cohort of promoters (10, 12, 23, 51). Meanwhile, the LetA/LetS (LetA/S) two-component system (Legionella transmission activator and sensor, respectively) activates expression of two small regulatory RNAs, RsmY and RsmZ (33, 52, 56), which then bind to CsrA to relieve its repression of the transmission or postexponential (PE) traits (27, 28, 46). Together with the alternative sigma factors and other regulatory proteins, the LetA/LetS system induces traits that enable efficient host transmission and survival in the environment, including cytotoxicity, motility, pigment production, infectivity, and lysosome evasion (23, 59).
For most two-component systems, the physiological stimulus that activates the signal transduction pathway has remained elusive. Although the alarmone ppGpp is known to coordinate L. pneumophila differentiation when either amino acid or fatty acid biosynthesis is compromised (17, 22, 27), a precise signal that triggers LetS autophosphorylation has yet to be identified. For the two-component systems where the environmental cues are known, it appears that multiple inputs can induce the phosphorelay (9). By analogy, we predict that a variety of stimuli activate LetA/LetS and, likewise, L. pneumophila differentiation.
Whereas conventional two-component systems require a single phosphorylation event to induce a response, the L. pneumophila LetA/LetS system belongs to a family of signal-transducing proteins that use a multistep phosphorelay to regulate their response pathways. The prototype for this unorthodox family of signaling molecules is the Bordetella BvgA/BvgS system, which employs a four-step relay requiring consecutive phosphorylation of His-Asp-His-Asp residues (65, 67). BvgS is a polydomain sensor protein whose large periplasmic domain is linked by a membrane-spanning region to three cytoplasmic signaling domains (13). BvgA is the cytoplasmic activator kinase that, upon phosphorylation, gains affinity for Bvg-regulated promoters (13, 14). Upon receiving an appropriate signal, BvgS autophosphorylates on a conserved histidine residue and then sequentially transfers the phosphoryl group along the relay, culminating with BvgA activation (66). It has been proposed that the complexity of the BvgA/BvgS signaling mechanism enables Bordetella to express a spectrum of traits according to local conditions (15, 16, 60). In support of this model, Bordetella alternates between at least three distinct phenotypic phases in response to various external stimuli (15, 35). Cotter and Miller deduced that the BvgA/BvgS system regulates the amount of phosphorylated BvgA (BvgA∼P) present in the cell (15). The level of BvgA∼P, together with the inherent binding affinity of each Bvg-regulated promoter, enables Bordetella to control the temporal expression of different classes of genes and, likewise, its different phenotypic states (14).
Apart from the well-studied Bordetella system, other members within this family of two-component systems have not been analyzed to discern whether they also exhibit rheostat-like behavior that broadens their spectrum of phenotypic states. Sequence analysis indicates that the three predicted signaling domains of LetS are highly homologous to the analogous regions of Bordetella bronchiseptica BvgS, and the domain architecture is also comparable. Therefore, we exploited LetA/LetS to test whether the two-component regulatory system confers rheostat control in L. pneumophila. For this purpose, we constructed single amino acid substitutions in LetS and analyzed the mutants' phenotypic and transcriptional profiles. Our data indicate that, although their downstream circuitries differ, the L. pneumophila LetA/LetS two-component system resembles Bordetella BvgA/BvgS by functioning as a rheostat that can fine-tune the bacteria's virulence traits, which may augment versatility and fitness.
MATERIALS AND METHODS
Bacterial strains and culture.
L. pneumophila Lp02 (thyA hsdR rpsL; MB110) is a virulent thymine auxotroph derived from the Philadelphia-1 clinical isolate (5) and was the parental strain for all constructed mutants (Table 1). To construct a letS (lpg1912) insertion mutant that lacks the plasmid pflaG (which contains the promoter for the flagellin gene, flaA, fused to green fluorescent protein [GFP]), the letS locus containing the transposon insertion was amplified from MB417 and transferred to Lp02 by natural competence, resulting in strain MB416 (28). Bacteria were cultured at 37°C in 5-ml aliquots of N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract (AYE) broth and supplemented with 100 μg/ml thymidine when necessary. Cultures having an optical density at 600 nm (OD600) of 3.4 to 4.5 were defined as postexponential; within each experiment, similar culture densities were used to analyze strain phenotypes. To enumerate CFU, L. pneumophila cells were plated on ACES-buffered charcoal-yeast extract agar supplemented with 100 μg/ml thymidine (CYET) and incubated at 37°C. For constructing amino acid substitutions in letS, the semidefined medium CAA was prepared as described previously (43), and thymidine was added to 100 μg/ml (CAAT medium). For solid CAAT medium, agar was added to 15 mg/ml, starch to 5 mg/ml, and trimethoprim to 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Laboratory collection |
| XL10-Gold | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F[′] proAB lacIqZΔM15 Tn10(Tetr) Amy Camr] | Stratagene |
| MB596 | DH5α pletS | This work |
| MB597 | DH5α pletST311M | This work |
| MB598 | DH5α pletS::thyA | This work |
| MB610 | DH5α pletSH307Q | This work |
| MB540 | DH5α pBluescript KS+ with thyA | Laboratory collection |
| L. pneumophila | ||
| MB110 | Lp02 wild type; thyA hsdR rpsL | 5 |
| MB355 | Lp02 pflaG | 27 |
| MB416 | letS-36::Kan | 28 |
| MB417 | letS::kan pflaG | 28 |
| MB599 | letS::thyA | This work |
| MB600 | letS(T311M) | This work |
| MB605 | letS(T311M) pflaG | This work |
| MB611 | letS(H307Q) | This work |
| Plasmids | ||
| pGEM-T | Multiple cloning site within coding region of β-lactamase α fragment linearized with single-T overhangs; 3 kb; Ampr | Promega |
| pflaG | 150-bp flaA promoter fragment fused to GFP; encodes thymidylate synthetase; 10.5 kb; Ampr | 27 |
| pletS | pGEM-T containing 3.1-kb letS fragment PCR amplified from Lp02 chromosome and ligated into T overhangs; 6.1 kb; Ampr | This work |
| pletST311 M | pletS with a ACC to ATG change; 6.1 kb; Ampr | This work |
| pletSH307Q | pletS with a CAT to CAA change; 6.1 kb; Ampr | This work |
| pletS::thyA | pletS with 1.8 kb thyA fragment inserted into EcoRI site at base 436 of letS; 7.9 kb; Ampr | This work |
| pMB540 | pBluescript KS+ with thyA | Laboratory collection |
Construction of letS chromosomal substitutions.
The 3.1-kb letS locus was amplified from Lp02 genomic DNA using primers LetS F and LetS R (Table 2); the PCR fragment was purified and ligated into pGEM-T (Promega), and the resulting plasmid was designated pletS (MB596). Nucleotide substitutions were introduced into the letS open reading frame (ORF) in pletS using a QuikChange XL site-directed mutagenesis kit (Stratagene). A glutamate was substituted for the histidine at amino acid 307 by changing CAT to CAA with primers LetS Mut His F and LetS Mut His R (Table 2), resulting in the plasmid pletSH307Q (MB610). To substitute methionine for the threonine at residue 311 of letS, ACC was changed to ATG using primers LetS Mutagenesis F and LetS Mutagenesis R (Table 2), resulting in pletST311M (MB597). Synthesis of plasmid DNA, template digestion, and transformations into Escherichia coli XL10-Gold were performed according to the manufacturer's protocols. Mutagenesis was verified by sequencing the letS locus using the primer LetS Mut Seq F (Table 2).
TABLE 2.
Primers for PCR and RT-PCR
| Primer function and name | Sequencea |
|---|---|
| PCR | |
| LetS F | 5′-AATAATGCAGTCCTTACCC-3′ |
| LetS R | 5′-TGGATGACACCACAAGC-3′ |
| LetS Mut. His F | 5′-TTATTGCCAACATGAGTCAAGAAATTCGTACCCCAATGAATGGC-3′ |
| LetS Mut. His R | 5′-GCCATTCATTGGGGTACGAATTTCTTGACTCATGTTGGCAATAA-3′ |
| LetS Mutagenesis F | 5′-CATGAGTCATGAAATTCGTATGCCAATGAATGGCGTGATTGG-3′ |
| LetS Mutagenesis R | 5′-CCAATCACGCCATTCATTGGCATACGAATTTCATGACTCATG-3′ |
| LetS Mut Seq F | 5′-CGA TTG CGT CGA AGT ATG-3′ |
| RT-PCR | |
| CsrA_RT_F | 5′-TTTGACTCGGCGTATAGGTG-3′ |
| CsrA_RT_R | 5′-TTCCTAAGCGAACTTGATTGC-3′ |
| RsmY_RT_F | 5′-ATGGATATGTCTGACAGGAAGTC-3′ |
| RsmY_RT_R | 5′-ATTAGAGAATAAGTGCTGCATCC-3′ |
| RsmZ_RT_F | 5′-TGGATATGAGTCGTGCAAATGG-3′ |
| RsmZ_RT_R | 5′-GACTCAGCCCTGGCTTTTC-3′ |
Underlined nucleotides indicate site changes.
The gene encoding thymidylate synthetase was excised from pMB540 via EcoRI digestion and subcloned into the EcoRI site of pletS, which resulted in the insertion mutant pletS::thyA (MB598). The newly interrupted gene was amplified by PCR using primers LetS F and LetS R (Table 2), transferred to Lp02 by natural competence (61), and selected for growth in the absence of thymidine (MB599). Then, the point mutants were amplified from their respective plasmids, pletSH307Q or pletST311M, using primers LetS F and LetS R (Table 2), and the PCR fragments were purified (Qiaquick PCR purification kit; Qiagen). Approximately 50 μl of each PCR product was transferred to a 1-in. patch of MB599 on CYET medium. Following a 2-day incubation at 30°C, the patches of MB599 containing the PCR products were scraped off the CYET plates with a 1-ml pipette tip, and the cells were resuspended in 700 μl of CAA medium. Several dilutions of the resuspension were plated onto solid CAAT medium with or without trimethoprim. Recombinants were selected for growth on medium containing trimethoprim and confirmed both by their thymidine requirement and by sequencing of the letS locus with the LetS Mut Seq F primer (Table 2). The resulting L. pneumophila chromosomal substitution mutants, letS(T311M) and letS(H307Q), were designated MB600 and MB611, respectively.
Sequence and protein analysis.
The membrane-spanning regions of LetS were predicted using Kyte-Doolittle hydropathy plots (34). To predict the protein domains that are present in LetA and LetS, amino acid sequences were analyzed using the Conserved Domain Database (38). For alignments of the two-component sensor kinases, amino acid sequences from the following were aligned using T-Coffee (6, 12, 19, 21, 39, 47, 48, 62, 72): Acinetobacter baumannii ATCC 17978 GacS (A1S_0574); B. bronchiseptica RB50 BvgS (BB2995); Coxiella burnetii RSA 331 GacS (COXBURSA331_A1160) (GenBank accession number CP000890); E. coli K-12 substrains MG1655 ArcB (b3210), BarA (b2786), EvgS (b2370), and TorS (b0993); Klebsiella pneumoniae subsp. pneumoniae MGH 78578 BarA (KPN_03128; accession number CP000647); L. pneumophila subsp. pneumophila Philadelphia 1 LetS (lpg1912); Pseudomonas aeruginosa PAO1 GacS (PA0928); Salmonella enterica serovar Typhimurium LT2 BarA (STM2958); Shigella flexneri 2a 2457T BarA (S2993); Vibrio cholerae 0395 BarA (VC0395_A2032; accession number CP000627); and Yersinia pestis KIM BarA (y0808). Locus tags are listed in parentheses following each LetS homologue. To determine the percent identity and similarity between L. pneumophila LetS and B. bronchiseptica BvgS amino acid sequences, the GeneStream align program was used (49).
Macrophage culture.
Bone marrow-derived macrophages were isolated from femurs of A/J mice (Jackson Laboratory) and cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (RPMI-FBS medium; Gibco BRL) as described previously (63). After a 7-day incubation in L-cell supernatant-conditioned medium, macrophages were plated at 2.5 × 105/well in 24-well plates for infectivity and degradation assays and at 5 × 104/well in 96-well plates for cytotoxicity assays.
Infectivity.
To ascertain the degree to which L. pneumophila cells bind, enter, and survive inside macrophages, PE-phase bacteria were cocultured with macrophages at a 1:1 ratio in duplicate. The cells were centrifuged at 400 × g for 10 min at 4°C and then incubated an additional 2 h at 37°C. To remove extracellular bacteria, the infected monolayers were washed three times with fresh RPMI-FBS medium. Macrophages were mechanically lysed in 1 ml of 1× PBS, the lysate was plated onto CYET medium, and the cell-associated bacteria were enumerated (44). Infectivity was expressed as follows: (number of cell-associated CFU at 2 h/number of CFU added at 0 h) × 100 (2, 8).
Cytotoxicity.
To measure contact-dependent cytotoxicity of L. pneumophila for macrophages, PE-phase bacteria suspended in RPMI-FBS medium were added to macrophages at various multiplicities of infection (MOIs) in triplicate. After centrifugation at 400 × g for 10 min at 4°C (44), cells were incubated for 1 h at 37°C. To assess macrophage viability, the infected monolayers were incubated for 6 to 12 h with RPMI-FBS medium that contained 10% alamarBlue (Trek Diagnostic Systems), and then reduction of the colorimetric dye was measured by spectrophotometry and calculated as described previously (27, 44). In each experiment, experimental samples are compared to E- and PE-phase WT L. pneumophila negative and positive reference samples, as some variability that we do not understand can occur.
Lysosomal degradation.
The percentage of microbes that were intact following a 2-h incubation in macrophages was determined by fluorescence microscopy. Briefly, cells plated onto coverslips in a 24-well plate were infected with PE-phase L. pneumophila at an MOI of ∼1. Following centrifugation at 400 × g for 10 min at 4°C, the cells were incubated at 37°C for 2 h. After uninternalized bacteria were removed by washing with RPMI-FBS medium, the macrophages were fixed, permeabilized, and stained for L. pneumophila as described above, and duplicate coverslips were scored for intact rods versus degraded particles (3, 46).
Sodium sensitivity.
Sodium sensitivity was determined by plating 10-fold serial dilutions of PE broth cultures in 1× PBS onto CYET agar containing or lacking 100 mM NaCl. Following a 6-day incubation at 37°C, CFU were enumerated, and the percentage of sodium-sensitive bacteria was calculated as described previously (8).
Pigmentation.
To quantify pigment accumulation, 1-ml samples were obtained from broth cultures maintained in the PE phase for 5 days at 37°C. The aliquots were centrifuged at 16,000 × g for 10 min, and supernatants were measured at the OD550 (46).
Motility.
To qualitatively assess motility, 10-μl wet mounts of broth-grown L. pneumophila were prepared and immediately examined by phase-contrast microscopy. The estimated percentage of motility was based on at least three independent observations of fields that contained several hundred microbes.
Flow cytometry.
To monitor the promoter activity for an entire letS(T311M) population of cells, MB600 was transformed with pflaG, which contains the promoter for the flagellin gene, flaA, fused to a GFP reporter (27). The resulting strain, MB605, was cultured in AYE medium; at the designated optical densities, samples were centrifuged at 5,900 × g and washed in 1× PBS to remove impurities, and the cells were normalized to 5 × 105 in 1× PBS. Total GFP fluorescence was analyzed using a BD FACSAria cell sorter. PE-phase MB355 and MB417 cells were used as positive and negative controls, respectively.
Statistical analyses for phenotypic assays.
To calculate P values for infectivity, lysosomal degradation, sodium sensitivity, and pigmentation assays, one-way analysis of variance (ANOVA) was used for at least three independent samples.
RNA isolation, RNA labeling, and microarray hybridization.
WT and letS(T311M) mutants were cultured on an orbital shaker at 37°C to an OD600 of either 2 or 3 in 500 ml of AYE medium containing 100 μg/ml thymidine. Next, 10-ml aliquots were centrifuged at 6,000 × g for 2 min at 4°C, the culture supernatants were discarded, and the pellets were flash frozen and stored at −80°C. Total RNA was extracted using TRIzol (Invitrogen) as described previously (42). The RNA was reverse transcribed and labeled with Cy3 or Cy5 according to the manufacturer's instructions (Amersham Biosciences). The microarrays were designed to contain gene-specific 70-mer oligonucleotides based on all predicted genes within the genome of L. pneumophila strain Paris (CR628336) and its plasmid (CR628338) (7). In addition, genes specific to L. pneumophila strain Philadelphia-1 (AE017354) and strain Lens (CR628337) and its plasmid (CR628339) were added to the microarrays, as described previously (7). Hybridizations were performed following the manufacturer's recommendations (Corning) using 250 pmol of Cy3- and Cy5-labeled cDNA. Slides were scanned on a GenePix 4000A scanner (Axon Instruments), and the laser power and photomultiplier tube (PMT) were adjusted to balance the two channels. The resulting files were analyzed using Genepix Pro, version 5.0, software. Spots were excluded from analysis if they contained high background fluorescence, slide abnormalities, or weak intensity.
Data and statistical analysis for microarrays.
Data normalization and differential analysis were conducted using the R software package (http://www.R-project.org). Background subtraction was not performed, but a careful graphical examination of all the slides was done to ensure a homogeneous, low-level background in both channels. A lowess normalization (76) was performed on a slide-by-slide basis (BioConductor package marray [http://bioconductor.org/packages/2.2/bioc/html/marray.html]). Differential analysis was carried out separately for each comparison between the two time points, using the VM method (VarMixt package [18]), together with the Benjamini and Yekutieli P value adjustment method (53). If not stated otherwise, only differently expressed genes with 2-fold change were taken into consideration. Empty and flagged spots were excluded from the data set, and only genes without missing values for the comparison of interest were analyzed. The complete data set is available at http://genoscript.pasteur.fr in a MIAME (minimum information about a microarray experiment)-compliant public database maintained at the Institut Pasteur.
Real-time PCR.
Transcriptional analysis of RsmY and RsmZ was performed at cDNA concentrations ranging from 0.1 pg to 100 pg as described previously (7), and primers used are reported in Table 2. Two biological replicates of the WT and the letS(T311M) mutant grown to OD600s of 2 and 3 were analyzed, and RsmY and RsmZ transcripts were quantified twice from each sample. As a control, csrA transcripts were quantified in duplicate in the same WT and letS(T311M) samples. Values of relative induction were calculated by groupwise comparison of the letS(T311M) transcripts versus WT transcripts at both cell densities (50). All values reported are significant to a P value of <0.001.
RESULTS
Architecture of the LetA/LetS signal transduction system.
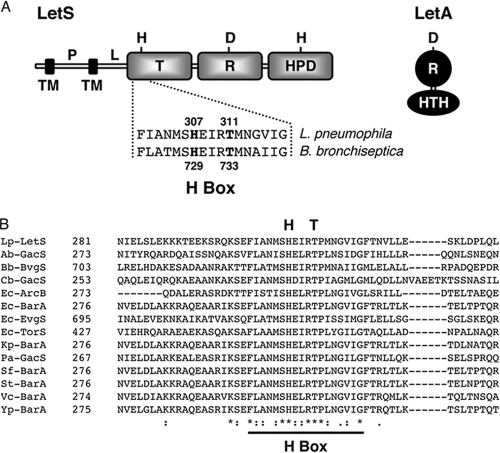
Based on protein sequence, the L. pneumophila LetA/LetS two-component system belongs to the family of signaling molecules that use a polydomain sensor to activate or repress their target genes. LetS is a 103-kDa sensor kinase that is likely localized to the membrane by two transmembrane regions (Fig. 1A). LetS is predicted to have three cytoplasmic signaling domains, namely, a transmitter, a receiver, and a histidine phosphotransfer domain (Fig. 1A). The cognate response regulator, LetA, is a 43-kDa protein that contains a receiver domain and a helix-turn-helix motif (Fig. 1A). By analogy to proteins of similar structure, it is predicted that, in response to a signal input, LetS autophosphorylates on a histidine residue and then sequentially transfers the phosphoryl group to an aspartic acid in the receiver domain, to a second histidine in the histidine phosphotransfer domain, and finally to an aspartic acid located in the receiver domain of LetA. Overall, LetS is only 18% identical to B. bronchiseptica BvgS at the amino acid level. However, within the region that contains the primary phosphorylation site, known as the H box, LetS is 78% identical and 89% similar to BvgS.
FIG. 1.
The L. pneumophila LetA/LetS two-component system. (A) Domain architecture of the LetA/LetS two-component system. LetS, a 103-kDa sensor protein, is likely tethered to the inner membrane by two transmembrane (TM) domains at its N terminus. The periplasmic (P) domain is connected via a linker (L) region to three cytoplasmic signaling domains, a transmitter (T), receiver (R), and histidine phosphotransfer domain (HPD). LetA is a 43-kDa activator kinase that contains a receiver (R) domain and a helix-turn-helix motif (HTH). It is predicted that, upon receiving a signal, LetS autophosphorylates on a conserved histidine residue, and then the phosphate is sequentially transferred to aspartic acid and histidine residues in LetS and finally to an aspartic acid in LetA. A histidine-to-glutamine substitution at amino acid 307 of LetS abolishes LetS activity, while a threonine-to-methionine substitution at position 311 creates a strain with sluggish transcriptional and phenotypic profiles. (B) Sequence alignment of the L. pneumophila LetS H-box region with related sensor kinases. Amino acid alignments were produced using T-Coffee. Dashes represent gaps introduced to optimize sequence alignments. Asterisks indicate identical residues while the colon and period represent conserved and semiconserved amino acids, respectively. The H-box region is underlined, and the primary histidine and conserved threonine residues are displayed above the alignment. Lp, L. pneumophila; Ab, A. baumannii; Bb, B. bronchiseptica; Cb, C. burnetii; Ec, E. coli; Kp, K. pneumoniae; Pa, P. aeruginosa; Sf, S. flexneri; St, S. Typhimurium; Vc, V. cholerae; Yp, Y. pestis.
A threonine residue near the autophosphorylation site is conserved among the polydomain sensor kinases.
Since LetS belongs to a family of two-component systems, we postulated that other sensor kinases within this unorthodox class of signaling molecules might have comparable domain architecture and, likewise, might employ similar regulatory mechanisms. Amino acid sequences from the following organisms were aligned using T-Coffee (47): L. pneumophila LetS; A. baumannii GacS; B. bronchiseptica BvgS; C. burnetii GacS; E. coli ArcB, BarA, EvgS, and TorS; K. pneumoniae BarA; P. aeruginosa GacS; S. Typhimurium BarA; Shigella flexneri BarA; V. cholerae BarA; and Y. pestis BarA. Indeed, the H-box regions of all the sensor proteins analyzed are remarkably similar. Moreover, the primary histidine residue, which is the proposed site for autophosphorylation, is conserved (Fig. 1B). Importantly, the threonine residue, which, when replaced with methionine enables Bordetella to stably display an intermediate class of genes and phenotypes (15), is also conserved among all family members analyzed (Fig. 1B). Therefore, we used LetS as a tool to test whether other multidomain sensors within this family of two-component systems might exhibit rheostat-like behavior to customize their traits when they are challenged by environmental stresses and fluctuations.
Histidine 307 of LetS is critical for LetA/LetS activity.
In response to a stimulus, the sensor protein of a microbial two-component system autophosphorylates on a histidine residue using ATP as a phosphate donor. In B. bronchiseptica, a histidine-to-glutamine substitution at amino acid 729 of BvgS abolishes its autophosphorylation as well as BvgA activation (66). Sequence alignments between the L. pneumophila and B. bronchiseptica sensor kinases predict that histidine 307 of LetS is the initial site of phosphorylation (Fig. 1). To test this model, we substituted a glutamine residue for histidine 307 of LetS. After verifying the unmarked, chromosomal point mutant, designated letS(H307Q), the mutant strain was analyzed for its expression of PE-phase phenotypes using WT L. pneumophila and an letS transposon insertion mutant (allele letS-36) as positive and negative controls, respectively.
For every trait examined, letS(H307Q) bacteria resembled the null mutant strain. After a 2-h incubation with macrophages, less than 5% of the L. pneumophila of letS(H307Q) and letS null mutant bacteria in the inoculum remained cell associated, whereas more than 15% of the WT bacteria did (Fig. 2A). In addition, histidine 307 of LetS was essential for both flagellin- and contact-dependent cell death of macrophages (Fig. 2B) (44). Likewise, phase-contrast microscopy indicated that letS(H307Q) mutants were completely defective for motility (data not shown). Using immunofluorescence microscopy to analyze the morphology of intracellular bacteria following 2 h of incubation within macrophages, we determined that, similar to the letS insertion mutant, only 40% of letS(H307Q) mutant cells resisted lysosomal degradation, whereas 80% of WT cells remained intact (Fig. 2C). Furthermore, histidine 307 of LetS was necessary for both salt sensitivity (Fig. 2D), a phenotype that reflects activity of the Dot/Icm T4SS (8, 55, 70), and for production of a soluble melanin-like pigment that accumulates in the PE phase (Fig. 2E) (71, 73, 75). Taken together, these genetic data are consistent with the model that histidine 307 is the autophosphorylation site of LetS and that the residue is critical for LetA/LetS activity.
FIG. 2.
Histidine 307 of LetS is required for the expression of PE-phase phenotypes. (A) The percentage of PE-phase bacteria that were viable and associated with macrophages after a 2-h incubation at an MOI of 1 is shown. (B) The fraction of viable macrophages was assessed by the reduction of the colorimetric dye alamarBlue after coculture for 1 h with PE-phase WT (circles), letS (squares), or letS(H307Q) (triangles) bacteria over a range of MOIs. Shown is a representative graph from three independent experiments preformed in triplicate. (C) The percentage of PE-phase bacteria that remained intact following a 2-h incubation with macrophages at an MOI of 1 was determined by fluorescence microscopy. (D) The percentage of sodium-resistant bacteria was determined after PE-phase cultures were plated on medium with or without 100 mM NaCl. (E) Soluble pigment in culture supernatants of WT, letS, and letS(H307Q) bacteria was quantified after a 5-day incubation period. For bar graphs in panels A, C, D, and E, the means ± standard deviations from duplicate samples in three independent experiments are displayed. Asterisks indicate statistically significant differences (P < 0.01) compared to WT PE-phase bacteria.
An amino acid substitution at position 311 of LetS unveils a hierarchy among PE phenotypes.
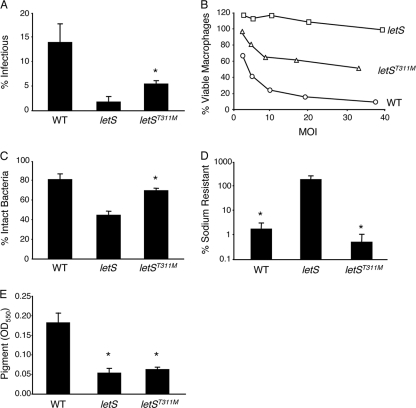
The paradigm in Bordetella suggests that the BvgA/BvgS system equips the bacterium with a rheostat to customize its expression profile. In support of this model, seminal work from Cotter and Miller demonstrated that a single amino acid substitution at residue 733 of B. bronchiseptica BvgS created a mutant that stably displayed intermediate phenotypes (15). To test whether this feature is common to other family members, a methionine was substituted for the corresponding threonine at position 311 in the cytoplasmic transmitter domain of LetS, four residues from the proposed autophosphorylation site (Fig. 1A). After sequence verification, the letS(T311M) mutant was analyzed for a panel of PE phenotypes.
Depending on the trait, the impact of the letS(T311M) mutation was complete, partial, or inconsequential. For example, the point mutant was intermediate for several PE-phase phenotypes, including entry and survival in macrophages (Fig. 3A), contact-dependent cytotoxicity (Fig. 3B), and evasion of lysosomal degradation (Fig. 3C). Likewise, only approximately 25 to 40% of letS(T311M) mutants were motile, as indicated by microscopic analysis (data not shown). Surprisingly, the letS(T311M) mutants were identical to WT L. pneumophila in the PE phase with regard to their sodium sensitivity (Fig. 3D), but the letS(T311M) mutants resembled letS null bacteria for pigment production (Fig. 3E). Therefore, these phenotypic data support the model that the LetA/LetS system enables the bacteria to display a spectrum of traits. Moreover, the profile of letS(T311M) mutants revealed a hierarchy with regard to the expression of particular L. pneumophila PE-phase phenotypes since some traits were sensitive to the mutation, but others were not.
FIG. 3.
A strain containing a threonine-to-methionine substitution at position 311 of LetS uncovers a hierarchy among the PE traits. (A) The number of viable PE bacteria associated with macrophages following a 2-h incubation at an MOI of 1 was calculated from lysates. The means ± standard deviations from duplicate samples in three independent experiments are displayed. Asterisks indicate statistically significant differences (P < 0.01) compared to WT microbes and (P < 0.05) compared to letS null bacteria. (B) Macrophage viability was measured by the reduction of the dye alamarBlue after infection for 1 h with PE-phase WT, letS, and letS(T311M) microbes at the MOIs shown. A representative graph from three independent experiments performed in triplicate is displayed. (C) The percentage of intact bacteria 2 h after macrophages were infected with PE-phase cultures at an MOI of 1 was quantified by fluorescence microscopy. Shown are the means from three independent experiments performed in duplicate. Error bars represent standard deviations, and asterisks indicate a statistically significant difference (P < 0.01) compared with WT and letS PE-phase bacteria. (D) The percentage of sodium-resistant CFU was quantified by plating PE-phase bacteria on CYET medium containing or lacking 100 mM NaCl. The means ± standard deviations from duplicate samples in three independent experiments are displayed, and the asterisks indicate significant differences (P < 0.01) in comparison to letS PE-phase cultures. (E) Pigment produced by WT, letS, and letS(T311M) cells cultured for 5 days was measured from supernatants at the OD550. Shown are the means from three independent experiments. Error bars indicate standard deviations, and asterisks indicate statistically significant differences (P < 0.01) compared to WT bacteria.
Promoter analysis demonstrates that the letS(T311M) mutant has a kinetic delay.
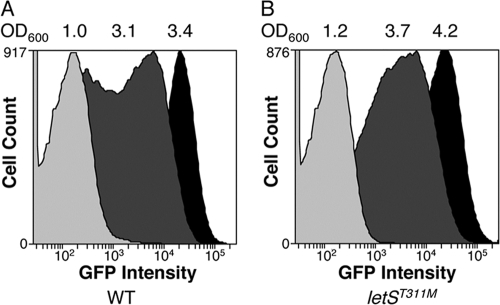
The PE phenotypes of infectivity, cytotoxicity, and lysosome evasion all depend on motility (44). Since the letS(T311M) mutant was intermediate for each of these traits, we predicted that, at the cell density analyzed, either all cells in the sample express the flagellin promoter at a weaker level than the WT, or the timing of flagellin expression is altered in letS(T311M) cells. To help distinguish between these two models, we monitored promoter expression by transforming the WT and the letS(T311M) mutant with a reporter construct that contained a transcriptional fusion of the flagellin promoter flaA fused to gfp. Flow cytometry indicated that at an OD600 of 1.0, the majority of cells in WT cultures had low levels of flaA expression (Fig. 4A). At the transition between the E and PE phases (OD600 of ≈3.0), two populations of cells were present in WT samples: one had low flaA promoter activity, and another showed robust induction of the flagellin promoter (Fig. 4A). Once WT L. pneumophila reached the PE phase (OD600 of 3.4), every cell in the population induced flaA to high levels (Fig. 4A). It was striking that flaA promoter activity was delayed in the letS(T311M) mutant compared to WT L. pneumophila (Fig. 4B). For example, cells in the letS(T311M) mutant population did not highly induce the flagellin promoter until an OD600 of 4.2, a density significantly higher than the OD600 of 3.4 where WT cultures fully activated flaA expression (compare Fig. 4A and B). Thus, every cell in the letS(T311M) population is able to activate the flaA promoter to WT levels, but the mutants have a kinetic defect.
FIG. 4.
Flow cytometry indicates that eventually every letS(T311M) cell in the population activates the flaA promoter to a similar level. To determine what percentage of WT and letS(T311M) populations expressed the pflaG reporter, GFP fluorescence was monitored via flow cytometry at the three culture densities (OD600) noted. Shown are representative curves from one experiment; similar results were obtained in three separate experiments.
Transcription of rsmY and rsmZ is also delayed in the letS(T311M) mutant.
The recently identified noncoding RNAs RsmY and RsmZ are critical components of the L. pneumophila differentiation circuitry (34, 52, 56). As described for orthologous systems (37), the LetA/LetS two-component regulatory system directly induces expression of RsmY and RsmZ, two regulatory RNAs that bind CsrA protein to relieve its repression of transmissive-phase genes (46, 52, 56). One indication that LetA/LetS governs L. pneumophila differentiation predominantly by inducing RsmY and RsmZ production in the PE phase (52, 56) is the similar transcriptional profiles of letA, letS, and rsmY rsmZ null mutant L. pneumophila bacteria (56). Since flow cytometry analyses determined that the letS(T311M) mutants exhibit a kinetic delay in expression of the transmissive-phase gene flaA (Fig. 4), we tested the prediction that, compared to WT L. pneumophila, letS(T311M) mutant induction of rsmY and rsmZ expression in the PE phase would be delayed.
Indeed, when cultured to OD600 of 2, letS(T311M) bacteria contained 20- and 50-fold less RsmY and RsmZ RNA, respectively, than the WT, as determined by reverse transcription-PCR (RT-PCR). However, at a later growth phase (OD600 of 3), the difference between letS(T311M) mutant and WT bacteria had diminished, as the RsmY and RsmZ RNAs were only 6- and 3-fold lower, respectively. Thus, kinetic analysis of the PE-phase induction of both the regulatory RNAs and the transmissive-phase flaA gene (Fig. 4) suggests that a sluggish transcriptional response accounts for the altered transmissive profile of letS(T311M) mutants (Fig. 3).
Transcriptome analysis indicates that the letS(T311M) mutant has a delayed transmissive profile.
RsmY and RsmZ directly relieve CsrA repression of numerous transmissive traits and genes (25, 46, 52, 56), including several that are poorly expressed by letS(T311M) mutants (Fig. 3 and 4). Therefore, we tested the prediction that their delayed transmissive-phase gene expression extends beyond the flaA, rsmY, and rsmZ promoters. For this purpose, the transcription profiles of letS(T311M) and WT L. pneumophila were compared at two different stages of growth via microarrays using chips bearing 70-mer oligonucleotides that represent each gene in the Paris, Lens, and Philadelphia-1 strains.
The data displayed in Table 3 indicate that a kinetic hierarchy exists among the LetA/LetS-regulated genes. By this model, genes that are expressed poorly by the letS(T311M) mutant at OD600s of both 2 and 3 likely represent factors that the LetA/LetS system activates throughout the PE phase (e.g., lpg0012) (Table 3). On the other hand, genes that are poorly expressed by the letS(T311M) mutant compared to WT only at an OD600 of 2 represent factors that L. pneumophila normally induces early during the PE transition. Several class II flagellar genes displayed this pattern (flgDEFGHIJKL [lpg1218-26]) (Table 3). At an OD600 of 2, letS(T311M) mutants contained less of each flagellar class II RNA than the WT; however, by an OD600 of 3 the WT and mutant RNA levels were indistinguishable (Table 3). Two more class II flagellar genes were also repressed in the letS(T311M) mutant, fliHG (lpg1758-9), although the level of repression was slightly below the cutoff value of 2.0 (−1.7 and −1.8, respectively) (data not shown). In contrast to PE-phase WT and letS(T311M) mutant L. pneumophila bacteria, null mutants of letA and letS fail to induce expression of these class II flagellar genes (56).
TABLE 3.
Selected subset of genes that are delayed in the letS(T311M) mutant compared to the WT
| Family | Gene IDa | Gene name | Paris strain ID | Lens strain ID | Description | Fold change at the indicated OD600 valueb |
|
|---|---|---|---|---|---|---|---|
| 2 | 3 | ||||||
| Regulation | lpg0277 | lpp0351 | lpl0329 | Regulatory protein (EAL domain) | −3.2 | — | |
| lpg1168 | lpp1170 | lpl1176 | Regulatory protein (GGDEF and EAL domains) | −3.4 | — | ||
| lpg2457 | lpp2523 | lpl2376 | Two-component response regulator (crystallized) | −2.4 | — | ||
| lpg2732 | lqsR | lpp2788 | lpl2657 | LqsR response regulator | −2.5 | — | |
| lpg0586 | lpp0636 | lpl0620 | Putative transcriptional regulator | — | −5.4 | ||
| lpg1114ac | lpp1115 | lpl1119 | KaiB-like circadian clock protein | — | −2.1 | ||
| lpg1115 | kaiC2 | lpp1116 | lpl1120a | Putative circadian clock protein KaiC | — | −2.5 | |
| lpg1577 | rpoE | lpp1535 | lpl1448 | Sigma factor RpoE (σ24) | — | −3.9 | |
| lpg1796 | lpp1760 | lpl1760 | LysR family transcriptional regulator | — | −2.0 | ||
| lpg2132 | lpp2071 | lpl2061 | Regulatory protein (GGDEF domain) | — | −2.5 | ||
| lpg2145 | lpp2083 | lpl2073 | Putative two-component response regulator | — | −2.9 | ||
| lpg2146 | stuC | lpp2084 | lpl2074 | Sensor histidine kinase | — | −2.8 | |
| lpg2181 | arcB | lpp2133 | lpl2108 | Putative histidine kinase/response regulator | — | −2.5 | |
| lpg2524 | LuxR family transcriptional regulator | — | −2.9 | ||||
| Flagellum biosynthesis | lpg1218 | flgD | lpp1226 | lpl1226 | Flagellar basal-body rod modification protein | −2.4 | — |
| lpg1219 | flgE | lpp1227 | lpl1227 | Flagellar hook protein | −6.1 | — | |
| lpg1220 | flgF | lpp1228 | lpl1228 | Flagellar biosynthesis protein | −4.3 | — | |
| lpg1221 | flgG | lpp1229 | lpl1229 | Flagellar biosynthesis protein | −2.1 | — | |
| lpg1222 | flgH | lpp1230 | lpl1230 | Flagellar L-ring protein precursor | −2.3 | — | |
| lpg1223 | flgI | lpp1231 | lpl1231 | Flagellar P-ring protein precursor | −2.9 | — | |
| lpg1224 | flgJ | lpp1232 | lpl1232 | Flagellar biosynthesis protein | −2.6 | — | |
| lpg1225 | flgK | lpp1233 | lpl1233 | Flagellar hook-associated protein | −4.9 | — | |
| lpg1226 | flgL | lpp1234 | lpl1234 | Flagellar hook-associated protein | −4.4 | — | |
| lpg1340 | flaA | lpp1294 | lpl1293 | Flagellin | — | −4.0 | |
| Type IV pilus | lpg0627 | pilE | lpp0681 | lpl0664 | Type IV pilin | — | −2.6 |
| lpg0628 | lpp0682 | lpl0665 | Type IV fimbrial biogenesis PilY1-related protein | — | −3.4 | ||
| lpg0629 | lpp0683 | lpl0666 | Tfp pilus assembly protein PilX | — | −2.9 | ||
| lpg0631 | lpp0685 | lpl0668 | Type IV fimbrial biogenesis protein PilV | — | −3.8 | ||
| lpg0632 | lpp0686 | lpl0669 | Type IV fimbrial pilin related protein | — | −7.5 | ||
| Virulence | lpg0910 | enhA2 | lpp0972 | lpl0942 | Similar to enhanced entry protein EnhA | — | −3.7 |
| lpg1355 | sidG | lpp1309 | SidG; substrate of the Dot/Icm T4SS | — | −2.9 | ||
| lpg1386 | enhA3 | lpp1341 | lpl1337 | Similar to enhanced entry protein EnhA | — | −3.8 | |
| lpg2157 | sdeA | lpp2096 | lpl2085 | SdeA; substrate of the Dot/Icm T4SS | — | −3.1 | |
| lpg2862 | legC8 | Cytotoxic glucosyltransferase | — | −4.3 | |||
| Eukaryotic-like | lpg0625 | lpp0679 | lpl0662 | Similar to unknown eukaryotic proteins | — | −2.8 | |
| lpg1158 | lpp1160 | Some similarity with eukaryotic proteins | — | −3.0 | |||
| lpg1491 | lpp1447 | Some similarity with eukaryotic proteins | — | −3.0 | |||
| Putatively involved in PHB synthesis | lpg0560 | phaB1 | lpp0620 | lpl0603 | Acetoacetyl-coenzyme A reductase | −2.8 | — |
| lpg1059 | phaB3 | lpp2322 | lpl1056 | Acetoacetyl-coenzyme A reductase | −2.2 | — | |
| Unknown | lpg0012 | lpp0012 | lpl0012 | Unknown | −3.2 | −2.6 | |
| lpg1174ac | lpp1177 | lpl1183 | Unknown | −5.8 | −8.5 | ||
| lpg0741 | lpp0806 | lpl0777 | Unknown | — | −5.5 | ||
| lpg1895 | lpp1864 | lpl1859 | Unknown | — | −6.0 | ||
| lpg2569 | Unknown | — | −5.9 | ||||
| lpg2803 | lpp2849 | lpl2718 | Unknown | — | −6.4 | ||
ID, identifier.
—, no statistically significant difference in expression was detected between WT and letS(T311M) mutant bacteria.
These genes were not predicted in the Philadelphia-1 strain. Their identifiers therefore became that of the gene located upstream on the chromosome with the addition of the letter “a.” For exact location and orientation of these genes, see Table S2 in the supplemental material.
In total, the mRNAs of 41 genes were significantly reduced in the letS(T311M) mutant compared to WT at an OD600 of 2 (see Table S1 in the supplemental material). Of these genes, 20 had been identified as “early transmissive traits” during L. pneumophila growth in Acanthamoeba castellanii (7). Genes critical for polyhydroxybutyrate (PHB) synthesis (phaB1 [lpg0560] and phaB3 [lpg1059]) were poorly expressed by the mutant strain at an OD600 of 2 (Table 3; see also Table S1). In addition, expression of several regulatory genes was reduced in the letS(T311M) mutant compared to the WT, including two genes that are predicted to contain EAL domains (lpg0277 and lpg1168), a two-component response regulator (lpg2457), and the response regulator lqsR (lpg2732) (Table 3; see also Table S1). Previous microarray experiments revealed that lqsR regulates the expression of genes involved in virulence, motility, and cell division, consistent with a role for LqsR in the transition from the E to the PE phase. However, lqsR expression is also dependent on RpoS and, to a lesser extent, LetA (64). Therefore, the letS(T311M) mutation may affect expression of lqsR and other early transmissive-phase genes either directly or indirectly.
At an OD600 of 3, shortly after the broth cultures entered PE phase, the expression of 87 genes was significantly reduced in the letS(T311M) mutant compared to WT L. pneumophila (see Table S1 in the supplemental material). Among this group are 13 of the 23 late transmissive-phase genes identified for WT L. pneumophila cultured in A. castellanii (7) (Table 3), including the sigma factor gene rpoE, putative transcription factor lpp0636, type IV fimbrial pilin-related genes lpg0632 and lpp0686, and the virulence-associated enhA3 locus. Nearly 50 of these genes encode unknown functions, and most lack similarity with any other protein or domain stored in the publicly available databases (see Table S1). Among the known genes, expression of several that are involved in L. pneumophila virulence were reduced at an OD600 of 3 in the letS(T311M) mutant, including those encoding substrates of the Dot/Icm T4SS sdeA (lpg2157) and sidG (lpg1355), enhanced entry protein enhA2 (lpg0910), and the newly identified cytotoxic glycosyltransferase legC8 (lpg2862) (4). The expression of several regulatory proteins was also clearly affected by the letS(T311M) mutation. In addition to the PE-specific sigma factor rpoE (lpg1577) (7), which was reduced 4-fold in the letS(T311M) mutant compared to WT bacteria (Table 3; see also Table S1), mRNAs for the GGDEF protein encoded by lpg2132, as well as several members of putative two-component systems (lpg2145, stuC [lpg2146], and arcB [lpg2181]) were significantly diminished in letS(T311M) bacteria (Table 3; see also Table S1).
One especially informative class of genes was the flagellar regulon. Expression of flaA (lpg1340) was decreased 4-fold in the letS(T311M) mutant compared to expression in the WT at an OD600 of 3. It is important to note that flaA, which encodes flagellin, is located at the bottom of the flagellar hierarchy and is commonly referred to as a class IV gene in the flagellar biosynthesis cascade (1, 59). Consistent with the sluggish flaA expression (Fig. 4 and Table 3; see also Table S1 in the supplemental material), expression of the upstream flagellar class II genes was diminished in the letS(T311M) mutant at an OD600 of 2 but not at the later time point when the OD600 reached 3 (Table 3; see also Table S1). Unlike PE-phase WT and letS(T311M) L. pneumophila bacteria, letA and letS null mutants are defective for induction of the flagellar regulon (56). Therefore, both our transcriptional profiling and flow cytometry data indicate that the letS(T311M) mutant is delayed in inducing the flagellar regulon.
DISCUSSION
The L. pneumophila LetA/LetS two-component system belongs to a family of signaling molecules that encode a four-step phosphorelay to activate or repress their target genes. The archetype for this family of two-component systems, the Bordetella BvgA/BvgS system, exhibits rheostat-like behavior. Likewise, in stark contrast to an on/off switch, the LetA/LetS system enables L. pneumophila to display a continuum of phenotypic phases to regulate its cohort of genes. To investigate whether the BvgA/BvgS paradigm applies to other family members, we analyzed the LetA/LetS system of L. pneumophila. By sequence analysis, we demonstrated that the H-box regions of the sensor kinases are highly conserved among all family members (Fig. 1B). Moreover, both the primary histidine residues and the threonine residues located four amino acids downstream of the autophosphorylation sites are also conserved (Fig. 1B). Using the letS(T311M) mutant as a tool, transcriptional and phenotypic analyses indicated that LetS permits Legionella to express a variety of phenotypic profiles, a versatility that may increase its ability to combat the stresses and challenges in its local environment. Based on the sequence homology within this family of two-component systems, we predict that rheostat-like behavior is widely used by microbes to confer versatility and enhance overall fitness.
Our data revealed a hierarchy among LetA/LetS-regulated genes and phenotypes. For example, the letS(T311M) mutant was similar to WT L. pneumophila with respect to salt sensitivity (Fig. 3D). This phenotype depends on the expression of the Dot/Icm T4SS and is thought to reflect a large pore formed by the apparatus that allows sodium ions to enter the bacterial cell (8, 55, 70). In support of this model, dot/icm genes were similar in the letS(T311M) mutant and WT bacteria based on microarray analysis, thus corroborating our phenotypic data (Fig. 3D and data not shown).
Unlike sodium sensitivity, expression levels of several other L. pneumophila PE-phase phenotypes by the letS(T311M) mutant pattern fell between those of WT and letS null bacteria. In particular, the point mutant was intermediate for its entry and survival in macrophages, cytotoxicity, and avoidance of the lysosomal compartment (Fig. 3A to C). Moreover, microscopic examination of the letS(T311M) mutant at various points during the L. pneumophila growth phase indicated that, at any given time, less than half of the population of the point mutant cells were motile (data not shown). Previous studies indicated that infectivity, cytotoxicity, and lysosomal degradation are all largely dependent upon motility (44). Thus, the intermediate phenotype displayed by the point mutant in each of these assays underscores the interdependency of this set of traits.
To assemble the flagellum, L. pneumophila requires a four-tiered regulatory cascade in which the expression and timing of each component must be tightly controlled (11, 59). The behavior of the letS(T311M) mutant illustrates that the precise coordination of the flagellar regulon is essential for constructing a functional apparatus. Although every cell in the letS(T311M) population eventually induces the promoter for flagellin to WT levels, as judged by flow cytometry data (Fig. 4), only a subset of the letS(T311M) mutant population becomes motile (data not shown). Furthermore, microarray data determined that, at an OD600 of 2, mRNAs for the flagellar class II genes (flgDEFGHIJKL [lpg1218-26]) were diminished in the letS(T311M) mutant compared to those of WT L. pneumophila (Table 3; see also Table S1 in the supplemental material). At a later growth phase (OD600 of 3), this defect disappeared; instead, expression of the flagellar class IV gene flaA was reduced in the letS(T311M) mutant (Table 3; see also Table S1). This pattern is distinct from those of null mutant letA and letS L. pneumophila bacteria, which are defective for induction of the flagellar regulon in the PE phase (56). Accordingly, we infer that the motility defect of the letS(T311M) mutant is the result of a kinetic defect that disrupts coordination of the flagellar regulatory cascade.
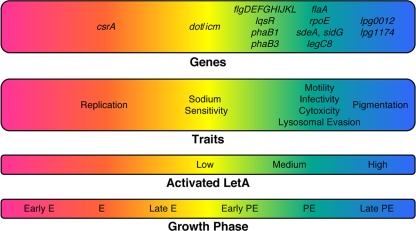
In Bordetella, the ability of the BvgA/BvgS system to regulate different classes of genes depends on the rate of the phosphorelay, the amount of BvgA∼P, and the binding affinities of BvgA protein for the promoter regions of Bvg-regulated genes (14). Accordingly, high rates of phosphate flowing through the relay lead to high levels of BvgA∼P, whereas lower rates of phospho-transfer lead to less phosphorylated activator kinase in the cell (14). It is predicted that the intermediate phase displayed in the Bordetella bvgS(T733M) mutant reflects a decrease in the intracellular concentration of BvgA∼P (20, 32, 74). We favor a similar model in which the transcriptional and phenotypic defects displayed by the letS(T311M) mutant are due to a slower or less efficient phosphorelay. In this scenario, the amount of phosphorylated LetA (LetA∼P) in the cell is significantly less than that of WT L. pneumophila, thereby altering the timing of the expression of LetA/LetS-regulated traits. Work from Sahr et al. indicates that the LetA/LetS system is inactive during the E phase of the L. pneumophila life cycle (56). Instead, genes required for replication are induced, including genes for DNA replication and protein synthesis, and also the global repressor of PE phenotypes csrA (Fig. 5) (25, 46). It is predicted that a signal generated at the onset of the PE phase triggers the LetA/LetS system (56). We postulate that lower levels of LetA∼P are sufficient to activate genes of the Dot/Icm T4SS and, likewise, induce sodium sensitivity since the letS(T311M) mutant was transcriptionally and phenotypically similar to WT L. pneumophila for these traits (Fig. 3D and 5; also data not shown). Perhaps more LetA∼P is needed to induce the flagellar cascade since the letS(T311M) mutant was delayed in transcription of the flagellar genes (Fig. 4 and 5 and Table 3; see also Table S1 in the supplemental material) and also intermediate for each of the motility-dependent phenotypes (Fig. 3A to C and 5). Finally, high levels of LetA∼P are likely required for pigmentation, because the letS(T311M) mutant never accumulated detectable levels of the soluble pigment (Fig. 3E and 5). Likewise, we infer that higher levels of LetA∼P are needed to transcribe lpg0012 and lpg1174 since the mRNA of each was significantly diminished in the letS(T311M) mutant compared to WT bacteria at OD600s of both 2 and 3 (Table 3 and Fig. 5; see also Table S1). Taken together, our transcriptional and phenotypic data support the rheostat model of regulation (14), whereby a two-component system can equip a bacterium with a mechanism to fine-tune its expression of gene hierarchies.
FIG. 5.
Model of the LetA/LetS system as a rheostat to fine-tune its phenotypic profile. When L. pneumophila cells are in the E phase of growth, the LetA/LetS system is likely inactive. At this time point, genes that are essential for L. pneumophila replication are induced, for example, the posttranscriptional regulator csrA. When LetS receives an appropriate signal, the sensor kinase autophosphorylates, and as phosphate flows through the relay, the amount of LetA∼P accumulates. It is predicted that the amount of LetA∼P required to activate the Dot/Icm T4SS and sodium sensitivity is low, as letS(T311M) mutants are similar to the WT for these traits. Perhaps intermediate levels of LetA∼P are required to activate the PE-phase phenotypes of infectivity, cytotoxicity, and lysosomal avoidance as letS(T311M) mutants exhibit intermediate phenotypes compared to WT and letS null bacteria. Also, intermediate levels of LetA∼P are likely needed to induce motility since a kinetic defect was observed in genes of the flagellar cascade for the letS(T311M) mutant. Presumably, substantial levels of LetA∼P are required for pigmentation as letS(T311M) mutants do not accumulate high levels of the soluble pigment. Likewise, high levels of LetA∼P are probably required to induce lpg0012 and lpg1174 since the letS(T311M) mutant was repressed for these genes at OD600s of both 2 and 3 compared to WT L. pneumophila.
While work in Bordetella indicates that the Bvg intermediate phase is mediated by lower levels of BvgA∼P (20, 32, 74), we have not ruled out the formal possibility that the sluggish transcriptional and phenotypic profiles observed in the L. pneumophila letS(T311M) mutant are due to alterations in protein stability. Attempts to analyze protein levels and phosphorylation through overexpression and purification of epitope-tagged WT LetS and the LetS(T311M) mutant proteins were unsuccessful (data not shown). In particular, both strain and plasmid variants arose at high frequencies. The instability we observed is in accordance with molecular analysis indicating that spontaneous mutations commonly arise in a homopolymeric tract of thymines present in LetS, which frequently generates truncated proteins (C. Buchrieser et al., personal communication). Moreover, this phenomenon is not unique to the L. pneumophila LetA/LetS system. Studies of the analogous GacA/GacS two-component system in Pseudomonas indicate that spontaneous point mutations, deletions, and DNA rearrangements in either the sensor or the activator kinases lead to colony phase variation (68). However, based on the striking sequence and functional similarities displayed by the LetA/LetS and BvgA/BvgS systems, we favor the model that the transcriptional and phenotypic profiles controlled by the L. pneumophila two-component system are due to fluctuations in the rate of the phosphorelay rather than alterations in LetS protein levels (14).
Although the Bordetella BvgA/BvgS system has been an informative paradigm for this family of signaling molecules, significant differences in the architecture of the Bordetella and Legionella regulatory cascades exist. Namely, the Bordetella two-component system controls many classes of genes through differences in the binding affinities of BvgA∼P to variations in the consensus sequences of the Bvg-regulated promoter regions (14). However, no conserved LetA-dependent DNA-binding motifs could be identified upstream of any of the LetA/LetS-regulated genes (56). Instead, bioinformatic and biochemical data suggest that LetA binds only to a conserved palindromic sequence located upstream of the regulatory RNAs, RsmY and RsmZ, which work together with LetA/LetS to regulate the genes and phenotypes responsible for the PE phase in L. pneumophila (25, 31, 33, 46, 52, 57). Interestingly, while rsmY and rsmZ transcripts are reduced in ΔletA and ΔletS mutants in the late PE phase (OD of 4.3), their expression is not completely abolished; thus other factors might contribute to their regulation (55). We predict that the rate of the LetA/LetS phosphorelay affects the amount of LetA∼P, which in turn impacts the amount of RsmY and RsmZ transcribed and the ability of the small RNAs to titrate CsrA from its respective cohort of mRNAs (56). In support of this model, we demonstrated that the letS(T311M) mutant had less RsmY and RsmZ transcribed than WT L. pneumophila, presumably because less intracellular LetA∼P is present in letS(T311M) bacteria. By analogy to the Bordetella system, perhaps CsrA has different affinities for particular mRNAs. If so, the amount of RsmY and RsmZ would then affect the order in which mRNAs are relieved from CsrA repression. We envision that by having the LetA/LetS and Csr systems in tandem, L. pneumophila can adapt to stresses more quickly since mRNA transcripts for critical transmission traits would already have been generated. Unlike Bordetella, which lacks the Csr system, many other members within this family of two-component systems contain this additional layer of regulation, including A. baumannii, C. burnetii, E. coli, S. Typhimurium, P. aeruginosa, V. cholerae, K. pneumonia, S. flexneri, and Yersinia (29, 36, 37). Thus, the L. pneumophila LetA/LetS system can serve as a valuable alternative model for this large family of signaling molecules.
Supplementary Material
Acknowledgments
We thank Michael A. Bachman and Brenda Byrne for their technical assistance in developing the thymidylate synthetase/trimethoprim selection method.
This work was supported by the Cellular and Molecular Biology Training Program, the University of Michigan Rackham Predoctoral Fellowship, the University of Michigan Rackham Graduate Student Research Grant, the Network of Excellence “Europathogenomics” (LSHB-CT-2005-512061), and NIH grant 2 R01 AI44212.
Editor: A. Camilli
Footnotes
Published ahead of print on 29 March 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Albert-Weissenberger, C., T. Sahr, O. Sismeiro, J. Hacker, K. Heuner, and C. Buchrieser. 2010. Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J. Bacteriol. 192:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72:2468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Belyi, Y., I. Tabakova, M. Stahl, and K. Aktories. 2008. Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J. Bacteriol. 190:3026-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mao, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 7.Bruggemann, H., A. Hagman, M. Jules, O. Sismeiro, M. A. Dillies, C. Gouyette, F. Kunst, M. Steinert, K. Heuner, J. Y. Coppee, and C. Buchrieser. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8:1228-1240. [DOI] [PubMed] [Google Scholar]
- 8.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calva, E., and R. Oropeza. 2006. Two-component signal transduction systems, environmental signals, and virulence. Microb. Ecol. 51:166-176. [DOI] [PubMed] [Google Scholar]
- 10.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 11.Chevance, F. F., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 13.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11:367-373. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. A., and J. F. Miller. 1997. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol. Microbiol. 24:671-685. [DOI] [PubMed] [Google Scholar]
- 16.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalebroux, Z. D., R. L. Edwards, and M. S. Swanson. 2009. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol. Microbiol. 71:640-658. [DOI] [PubMed] [Google Scholar]
- 18.Delmar, P., S. Robin, and J. J. Daudin. 2005. VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics 21:502-508. [DOI] [PubMed] [Google Scholar]
- 19.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deora, R., H. J. Bootsma, J. F. Miller, and P. A. Cotter. 2001. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol. Microbiol. 40:669-683. [DOI] [PubMed] [Google Scholar]
- 21.Dorsey, C. W., A. P. Tomaras, and L. A. Actis. 2002. Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68:6353-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards, R. L., Z. D. Dalebroux, and M. S. Swanson. 2009. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol. Microbiol. 71:1190-1204. [DOI] [PubMed] [Google Scholar]
- 23.Edwards, R. L., and M. S. Swanson. 2007. Regulation of the L. pneumophila life cycle, p. 95-111. In P. Hoffman, T. Klein, and H. Friedman (ed.), Legionella pneumophila: pathogenesis and immunity. Springer, Berlin, Germany.
- 24.Fernandez-Moreira, E., J. H. Helbig, and M. S. Swanson. 2006. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect. Immun. 74:3285-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpression of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 26.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 28.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 29.Heroven, A. K., K. Bohme, M. Rohde, and P. Dersch. 2008. A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68:1179-1195. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Invest. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovel-Miner, G., S. Pampou, S. P. Faucher, M. Clarke, I. Morozova, P. Morozov, J. J. Russo, H. A. Shuman, and S. Kalachikov. 2009. σS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J. Bacteriol. 191:2461-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, A. M., P. E. Boucher, C. L. Williams, S. Stibitz, and P. A. Cotter. 2005. Role of BvgA phosphorylation and DNA binding affinity in control of Bvg-mediated phenotypic phase transition in Bordetella pertussis. Mol. Microbiol. 58:700-713. [DOI] [PubMed] [Google Scholar]
- 33.Kulkarni, P. R., X. Cui, J. W. Williams, A. M. Stevens, and R. V. Kulkarni. 2006. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 34:3361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 35.Lacey, B. W. 1960. Antigenic modulation of Bordetella pertussis. J. Hyg. (Lond.) 58:57-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lapouge, K., M. Schubert, F. H.-T. Allain, and D. Haas. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behavior. Mol. Microbiol. 67:241-253. [DOI] [PubMed] [Google Scholar]
- 37.Lucchetti-Miganeh, C., E. Burrowes, C. Baysse, and G. Ermel. 2008. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154:16-29. [DOI] [PubMed] [Google Scholar]
- 38.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 40.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory diseases. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 41.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 187:1527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 43.Mintz, C. S., J. Chen, and H. Shuman. 1988. Isolation and characterization of auxotrophic mutants of Legionella pneumophila that fail to multiply in human monocytes. Infect. Immun. 56:1449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molofsky, A. B., L. M. Shetron-Rama, and M. S. Swanson. 2005. Components of the Legionella pneumophila flagellar regulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect. Immun. 73:5720-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 46.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 47.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 48.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 49.Pearson, W. R., T. Wood, Z. Zhang, and W. Miller. 1997. Comparison of DNA sequences with protein sequences. Genomics 46:24-36. [DOI] [PubMed] [Google Scholar]
- 50.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potrykus, K., and M. Cashel. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35-51. [DOI] [PubMed] [Google Scholar]
- 52.Rasis, M., and G. Segal. 2009. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol. Microbiol. 72:995-1010. [DOI] [PubMed] [Google Scholar]
- 53.Reiner, A., D. Yekutieli, and Y. Benjamini. 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19:368-375. [DOI] [PubMed] [Google Scholar]
- 54.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663-674. [DOI] [PubMed] [Google Scholar]
- 55.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahr, T., H. Bruggemann, M. Jules, M. Lomma, C. Albert-Weissenberger, C. Cazalet, and C. Buchrieser. 2009. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol. Microbiol. 72:741-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauer, J.-D., M. A. Bachman, and M. S. Swanson. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. U. S. A. 102:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Segal, G., M. Purchell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U. S. A. 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinert, M., K. Heuner, C. Buchrieser, C. Albert-Weissenberger, and G. Glockner. 2007. Legionella pathogenicity: genome structure, regulatory networks and the host cell response. Int. J. Med. Microbiol. 297:577-587. [DOI] [PubMed] [Google Scholar]
- 60.Stockbauer, K. E., B. Fuchslocher, J. F. Miller, and P. A. Cotter. 2001. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol. Microbiol. 39:65-78. [DOI] [PubMed] [Google Scholar]
- 61.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 63.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tiaden, A., T. Spirig, S. S. Weber, H. Bruggemann, R. Bosshard, C. Buchrieser, and H. Hilbi. 2007. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell. Microbiol. 9:2903-2920. [DOI] [PubMed] [Google Scholar]
- 65.Uhl, M. A., and J. F. Miller. 1996. A new type of phosphodonor in two-component signal transduction systems: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 15:1028-1036. [PMC free article] [PubMed] [Google Scholar]
- 66.Uhl, M. A., and J. F. Miller. 1994. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc. Natl. Acad. Sci. U. S. A. 91:1163-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uhl, M. A., and J. F. Miller. 1996. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J. Biol. Chem. 271:33176-33180. [DOI] [PubMed] [Google Scholar]
- 68.van den Broek, D., T. Chin-A-Woeng, G. Bloemberg, and B. Lugtenberg. 2005. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J. Bacteriol. 187:593-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 70.Vogel, J. P., C. Roy, and R. R. Isberg. 1996. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann. N. Y. Acad. Sci. 797:271-272. [DOI] [PubMed] [Google Scholar]
- 71.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiater, L. A., A. B. Sadosky, and H. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903dIIlacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 74.Williams, C. L., P. E. Boucher, S. Stibitz, and P. A. Cotter. 2005. BvgA functions as both an activator and a repressor to control Bvgi phase expression of bipA in Bordetella pertussis. Mol. Microbiol. 56:175-188. [DOI] [PubMed] [Google Scholar]
- 75.Wintermeyer, E., M. Flügel, M. Ott, M. Steinert, U. Rdest, K. Mann, and J. Hacker. 1994. Sequence determination and mutational analysis of the lly locus of Legionella pneumophila. Infect. Immun. 62:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.