Abstract
Bordetella avium causes bordetellosis in birds, a disease similar to whooping cough caused by Bordetella pertussis in children. B. avium agglutinates guinea pig erythrocytes via an unknown mechanism. Loss of hemagglutination ability results in attenuation. We report the use of transposon mutagenesis to identify two genes required for hemagglutination. The genes (hagA and hagB) were adjacent and divergently oriented and had no orthologs in the genomes of other Bordetella species. Construction of in-frame, unmarked mutations in each gene allowed examination of the role of each in conferring erythrocyte agglutination, explanted tracheal cell adherence, and turkey poult tracheal colonization. In all of the in vitro and in vivo assays, the requirement for the trans-acting products of hagA and hagB (HagA and HagB) was readily shown. Western blotting, using antibodies to purified HagA and HagB, revealed proteins of the predicted sizes of HagA and HagB in an outer membrane-enriched fraction. Antiserum to HagB, but not HagA, blocked B. avium erythrocyte agglutination and explanted turkey tracheal ring binding. Bioinformatic analysis indicated the similarity of HagA and HagB to several two-component secretory apparatuses in which one product facilitates the exposition of the other. HagB has the potential to serve as a useful immunogen to protect turkeys against colonization and subsequent disease.
Bordetella avium is the causative agent of bordetellosis, an avian upper respiratory tract disease to which commercially raised turkeys are particularly susceptible (10). As with other pathogenic species of the Bordetella genus (e.g., B. pertussis and B. bronchiseptica), B. avium binds preferentially to ciliated tracheal epithelial cells (1, 25, 34). Subsequent death of the ciliated cells is thought to contribute to the clinical signs associated with bordetellosis (e.g., coughing and oculonasal discharge [10]). In addition, infected turkeys are more susceptible to secondary infections with other pathogens such as Escherichia coli (3, 10, 26).
As with many medically important bacteria, B. avium has the ability to agglutinate erythrocytes from certain animal species (2, 23). B. avium mutants that are hemagglutination negative are attenuated in experimental infections in turkey poults and impaired in their ability to bind to explanted turkey tracheal rings in vitro (33). In B. avium, the ability to cause hemagglutination is not associated with particular attachment organelles such as pili (11). However, an exposed cell surface factor required for hemagglutination was detected by Moore and Jackwood (22), who employed monoclonal antibodies and periodate treatment to infer that the hemagglutinin is a cell surface carbohydrate (associated with a 41-kDa protein).
A large (ca. 220-kDa) secreted protein, filamentous hemagglutinin (FHA), that mediates hemagglutination is produced by the two best-described Bordetella species (B. pertussis and B. bronchiseptica) (5, 18). B. avium encodes a product very similar to FHA, but its loss (via a fhaB mutation), while dramatically attenuating, does not cause the loss of hemagglutinating ability (31). The property of hemagglutination in B. avium is thus conferred by a mechanism that is unique to, and important for, the normal pathogenesis of this Bordetella species.
In a prior study, we associated the loss of hemagglutination with attenuation in turkey poults in a large screen of B. avium transposon insertion mutants (33). In that study, the insertions associated with hemagglutination loss were not mapped. Consequently, the number of genes involved and the nature of their putative products were not uncovered. Here we report the identification of two genes whose trans-acting products are required for hemagglutination. The genes (hagA and hagB) were adjacent and divergently oriented and had no orthologs in the sequenced Bordetella genomes. Construction of in-frame, unmarked mutations in each gene allowed examination of each product's characteristics. The hagB product (HagB) was directly required for hemagglutination and explanted tracheal ring binding, since antiserum to purified HagB, but not purified HagA, blocked these activities. Bioinformatic predictions that products orthologous to HagA are often involved in proper localization of an active component were compatible with our biochemical and genetic findings.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains and plasmids employed in this study are listed in Table 1. Brain heart infusion (BHI) (Difco) was used under B. avium growth conditions previously described (33). Antibiotics were added at the concentrations reported by Spears et al. (32). All E. coli strains were grown in Luria (L) broth or agar (21) at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| 197N | Parental B. avium strain; Strs Nalr Kms Hag+ | 33 |
| 197N2 | 197N; spontaneous Strr mutant | This study |
| P206b | 197N except hagA::TnphoA206 Kmr Hag− | This studyb |
| P207a | 197N except hagA::TnphoA207 Kmr Hag− | This study |
| G145 | 197N except hagA::TnlacZ145 Kmr Hag− | This studyc |
| P225a | 197N except hagA::TnphoA225 Kmr Hag− | This study |
| P218a | 197N except hagA::TnphoA218 Kmr Hag− | This study |
| P215b | 197N except hagA::TnphoA215 Kmr Hag− | This study |
| P208a | 197N except hagB::TnphoA208 Kmr Hag− | This study |
| P212a | 197N except hagB::TnphoA212 Kmr Hag− | This study |
| P205a | 197N except hagB::TnphoA205 Kmr Hag− | This study |
| P201a | 197N except hagB::TnphoA201 Kmr Hag− | This study |
| PAS666 | 197N2 except ΔhagA Kms Hag− | This study |
| PAS667 | 197N2 except ΔhagB Kms Hag− | This study |
| S17.1 λ pir | E. coli conjugation donor; Strr Nals Kms | 9 |
| HB101 | E. coli cloning strain | Laboratory collection |
| DH5α | E. coli cloning strain | Invitrogen |
| M13/pREP4 | E. coli cloning strain containing plasmid pREP4 | Qiagen |
| Plasmids | ||
| pLAFR5 | Broad-host-range cloning vector | 16 |
| pUC19 | E. coli cloning vector | 36 |
| pCR2.1TOPO | E. coli cloning plasmid; Kmr Apr | Invitrogen |
| pQE30 | His tag cloning vector | Qiagen |
| pQE80-L | His tag cloning vector | Qiagen |
| pKAS46 | oriR6K oriT rpsL Apr Kmr | 30 |
| phagA | pLAFR5 hagA | This study |
| phagB | pLAFR5 hagB | This study |
| pΔhagA | pTOPO ΔhagA | This study |
| pΔhagB | pTOPO ΔhagB | This study |
| pKASΔhagA | pKAS46 ΔhagA Kmr | This study |
| pKASΔhagB | pKAS46 ΔhagB Kmr | This study |
| pΩhagAhis | pQE30 ΩhagAhis | This study |
| pΩhagBhis | pQE80-L ΩhagBhis | This study |
Abbreviations: Hag, phenotypic characteristic of hemagglutination; Strr, streptomycin resistant; Nalr, nalidixic acid resistant; Nals, nalidixic acid sensitive; Kms, kanamycin sensitive; Kmr, kanamycin resistant; Apr, ampicillin resistant. Sequences for each hagA and hagB mutant allele are available at GenBank, with accession numbers assigned as follows: hagA::TnphoA206, FJ001329; hagA::TnphoA207, FJ001327; hagA::TnlacZ145, FJ001328; hagA::TnphoA225, FJ001330; hagA::TnphoA218, FJ001331; hagA::TnphoA215, FJ001332; hagB::TnphoA208, FJ001333; hagB::TnphoA212, FJ001334; hagB::TnphoA205, FJ001335; hagB::TnphoA201, FJ001336; ΔhagA, GQ240879; and ΔhagB, GQ240878.
This strain has been previously reported (33), but the map location of the lesion conferring the Hag− phenotype was not known.
The strain has been previously reported (28), but the map location of the lesion conferring the Hag− phenotype was not known.
Transposon mapping.
B. avium hemagglutination-negative insertion mutants were identified in a prior study by screening insertion mutant libraries for isolates that had lost the ability to agglutinate guinea pig erythrocytes (33). To locate the transposons in hemagglutination-negative mutants, chromosomal DNA from each mutant was prepared using the Qiagen DNeasy kit and then digested with NotI, which cuts once between the phoA and the neoR genes within the mini-Tn5 lacZ and phoA transposons (6). Digested chromosomal DNA was ligated into NotI-digested pUC19 vector and introduced into E. coli HB101 cells by transformation, selecting for kanamycin-resistant colonies (32). Using the resulting clones, primers unique to the distal end of the neoR sequence of Tn5 (ACTTGTGTATAAGAGTCAG) and pUC19 forward (TTGTAAAACGACGGCCAGTGA) or reverse (CAGGAAACAGCTATGACCATG) primers were used to obtain a partial sequence of the inserted DNA and pinpoint the insertion site. The clones were sequenced at the UNC-CH Automated DNA Sequencing Facility on a model 377 DNA sequencer (Perkin-Elmer, Applied Biosystems Division) using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystems Division).
Obtaining the hagA and hagB genes.
In order to clone the hagA gene, a DNA fraction enriched for hagA was identified by Southern blot analysis of HindIII-digested chromosomal B. avium 197N DNA using a digoxigenin (DIG) PCR probe consisting of portion of the hagA gene adjacent to a Tn5 insertion. Colony blot hybridization and restriction endonuclease mapping of clones harboring HindIII-digested DNA revealed that hagA resided on a 7-kb DNA fragment (32). This DNA fragment was introduced into the B. avium replication-competent vector pLAFR5, and the construct was designated phagA. The hagB gene was obtained by amplifying a 4,508-bp chromosomal DNA fragment from strain 197N (BA3064372 to BA3068880) that was subsequently cloned into pLAFR5 with the aid of PstI restriction endonuclease sites that had been incorporated into the primers AACTGCAGGCATGCAAATTGACACCCAATTC and AACTGCAGCGAGTAACTGGGACCTGCCAT. This construct was designated phagB.
Deletion mutation construction.
To construct an in-frame hagA deletion, the full-length hagA gene (BA3062538 to BA3064682) was cloned following PCR amplification using primers AGCTTTGACGCAGATCGGAAA and GCCGTGTTTGTGATGACTACG into the pCR2.1TOPO vector (Invitrogen). Restriction enzymes BbsI and BsiWI were used to remove all but 261 and 237 nucleotides on the 3′ and 5′ ends of the gene, respectively. Recircularization of the plasmid via ligation of the blunt ends restored the proper reading frame in the truncated gene.
To construct the in-frame hagB deletion, two primer pairs were used to amplify a 304-bp fragment and a 301-bp fragment on the 5′ and 3′ ends of the gene (BA3064793 to BA3065096 for the 5′ end and BA3068538 to BA306838 for the 3′ end). The primers employed were GACTCAGCGCCATGAAACACC and GGGCCTCGTTGCGTTTTACC for the 5′ fragment and CAGCGGCCGTGATTACACC and TGCACGGTAATGATGCAACG for the 3′ fragment. The 5′ and 3′ fragments were cloned individually into the pCR2.1TOPO vector (Invitrogen) and then were subsequently fused together by isolating the fragments excised via the adjacent EcoRV and BamHI sites in the pCR2.1TOPO vector. A brief ligation of the fragments enriched for the desired deletion product because of the high reactive efficiency of the BamHI overhanging ends. The ligation mix was subjected to PCR amplification using the original external primers then the amplicons cloning into the pCR2.1TOPO vector. Selected constructs were sequenced to confirm the predicted extent of the deletion and retention of an open reading frame. DNA sequencing revealed the deletion of 1,308 nucleotides (BA3062724 to BA3064032) from hagA and 3,439 nucleotides (BA3065097 to BA3068536) from hagB. For both hagA and hagB, restriction enzymes Asp718 and NotI were used to subclone the gene fragments into the allelic exchange vector pKAS46 (30).
Allelic replacements.
The in-frame deletion constructs (ΔhagA and ΔhagB) were introduced into the chromosome of strain 197N2 via homologous recombination. The mutant alleles were introduced first into E. coli S17.1 λpir (9) by transformation and then into B. avium via conjugation. For conjugation, the E. coli donor and B. avium recipient were concentrated from overnight growth (in L broth and BHI, respectively) to give ca. 1010 CFU of each in 1.0 ml L broth, where mating took place for 2 h at 37°C. Exconjugants were selected on L agar plates containing kanamycin and nalidixic acid. Individual Nalr Kmr colonies were picked and restruck on the same medium, and isolated colonies were patched onto medium containing no selective antibiotics. Growth from such patches was struck onto L agar containing streptomycin, and individual colonies were picked and scored for hemagglutination and kanamycin sensitivity. Hemagglutination-negative (Hag−) Kms individuals were subsequently tested for the deletion by sequencing PCR amplicons generated from amplification of the region of the mutant alleles predicted to contain the unmarked deletion, using the same primers utilized in constructing the deletion plasmids. Two isolates were chosen and designated PAS666, carrying the hagA deletion (ΔhagA), and PAS667, carrying the hagB deletion (ΔhagB).
Histidine fusion constructions.
Segments of the hagA and hagB genes were introduced into the histidine fusion vectors pQE30 and pQE80-L, respectively, so as to produce products that lacked their predicted amino-terminal signal sequences and had a series of six histidine amino acids fused in the amino-terminal end. The mature (processed) HagA and HagB products were predicted from probable signal sequence length (Signal P version 2; http://www.cbs.dtu.dk/services/SignalP-2.0/). The final products (HagAHis and HagBHis) were designed to contain, in the case of HagAHis, the 26th amino acid at nucleotide position BA3064223 and end with the stop codon at position BA3062687. For HagBHis, the product was designed to begin with the 38th amino acid residue at nucleotide position BA30640904 and end with the stop codon at position BA3068858. The predicted products were not anticipated to be exported due to the elimination of the predicted signal sequence. PCR amplicons were generated using B. avium 197N chromosomal DNA (32) and DNA primers containing BamHI and PstI restriction endonuclease sites. The sites were added in order to facilitate amplicon introduction into the multiple cloning site of pQE30 and pQE80-L. The manufacturer's instructions were followed regarding the cloning of amplicons and the introduction of the resulting plasmids (pΩhagAhis and pΩhagBhis) into E. coli M15/pREP4 and DH5α, respectively (hagA primers, GCGCGGATCCCAAAGTATAGGGAATGAGATTAATCG and GCGCAAGCTTTTAGAACTGAGCTTGCAGCG; hagB primers, GCGCGGATCCCAAGTCGTGCCCACCAATG and GCGCCTGCAGTCAAGTCGGTTTTTTGGGCGTATG). E. coli transformants were selected, and representatives having plasmids with inserts of the proper size were identified as containing in-frame insertions by DNA sequencing.
Isolation of HagAHis and HagBHis.
For both fusion proteins, isolation was accomplished essentially by employing the cleared lysate protocol described in the Qiagen manual (23a). In this protocol, induction, lysis, denaturation, and Ni+2 column chromatography are described. Different techniques were used to recover HagAHis and HagBHis from column eluates. HagAHis recovery was low using filtration techniques to remove urea and salts. Consequently, dialysis against phosphate-buffered saline (PBS) was employed. In some preparations, a small amount of precipitate was seen following dialysis. The precipitate was routinely eliminated by adding SDS to a final concentration of 0.12%. In the case of HagBHis column eluates, filtration (Amicon Ultra-4 concentrators [Millipore]; 30,000-molecular-weight cutoff) produced adequate recovery levels (a precipitate was not observed). Protein concentrations were estimated following Coomassie blue staining of SDS-polyacrylamide gels (12% acrylamide) (17). The samples were diluted with PBS to achieve a protein concentration of ca. 50 μg/ml purified protein, and 0.5 ml was used for immunization.
Preparation of polyclonal antisera.
Four female New Zealand White rabbits (certified Bordetella free) were used to generate polyclonal HagAHis- and HagBHis-specific antisera (2 rabbits for each antigen) using standard techniques. Complete Freund's adjuvant was employed for the immunizing dose, with boosts in incomplete Freund's adjuvant at 4-week intervals. After 12 weeks, the rabbits were exsanguinated. Antisera collected over the 12-week period showed increasing reactivity compared to nonimmune sera (taken prior to immunization). All procedures were carried out in full compliance with federal guidelines and institutional policies.
Hemagglutination and tracheal ring binding.
Insertion libraries were screened initially for hemagglutination-negative mutants by mixing a small amount of plate-grown bacteria into ca. 20 μl of PBS containing 2 to 4% settled guinea pig erythrocytes with a sterile toothpick. Quantitative hemagglutination titers were determined in microtiter plates as previously described (33). Tracheal ring attachment assays were performed as previously described (33). Briefly, a constant number of explanted tracheal rings (typically three) from embryonic turkeys were incubated in individual wells containing tissue culture medium and various concentrations of B. avium. Comparisons of adherent bacterial numbers were made after extensive washing with PBS and detergent treatment to solubilize the epithelial cells and release the adherent bacteria. Binding efficiency (bacteria bound divided by bacteria added) was used to compare the binding abilities of the strains employed.
Western blot analysis.
Outer membrane and other cellular fractions were prepared using the method of Hellwig and Arp (8). Approximately 25 μg of protein from each fraction was separated on a 4 to 20% denaturing polyacrylamide gradient gel and a Western blot performed following the instructions of the gel manufacturer (Invitrogen), using primary antiserum raised against purified HagAHis or HagBHis. The secondary antibody was horseradish peroxidase-conjugated mouse anti-rabbit IgG (Sigma), and the substrate was diaminobenzidine. Gels were silver stained according to manufacturer's directions (GE Healthcare, kit 17-1150-01).
Infectious dose determinations.
The 50% infectious dose (ID50) measurement for each mutant was performed in turkey poults obtained from North Carolina State University Poultry Unit II as previously described (33), and the results were analyzed by the methods of Reed and Muench (24).
Statistical methods.
The standard deviation of the mean was calculated with the aid of the Microsoft Excel STDEV function. The standard error was calculated as the standard deviation divided by the square root of the number of experiments. The statistical significance of median differences between two groups was determined using the Mann-Whitney rank sum test with the aid of Minitab statistical analysis software (release 14). In all cases, a P value of <0.05 was considered significant. DNA sequences were analyzed for open reading frames using Orf Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). Amino acid sequences were used for comparison to known proteins in NCBI databases using protein BLAST (7), PsiPred (14, 20), and Conserved Domain Database (19) analyses.
Nucleotide sequence accession numbers.
Accession numbers for all insertion and deletion mutations are noted in Table 1. In some cases, positions of insertion and deletion mutations are additionally noted by their positions in the B. avium 197N genome sequence (GenBank accession number AM167904). For insertion mutations, the genomic position number is the first base in the coding region toward the 3′ end of the respective gene, beyond where the transposon was inserted. For deletion mutations, the gene sequence is shown with the deleted bases omitted and the breakpoint noted.
RESULTS
Two genes are required for B. avium hemagglutination.
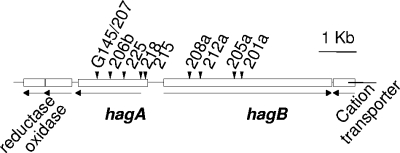
A screen of our miniTn5 insertion mutant banks for hemagglutination-negative (Hag−) mutants resulted in the isolation of 20 mutants, of which at least 10 were unique (Fig. 1). Initial DNA cloning adjacent to the transposon insertions and sequencing, combined with information derived from the complete chromosome sequence (27), revealed that the insertions resided in two separate but adjacent genes that were unique to B. avium, which we named hagA and hagB. All 20 mutants identified as Hag− had insertions that interrupted either hagA or hagB.
FIG. 1.
Organization of the hagA-hagB region. Divergent transcription is noted by arrows. Independent insertion mutations that define the genes are denoted by triangles. Open reading frames adjacent to hagA and hagB are noted by their predicted putative products. The exact locations of the individual insertions (given alphanumeric designations in the figure) are matched with the B. avium 197N genome (27) coordinates (BA) as follows: P207/G145, BA3063020; P206b, BA3063348; P225, BA3063666; P218, BA3064152; P215b, BA3064256; P208a, BA3065210;P212a, BA3065504; P205a, BA3066299; and P201a, BA3066509.
A bioinformatic analysis of the hagA and hagB genes revealed no orthologs within the genus Bordetella. However, certain similarities between the predicted hagA and fhaC product from B. avium were noted (31). FhaC is well known for its role in facilitating the export of filamentous hemagglutinin (FhaB) in B. pertussis and B. bronchiseptica (13), which suggested that HagA might serve a similar function for HagB (27). Because transposon insertions have been known to complicate an analysis of the loss of gene function, we first created in-frame, unmarked deletion mutations in hagA and hagB.
Unmarked, in-frame deletion mutations in either hagA or hagB result in a loss of hemagglutination and are complemented in trans.
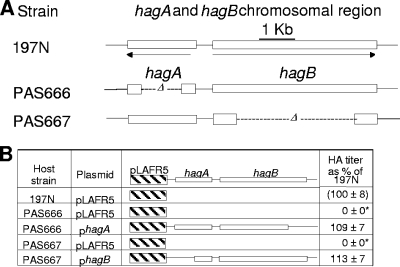
Unmarked, in-frame deletion mutations were created in hagA and hagB via allelic exchange in a streptomycin-resistant version of 197N (197N2). The resulting lesions eliminated 60% and 85% of the coding regions of hagA and hagB, respectively (Fig. 2A). As expected, hemagglutination assays revealed that the deletions created a Hag− phenotype indistinguishable from that caused by representative miniTn5 mutations in hagA and hagB (data not shown). Also, both deletion mutations could be fully complemented in trans following introduction of the broad-host-range vector pLAFR5 carrying the appropriate parental allele (Fig. 2B). We note that here, in the interest of continuity, 197N (rather than 197N2) was used as a point of positive (wild-type) comparison. The abilities of 197N and 197N2 to hemagglutinate and colonize turkey poult tracheas were indistinguishable (our unpublished observations).
FIG. 2.
(A) Locations of hagA and hagB in-frame deletion mutations in strains PAS666 and PAS667 (PAS666 is hagA and PAS667 is hagB). The deletion limits are denoted by their alphanumeric coordinates in the B. avium 197N genome sequence (27) in the text. (B) Hemagglutination complementation effected by specific plasmids encoding the parental alleles diagrammed. Hemagglutination titer is shown as a percentage of 197N after a log2 transformation of the reciprocal titers. Values that differed significantly from those for 197N are denoted by an asterisk.
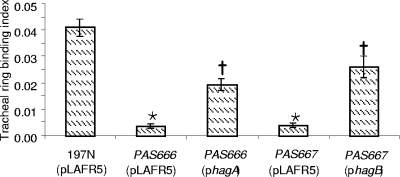
Complementation restores tracheal ring binding in hagA and hagB deletion mutants.
Strains PAS666 and PAS667 were defective in their ability to bind to explanted tracheal rings from embryonic turkeys. Complementation in trans was effected (Fig. 3) with the same plasmids used to restore the Hag+ phenotype in PAS666 and PAS667 mutants. The level of tracheal ring binding shown by the complemented mutants was clearly distinguishable from that of the uncomplemented mutants and was approximately half of that shown by strain 197N harboring the pLAFR5 vector plasmid. Why complementation was not as complete for tracheal binding as for hemagglutination (Fig. 2) is not known.
FIG. 3.
Effect of complementing plasmids on the tracheal ring binding index of the ΔhagA and ΔhagB mutant strains (PAS666 and PAS667, respectively, complemented by the plasmids bearing the denoted hagA or hagB alleles). The binding index is a measure of the number of bacteria bound to a tracheal ring normalized to the bacteria added (33). The column height indicates the average for at least seven separate tracheal rings. The error bars denote the standard errors of the means. Asterisks denote values that are distinguishable from the parental and complemented mutant values. Crosses indicate a statistical difference between the parent and mutant.
Complementation restores turkey poult colonization ability in hagA and hagB deletion mutants.
As was found in our earlier studies of selected Hag− insertion mutants (33), the hagA and hagB deletion mutants (PAS666 and PAS667, respectively) were attenuated in turkey poults. In the present study, Hag+ complemented mutants were employed and were as effective as strain 197N harboring the vector plasmid (pLAFR5) in colonization measurements (Table 2).
TABLE 2.
Virulence of B. avium Hag mutants
| Strain | Hag genotype | Hag phenotype | ID50 × 10−6a | ID50 statistically different from that of 197Nb |
|---|---|---|---|---|
| 197N | Wild type | + | 2 ± 3 | NA |
| PAS666 | ΔhagA | − | >35 | + |
| PAS667 | ΔhagB | − | >40 | + |
| 197N/pLAFR5 | Wild type/vector | + | 7 | − |
| PAS666/pLAFR5 | ΔhagA/vector | − | >40 | + |
| PAS667 /pLAFR5 | ΔhagB/vector | − | >30 | + |
| PAS666/phagA | ΔhagA/phagA | + | 8 | − |
| PAS667/phagB | ΔhagB/phagB | + | 2 ± 2 | − |
Fifty percent infectious dose (ID50) values were determined as described previously (29). The values indicate the dose needed to recover the inoculated strain from the tracheas of 50% of turkeys at 2 weeks postinoculation. The “greater than” sign (>) indicates that no isolates were recovered from colonized turkeys exhibiting the inoculated Hag phenotype at any dose given. The numeric value shown after the > indicates the lowest possible ID50 achievable (i.e., it represents the ID50 value generated if all birds were colonized at a dose 1 order of magnitude higher than the highest dose employed in the actual experiment). The standard deviation of the mean ID50 is indicated. For the parental strain, the ID50 values from seven independent and identical experiments were averaged. For PAS667/phagB, the values from two experiments were averaged.
The statistical analysis was performed using the log10 ID50 values shown in the table. +, significant difference; −, no difference (P > 0.05). The Z test was employed as described previously (29) in those cases where a single ID50 determination was performed. NA, not applicable.
Inoculated turkeys were scored as colonized if the inoculated strain was recovered from tracheal swabs after a 2-week incubation. The percentage of culture-positive birds in a group receiving a specified dose was used to determine the ID50 (27). In checking individual isolates from culture-positive birds, we did not recover any phenotypically Hag− mutants from any turkey inoculated with the uncomplemented hagA or hagB mutants. Rather, Hag+ isolates were often recovered from these birds. Such isolates still possessed the relevant hagA or hagB lesion (tested by PCR analysis). These phenotypically Hag+ isolates had evidently acquired a secondary lesion (or lesions) that conferred a stable hemagglutinating ability superficially similar to that of the parental strain. Such pseudorevertants were noted and reported in earlier work in which transposon mutants (Fig. 1) were employed (33). The genetic basis for the pseudoreversion has not been uncovered, but cryptic hagA- and hagB-like genes have been identified (27). These may undergo activation via mutation. Birds colonized by such pseudorevertants were scored as not colonized by the inoculated (Hag−) strain, in keeping with earlier practices (33).
Evidence for a direct role for HagB in hemagglutination and tracheal ring binding.
Bioinformatic data suggest that the HagA and HagB products could constitute a two-component secretion system in which one protein is necessary for the proper presentation of the other (27). This model predicts that both would be outer envelope proteins but that only one would be directly involved in erythrocyte binding. This possibility was investigated by obtaining and employing HagA- and HagB-specific antisera.
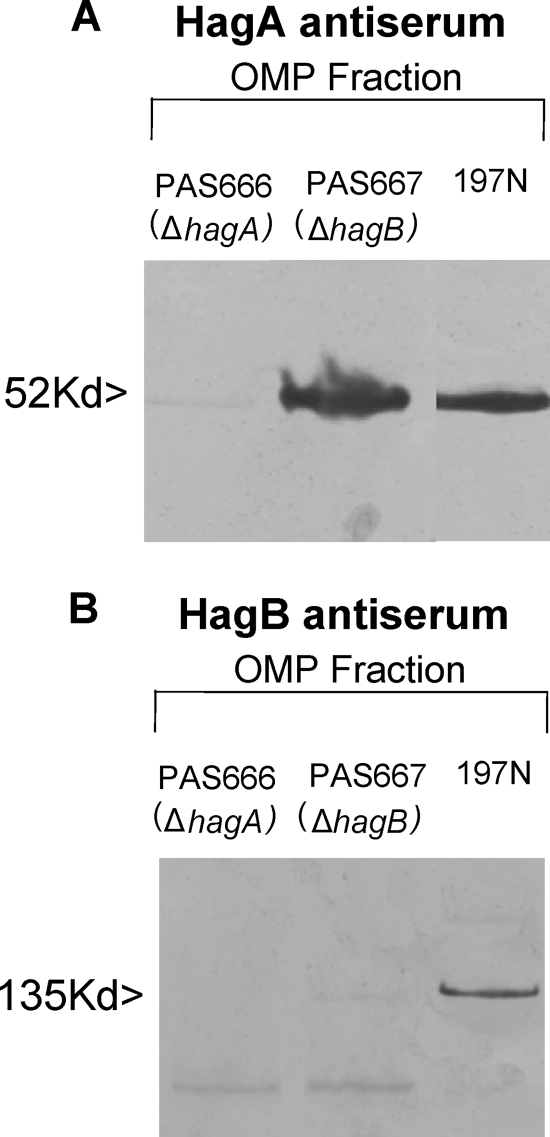
Western blot analysis of a fraction enriched in outer envelope material (OMP fraction) revealed the presence of proteins of the predicted sizes of HagA and HagB in strain 197N and the absence of these reactive products in the respective mutants (Fig. 4A and B). Additionally, silver-stained gels of preparations identical to those shown on the Western blots clearly revealed the absence of a band migrating identically to HagA in the hagA mutant. Obscuring material migrating in the HagB region precluded a comparable supporting statement for the hagB mutant (data not shown). Interestingly, the HagA product was readily detectable in the hagB mutant (Fig. 4A). However, we failed to detect the HagB product in the hagA mutant in the OMP fraction (Fig. 4B) or in other cellular fractions (data not shown).
FIG. 4.
Western blots of the outer membrane-enriched fractions (OMP) from the indicated strains using polyclonal HagA antiserum (A) or polyclonal HagB antiserum (B).
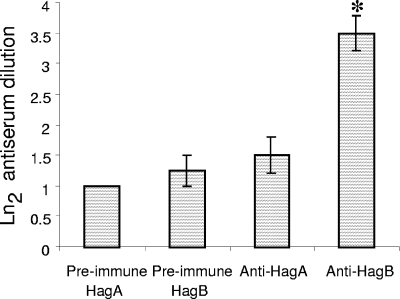
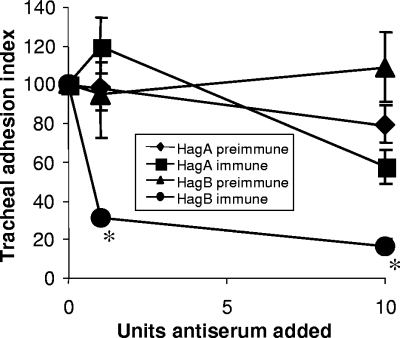
Antisera were also employed to inhibit erythrocyte agglutination. Antiserum specific to HagB, but not to HagA, was capable of significantly reducing erythrocyte agglutination by strain 197N (Fig. 5). Similarly, tracheal ring binding by strain 197N was significantly reduced by HagB, but not HagA, antiserum (Fig. 6). We inferred from these results that the HagB product was surface exposed and directly involved in the erythrocyte and tracheal ring attachment process. A direct role for HagA in attachment was not supported by the test applied.
FIG. 5.
Hemagglutination inhibition by antiserum raised against HagA or HagB. Dilutions of antisera were added to microtiter wells containing strain197N and erythrocytes added as described in the text. The last dilution capable of inhibiting agglutination was noted. Four replicates were performed. An asterisk denotes a difference in inhibitory titer from the preimmune serum.
FIG. 6.
Effect of the addition of 1 or 10 units of polyclonal HagA or HagB antiserum on the ability of strain 197N to bind to turkey embryonic tracheal rings in vitro. Tracheal binding inhibition is shown as the binding index normalized to 100% in the absence of antiserum (0 units) and binding inhibition as the percentage inhibition. The results show the average of 10 separate determinations. Values that are statistically distinguishable from that for the preimmune antiserum control are denoted by asterisks.
DISCUSSION
B. avium agglutinates guinea pig erythrocytes, and loss of this property via mutation results in attenuation (2). However, the mechanism by which hemagglutination is carried out remains ill defined. Here we show that the characteristics of hemagglutination, explanted tracheal ring binding, and turkey poult colonization are all linked in their requirement for the products of the hagA and hagB genes. Further, we present evidence for the direct involvement of HagB in erythrocyte agglutination and tracheal ring binding.
We discovered the hagA and hagB genes in the course of characterizing 20 hemagglutination-negative miniTn5 insertion mutants and report here the locations of 10 insertions that were demonstrably unique. All isolated mutants had lesions in either hagA or hagB. The hagA and hagB genes were transcriptionally divergent and had no orthologs in any other published Bordetella genomes (27).
To better characterize the HagA and HagB products, we constructed in-frame, unmarked deletions in hagA and hagB. As expected, the deletion mutants were hemagglutination negative. Additionally, complementation experiments revealed that each gene produced a unique trans-acting product required for hemagglutination, explanted tracheal ring binding, and turkey poult tracheal colonization. Part of the success of the complementations, especially those performed in vivo (in the absence of antibiotic-contributed selective pressure for plasmid maintenance), was likely due to the stability of the pLAFR5 vector, a property noted previously (32).
In order to obtain evidence for the direct involvement of one or both of the HagA and HagB products in attachment, we generated polyclonal HagA and HagB antisera. The antisera raised were each demonstrably specific for proteins of the predicted size of HagA or HagB present in our outer envelope-enriched fraction in Western blots. However, antiserum raised to HagA did not block hemagglutination or impede explanted tracheal ring attachment by strain 197N any differently than preimmune serum, even though we could additionally confirm (using immunoblots) the reactivity of the HagA antiserum with the isolated immunizing protein (our unpublished observations). In contrast to the HagA antiserum, the HagB antiserum inhibited both hemagglutination and tracheal ring binding.
We inferred from the foregoing that HagB directly binds a component or components common to the surface of guinea pig erythrocytes and explanted turkey tracheal cells. We found no support for a direct role for HagA in receptor binding. However, our results do not rule out the possibility that HagA is also directly involved, because the ability of each protein to elicit antibodies that specifically block attachment could vary significantly. This could be due to differences in sensitivity to their isolation procedures or other factors unique to each protein.
Bioinformatic analysis places HagA and HagB in a family of two-component secretory pairs (12, 15, 27). With regard to HagA, recent analysis of B. pertussis FhaC (a protein required for the export of filamentous hemagglutinin, FhaB), has reinforced predicted structural similarities between HagA and FhaC (4). Also, portions of HagA show characteristics of proteins that facilitate exposition of hemagglutinins and hemolysins from other microorganisms (4, 35). With regard to HagB, bioinformatic data indicate that strong similarities exist between portions of FhaB and other hemagglutinins from bacteria outside the bordetellae (4, 13, 35).
Our complementation experiments told us that HagB was synthesized in the hagA mutant and vice versa. However, we did not detect HagB in our hagA mutant outer envelope-enriched fraction. One way to explain this observation is that HagA was necessary for the retention or stabilization of the HagB product. If that is the case, there was no reciprocity in this relationship because HagB was not required for the stability or retention of HagA. Consistent with the bioinformatic inferences, HagA may act to facilitate the proper localization and presentation of HagB via direct interactions characteristic of two-component regulators. However, additional means will need to be employed to unequivocally place HagA and HagB in the same or different membranes and subsequently address the possibility that HagA directly interacts with HagB (e.g., coimmunoprecipitation). Also, tracheal and erythrocyte competitive binding studies with purified HagA and HagB could help reinforce the present conclusions regarding the different roles of the proteins determined here with antisera.
We note that the results presented here (and additional unpublished observations) did not lend support to the earlier findings of Moore and Jackwood (22), who proposed that hemagglutination in B. avium is associated with a 41-kDa protein, as well as a periodate-sensitive component. However, our results do not rule out the possibility that there are additional factors required for hemagglutination that were not detected by the mutagenic methods we employed to discover HagA and HagB.
In B. avium, hemagglutination is essential for turkey poult colonization (33). In no other member of the bordetellae is the loss of hemagglutinating ability so profoundly attenuating. The HagB hemagglutinin may prove to be a beneficial adjunct in vaccine formulations aimed at preventing tracheal colonization. Targeting this molecule could reduce the level of subclinical carriage and reduce disease incidence in commercial turkey operations. In addition, the surface location of HagB may make this product a good candidate to expose foreign antigens as gene fusions. This measure could facilitate the use of B. avium as a live oral poultry vaccine platform to protect poultry against other infectious agents.
Acknowledgments
We thank Denarra Nevels and Kelly Prescott for technical assistance.
This work was supported by USDA grants 950-934, 99-35204-7734, and 02-35204-12236 to P.E.O., by College of Liberal Arts, Drew University, funding to L.M.T., and by NIH grant R15A157382-01 to L.M.T. and D.M.M.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Arp, L. H., and N. F. Cheville. 1984. Tracheal lesions in young turkeys infected with Bordetella avium. Am. J. Vet. Res. 45:2196-2200. [PubMed] [Google Scholar]
- 2.Arp, L. H., R. D. Leyh, and R. W. Griffith. 1988. Adherence of Bordetella avium to tracheal mucosa of turkeys: correlation with hemagglutination. Am. J. Vet. Res. 49:693-696. [PubMed] [Google Scholar]
- 3.Barnes, H. J., and M. S. Hofstad. 1978. Factors involved in respiratory disease of turkeys in Iowa. J. Am. Vet. Med. Assoc. 173:889-897. [Google Scholar]
- 4.Clantin, B., A. S. Delattre, P. Rucktooa, N. Saint, A. C. Meli, C. Locht, F. Jacob-Dubuisson, and V. Villeret. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317:957-961. [DOI] [PubMed] [Google Scholar]
- 5.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gish, W., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]
- 8.Hellwig, D. H., and L. H. Arp. 1990. Identification of Bordetella avium antigens recognized after experimental inoculation in turkeys. Am. J. Vet. Res. 51:1188-1191. [PubMed] [Google Scholar]
- 9.Hoffmann, A., T. Thimm, M. Droge, E. R. B. Moore, J. C. Munch, and C. C. Tebbe. 1998. Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola). Appl. Environ. Microbiol. 64:2652-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackwood, M. W., and Y. M. Saif. 2003. Bordetellosis, p. 705-718. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougal, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames, IA.
- 11.Jackwood, M. W., and Y. M. Saif. 1987. Pili of Bordetella avium: expression, characterization, and role in in vitro adherence. Avian Dis. 31:277-286. [PubMed] [Google Scholar]
- 12.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 13.Jacob-Dubuisson, F., B. Kehoe, E. Willery, N. Reveneau, C. Locht, and D. A. Relman. 2000. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology 146:1211-1221. [DOI] [PubMed] [Google Scholar]
- 14.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 15.Kajava, A. V., N. Cheng, R. Cleaver, M. Kessel, M. N. Simon, E. Willery, F. Jacob-Dubuisson, C. Locht, and A. C. Steven. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279-292. [DOI] [PubMed] [Google Scholar]
- 16.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35:D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. F. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 22.Moore, K. M., and M. W. Jackwood. 1994. Production of monoclonal antibodies to the Bordetella avium 41-kilodalton surface protein and characterization of the hemagglutinin. Avian Dis. 38:218-224. [PubMed] [Google Scholar]
- 23.Moore, K. M., M. W. Jackwood, T. P. Brown, and D. W. Dreesen. 1994. Bordetella avium hemagglutination and motility mutants: isolation, characterization, and pathogenicity. Avian Dis. 38:50-58. [PubMed] [Google Scholar]
- 23a.Qiagen, Inc. 2003. The QIAexpressionist handbook, 5th ed. Qiagen, Inc., Valencia, CA.
- 24.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:293-299. [Google Scholar]
- 25.Rhea, L. J. 1915. The comparative pathology of the tracheal and bronchial lesions produced in man by B. pertussis (whooping cough) and those produced in dogs by B. bronchiseptica (canine distemper). J. Med. Res. 32:471-474. [PMC free article] [PubMed] [Google Scholar]
- 26.Saif, Y. M., P. D. Moorhead, R. N. Dearth, and D. J. Jackwood. 1980. Observations on Alcaligenes faecalis infection in turkeys. Avian Dis. 24:665-684. [PubMed] [Google Scholar]
- 27.Sebaihia, M., A. Preston, D. J. Maskell, H. Kuzmiak, T. D. Connell, N. D. King, P. E. Orndorff, D. M. Miyamoto, N. R. Thomson, D. Harris, A. Goble, A. Lord, L. Murphy, M. A. Quail, S. Rutter, R. Squares, S. Squares, J. Woodward, J. Parkhill, and L. M. Temple. 2006. Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J. Bacteriol. 188:6002-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelton, C. B., D. R. Crosslin, J. L. Casey, S. Ng, L. M. Temple, and P. E. Orndorff. 2000. Discovery, purification, and characterization of a temperate transducing bacteriophage for Bordetella avium. J. Bacteriol. 182:6130-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shelton, C. B., L. M. Temple, and P. E. Orndorff. 2002. Use of bacteriophage Ba1 to identify properties associated with Bordetella avium virulence. Infect. Immun. 70:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 31.Spears, P. A., L. M. Temple, D. M. Miyamoto, D. J. Maskell, and P. E. Orndorff. 2003. Unexpected similarities between Bordetella avium and other pathogenic bordetellae. Infect. Immun. 71:2591-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spears, P. A., L. M. Temple, and P. E. Orndorff. 2000. A role for lipopolysaccharide in turkey tracheal colonization by Bordetella avium as demonstrated in vivo and in vitro. Mol. Microbiol. 36:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Temple, L. M., A. A. Weiss, K. E. Walker, H. J. Barnes, V. L. Christensen, D. M. Miyamoto, C. B. Shelton, and P. E. Orndorff. 1998. Bordetella avium virulence measured in vivo and in vitro. Infect. Immun. 66:5244-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuomanen, E. I., and J. O. Hendley. 1983. Adherence of Bordetella pertussis to human respiratory epithelial cells. J. Infect. Dis. 148:125-130. [DOI] [PubMed] [Google Scholar]
- 35.Willems, R. J., C. Geuijen, H. G. van der Heide, G. Renauld, P. Bertin, W. M. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]