Abstract
Chlamydia trachomatis strains are obligate intracellular human pathogens that share near genomic synteny but have distinct infection and disease organotropisms. The genetic basis for differences in the pathogen-host relationship among chlamydial strains is linked to a variable region of chlamydial genomes, termed the plasticity zone (PZ). Two groups of PZ-encoded proteins, the membrane attack complex/perforin (MACPF) domain protein (CT153) and members of the phospholipase D-like (PLD) family, are related to proteins that modify membranes and lipids, but the functions of CT153 and the PZ PLDs (pzPLDs) are unknown. Here, we show that full-length CT153 (p91) was present in the elementary bodies (EBs) of 15 C. trachomatis reference strains. CT153 underwent a rapid infection-dependent proteolytic cleavage into polypeptides of 57 and 41 kDa that was independent of de novo chlamydial protein synthesis. Following productive infection, p91 was expressed during the mid-developmental cycle and was similarly processed into p57 and p41 fragments. Infected-cell fractionation studies showed that insoluble fractions contained p91, p57, and p41, whereas only p91 was found in the soluble fraction, indicating that unprocessed CT153 may be secreted. Finally, CT153 localized to a distinct population of reticulate bodies, some of which were in contact with the inclusion membrane.
Chlamydia trachomatis is a Gram-negative obligate intracellular pathogen that is the cause of trachoma and sexually transmitted infections in humans. Chlamydiae have a unique biphasic developmental cycle in which the infectious, metabolically inactive elementary body (EB) invades host cells and remains restricted within a membrane-bound vacuole termed an inclusion. EBs transform into metabolically active, noninfectious reticulate bodies (RBs) that grow and replicate by binary fission. RBs undergo a secondary differentiation into EBs, which are released from the host cell to initiate another round of infection (35).
C. trachomatis depends upon the eukaryotic host for a number of metabolic intermediates and captures host-derived lipids via multiple routes during the intracellular phase of its developmental cycle (24, 59). Chlamydia induces Golgi compartment fragmentation (25) and intercepts Golgi compartment-derived exocytic vesicles (11, 21), whereby the pathogen acquires cholesterol, sphingomyelin, and possibly other nutrients. Chlamydial inclusions sequester lipid droplet (LD) organelles from host cells (14) and fuse with multivesicular bodies (MVBs), which serve as a primary source for sphingomyelin and lysobisphosphatidic acid (3, 46). The inclusion membrane protein IncA (14), chlamydial Lda proteins (32), and the phospholipase D (PLD) paralogs (37) have all been implicated in lipid acquisition, but the specific mechanisms by which chlamydiae target and assimilate lipids remain largely unknown.
The genomes of many chlamydial strains are fully sequenced (1, 13, 29, 44, 45, 50, 52, 55, 56). Comparative genomic analyses have strongly influenced our understanding of the molecular basis of the pathogenic variability in the Chlamydiaceae and have provided important insights into common and species-variable virulence factors. C. trachomatis and Chlamydia muridarum, human and murine pathogens, respectively, have more than 99%-orthologous gene contents (44). C. trachomatis serovariants that cause trachoma or sexually transmitted diseases have more than 99.5% overall nucleic acid sequence identity (13, 30). Thus, Chlamydia contains a highly conserved core genome that mediates genus-common functions such as metabolism and cell division. Major genomic differences fall into two highly variable gene families: ompA, which encodes the major outer membrane protein (MOMP), and polymorphic membrane protein genes (pmp's), which encode type V autotransporters (13, 29). A third region of genomic variability is near the replication terminus and is termed the plasticity zone (PZ) (44). Genes in the PZ encode strain- and species-variable alleles that have been implicated in in vivo pathogenic diversity by mechanisms involving immune avoidance (10, 34, 39).
Genetic diversity in the PZ reflects an array of chlamydial virulence factors that have evolved to counteract or evade species-specific immune effectors in chlamydial host organisms. For example, PZ genes encoding the tryptophan biosynthesis operon and chlamydial cytotoxins correlate with in vivo infection tropisms and immune evasion strategies (6, 10, 12, 34, 38, 39). The expanded family of genes that encode PZ phospholipase Ds (pzPLDs), which are putative lipid-modifying enzymes, may play an important role in chlamydial survival late in the developmental cycle (37). pzPLDs contain an HKD motif (42, 52) similar to those seen in lipid-hydrolyzing enzymes and have been suggested to function in chlamydial lipid modification or metabolism (37). Supporting this hypothesis, CT156/Lda1, a pzPLD, was recently shown to localize to cytosolic-neutral, lipid-rich structures adjacent to the inclusion membrane (32).
CT153 gene orthologs are conserved in all sequenced C. trachomatis genomes and are located immediately upstream from the pzPLD genes, suggesting that the proteins have concomitant functions (13, 41, 44, 52, 55). C-terminal amino acid residues 427 to 621 of CT153 share homology with the membrane attack complex/perforin (MACPF) domain (41). The MACPF domain of human perforin and complement 9 contains membrane-spanning regions that map to two amphipathic α-helices that form a helix-loop-helix functional motif (40). The crystal structures of prokaryotic and eukaryotic MACPF domains demonstrated that the MACPF domain is similar to the pore-forming, cholesterol-dependent cytolysins (CDC) of Gram-positive bacteria (22, 47). A characteristic shared by members of the MACPF/CDC superfamily is the ability of secreted monomers to convert into membrane-inserted oligomers that form lytic pores in membranes or function in nonlytic membrane interactions (48). MACPF domain-containing proteins from Toxoplasma and Plasmodium form pores to facilitate parasite egress from cells (28) and disrupt membranes to allow the parasite to transverse host cells (26, 27), respectively. Listeria monocytogenes listeriolysin O, a secreted cytolysin, and listerial phospholipases mediate pathogen escape from the nascent membrane-bound vacuole and disrupt host membranes during intercellular dissemination (15, 51). Thus, both the presence of an MACPF domain in CT153 and its linkage with the putative lipid-modifying pzPLD proteins suggest the hypothesis that CT153 interacts with host cell membranes (44). There are limited reports on the biological characterization of CT153. Transcriptome analysis of the C. trachomatis developmental cycle showed CT153 to be a gene expressed during the mid-developmental cycle (7), an expression pattern that correlates with RB replication.
Here, we provide the first biological characterization of CT153. We show that CT153 is expressed by all C. trachomatis serovars, undergoes proteolytic processing, and is associated with both the insoluble and soluble fractions of chlamydia-infected cells. CT153 localized to a distinct population of RBs located at the inclusion membrane interface and to atypical large RBs within the inclusion lumen. Lastly, we show that CT153 expression is correlated with the ability of chlamydiae to accumulate intrainclusion LDs in a strain-specific manner.
MATERIALS AND METHODS
Chlamydial strains and propagation.
C. trachomatis serovars A/Har-13, B/TW-5/OT, Ba/Ap-2, C/TW-3/OT, D/UW-3/Cx, E/Bour, F/IC-Cal-3, G/UW-524/Cx, H/UW-4/Cx, I/UW-12/Ur, J/UW-36/Cx, K/UW-31/Cx, L1/LGV-440, L2/LGV-434, and L3/LGV-404, C. muridarum strain Weiss (MoPn), and Chlamydia caviae strain guinea pig inclusion conjunctivitis (GPIC) were propagated and purified from HeLa 229 cells as previously described (8). Total protein concentrations of the purified EBs were measured by bicinchoninic acid assay (Thermo Fisher Scientific, Rockford, IL).
Cloning, expression, and purification of CT153.
The CT153 allele from C. trachomatis serovar D genomic DNA was amplified by PCR using primers CT153A (5′-CACCATGACTAAGCCTTCTTTCTTATACG-3′) and CT153B (5′-ATAACCTGAAGATTTTTTAATAATAAAGATAGC-3′). The resulting PCR product was cloned into pENTR/SD/D-TOPO (Invitrogen, Carlsbad, CA) and was subcloned into pEXP2, a prokaryotic expression vector containing a C-terminal six-histidine tag and a 14-amino-acid V5 epitope tag for purification and detection (Invitrogen, Carlsbad, CA). The final vector was confirmed by restriction digest and by DNA sequencing. BL21(DE3) Escherichia coli cells (Invitrogen, Carlsbad, CA) were transformed and induced with isopropyl-β-d-thiogalactopyranoside during mid-log-phase growth. Cultures were harvested by centrifugation 12 h after protein expression was induced. Cell pellets were suspended in phosphate-buffered saline (PBS), pH 7.2, extracted by sonication, and centrifuged to obtain insoluble and soluble fractions. Recombinant CT153 (rCT153) was batch purified from the soluble fractions by immobilized metal affinity chromatography (IMAC) using cobalt resin according to the manufacturer's protocol for nondenaturing purification (BD Biosciences, San Jose, CA) and was eluted from the column using a step gradient of increasing imidazole concentrations.
Generation of anti-CT153 antibodies.
Pooled elution fractions from IMAC analyses were concentrated, separated by SDS-PAGE, and stained with GelCode blue (Thermo Fisher Scientific, Rockford, IL). The predominant eluted polypeptides were excised, digested with trypsin, and subjected to matrix-assisted laser desorption ionization-time of flight (MALDI) mass spectrometry (MS) analysis to confirm their identities (Scripps Center for Mass Spectrometry, La Jolla, CA). The Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases approved all animal experiments. Six 8- to 12-week-old female BALB/c mice were subcutaneously immunized with 2 μg of purified rCT153 emulsified in 0.2 ml of Ribi adjuvant system (Corixa, Hamilton, MT) as previously described (37). Sera were collected, antibody (Ab) specificity was confirmed by Western blotting of purified EBs, and titers were determined by enzyme-linked immunosorbent assay using rCT153.
SDS-PAGE and Western blotting.
Equivalent amounts of protein from density gradient (DG)-purified EBs from 15 C. trachomatis reference serovars, C. muridarum, and C. caviae were boiled for 10 min in Laemmli buffer and were separated on 10% or 4 to 15% Criterion precast gels (Bio-Rad, Hercules, CA). Proteins were electrophoretically transferred to 0.2-μm nitrocellulose membranes in sodium phosphate buffer at 100 V for 35 min. Membranes were blocked at room temperature (RT) for 2 h in PBS containing 3% bovine serum albumin (BSA), 0.05% Tween 20, and 0.02% NaN3 and then incubated with designated primary Abs at RT overnight. Blots were washed with PBS containing 3% BSA and 0.05% Tween 20 and then incubated at RT for 2 h with goat anti-mouse horseradish peroxidase-conjugated secondary Ab (MP Biomedicals, Solon, OH). The blots were washed with PBS containing 0.05% Tween 20, rinsed with PBS, and then developed with 4-chloro-1-naphthol and H2O2.
Temporal kinetics of CT153 processing and expression in infected cells.
HeLa 229 cells were grown in 6-well plates and infected with C. trachomatis serovar D or mock infected in duplicate using a multiplicity of infection (MOI) of 1 or 1,000 in the presence or absence of 1 μg/ml rifampin and 5 μg/ml chloramphenicol. Monolayers were washed with Hanks balanced salt solution and suspended in 300 μl of Laemmli buffer warmed to 56°C at the times postinfection designated in Fig. 2 and 3. Samples were subjected to SDS-PAGE and Western blotting using anti-CT153, anti-MOMP (B-B5b), or anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, MA).
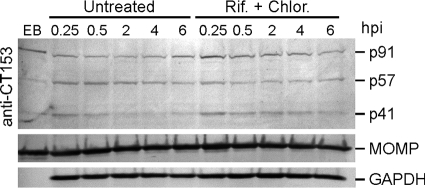
FIG. 2.
Processing of CT153 is independent of de novo chlamydial protein synthesis. HeLa 229 cells were infected with C. trachomatis at an MOI of 1,000 in the absence or presence of rifampin (Rif.) and chloramphenicol (Chlor.) and harvested at the hour postinfection (hpi) indicated. Samples were analyzed by SDS-PAGE and Western blotting using anti-CT153, -MOMP, or -GAPDH Abs. p91 was the predominant polypeptide in EBs; reactivity against p57 and p41 was observed but at a lesser intensity (EB lane). Following infection of untreated and antibiotic-treated cells, the p57- and p41 polypeptides were observed over the entire 6-h incubation period. Note that the p41 signal diminishes with time in the untreated but not the antibiotic-treated cells.
FIG. 3.
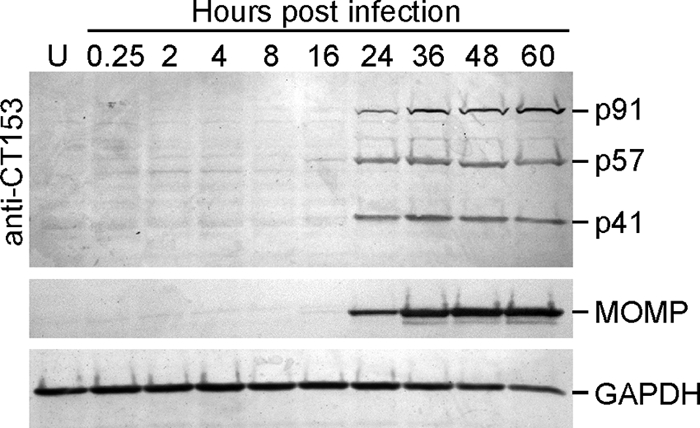
CT153 is expressed during the mid-developmental cycle and is processed to 57-kDa and 41-kDa polypeptides. C. trachomatis-infected HeLa 229 cells were harvested at the times indicated, and cell lysates were separated by SDS-PAGE, blotted to nitrocellulose, and probed with Abs against CT153, MOMP, or GAPDH. Full-length CT153 was detected during the middle to late development cycle, and immunoreactive polypeptides of 57 kDa and 41 kDa in equimolar ratios indicated processed forms of CT153. U, uninfected.
Preparation of cellular fractions.
C. trachomatis serovar L2-infected L929 suspension cultures were harvested by centrifugation at 0, 12, 24, 30, and 36 h postinfection and lysed by Dounce homogenization, and cellular fractionation was performed as previously described (9, 18, 53). Insoluble and soluble fractions were analyzed by Western blotting using anti-CT153, anti-MOMP, or anti-chlamydial protease-like activity factor (CPAF) (16) Abs.
Confocal microscopy.
HeLa 229 cells were grown on coverslips, infected with C. trachomatis serovar D at an MOI of 0.5, fixed at different times postinfection in 2% paraformaldehyde (PFA), and permeabilized with 0.2% saponin. Indirect immunofluorescence was performed using anti-CT153 mouse Abs and anti-EB rabbit serum, followed by Alexa Fluor-conjugated secondary Abs (Molecular Probes, Eugene, OR). Coverslips were washed, stained with DRAQ5 (Vinci-Biochem, Vinci, Italy), and mounted in Mowiol (Calbiochem, La Jolla, CA). Images were collected using a Yokogawa spinning disk head mounted on a Nikon Eclipse TE2000-S microscope with a 60× and 1.4-numerical-aperture (NA) oil immersion objective (Nikon, Tokyo, Japan). Fluorescence was detected using a Photometrics Cascade II:512 camera (Princeton Instruments, Trenton, NJ). Images were processed using Adobe Photoshop.
Transmission electron microscopy.
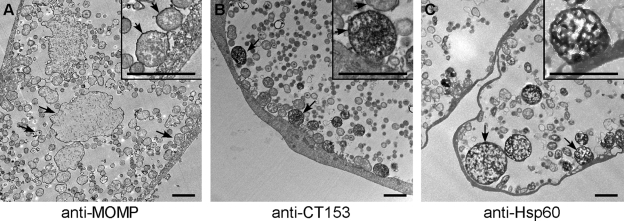
C. trachomatis serovar D-infected HeLa 229 cells were fixed in 2% PFA, labeled with anti-CT153 Abs and peroxidase-conjugated secondary Ab, and prepared for transmission electron microscopy (TEM) as previously described (7). Anti-MOMP and anti-Hsp60 were used as controls for chlamydial outer membrane and cytosolic staining patterns, respectively. Sections were examined at 80 kV on a Hitachi H75000 transmission electron microscope. Images were captured using an Advantage high-resolution charge-coupled-device camera.
MVB inhibition assays and LD staining.
HeLa 229 cells were grown on coverslips and infected with C. trachomatis or C. caviae, and 1, 5, or 10 mM 3-methyladenidne (3-MeA) was added to the culture medium at 1 h postinfection, as previously described (3). The effect of the MBV inhibitor was determined by titrating recoverable inclusion-forming units (IFU) on monolayers of HeLa 229 cells. C. trachomatis- or C. caviae-infected HeLa cells were fixed in 3% PFA plus 0.025% glutaraldehyde at 36 or 28 h postinfection, respectively. Monolayers were washed, stained with DRAQ5 and BODIPY 495/503 (Molecular Probes, Eugene, OR) as previously described (32), and analyzed by confocal microscopy.
RESULTS
CT153 is expressed by all C. trachomatis serovars.
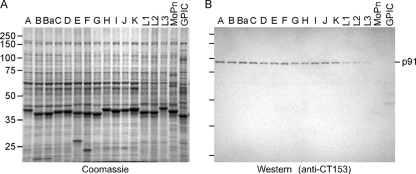
Highly conserved CT153 gene orthologs are present in the genomes of all sequenced C. trachomatis strains, including serovars A, D, and L2 (13, 52, 55); however, it is unknown if CT153 gene orthologs are present in the unsequenced human serovars. We analyzed EBs from chlamydial reference strains by SDS-PAGE (Fig. 1A) and by Western blotting with anti-CT153 Abs (Fig. 1B) to determine if CT153 was broadly conserved among C. trachomatis serovars. A protein corresponding to the predicted mass of CT153 (91 kDa) that was unique to C. trachomatis serovars was not discernible following Coomassie brilliant blue (CBB) staining of EB lysates (Fig. 1A). The inability to identify p91 with CBB indicated that it was not a highly abundant protein. Anti-CT153 Abs reacted with a single polypeptide of 91 kDa (p91) in lysates of EBs from all 15 human C. trachomatis reference serovars, as determined by Western blotting (Fig. 1B). The amount of CT153, as judged by the intensities of the Western blot bands, was greater in lysates from trachoma (Fig. 1B, lanes A to K) than in lymphogranuloma venereum (Fig. 1B, lanes L1 to L3) biovar EBs. Anti-CT153 Abs failed to recognize the C. muridarum CT153 ortholog, TC0431, although these proteins share 70% amino acid identity. This could indicate that TC0431 does not localize to the EB or that the immunodominant portion(s) of CT153 and TC0431 resides in regions of amino acid diversity. Anti-CT153 Abs were nonreactive with EBs from the C. caviae strain, which is consistent with the absence of a CT153 gene ortholog in this strain (45).
FIG. 1.
CT153 is conserved among C. trachomatis serovars. Equal amounts of protein from DG-purified EBs from C. trachomatis serovars (lanes A to L3), C. muridarum (lane MoPn), and C. caviae (lane GPIC) were separated by SDS-PAGE, blotted to nitrocellulose, and probed with anti-CT153 Abs. (A) CT153 was not identifiable following CBB staining of electrophoresed polypeptides, indicating that it is not an abundant protein component of the organism. (B) Western blotting showed that anti-CT153 Abs reacted with a single 91-kDa polypeptide common to all C. trachomatis serovars (lanes A to L3). Anti-CT153 failed to detect the CT153 ortholog TC0431 in C. muridarum (lane MoPn) and did not react with C. caviae (lane GPIC).
Processing of CT153 is independent of de novo chlamydial protein synthesis.
The association of p91 with EBs prompted us to examine CT153 in the context of early interactions of EBs with host cells. HeLa cell monolayers were infected with C. trachomatis at an MOI of 1,000 in the absence or presence of prokaryotic RNA and protein synthesis inhibitors and were analyzed by Western blotting (Fig. 2). Full-length p91 was the predominant immunoreactive protein in purified EBs. Following infection of untreated and treated cells, anti-CT153 Abs exhibited reactivity against p91 and polypeptides of 57 kDa and 41 kDa. The combined mass of these lower-molecular-weight polypeptides was similar to that of full-length CT153, indicating that p91 was proteolytically processed into p57 and p41. Cleavage of p91 occurred as early as 15 min postinfection. The p57 and p41 proteins were observed in antibiotic-treated cultures throughout the 6-h incubation period. These results demonstrated that the proteolytic processing of p91 occurred in the absence of de novo chlamydial protein synthesis. Unlike with p91and p57, the intensity of the p41 signal diminished with time in the untreated infected cells, indicating that the instability of p41 required de novo chlamydial protein synthesis.
CT153 is expressed during the mid-developmental cycle and is processed to 57-kDa and 41-kDa polypeptides.
Temporal Western blot analysis of C. trachomatis-infected cells was performed to examine the kinetics of de novo CT153 expression and proteolytic processing (Fig. 3). Anti-MOMP and anti-GAPDH Abs were used as chlamydial and cellular controls, respectively. MOMP expression was first detected by the mid-developmental cycle (24 h); GAPDH levels remained constant at all time points examined. Anti-CT153 Abs detected p91 by 24 h postinfection, and the amount of protein progressively increased until the end of the developmental cycle (Fig. 3). This is consistent with results of a previous transcriptome analysis of the C. trachomatis developmental cycle that shows that de novo expression of the CT153 gene mRNA initiates 8 h postinfection, peaks by 24 h postinfection, and remains elevated throughout the remainder of the cycle (7). The anti-CT153 Abs were immunoreactive with p57 and p41 at similar intensities at the same time points, indicating that the processed forms of CT153 were stable during the middle to late developmental cycle.
CT153 is found in the insoluble and soluble fractions of infected cells.
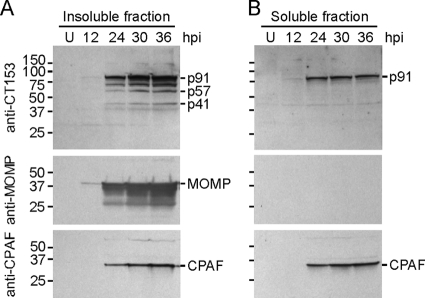
Many MACPF/CDC superfamily proteins are secreted into infected host cells or into the extracellular environment (48, 57). Insoluble and soluble fractions of C. trachomatis-infected cells were analyzed by Western blotting using anti-CT153 Abs to examine the solubility of CT153 (Fig. 4). Anti-MOMP and -CPAF Abs were used as positive controls for insoluble and soluble chlamydial proteins, respectively. Both of these proteins were first detected during the middle to late developmental cycle. MOMP was present only in the insoluble fractions, and CPAF was detected in both the insoluble and soluble fractions (Fig. 4), as previously described (53). Consistent with the kinetic study, anti-CT153 Abs recognized full-length p91, p57, and p41 by 24 h postinfection in the insoluble fractions (Fig. 4A). These results support the conclusion that insoluble forms of CT153 were associated with chlamydial organisms or with membranes. Full-length p91 was detected at the same time points as CPAF in the soluble fractions (Fig. 4B). There are probable explanations for these results: (i) fragile RBs (54) lysed during the homogenization process and released p91 into the soluble fraction or (ii) CT153 was secreted from organisms into the inclusion lumen or host cytosol, similarly to other soluble chlamydial proteins (31, 53, 60).
FIG. 4.
CT153 distributes into insoluble and soluble fractions of homogenized infected cells. Western blots of insoluble (A) and soluble (B) fractions of C. trachomatis-infected L929 cells at various hours postinfection were probed with anti-CT153 Abs. (A) Insoluble fractions contained p91, p57, and p41. The CT153 Ab reacted with an ∼70-kDa polypeptide that may represent an intermediate processed form. (B) Only p91 was detected in soluble fractions. The lower-molecular-weight band in the soluble fractions was nonspecific, as it was also detected in uninfected cells. Monoclonal Abs against MOMP and CPAF were used for positive controls for insoluble and soluble proteins, respectively.
Confocal microscopy of CT153 expression in chlamydia-infected cells.
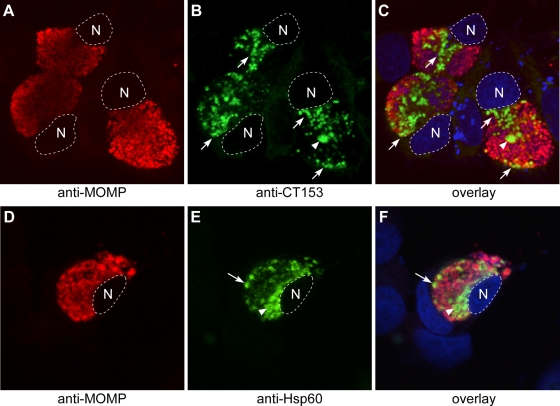
C. trachomatis-infected cells were immunostained with anti-CT153 Abs to localize the protein. Infected cells were also stained with anti-MOMP and anti-Hsp60 monoclonal Abs as positive controls for outer membrane and cytoplasmic staining, respectively. Characteristically, anti-MOMP uniformly and specifically reacted with the outer membranes of RB and EB developmental forms, and immunofluorescence was distributed throughout the contents of mature inclusions (Fig. 5A, C, D, and F). In contrast, organisms stained with anti-CT153 showed a nonuniform staining pattern that labeled only what appeared to be RB developmental forms (Fig. 5B and C, arrows) and a minor population of very large particles (Fig. 5B and C, arrowheads). Organisms stained with anti-CT153 were commonly observed along the inner leaflet of the inclusion membrane and tended to polarize next to nuclear regions within inclusions (Fig. 5B and C, arrows). Organisms stained with anti-CT153 did not exhibit discernible outer membrane staining at this level of resolution but localized to the chlamydial cytoplasm. Despite a rigorous examination of infected cells, we were not able to detect CT153 in either the host cell cytosol or cytoplasmic membrane. Hsp60 staining (Fig. 5E and F) was similar to CT153 staining (Fig. 5B and C) and to that previously reported for pzPLD CT155 (37).
FIG. 5.
Confocal microscopy of CT153 expression in chlamydia-infected cells. C. trachomatis-infected HeLa cells were fixed late in the developmental cycle and were immunolabeled with anti-MOMP and anti-CT153 (A to C) or anti-Hsp60 (D to F) Abs. (A to C) CT153 was distinct from MOMP and localized to RBs lining the inclusion membrane and to regions of the inclusion near the nucleus (arrows) and to large RB-like structures that may contact the inclusion membrane at points beyond the confocal plane (arrowheads). (D to F) Like CT153, Hsp60 labeled RBs adjacent to the inclusion membrane (arrows) and larger RBs (arrowheads). CT153 and Hsp60 staining was not a fixation artifact, because identical staining patterns were observed in methanol-fixed cells (data not shown), and this pattern of staining has been independently observed for Hsp60 and CT155 by other investigators (2, 4, 25, 37). The nucleus is outlined by a white dotted line and is stained blue in the overlay. N; host cell nucleus. Single representative confocal sections are shown.
CT153 localizes to distinct RB developmental forms.
Immunoelectron microscopy was performed on infected cells to more precisely define the developmental forms reactive with anti-CT153 (Fig. 6). Anti-MOMP Abs reacted strongly with the outer membranes but not with the cytosols of all chlamydial developmental particles, including the very large morphologically aberrant forms within the lumen of the inclusion (Fig. 6A). In contrast, and consistent with our confocal findings, CT153 Abs labeled a distinct population of RBs that were predominantly positioned adjacent to the inner leaflet of the inclusion membrane (Fig. 6B, arrows) and to the less abundant population of large atypical RB particles described above (data not shown). CT153 staining of the RB outer membrane was suggested (Fig. 6B, inset) but was not clearly discernible due to the intense cytoplasmic staining. Anti-Hsp60 staining was similar to anti-CT153 staining in being restricted to a subpopulation of RBs with strong cytoplasmic localization (Fig. 6C). Likewise, it was difficult to discern specific anti-Hsp60 outer membrane staining due to its cytoplasmic labeling. The differential staining of RBs and large aberrant RB forms with CT153 and Hsp60 was most pronounced in mature, late-developmental-cycle inclusions (30 to 48 h).
FIG. 6.
Immunoelectron microscopy localizes CT153 to distinct RB developmental forms. C. trachomatis-infected HeLa cells were fixed late in the developmental cycle, immunolabeled with anti-MOMP (A), anti-CT153 (B), or anti-Hsp60 (C), and prepared for TEM. (A) MOMP localized to the outer membranes of EBs, RBs, and large RBs (arrows) that were observed in mature inclusions. The arrows in the inset point to the dense immunoreactivity of the outer membrane. (B) CT153 stains RBs that are located primarily at the RB-inclusion membrane interface (arrows). The arrows in the inset show that the intensity of cell wall staining is greater in organisms that exhibit cytosolic CT153 labeling than in organisms that do not express CT153 in the cytosol. (C) Hsp60 stained similar RB developmental forms. Scale bars, 2 μm.
Effect of MVB inhibitors on chlamydial growth and LD staining.
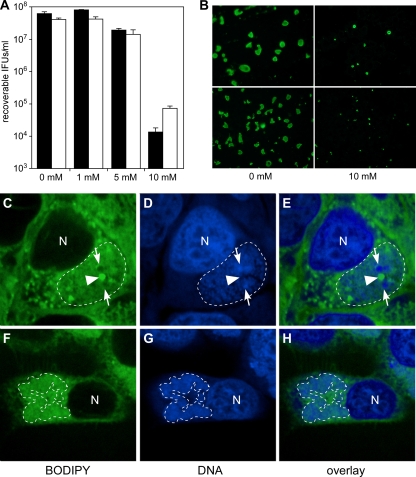
The fusion of MVBs with chlamydial inclusions is necessary for optimal C. trachomatis growth (3). The effect of MVB inhibition on chlamydial development was tested with C. trachomatis and C. caviae strains that possess and lack CT153, respectively (Fig. 7). There was a 3-log reduction in the number of C. trachomatis and C. caviae IFU recoverable in the presence of 10 mM 3-MeA (Fig. 7A), which was accompanied by a marked reduction in inclusion size (Fig. 7B). The inhibitory effect of 3-MeA on C. trachomatis is consistent with the work of Beatty (3). Our results show that C. caviae growth was also inhibited by 3-MeA, indicating that this strain requires normal MVB maturation for survival. These data suggest that interactions with MVBs may be important for the development of all chlamydial species. Thus, there was not a correlation between strain sensitivity to 3-MeA growth inhibition and retention of CT153. In contrast, we observed differences between the strains when inclusions were analyzed for LD distribution. LD staining in C. trachomatis (CT153-positive)- and C. caviae (CT153-negative)-infected cells was distinct (Fig. 7C to H). In C. trachomatis-infected cells, LDs were redistributed in the host cytosol and were detected inside inclusions (Fig. 7C to E), an observation consistent with previous reports (14, 32). There was a definite association between large chlamydial developmental forms and intrainclusion sequestered LDs. In contrast, neutral lipid staining inside C. caviae inclusions was weak and dispersed, and no prominent droplet-like structures were detected (Fig. 7F to H). Thus, the occurrence of intrainclusion LDs correlates with the expression of CT153. These findings suggest that the degrees of chlamydial interaction with LDs vary among chlamydial species.
FIG. 7.
Effect of MVB inhibitors on chlamydial growth and LD staining. C. trachomatis- or C. caviae-infected HeLa cells were treated with 3-MeA and assayed for growth. (A) The recovery of infectious C. trachomatis and C. caviae organisms from infected cells treated with 3-MeA was decreased 3 logs. Black bars, C. trachomatis; white bars, C. caviae. (B) Immunofluorescence microscopy of infected cells using MOMP-specific Abs showed that the inclusion size of C. trachomatis (top panels) and C. caviae (bottom panels) was reduced in the presence of an MVB inhibitor. C. trachomatis (C to E)- and C. caviae (F to H)-infected HeLa cells were labeled with BODIPY 495/503 to compare neutral lipid distributions. (C to E) C. trachomatis inclusions contained large intrainclusion LDs (arrowheads) adjacent to large RB-like structures (arrows). (F to H) Neutral lipid staining was weak and dispersed in C. caviae inclusions. The white dotted lines represent the boundaries of inclusions. DNA is labeled blue. N, host cell nucleus. Single representative confocal sections are shown.
DISCUSSION
Species-variable alleles in the PZs of chlamydiae play important roles in chlamydial disease diversity and immune evasion (6, 10, 12, 34, 38, 39). Here, we provide the first biological characterization of the PZ MACPF domain family protein CT153. We show that CT153 is present in EBs of all 15 human C. trachomatis reference serovars as a full-length p91 polypeptide, indicating a species-common function. The p91 polypeptide was rapidly cleaved (15 min) into immunoreactive p57 and p41 fragments, suggesting a possible role for CT153 in early EB host cell interactions. It is unknown if proteolytic processing is required for CT153 function, as some MACPF/CDC proteins require cleavage for activation (58) while others do not (49). The early infection-dependent processing of CT153 occurs independently of chlamydial de novo protein synthesis, implicating either host-mediated proteolytic cleavage or autoproteolysis. The p91 and p57 polypeptides are present in both untreated and antibiotic-treated infected cells for up to 6 h postinfection, and the p41 fragment is less immunoreactive at these time points. The significance of these observations is unclear, but they suggest distinct functions for full-length p91 and its processed peptide fragments. Collectively, our findings implicate CT153 as a potential virulence factor that could function in early events of C. trachomatis infection, possibly by interacting with host cell membranes. Anti-CT153 Abs failed to react with EBs by immunodot blotting and were nonneutralizing, suggesting that p91 is not a surface-exposed antigen (data not shown). However, these findings do not exclude a possible secretory mechanism for mediating p91-host cell interactions.
CT153 was expressed during the mid-developmental cycle, and levels of p91 increased later during the growth cycle, a time when RBs actively differentiate into EBs. This finding is consistent with our observation that full-length CT153 is the predominate polypeptide present in purified EBs. CT153 exhibited proteolytic processing that resulted in p57 and p41 fragments similar to those detected following early infection. Members of the MACPF/CDC superfamily are secreted proteins that transition from soluble monomers to oligomeric membrane-spanning pores (48, 57). In addition to playing roles in parasite (26) and protein translocation (33) across membranes, the proteins exhibit cytolytic activity in vitro (19, 23). Recombinant p91 was not active in red blood cell hemolytic assays (data not shown). The hemolytic activity of CDCs can be species specific (36) and sensitive to the cholesterol content of the target membrane (20); therefore, these results are inconclusive, as our assays may have been limited by suboptimal conditions or by the use of recombinant p91. Western blotting of cellular fractions from C. trachomatis-infected cells indicated that p57 and p41 localized to insoluble membrane fractions. Interestingly, p91 was detected in both the insoluble and soluble fractions of infected cells. The solubility characteristics of p91 are similar to those of CPAF, a well-studied chlamydial protease that is secreted into the host cell cytosol by an unknown mechanism (60). One interpretation of our results is that full-length p91 is similarly secreted and undergoes processing after secretion or once inserted into a target membrane. Further biochemical and membrane fractionation studies are needed to test this hypothesis and better define CT153 interactions with host cell membranes.
CT153 was expressed in a distinct population of RBs, some of which were closely positioned next to the inclusion membrane. Confocal microscopy and TEM showed that CT153 expression in RBs was indistinguishable from that of Hsp60 (Fig. 5 and 6). Hsp60 is a chaperonin (17) that exhibits increased expression when chlamydiae respond to physiological stress (5, 43). Thus, RBs expressing CT153 may have specialized metabolic activity, supporting a hypothesis that there may be unique developmental forms with important functions in chlamydial intracellular growth. The similarities in Ab staining patterns between CT153 and the pzPLD CT155 (37) are striking and suggest that these proteins are functionally related. We also observed that C. trachomatis RBs expressing CT153 were closely associated with intrainclusion LDs. Metabolically active RBs in close proximity with the inclusion membrane could be poised for physical interactions with the host cell environment, including responding to signals in the eukaryotic cytosol, facilitating nutrient uptake, or secreting proteins. Each of these activities could be mediated by a chlamydial membrane-spanning pore protein. Thus, these proteins might participate in a lipid acquisition or modification pathway unique to C. trachomatis. This hypothesis is supported by the correlation between intrainclusion LD and a C. trachomatis strain that expresses both CT153 and the pzPLDs. Unfortunately, we were unable to localize CT153 within the host cytosol, in host membranes, or in chlamydial membranes by immunofluorescent confocal microscopy or immunoelectron microscopy. These findings do not rule out a pore-forming function for CT153 in chlamydia-host cell interactions, as these assays may be too insensitive to detect the small quantities of protein that possibly associate with membranes in a specialized and restricted manner.
In summary, we show that full-length CT153 (p91) is present in EBs of all human-pathogenic C. trachomatis strains and is proteolytically processed immediately following infection of host cells. Our findings show that CT153 is expressed by RBs closely associated with the inclusion membrane and with intrainclusion LDs and exists in both insoluble and soluble forms. The function of CT153 is unknown, but the gene encoding it is conserved in human strains, implicating CT153 as a potentially important protein in the pathogenesis of human infection and disease. We propose that CT153 is activated by proteolytic processing and, in conjunction with the pzPLDs, plays an important role in the acquisition or modification of host-derived lipids. Interestingly, reactivation of C. trachomatis from a gamma interferon-induced persistent state is blocked by primary alcohols, which are potent inhibitors of PLDs, implicating a role for the pzPLDs in this process (37). In this context, CT153 expression of persistently infected cells is worthy of further study, as a recent report shows that recovery from persistence requires incorporation of host lipids into chlamydial membranes (46). In future studies, we hope to address the function of CT153 during normal and persistent infection.
Acknowledgments
We thank Sunia Trauger at the Scripps Center for Mass Spectrometry at the Scripps Research Institute for MALDI-TOF MS analysis, Guangming Zhong at the University of Texas Health Sciences Center at San Antonio for the kind gift of anti-CPAF antibodies, Debbie Crane and John Carlson for technical assistance, and Kelly Matteson and Anita Mora for editorial and graphical assistance.
This research was supported by the Intramural Research Program of the NIH, NIAID.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 29 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Azuma, Y., H. Hirakawa, A. Yamashita, Y. Cai, M. A. Rahman, H. Suzuki, S. Mitaku, H. Toh, S. Goto, T. Murakami, K. Sugi, H. Hayashi, H. Fukushi, M. Hattori, S. Kuhara, and M. Shirai. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 3.Beatty, W. L. 2006. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119:350-359. [DOI] [PubMed] [Google Scholar]
- 4.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect. Immun. 62:4059-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. U. S. A. 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U. S. A. 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell, H. D., C. C. Kuo, and G. E. Kenny. 1975. Antigenic analysis of chlamydiae by two-dimensional immunoelectrophoresis. I. Antigenic heterogeneity between C. trachomatis and C. psittaci. J. Immunol. 115:963-968. [PubMed] [Google Scholar]
- 10.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Invest. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carabeo, R. A., D. J. Mead, and T. Hackstadt. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U. S. A. 100:6771-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson, J. H., S. Hughes, D. Hogan, G. Cieplak, D. E. Sturdevant, G. McClarty, H. D. Caldwell, and R. J. Belland. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72:7063-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson, J. H., S. F. Porcella, G. McClarty, and H. D. Caldwell. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect. Immun. 73:6407-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchiaro, J. L., Y. Kumar, E. R. Fischer, T. Hackstadt, and R. H. Valdivia. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U. S. A. 105:9379-9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cossart, P., M. F. Vicente, J. Mengaud, F. Baquero, J. C. Perez-Diaz, and P. Berche. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong, F., J. Sharma, Y. Xiao, Y. Zhong, and G. Zhong. 2004. Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect. Immun. 72:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis, R. J., and S. M. van der Vies. 1991. Molecular chaperones. Annu. Rev. Biochem. 60:321-347. [DOI] [PubMed] [Google Scholar]
- 18.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1987. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giddings, K. S., A. E. Johnson, and R. K. Tweten. 2003. Redefining cholesterol's role in the mechanism of the cholesterol-dependent cytolysins. Proc. Natl. Acad. Sci. U. S. A. 100:11315-11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 22.Hadders, M. A., D. X. Beringer, and P. Gros. 2007. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science 317:1552-1554. [DOI] [PubMed] [Google Scholar]
- 23.Hadding, U., and H. J. Muller-Eberhard. 1969. The ninth component of human complement: isolation, description and mode of action. Immunology 16:719-735. [PMC free article] [PubMed] [Google Scholar]
- 24.Hatch, G. M., and G. McClarty. 1998. Phospholipid composition of purified Chlamydia trachomatis mimics that of the eucaryotic host cell. Infect. Immun. 66:3727-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuer, D., A. R. Lipinski, N. Machuy, A. Karlas, A. Wehrens, F. Siedler, V. Brinkmann, and T. F. Meyer. 2009. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature 457:731-735. [DOI] [PubMed] [Google Scholar]
- 26.Ishino, T., Y. Chinzei, and M. Yuda. 2005. A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell. Microbiol. 7:199-208. [DOI] [PubMed] [Google Scholar]
- 27.Kadota, K., T. Ishino, T. Matsuyama, Y. Chinzei, and M. Yuda. 2004. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc. Natl. Acad. Sci. U. S. A. 101:16310-16315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kafsack, B. F., J. D. Pena, I. Coppens, S. Ravindran, J. C. Boothroyd, and V. B. Carruthers. 2009. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science 323:530-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 30.Kari, L., W. M. Whitmire, J. H. Carlson, D. D. Crane, N. Reveneau, D. E. Nelson, D. C. Mabey, R. L. Bailey, M. J. Holland, G. McClarty, and H. D. Caldwell. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J. Infect. Dis. 197:449-456. [DOI] [PubMed] [Google Scholar]
- 31.Kleba, B., and R. S. Stephens. 2008. Chlamydial effector proteins localized to the host cell cytoplasmic compartment. Infect. Immun. 76:4842-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, Y., J. Cocchiaro, and R. H. Valdivia. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646-1651. [DOI] [PubMed] [Google Scholar]
- 33.Madden, J. C., N. Ruiz, and M. Caparon. 2001. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in Gram-positive bacteria. Cell 104:143-152. [DOI] [PubMed] [Google Scholar]
- 34.McClarty, G., H. D. Caldwell, and D. E. Nelson. 2007. Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10:47-51. [DOI] [PubMed] [Google Scholar]
- 35.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagamune, H., C. Ohnishi, A. Katsuura, K. Fushitani, R. A. Whiley, A. Tsuji, and Y. Matsuda. 1996. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun. 64:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, D. E., D. D. Crane, L. D. Taylor, D. W. Dorward, M. M. Goheen, and H. D. Caldwell. 2006. Inhibition of chlamydiae by primary alcohols correlates with the strain-specific complement of plasticity zone phospholipase D genes. Infect. Immun. 74:73-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, D. E., L. D. Taylor, J. G. Shannon, W. M. Whitmire, D. D. Crane, G. McClarty, H. Su, L. Kari, and H. D. Caldwell. 2007. Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cell. Microbiol. 9:2289-2298. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, D. E., D. P. Virok, H. Wood, C. Roshick, R. M. Johnson, W. M. Whitmire, D. D. Crane, O. Steele-Mortimer, L. Kari, G. McClarty, and H. D. Caldwell. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peitsch, M. C., P. Amiguet, R. Guy, J. Brunner, J. V. Maizel, Jr., and J. Tschopp. 1990. Localization and molecular modelling of the membrane-inserted domain of the ninth component of human complement and perforin. Mol. Immunol. 27:589-602. [DOI] [PubMed] [Google Scholar]
- 41.Ponting, C. P. 1999. Chlamydial homologues of the MACPF (MAC/perforin) domain. Curr. Biol. 9:R911-R913. [DOI] [PubMed] [Google Scholar]
- 42.Ponting, C. P., and I. D. Kerr. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raulston, J. E. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson, D. K., L. Gu, R. K. Rowe, and W. L. Beatty. 2009. Inclusion biogenesis and reactivation of persistent Chlamydia trachomatis requires host cell sphingolipid biosynthesis. PLoS Pathog. 5:e1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosado, C. J., A. M. Buckle, R. H. Law, R. E. Butcher, W. T. Kan, C. H. Bird, K. Ung, K. A. Browne, K. Baran, T. A. Bashtannyk-Puhalovich, N. G. Faux, W. Wong, C. J. Porter, R. N. Pike, A. M. Ellisdon, M. C. Pearce, S. P. Bottomley, J. Emsley, A. I. Smith, J. Rossjohn, E. L. Hartland, I. Voskoboinik, J. A. Trapani, P. I. Bird, M. A. Dunstone, and J. C. Whisstock. 2007. A common fold mediates vertebrate defense and bacterial attack. Science 317:1548-1551. [DOI] [PubMed] [Google Scholar]
- 48.Rosado, C. J., S. Kondos, T. E. Bull, M. J. Kuiper, R. H. Law, A. M. Buckle, I. Voskoboinik, P. I. Bird, J. A. Trapani, J. C. Whisstock, and M. A. Dunstone. 2008. The MACPF/CDC family of pore-forming toxins. Cell. Microbiol. 10:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakurai, N., J. Kaneko, Y. Kamio, and T. Tomita. 2004. Cloning, expression, and pore-forming properties of mature and precursor forms of pleurotolysin, a sphingomyelin-specific two-component cytolysin from the edible mushroom Pleurotus ostreatus. Biochim. Biophys. Acta 1679:65-73. [DOI] [PubMed] [Google Scholar]
- 50.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 53.Swanson, K. A., L. D. Taylor, S. D. Frank, G. L. Sturdevant, E. R. Fischer, J. H. Carlson, W. M. Whitmire, and H. D. Caldwell. 2009. Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect. Immun. 77:508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura, A., A. Matsumoto, G. P. Manire, and N. Higashi. 1971. Electron microscopic observations on the structure of the envelopes of mature elementary bodies and developmental reticulate forms of Chlamydia psittaci. J. Bacteriol. 105:355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson, N. R., M. T. Holden, C. Carder, N. Lennard, S. J. Lockey, P. Marsh, P. Skipp, C. D. O'Connor, I. Goodhead, H. Norbertzcak, B. Harris, D. Ormond, R. Rance, M. A. Quail, J. Parkhill, R. S. Stephens, and I. N. Clarke. 2008. Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeno-Tarraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tweten, R. K. 2005. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 73:6199-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uellner, R., M. J. Zvelebil, J. Hopkins, J. Jones, L. K. MacDougall, B. P. Morgan, E. Podack, M. D. Waterfield, and G. M. Griffiths. 1997. Perforin is activated by a proteolytic cleavage during biosynthesis which reveals a phospholipid-binding C2 domain. EMBO J. 16:7287-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wylie, J. L., G. M. Hatch, and G. McClarty. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179:7233-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]