Abstract
Swarming motility by the urinary tract pathogen Proteus mirabilis has been a long-studied but little understood phenomenon. On agar, a P. mirabilis colony grows outward in a bull's-eye pattern formed by consecutive waves of rapid swarming followed by consolidation into shorter cells. To examine differential gene expression in these growth phases, a microarray was constructed based on the completed genome sequence and annotation. RNA was extracted from broth-cultured, swarming, and consolidation-phase cells to assess transcription during each of these growth states. A total of 587 genes were differentially expressed in broth-cultured cells versus swarming cells, and 527 genes were differentially expressed in broth-cultured cells versus consolidation-phase cells (consolidate). Flagellar genes were highly upregulated in both swarming cells and consolidation-phase cells. Fimbriae were downregulated in swarming cells, while genes involved in cell division and anaerobic growth were upregulated in broth-cultured cells. Direct comparison of swarming cells to consolidation-phase cells found that 541 genes were upregulated in consolidate, but only nine genes were upregulated in swarm cells. Genes involved in flagellar biosynthesis, oligopeptide transport, amino acid import and metabolism, cell division, and phage were upregulated in consolidate. Mutation of dppA, oppB, and cysJ, upregulated during consolidation compared to during swarming, revealed that although these genes play a minor role in swarming, dppA and cysJ are required during ascending urinary tract infection. Swarming on agar to which chloramphenicol had been added suggested that protein synthesis is not required for swarming. These data suggest that the consolidation phase is a state in which P. mirabilis prepares for the next wave of swarming.
Proteus mirabilis, a member of the Enterobacteriaceae and an opportunistic pathogen, is especially problematic as a urinary tract pathogen in catheterized patients, those with spinal cord injury, or those with anatomical abnormality of the urinary tract (reviewed in reference 16). This urease-positive bacterium causes an increase in urinary pH and the production of kidney and bladder stones (25, 39). In addition, urinary catheters become encrusted and even blocked during P. mirabilis infections (45). As our population continues to age, resulting in larger numbers of catheterized patients in hospitals and nursing homes, this organism will likely be of special concern as an agent of nosocomial infections. Particularly worrisome is the more frequent appearance of multidrug-resistant strains of P. mirabilis (17, 47).
P. mirabilis was first characterized in 1885 by G. Hauser for its ability to swarm over agar surfaces, resulting in a characteristic bull's-eye pattern (reviewed in reference 65). During the swarm process, P. mirabilis differentiates into very long (>50 μm), multinucleate, highly motile hyperflagellated cells (65). At intervals, swarm cells slow down or cease movement and dedifferentiate into shorter rod-shaped cells in what is known as the consolidation phase. Repeated cycles of swarming and consolidation lead to the bull's-eye pattern. Reflecting this phenotype, P. mirabilis was named for the Greek god Proteus, who was able to change form at will to avoid questioning (65). Flagellum-mediated swarming, accompanied by cell elongation, has been observed in other bacterial species, including Escherichia coli, Salmonella enterica serovar Typhimurium, Serratia marcescens, Vibrio parahaemolyticus, and Pseudomonas aeruginosa (reviewed in references 22, 28, and 61). However, it should be noted that swarming by these species requires a much lower agar concentration and lacks the characteristic cyclic pattern of swarming and consolidation. Thus, swarming by these other species is analogous but not identical to the robust swarming activity of P. mirabilis.
Numerous studies have been conducted in an effort to understand how and why P. mirabilis swarms, yet despite some advances, much remains unknown. However, it has been definitively demonstrated that flagella (2, 8) and chemotaxis (12) are necessary for swarming. Polysaccharides (including lipopolysaccharides [LPS]) (10, 26), extracellular matrix components (57), and fatty acids (40) may also play roles in helping P. mirabilis to move across solid surfaces. Putrescine (59) and glutamine (3) represent signals involved in the initiation of swarming. Cell density may also play a role in the transition between swarming and consolidation phases (53) and in the initiation of swarming (11); however, a luxS mutant of P. mirabilis swarms normally (56). Mutagenesis and library screens have identified several regulators of swarming motility, including rsbA (11), ccmA (31), umoA, umoB, umoC, umoD (20), wosA (30), disA (58), and the Lon protease (14). Multiple virulence factors are also upregulated during swarming, including hemolysin, urease, and the Zap metalloprotease (4, 62).
In the clinical setting, swarming may present an issue for patients with indwelling urinary catheters. P. mirabilis swarms across the surface of both latex and silicone urinary catheters (35, 54), thus gaining access to the bladder. Rapid colonization of catheter surfaces, coupled with catheter encrustation resulting from urease production, makes P. mirabilis a particularly troublesome pathogen in the hospital setting.
The recent sequencing and annotation of the P. mirabilis HI4320 genome (52) allowed for the construction of a P. mirabilis microarray. In this report, the first use of microarrays to study P. mirabilis gene expression, we investigated the transcriptome of this organism during its swarming and consolidation phases and contrasted gene expression during these phases with expression during logarithmic-phase broth culture. Both swarming and consolidation phases were monitored in real time. Only the edges of colonies in swarming or consolidation phases, confirmed by both Gram staining and microscopic observation of motility in intact colonies, were collected so that only the youngest cells of the colony were analyzed. Selected genes identified by microarray analysis were mutated and assessed for their effect on swarm behavior and virulence in a mouse model of ascending urinary tract infection. The necessity of active protein synthesis during swarming was also investigated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. mirabilis HI4320 was isolated from the urine of an elderly, long-term catheterized woman (45). E. coli DH5α was used as the host strain for cloning of mutagenesis constructs. All strains and mutants were cultured at 37°C with aeration in nonswarm LB broth (1% tryptone, 0.5% yeast extract, 0.05% NaCl) or on LB medium solidified with 1.5% agar. Media were supplemented with chloramphenicol (20 μg/ml), ampicillin (50 μg/ml), or kanamycin (25 μg/ml) as necessary.
Swarm agar and collection of swarming P. mirabilis bacteria.
Swarming motility was assessed by spotting 5 μl late-logarithmic-phase bacterial culture (optical density at 600 nm [OD600] of 1.0) onto the center of an LB swarm plate (1% tryptone, 0.5% yeast extract, 1% NaCl, 1.5% agar) followed by overnight incubation at 30°C. P. mirabilis was assessed for swarm or consolidation behavior by morphology of the colony edge, motility (using an inverted microscope), and by Gram staining. In three representative experiments, the lengths of bacteria from swarm and consolidation-phase samples were measured. In all three experiments, more than 99% of the consolidation-phase cells (consolidate) were less than 10 μm in length, and 88 to 97% of the swarming-phase cells were greater than 10 μm in length. Bacteria from the outermost edge of a swarming or consolidating colony were scraped with a plastic loop into 1.5 ml RNA Protect (Qiagen); the colony perimeter from four 7.5-cm diameter plates was scraped for each RNA sample. P. mirabilis was also cultured to mid-logarithmic phase (OD600 = 0.8) in LB broth at 30°C; 1 ml of culture was added to 2 ml RNA Protect. RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions, except that 3 mg/ml lysozyme was added to cells for 15 min. DNA was digested using Turbo DNAfree DNase (Ambion).
P. mirabilis microarray.
A preliminary annotation of the incomplete HI4320 genome located 3,722 potential open reading frames (ORFs) in the chromosome. This annotation was used to design unique 70-mer oligonucleotide probes (Operon Technologies, Inc.) corresponding to each ORF. The completed genome has 3,693 annotated ORFs; thus, the microarray contains probes for some regions that are now annotated as pseudogenes or were incorporated into neighboring ORFs. Our analysis accounts for these minor modifications. This microarray lacks probes corresponding to the genes in the 36-kb plasmid that is present in P. mirabilis HI4320. Oligonucleotide probes were spotted in triplicate onto UltraGaps II glass slides (Corning) by Microarrays, Inc. RNA (4 μg), obtained from broth-cultured, swarm, or consolidation-phase P. mirabilis, was used as a template for cDNA synthesis and was labeled with either cyanine 3 or cyanine 5 dye (GE Healthcare) according to standard operating procedure (SOP) no. M007 from The Institute for Genomic Research (TIGR) (ftp://ftp.jcvi.org/pub/data/PFGRC/pdf_files/protocols/M007.pdf). Labeled cDNA was mixed and hybridized to an array slide by using TIGR SOP no. M008 (ftp://ftp.jcvi.org/pub/data/PFGRC/pdf_files/protocols/M008.pdf). The microarray was scanned using a ScanArray Express microarray scanner (Perkin Elmer) at 10-μm resolution and quantified using the ScanArray Express software. Five independent arrays were each analyzed for broth-cultured versus consolidate or swarm bacteria. Three independent arrays were analyzed for the direct comparison of consolidate to swarm bacteria.
Microarray analysis.
The total normalization algorithm (ScanArray Express; Perkin Elmer) was used to normalize microarray data. Median spot values for each triplicate probe were extracted using MIDAS (TIGR). For the arrays comparing broth-cultured P. mirabilis to swarm or consolidate, a gene was considered differentially regulated if it was greater than 2-fold changed in four or more of the arrays and with the median being greater than a 2-fold change. For the arrays directly comparing consolidate to swarm, genes were considered differentially regulated if they were up- or downregulated >2-fold in two of the three datasets. The complete microarray data sets can be accessed at GEO database number GSE17957.
qRT-PCR.
RNA from broth-cultured, swarming, or consolidating P. mirabilis bacteria was converted into cDNA by using the Superscript first-strand synthesis system (Invitrogen) according to the manufacturer's protocol. PCR with primers specific to the rpoA (RNA polymerase A) gene was performed on cDNA samples prepared with and without reverse transcriptase to confirm lack of genomic DNA contamination of the RNA preparations. Each quantitative reverse transcriptase PCR (qRT-PCR) was set up in duplicate and consisted of 30 ng cDNA template, 150 nM each primer, and 12.5 μl 2× SYBR green PCR master mix (Stratagene). Target genes were amplified using an Mx3000P thermal cycler (Stratagene). Melting curve analysis was used to confirm a lack of primer dimers or genomic DNA contamination of reagents. Data were normalized to rpoA, and analyzed by the threshold cycle (2−ΔΔCT) method (42). Four independently isolated cDNA samples were analyzed. Primer sequences are listed in Table S1 in the supplemental material.
Mutant construction.
A kanamycin resistance gene was inserted into dppA (PMI2847), oppB (PMI1474), cysJ (PMI2250), lrhA (PMI0629), and hexA (PMI1764) by using the TargeTron gene knockout system (Sigma) with a modification previously described for P. mirabilis (51). Briefly, a group II intron was reprogrammed according to the manufacturer's protocol to specifically insert into the target gene by mutagenic PCR. The retargeted intron was ligated into plasmid pACD4K-C or pACD4K-CloxP to create plasmids pMP206, pMP207, pMP208, pMP196, and pMP197 for dppA, oppB, cysJ, lrhA, and hexA, respectively. To construct the lrhA hexA double mutant, the lrhA mutant, HI4320 lrhA Ωkan, was electroporated with pQL123 (41), which encodes an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible cre recombinase. IPTG induction led to the removal of the kanamycin resistance cassette from the intron in lrhA, creating the mutant HI4320 lrhAΔkan. This mutant was then electroporated with pMP197 to the construct HI4320 lrhAΔkan hexAΩkan. For all mutants, growth in LB was measured using a Bioscreen C growth curve analyzer (Growth Curves Ltd.). Mutants were screened for swarming ability on LB swarm agar or on minimal A agar (9) containing glucose as a carbon source and supplemented with 1% tryptone (MinA-T). Swarm radii were measured 16.0 h after inoculation in five independent experiments. Statistical significance was evaluated using the two-tailed paired t test.
Mouse model of ascending UTI.
A modification (34) of the CBA/J mouse model of ascending urinary tract infection (UTI) has been described previously (27, 38). Briefly, overnight cultures of wild-type P. mirabilis HI4230 and the isogenic mutant strain (dppA, oppB, cysJ, lrhA, or hexA) were individually adjusted to an estimated density of 2 × 108 CFU/ml (OD600 = 0.2). Cultures of mutants were each mixed in a 1:1 ratio with the wild-type parent strain. Six- to 8-week-old female CBA/J mice were inoculated transurethrally with a 50-μl suspension of the mixture (i.e., 1 × 107 CFU) via a sterile polyethylene catheter, using an infusion pump (Harvard Apparatus). At 7 days postinoculation, the bladders, kidneys, and spleens of these mice were homogenized in 3 ml phosphate-buffered saline (PBS) by using an OMNI mechanical homogenizer (OMNI International) and plated onto LB agar or LB agar supplemented with kanamycin by using a spiral plater (Autoplate 4000; Spiral Biotech) to assess bacterial burden; urine was diluted in PBS and also plated. Colony counts were enumerated using a Qcount (Spiral Biotech). Wild-type HI4320 infection was determined by subtracting the number of colonies on the kanamycin plate from the number of colonies on the plain LB plate. Statistical significance was assessed using Wilcoxon's matched-pairs test. The University of Michigan University Committee on Use and Care of Animals approved all mouse protocols.
Swarming on agar containing antibiotic discs.
Chloramphenicol or kanamycin was added to 6-mm-diameter sterile blank paper discs (Becton, Dickinson and Company) in specified amounts and allowed to dry. Swarm plates were inoculated with P. mirabilis HI4320 as described above, and the antibiotic discs were placed approximately 1 cm from the inoculation point. As a control, 50 μl of P. mirabilis HI4320 at an OD600 of 1.0 was spread plated onto nonswarm LB agar (0.5 g/liter NaCl) to create a lawn. Antibiotic discs were placed on the plate to determine zones of inhibition. Plates were incubated at 30°C overnight.
RESULTS
P. mirabilis swarm and consolidate.
When cultured on swarm agar, P. mirabilis alternates with regular periodicity between an elongated, hyperflagellated “swarmer” form and a shorter “consolidation” form, resulting in a characteristic bull's-eye pattern (Fig. 1A). The swarm and consolidation phases can be distinguished as they are forming at the colony edge; consolidating colonies have a ragged edge (Fig. 1B) and have little visible motion when examined microscopically (×50), while actively swarming colonies have a smooth edge (Fig. 1C) and are in vigorous motion (see Fig. S1 in the supplemental material). Gram stains of consolidate or swarm from the colony edge show the distinct cell morphologies corresponding to these well-defined phases (Fig. 1D and E). Using a cell length of greater than 10 μm to define swarm cells (14, 33, 44), more than 99% (298/300) of the bacteria shown in Fig. 1D were in the consolidation phase, while 88% (120/137) of the cells shown in Fig. 1E had the swarmer morphology.
FIG. 1.
(A) P. mirabilis swarm plate, with sample collection times indicated for microarray analysis. The edge of a newly forming consolidation ring forms finger-like projections (B) while the edge of an active swarm front is smooth (C). When bacteria are visualized by Gram staining from the edge of a consolidating colony (D), vegetative cells are found, while bacteria isolated from an active swarm edge display the elongated swarm morphology (E). In panel D, more than 99% of the bacteria are less than 10 μm in length, while in panel E, 88% of the bacteria are greater than 10 μm in length. The photographs in panels B and C were taken at ×50 magnification, and the photographs in panels D and E were taken at ×400 magnification. The size bar in panels D and E indicates 50 μm.
Microarray analysis of broth-cultured P. mirabilis and swarm or consolidate.
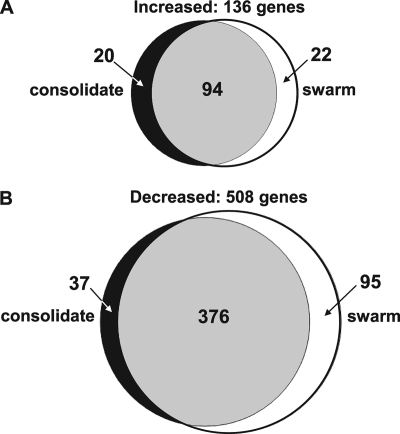
The transcriptome of broth-cultured P. mirabilis HI4320 was compared to consolidation- or swarming-phase P. mirabilis HI4320. For all agar-cultured samples, cell morphology was confirmed as swarm or consolidate by Gram staining. Bacteria for both swarm and consolidation samples were collected from the leading edge of the colony. Analysis of data from five arrays indicated that 587 genes (16% of the genome) were differentially expressed in broth-grown cells versus swarm cells, and 527 genes (14%) were differentially expressed in broth-grown cells versus consolidate. Most genes (470 of 644 [73%]) were found to be regulated similarly in swarm or consolidate cells compared to broth-grown cells (Fig. 2); that is, genes that were upregulated in consolidate compared to broth-cultured cells (Table 1; see also Table S2 in the supplemental material) also were likely to be upregulated in swarmer cells compared to broth-cultured cells (Table 2; see also Table S3 in the supplemental material). Genes that were upregulated on swarm agar compared to broth included numerous genes previously correlated with swarming behavior, such as flagella, the Zap metalloprotease, and hemolysin (4, 62). In fact, flaA (PMI1620), encoding flagellin, the major subunit of flagella, was among the most highly expressed genes in both consolidate (sixth) and swarm (third) cells. Fimbrial genes were upregulated in broth compared to either swarm or consolidate. See Tables S2, S3, S4, and S5 in the supplemental material for the complete list of genes differentially regulated at least 2-fold in broth compared to consolidate or swarm. See Tables S6, S7, and S8 in the supplemental material for lists of the most highly expressed genes in consolidate, swarm, and broth-cultured cells, respectively.
FIG. 2.
Venn diagrams showing the numbers of differentially regulated genes during broth culture compared to consolidation or swarming phases. (A) Genes with increased transcription on swarm agar (consolidation or swarming) compared to broth. (B) Genes with decreased transcription on swarm agar (consolidation or swarming) compared to broth.
TABLE 1.
Fifty genes most upregulated in consolidate compared to broth-cultured cells
| Gene | Annotation | Swarma | Fold change |
|---|---|---|---|
| PMI1629 | fliE flagellar hook-basal body complex protein FliE | Yes | 10.20 |
| PMI1654 | flgB flagellar basal body rod protein FlgB | Yes | 10.17 |
| PMI1618 | fliA RNA polymerase sigma factor for flagellar operon | Yes | 9.86 |
| PMI1961 | ccm putative membrane protein (Ccm1 protein) | Yes | 9.49 |
| PMI1649 | flgG flagellar basal body rod protein FlgG (distal rod protein) | Yes | 9.24 |
| PMI1653 | flgC flagellar basal body rod protein FlgC | Yes | 8.96 |
| PMI2898 | Putative amino acid ABC transporter, substrate-binding protein | Yes | 8.86 |
| PMI1650 | flgF flagellar basal body rod protein | Yes | 8.85 |
| PMI1631 | fliG flagellar motor switch protein FliG | Yes | 8.50 |
| PMI1637 | fliM flagellar motor switch protein FliM | Yes | 8.48 |
| PMI1636 | fliL flagellar protein FliL | Yes | 8.37 |
| PMI1621 | flaD flagellar hook-associated protein 2 | Yes | 8.19 |
| PMI1617 | fliZ putative alternative sigma factor regulatory protein | Yes | 8.19 |
| PMI1359 | Putative attachment invasion locus protein | Yes | 8.13 |
| PMI0182 | Putative transcriptional regulator (MrpJ homolog) | Yes | 7.90 |
| PMI1630 | fliF flagellar M-ring protein | No | 7.82 |
| PMI1651 | flgE flagellar hook protein FlgE | Yes | 7.76 |
| PMI1318 | nlpA lipoprotein 28 | No | 7.71 |
| PMI1638 | fliN flagellar motor switch protein FliN | Yes | 7.39 |
| PMI1442 | Hypothetical protein | Yes | 7.28 |
| PMI1647 | flgI flagellar P-ring protein precursor | Yes | 7.21 |
| PMI0876 | umoD upregulator of flagellar master operon | Yes | 7.18 |
| PMI1660 | flhB flagellar biosynthetic protein FlhB | No | 7.18 |
| PMI1671 | flhC flagellum biosynthesis transcription activator | No | 6.85 |
| PMI1624 | Conserved hypothetical protein | No | 6.82 |
| PMI3460 | Putative lipoprotein | Yes | 6.63 |
| PMI0808 | ddg cold-induced palmitoleoyl transferase | No | 6.62 |
| PMI2496 | rsmC rRNA small subunit methyltransferase C | Yes | 6.49 |
| PMI1908 | Putative colicin | No | 6.38 |
| PMI3036 | sodA superoxide dismutase (Mn) | Yes | 6.31 |
| PMI1248 | Putative cystathionine gamma-synthase | Yes | 6.09 |
| PMI1669 | motB chemotaxis motility protein B | Yes | 5.96 |
| PMI1657 | flgN flagellar synthesis protein FlgN | Yes | 5.92 |
| PMI1666 | cheD methyl-accepting chemotaxis protein | No | 5.76 |
| PMI0994 | Putative lipoprotein | Yes | 5.72 |
| PMI1645 | flgK flagellar hook associated protein 1 | Yes | 5.72 |
| PMI1661 | cheZ chemotaxis phosphatase | Yes | 5.65 |
| PMI1644 | flgL flagellar hook-associated protein 3 | Yes | 5.51 |
| PMI0272 | Probable transport protein | Yes | 5.41 |
| PMI1622 | fliS flagellar protein FliS | Yes | 5.40 |
| PMI1635 | fliK flagellar hook length control protein | Yes | 5.39 |
| PMI0044 | Putative outer membrane protein | No | 5.37 |
| PMI0842 | TonB-dependent receptor | Yes | 5.33 |
| PMI1670 | motA chemotaxis motility protein A | Yes | 5.29 |
| PMI1633 | fliI flagellum-specific ATP synthase | No | 5.18 |
| PMI0029 | exbB biopolymer transport protein | Yes | 5.03 |
| PMI1662 | cheY chemotaxis response regulator | Yes | 5.01 |
| PMI3384 | fklB FkbP-type peptidyl-prolyl cis-trans isomerase | No | 4.99 |
| PMI2808 | Methyl-accepting chemotaxis protein | Yes | 4.92 |
| PMI1835 | cysP sulfate/thiosulfate ABC transporter, thiosulfate-binding protein | Yes | 4.91 |
Genes that are also listed on Table 2.
TABLE 2.
Fifty genes most upregulated in swarm compared to broth-cultured cells
| Gene | Annotation | Consolidatea | Fold change |
|---|---|---|---|
| PMI1618 | fliA RNA polymerase sigma factor for flagellar operon | Yes | 10.11 |
| PMI1359 | Putative attachment invasion locus protein | Yes | 9.95 |
| PMI1629 | fliE flagellar hook-basal body complex protein FliE | Yes | 9.17 |
| PMI2898 | Putative amino acid ABC transporter, substrate-binding protein | Yes | 9.17 |
| PMI1654 | flgB flagellar basal body rod protein FlgB | Yes | 8.94 |
| PMI1617 | fliZ putative alternative sigma factor regulatory protein | Yes | 8.91 |
| PMI1653 | flgC flagellar basal body rod protein FlgC | Yes | 8.40 |
| PMI3460 | Putative lipoprotein | Yes | 8.21 |
| PMI2057 | hpmA hemolysin | No | 8.12 |
| PMI1438 | Putative membrane protein | No | 7.96 |
| PMI1650 | flgF flagellar basal body rod protein | Yes | 7.76 |
| PMI1637 | fliM flagellar motor switch protein FliM | Yes | 7.48 |
| PMI1442 | Hypothetical protein | Yes | 7.32 |
| PMI0993 | Putative lipoprotein | No | 7.21 |
| PMI1649 | flgG flagellar basal body rod protein FlgG (distal rod protein) | Yes | 7.19 |
| PMI1647 | flgI flagellar P-ring protein precursor | Yes | 6.96 |
| PMI1644 | flgL flagellar hook-associated protein 3 | Yes | 6.90 |
| PMI1961 | ccm putative membrane protein (Ccm1 protein) | Yes | 6.90 |
| PMI0182 | Putative transcriptional regulator (MrpJ homolog) | Yes | 6.78 |
| PMI1621 | flaD flagellar hook-associated protein 2 | Yes | 6.73 |
| PMI3036 | sodA superoxide dismutase (Mn) | Yes | 6.71 |
| PMI2496 | rsmC rRNA small subunit methyltransferase C | Yes | 6.65 |
| PMI1651 | flgE flagellar hook protein FlgE | Yes | 6.63 |
| PMI1636 | fliL flagellar protein FliL | Yes | 6.39 |
| PMI1638 | fliN flagellar motor switch protein FliN | Yes | 6.36 |
| PMI2808 | Methyl-accepting chemotaxis protein | Yes | 6.31 |
| PMI1631 | fliG flagellar motor switch protein FliG | Yes | 6.20 |
| PMI1669 | motB chemotaxis motility protein B | Yes | 6.16 |
| PMI0842 | TonB-dependent receptor | Yes | 6.16 |
| PMI0994 | Putative lipoprotein | Yes | 6.10 |
| PMI1622 | fliS flagellar protein FliS | Yes | 6.02 |
| PMI0187 | dsdAd-serine dehydratase | No | 6.00 |
| PMI1634 | fliJ flagellar protein FliJ | No | 5.89 |
| PMI1835 | cysP sulfate/thiosulfate ABC transporter, thiosulfate-binding protein | Yes | 5.74 |
| PMI1661 | cheZ chemotaxis phosphatase | Yes | 5.45 |
| PMI2385 | terC tellurite resistance protein | No | 5.42 |
| PMI1665 | tap methyl-accepting chemotaxis protein | No | 5.36 |
| PMI1645 | flgK flagellar hook-associated protein 1 | Yes | 5.34 |
| PMI2196 | Putative membrane-associated CAAX amino-terminal protease | No | 5.19 |
| PMI0876 | umoD upregulator of flagellar master operon | Yes | 5.17 |
| PMI1005 | Putative membrane protein | No | 5.11 |
| PMI1657 | flgN flagellar synthesis protein FlgN | Yes | 5.10 |
| PMI0272 | Probable transport protein | Yes | 5.07 |
| PMI1248 | Putative cystathionine gamma-synthase | Yes | 5.05 |
| PMI1635 | fliK flagellar hook length control protein | Yes | 5.00 |
| PMI1670 | motA chemotaxis motility protein A | Yes | 4.95 |
| PMI0276 | zapD type I secretion outer membrane protein | No | 4.94 |
| PMI1662 | cheY chemotaxis response regulator | Yes | 4.92 |
| PMI2383 | terE tellurite resistance protein | No | 4.88 |
| PMI0029 | exbB biopolymer transport protein | Yes | 4.83 |
Genes that are also listed on Table 1.
Microarray analysis of swarm compared to consolidate.
When the consolidate morphotype was compared directly to that of swarm, 541 genes were found to have transcription increased >2-fold in consolidate (see Table S9 in the supplemental material; the 50 most upregulated genes are listed in Table 3). Only nine genes had increased transcription in swarm cells compared to consolidate cells (Table 4).
TABLE 3.
Fifty genes most upregulated in consolidate compared to swarm
| Gene | Annotation | Fold change |
|---|---|---|
| PMI0785 | ompA outer membrane protein A | 9.09 |
| PMI1908 | Putative colicin | 8.30 |
| PMI1475 | oppA periplasmic oligopeptide-binding protein precursor | 8.10 |
| PMI1592 | fbaB fructose-bisphosphate aldolase class I | 7.86 |
| PMI0250 | Putative lipoprotein | 7.73 |
| PMI2019 | gcvP glycine dehydrogenase | 7.58 |
| PMI3037 | acs acetyl-coenzyme A synthetase | 7.52 |
| PMI2236 | rpoS RNA polymerase sigma factor RpoS | 7.48 |
| PMI2662 | chb secreted chitin-binding protein | 6.96 |
| PMI0437 | gltI glutamate/aspartate ABC transporter, substrate-binding protein | 6.91 |
| PMI3579 | Conserved hypothetical protein | 6.89 |
| PMI1056 | Putative TRAP-type transport system substrate-binding protein | 6.86 |
| PMI2114 | Putative sodium:alanine symporter | 6.82 |
| PMI1474 | oppB oligopeptide transport system permease protein | 6.70 |
| PMI1421 | ppsA phosphoenolpyruvate synthase | 6.69 |
| PMI1722 | Phage protein | 6.68 |
| PMI1615 | putP proline permease | 6.49 |
| PMI2246 | cysD sulfate adenylyltransferase subunit 2 | 6.37 |
| PMI2052 | coaE dephospho-CoA kinase | 6.36 |
| PMI2255 | Conserved hypothetical protein | 6.34 |
| PMI2254 | mdeA methionine gamma-lyase | 6.33 |
| PMI3177 | kbl 2-amino-3-ketobutyrate coenzyme A ligase | 6.28 |
| PMI1075 | Hypothetical protein | 6.27 |
| PMI0968 | Isocitrate dehydrogenase (NADP) (partial) | 6.20 |
| PMI3559 | accA3 putative bifunctional acetyl-/propionyl-coenzyme A carboxylase alpha chain | 6.17 |
| PMI3556 | Putative transmembrane transport protein | 6.16 |
| PMI0564 | gltA citrate synthase GltA | 6.11 |
| PMI0971 | Putative lipase | 6.03 |
| PMI1451 | uspG2 universal stress protein G | 6.02 |
| PMI3178 | tdh threonine 3-dehydrogenase | 6.01 |
| PMI2256 | Putative oxidoreductase | 5.99 |
| PMI1471 | oppF oligopeptide transport ATP-binding protein | 5.95 |
| PMI1473 | oppC oligopeptide transport system permease protein | 5.90 |
| PMI1171 | Conserved hypothetical protein | 5.77 |
| PMI1472 | oppD oligopeptide transport ATP-binding protein | 5.76 |
| PMI1616 | putA proline dehydrogenase/delta-1-pyrroline-5-carboxylate | 5.76 |
| PMI0360 | Putative general stress response protein | 5.69 |
| PMI1730 | rcsB two-component system response regulator, capsular synthesis | 5.62 |
| PMI2268 | Short-chain dehydrogenase | 5.60 |
| PMI0047 | Putative secreted 5'-nucleotidase | 5.54 |
| PMI2036 | Putative exported protein | 5.49 |
| PMI1737 | Putative lipoprotein | 5.49 |
| PMI3401 | sfsB sugar fermentation stimulation protein | 5.43 |
| PMI2890 | greB transcription elongation factor | 5.42 |
| PMI1509 | dadAd-amino acid dehydrogenase small subunit | 5.38 |
| PMI3701 | Putative sodium:dicarboxylate symporter | 5.37 |
| PMI0952 | Putative isocitrate dehydrogenase (NADP) (partial) | 5.34 |
| PMI3204 | sbp exported sulfate-binding protein | 5.33 |
| PMI3115 | umoA putative upregulator of flagellar operon | 5.32 |
| PMI3449 | Conserved hypothetical protein | 5.30 |
TABLE 4.
Genes upregulated >2-fold in swarm compared to consolidate
| Gene | Annotation | Fold change |
|---|---|---|
| PMI0875 | ndh NADH dehydrogenase | 4.67 |
| PMI0913 | cspA cold shock protein | 2.60 |
| PMI3584 | sugE quaternary ammonium compound resistance protein | 2.39 |
| PMI2309 | recB exodeoxyribonuclease V β-chain | 2.36 |
| PMI0807 | cspB major cold shock protein | 2.24 |
| PMI1140 | Rhs family protein | 2.24 |
| PMI0972 | cspG cold shock protein | 2.18 |
| PMI2667 | Putative mannosyl-glycoprotein endo-β-N-acetylglucosaminidase | 2.07 |
| PMI2732 | atfE fimbrial adhesin | 2.06 |
Genes encoding proteins involved in peptide uptake and amino acid uptake and biosynthesis were highly upregulated in consolidate compared to swarm. Genes encoding stress proteins, as well as the general stress response sigma factor RpoS, were also upregulated during consolidation, while three genes encoding cold shock proteins were upregulated during swarming. Numerous proteases, including Lon, which has previously been identified as active in swarm behavior (13, 14, 43), were upregulated during consolidation. In addition, genes involved in central metabolic pathways, including gluconeogenesis and the tricarboxylic acid (TCA) cycle, were upregulated during consolidation (see Table S6 in the supplemental material). Surprisingly, many flagellar genes were also upregulated during consolidation. Classes of genes found to be differentially regulated during swarming are summarized in Fig. 3.
FIG. 3.
Functional categories of genes differentially expressed during swarming or consolidation. The number of genes differentially regulated for each class is shown on the y axis.
Validation of microarray data by qRT-PCR.
To independently confirm data obtained by microarray analysis, qRT-PCR was conducted using cDNA templates obtained from consolidation-phase or swarming-phase P. mirabilis with primers corresponding to genes predicted to be up- or downregulated under each condition examined. Table 5 shows the results for genes predicted to be differentially regulated in consolidate compared to swarm. Nine of the 10 genes analyzed responded as predicted by the microarray.
TABLE 5.
Genes analyzed by qRT-PCRa
| Gene | Name | Annotation | Log2 fold change |
|
|---|---|---|---|---|
| Array | qRT-PCR | |||
| PMI0437 | gltI | Glutamate/aspartate ABC transport | 2.79 | 2.46 ± 0.72 |
| PMI0785 | ompA | Outer membrane protein A | 3.18 | 6.64 ± 0.59 |
| PMI0875 | ndh | NADH dehydrogenase | −2.22 | −2.11 ± 0.77 |
| PMI0913 | cspA | Cold shock protein | −1.38 | −2.00 ± 0.68 |
| PMI1475 | oppA | Oligopeptide ABC transporter | 3.02 | 2.80 ± 1.49 |
| PMI1722 | Phage protein | 2.74 | −1.77 ± 1.64 | |
| PMI1730 | rcsB | Capsular synthesis regulator | 2.49 | 0.16 ± 0.34 |
| PMI1908 | Putative colicin | 3.05 | 1.27 ± 1.11 | |
| PMI2250 | cysJ | Sulfite reductase (NADPH) flavoprotein | 2.09 | 3.08 ± 0.98 |
| PMI3037 | acs | Acetyl coenzyme A synthetase | 2.91 | 1.11 ± 0.40 |
Positive values are upregulated in consolidate compared to swarm; negative values are upregulated in swarm compared to consolidate.
Swarming-regulated genes are not necessarily required for swarming.
Because many genes involved in amino acid or peptide uptake were upregulated during consolidation, we hypothesized that these processes might play a role in swarm signaling. In particular, the entire opp oligopeptide permease operon, oppA2ABCDF, was upregulated greater than 5-fold in consolidate compared to swarm by microarray analysis (Table 3; see also Table S9 in the supplemental material), and this trend was confirmed for oppA by qRT-PCR (Table S5C). Therefore, three genes, dppA (dipeptide permease, PMI2847, upregulated 4.08-fold in consolidate compared to swarm), oppB (oligopeptide permease, PMI1474, upregulated 6.70-fold in consolidate compared to swarm), and cysJ (sulfite reductase flavoprotein subunit, PMI2250, upregulated 4.26-fold in consolidate compared to swarm) were inactivated by insertion of a kanamycin resistance cassette. Because each of the targeted genes is predicted to be involved with peptide uptake or amino acid biosynthesis, the mutant strains were tested for swarming on minimal A medium supplemented with 1% tryptone as a protein source (MinA-T). P. mirabilis swarms on MinA-T agar with a bull's-eye pattern of swarming and consolidation similar to that seen on LB swarm agar (Fig. 4, top panel), although the swarm edge is thinner on MinA-T. As on LB agar, P. mirabilis alternates between short consolidate and long swarmer forms on MinA-T agar (Fig. 4, center panel). Swarming initially proceeds more slowly on MinA-T but then advances at a rate comparable to that seen on LB agar (Fig. 4, bottom panel). Gene expression on MinA-T agar, analyzed by qRT-PCR, is similar to expression on LB agar; amino acid biosynthetic genes gltI and cysJ and the outer membrane protein OmpA are all upregulated during consolidate on MinA-T (fold changes of 2.02, 2.86, and 3.14, respectively, compared to swarm). However, the oligopeptide transporter gene oppA was not significantly upregulated during consolidation phase compared to swarm (fold change of 1.10). We hypothesized that if peptide and amino acid uptake contributed to swarm behavior, MinA-T agar might reveal subtle defects in swarming by peptide or amino acid uptake mutants. On this medium, all mutants tested had a defect in swarming (Fig. 5), with the most pronounced phenotype in the cysJ mutant. It should be noted that P. mirabilis HI4320 does not swarm on unaugmented minimal A agar. However, none of these mutants had a statistically significant defect in swarming behavior when tested on LB swarm agar (data not shown).
FIG. 4.
Comparison of swarming on LB and minimal A agar supplemented with 1% tryptone. Top panel, agar plates with characteristic bull's-eye P. mirabilis swarm colonies on LB agar (left) and MinA-T agar (right), 36 h postinoculation. Middle panel, Gram-stained cells collected during the consolidation (short form) and swarm (long form) phases from LB agar (left) and MinA-T agar (right). Consolidate was collected at 14 h postinoculation, and swarm was collected at 17 h postinoculation. All four Gram stain photos were recorded at ×400 magnification. Bottom panel, the swarm radius of colonies on LB (closed circles) and MinA-T (open circles) agar was measured hourly. Three plates were measured for each agar type. Error bars represent the standard errors of the means (SEM).
FIG. 5.
Swarming radii of P. mirabilis and isogenic mutants on MinA-T agar, 16 h postinoculation. *, P < 0.05; **, P < 0.01. Error bars represent SEM.
Sequencing and annotation of the P. mirabilis HI4230 genome (52) led to the observation that this species encodes two homologs of lrhA, a LysR family transcriptional regulator. These two genes, PMI0629 (lrhA) and PMI1764 (hexA), encode proteins with 54% amino acid sequence identity and 74% similarity to each other. Only one of the two homologs, hexA, was found to be upregulated in consolidate compared to swarm (3.60-fold); lrhA was not found to be differentially regulated greater than 2-fold under any of the conditions tested by microarray. To learn what roles lrhA and hexA might play in swarming, lrhA and hexA mutants and an lrhA hexA double mutant were tested for swarming on MinA-T agar (Fig. 5). While the lrhA mutant had only a modest decrease in swarming, the hexA and lrhA hexA mutants both swarmed significantly less than did the wild-type parent (P < 0.01). Interestingly, the double mutant had a slightly greater defect than did the hexA single mutant (P = 0.0318). On LB swarm agar, both the hexA and lrhA hexA mutants consistently had a decreased swarm radius compared to that of the wild-type parent, although these differences were not significant (data not shown).
The growth rates for all six mutants constructed for this study were also determined in LB broth. Only the oppB mutant had a modest growth deficiency in broth culture, both growing more slowly and not reaching the same density as that reached by the wild-type parent (data not shown).
Swarming-regulated genes contribute to in vivo fitness.
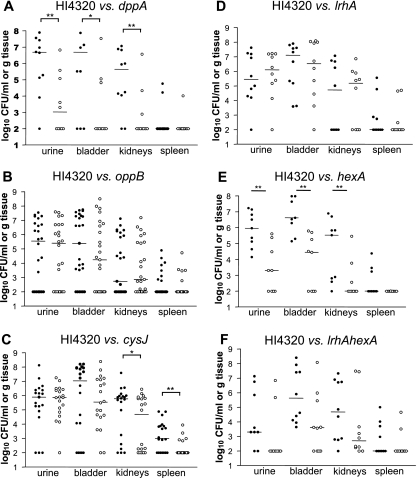
Virulence factors, including hemolysin and the Zap metalloprotease, are upregulated during culture on swarm agar (4, 62). To test whether the six mutants described above (dppA, oppB, cysJ, lrhA, hexA, and lrhA hexA) were attenuated in the ability to cause infection, we transurethrally inoculated mice with a 1:1 mixture of each mutant with the wild-type parent strain (total inoculum, 1 × 107 CFU/mouse). After 7 days, bacteria were enumerated in the urine, bladders, kidneys, and spleens (Fig. 6). The dppA, cysJ, and hexA mutants had significant colonization defects relative to the wild-type parent strain (Fig. 6A, C, and E), indicating that these genes contribute to fitness during infection of the urinary tract. Specifically, the dppA mutant was recovered in reduced numbers from the urine, bladder, and kidneys (P values of 0.0039, 0.0156, and 0.0078, respectively), the cysJ mutant was defective in colonization of the kidneys and spleen (P values of 0.0119 and 0.0052, respectively), and the hexA mutant was found in lower quantities in the urine, bladder, and kidneys (P values of 0.0039, 0.0039, and 0.0078, respectively). Likewise, the lrhA hexA mutant tended to be recovered in lower numbers from these same sites, although these differences were not significant compared to the wild type (P > 0.05). This suggests that cysteine biosynthesis or sulfate assimilation may be more important during infection of the upper urinary tract (i.e., pyelonephritis), while dipeptide uptake and regulation by HexA may be necessary throughout the urinary tract. In contrast, the oppB mutant survived at all sites in numbers similar to those of the wild-type parent (Fig. 6B) despite exhibiting a modest growth defect in LB broth culture. The lrhA mutant was also recovered in numbers similar to those of the wild-type parent (Fig. 6D).
FIG. 6.
In vivo cochallenge of wild-type P. mirabilis HI4320 with mutant strains. (A) dppA mutant cochallenge. (B) oppB mutant cochallenge. (C) cysJ mutant cochallenge. (D) lrhA mutant cochallenge. (E) hexA mutant cochallenge. (F) lrhA hexA mutant cochallenge. In all panels, closed circles represent wild-type CFU, and open circles represent mutant CFU. Numbers of actual input CFU/ml for each experiment were as follows: (A) HI4320, 1.51 × 108, and dppA mutant, 4.31 × 107; (B) HI4320, 1.79 × 108, and oppB mutant, 1.44 × 108; (C) HI4320, 1.57 × 108, and cysJ mutant, 2.31 × 108; (D) HI4320, 1.38 × 108, and lrhA mutant, 2.05 × 108; (E) HI4320, 1.36 × 108, and hexA mutant, 2.37 × 108; (F) HI4320, 1.60 × 108, and lrhA hexA mutant, 2.27 × 108. Bars indicate median values. The limit of detection is 102 CFU per ml or g tissue. *, P < 0.05; **, P < 0.01.
P. mirabilis swarms in the absence of protein synthesis.
Because 541 genes were upregulated in consolidate compared to swarm, but only nine genes were upregulated in swarm compared to consolidate, we hypothesized that P. mirabilis might be able to swarm in the absence of protein synthesis. To test this, the bacteriostatic antibiotic chloramphenicol, which interferes with protein synthesis by binding to the 50S subunit of the ribosome, was added to paper discs. P. mirabilis HI4320 was added to the center of swarm agar and allowed to dry. The chloramphenicol-infused discs were placed approximately 1 cm from the inoculation point, and the swarm colony was allowed to develop. As a control, P. mirabilis HI4320 was spread plated onto nonswarm LB agar to create a lawn and allowed to dry. Chloramphenicol-infused discs were placed on the lawn to determine concentrations of antibiotic that led to a zone of growth inhibition. In the presence of chloramphenicol, P. mirabilis swarmed more rapidly, though in a thinner, more disorganized manner (Fig. 7A, top left panel). However, P. mirabilis did not swarm in the presence of the bactericidal antibiotic kanamycin (Fig. 7A, bottom left panel). These results suggest that limited swarming behavior can occur in the absence of protein synthesis. Bacteria from the edge of the advancing swarm colony were examined by Gram staining for cell morphology (Fig. 7B). Swarm fronts within the area affected by the chloramphenicol-infused discs consistently contained bacteria with the elongated swarm morphology (Fig. 7B, top panel). In comparison, regions of the plate unaffected by the chloramphenicol discs alternated between swarm (Fig. 7B, middle panel) and consolidation (Fig. 7B, bottom panel) morphologies.
FIG. 7.
P. mirabilis swarming in the presence of antibiotics. (A) In the top panels, discs were infused with chloramphenicol (chl). In the bottom panels, discs were infused with kanamycin (kan). The panels on the left show swarm agar, while the panels on the right are nonswarm agar. On all plates the discs contain, clockwise from left, 2, 10, 20, or 50 μg antibiotic. (B) Gram stains from a representative chloramphenicol disc swarm plate (50 μg chl/disc). Top, swarm front next to the disc; middle, swarm front from a region of the plate unaffected by chloramphenicol; bottom, consolidation front from a region of the plate unaffected by chloramphenicol. Bar, 50 μm.
DISCUSSION
This is the first microarray analysis of the P. mirabilis transcriptome. When cultured on an agar surface, P. mirabilis elongates, upregulates its flagellar regulon, and undergoes swarming motility. This is followed by what was believed to be a resting stage, consolidation. However, this phase appears to represent a time of preparation for the next phase of swarming, marked by the upregulation of nutrient uptake systems, central metabolism (TCA cycle, gluconeogenesis, and glycerol metabolism), respiration, and cell wall synthetic enzymes that favor filamentation but discourage septation and division.
We first compared broth-cultured P. mirabilis directly to swarm or consolidate. Most genes that were differentially regulated in consolidate were similarly regulated in swarm, suggesting that this experimental approach was amenable for distinguishing the broth environment from the swarm agar environment, but is not as useful for pinpointing the changes that occur as P. mirabilis cycles from swarm to consolidate and back again. Thus, a second series of microarray experiments was conducted to compare swarm directly to consolidate.
The swarming phase of P. mirabilis is dynamic, with writhing motion easily detectable on the agar surface using light microscopy. Thus, it was initially surprising to find that 541 genes were upregulated during consolidation, yet only nine genes were upregulated during swarming in this direct comparison. Additionally, genes expected to be upregulated during swarming, especially those involved in flagellar synthesis, were instead either highly expressed during both swarm and consolidation phases or upregulated during the consolidation phase only. It should be noted, however, that P. mirabilis converts between the swarm and consolidation phases approximately every 2 h under the conditions used in this study. We hypothesize that the consolidation phase is a time where the bacteria are preparing for the next swarm cycle. That is, genes whose products are required during the swarming phase (i.e., flagella) should actually be upregulated during the previous consolidation phase, allowing time for proteins to be assembled and functional during the correct phase. Likewise, the activity of thousands of coordinated flagella during the swarming phase consumes considerable energy, and the consolidation phase may require the upregulation of genes involved in nutrient uptake (e.g., amino acid and peptide uptake systems) to replenish energy reserves. These observations are consistent with a previous report that indicated that swarm cells are metabolically less active than consolidate cells (7). However, our results, which revealed very high levels of flagellar gene expression through both swarming and consolidation phases, with highest expression during consolidation, contrast with previous work in which flhDC and flaA (called fliC in that report) varied in expression 30-fold by Northern blot analysis and, at lowest expression levels, had almost undetectable transcript (23). The major difference between these two studies is that the current study used bacteria taken from the edge of a swarming colony, while the earlier report used P. mirabilis spread evenly on an agar surface and followed transcript levels from bacteria washed from the agar surface for 5.5 h after seeding. That is, a 5.5-h-old seeded bacterium and a newly divided consolidation cell at the edge of a swarm colony may both have a short morphotype, but they are unlikely to be physiologically equivalent.
As one would predict, when P. mirabilis undergoes the radical transformation from an elongated, multinucleated cell to the short individual bacterium, there must be a significant remodeling of peptidoglycan and the cell wall. Indeed, genes involved in such remodeling are significantly upregulated in the consolidate compared to the swarmer. Genes upregulated during consolidation appear to prepare P. mirabilis for filamentation. For example, upregulation of minC (4.61-fold) and minD (2.36-fold) would produce an excess of MinCD, which prevents cell division and results in filamentation (18). Genes involved in chromosomal partitioning in the multinuclear swarmer cell, mukB, mukE, and mukF (46, 66), are all upregulated (2.31-, 3.26-, and 2.88-fold, respectively). Upregulation of mreC (2.75-fold) is consistent with maintenance of a rod-shaped swarmer cell (48). Finally, upregulation of mtgA, encoding a monofunctional biosynthetic peptidoglycan transglycosylase (19), would ensure glycan chain elongation in the developing swarmer cell. On the other hand, genes responsible for routine synthesis of peptidoglycan (ftsZ, ftsI, ftsL, murD, murE, murF, and mraY) are not altered in expression between swarmer and consolidate cells.
Genes differentially regulated during P. mirabilis swarming have interesting parallels and contrasts with similar studies conducted with swarming Salmonella (37, 63) and E. coli (32). For purposes of comparison, it should be noted that while P. mirabilis swarms on surfaces containing up to 2% agar, Salmonella and E. coli require a lower agar concentration (0.5 to 0.8%) and richer media (i.e., LB supplemented with glucose) to swarm. Swarming Salmonella and E. coli also lack the cyclic pattern of swarming and consolidation seen in P. mirabilis and instead swarm continuously to the agar edge (29, 37). Kim and Surette (37) examined protein expression in actively swarming Salmonella and found that amino acid biosynthetic pathways and central metabolism, particularly the TCA cycle, were increased compared to that in swimming samples; however, major outer membrane porins, including OmpA, were decreased during swarming. Interestingly, cysteine biosynthesis in particular was necessary for full swarm cell differentiation (60). In contrast, we found genes involved in all of these systems (amino acid biosynthesis, TCA cycle, as well as peptide and nonspecific uptake systems) were upregulated during consolidation. A microarray study by Wang et al. (63) analyzed gene expression in Salmonella on swarm agar with Salmonella on hard agar or in broth culture and found that gene expression patterns were more similar under the two agar-cultured conditions compared to broth culture; this is comparable to the finding that P. mirabilis has more genes expressed in common during swarm and consolidation than during exponential-phase broth culture. The same study also reported that genes involved in basic metabolism and cysteine biosynthesis were upregulated during swarming by Salmonella (63). A saturating knockout mutant screen of E. coli K-12 (32) also indicated the importance of the TCA cycle in swarming yet also identified glucose metabolism as a requirement for swarming in that species. In contrast, we did not find glucose metabolic genes upregulated during swarming by P. mirabilis, which does not require exogenous glucose for swarming.
Previous swarming reports also indicated that P. mirabilis is metabolically less active during swarming and that amino acid uptake increases during consolidation (7, 36). The microarray results from the current study support these findings. Numerous markers of metabolic activity were upregulated during consolidation, including peptide uptake, amino and nucleic acid biosynthesis, central metabolism, and aerobic respiration. Combining the microarray results with the ability of P. mirabilis to swarm in the absence of protein synthesis suggests that the swarming state is almost wholly consumed with flagellum-mediated motility. We propose that after this energy has been expended, P. mirabilis enters the consolidation phase, which is focused upon replenishing supplies. Perhaps the replenishing consolidation stage allows P. mirabilis to swarm under conditions that are not permissive for other species.
A particular concern when analyzing the transcriptome of a cyclical event is ensuring sample uniformity. Great effort was taken to confirm that the bacterial inputs were in the desired (swarm or consolidation) phase by observing the appearance and motion at the colony edge and by examining a portion of the input sample by Gram staining. The desired morphotype predominated in both consolidate (>99% of bacteria were <10 μm) and swarm (>88% bacteria were >10 μm) cells, respectively. One strength of this study is that only the edge of the bacterial colony was collected for RNA isolation; thus, only the youngest cells in the swarm colony were analyzed for both swarming and consolidation phases. Difficulties in obtaining pure samples of swarm or consolidate cells have been noted by others (7, 64), and older vegetative cells from the interior of the swarm colony have been used in the past for comparisons with swarm cells (1, 21). It is important to emphasize that in our study many genes previously known to be swarming regulated were also identified by microarray (e.g., flagella, fimbriae, and Lon and Zap proteases). Furthermore, our P. mirabilis HI4320 microarray has also been validated by comparing HI4320 transcripts from iron-restricted with iron-replete broth (S. D. Himpsl, M. M. Pearson, and H. L. T. Mobley, submitted for publication).
In this report, we found that a hexA mutant had a significant defect in swarming. In contrast, an earlier study (15) concluded that hexA (called lrhA in that report) had no effect on swarming. There are several possible explanations for the different outcomes. The first study examined swarming on LB agar incubated at 37°C, while the current report examined swarming on a minimal medium supplemented with tryptone, MinA-T agar, incubated at 30°C. Indeed, although we found that the hexA mutant tended to have reduced swarming motility on LB agar, the difference was not statistically significant (data not shown). It is also possible that, in the earlier study, the lrhA gene was able to complement the hexA mutant in the P. mirabilis strain used. The different results may also be a consequence of mutant construction. The hexA mutant in our study was constructed by inserting an intron between nucleotides 463 and 464 of hexA, resulting in the first 154 amino acids of the 305-amino-acid protein potentially being produced. Clemmer and Rather (15) used an internal cloned fragment of hexA on a suicide plasmid, which would result in the first 206 amino acids potentially being produced (i.e., the C-terminal 33% of the protein would be missing). Although this seems sufficient to ablate HexA function, we have deleted as much as the C-terminal third of another regulatory protein, MrpJ, and found that functional protein was still produced (50). Both hexA mutants could result in translated HexA protein with an intact helix-turn-helix domain; in addition, our mutant bears 57 of 151 amino acids of the predicted substrate-binding domain, while the other hexA mutant could produce 105/151 amino acids of this domain. As we cannot rule out the possibility that either hexA mutant is able to produce an active, truncated HexA protein, ideally, a complete hexA deletion would be tested for swarming ability in a future study.
P. mirabilis (4, 62) and other species (24, 49, 63) upregulate virulence factors during swarming, and in this report, more virulence factors in ascending UTI were identified. Thus, further analysis of the swarming transcriptome microarray data will likely be useful for identifying additional virulence factors for this and other pathogens capable of swarming. The association of swarming with virulence may seem to conflict with earlier work, in which P. mirabilis was found in the swarm morphotype only rarely in a mouse model of UTI (33). However, P. mirabilis swarms on urinary catheters (35, 54), which were not used in the previous study; this may represent a mechanism by which the bacterium gains access to the catheterized urinary tract, the bacterium's typical niche. It is not known how quickly P. mirabilis switches to the vegetative form once inside the urinary tract, and it is possible that swarming P. mirabilis cells are primed to be virulent as soon as they gain access to the bladder. The ability to prime genetic expression under a particular growth condition in preparation for entering a new environment has already been demonstrated for Vibrio cholerae, which expresses genes needed for survival in an aquatic environment during the later stages of intestinal infection (55). Urinary catheters may serve as a constant source of swarming P. mirabilis cells in the urinary tract. Further experimentation is needed to address this possibility.
The discovery that peptide and amino acid uptake are critical for P. mirabilis during UTI meshes with metabolic studies of uropathogenic E. coli (UPEC). P. mirabilis and other urinary tract pathogens are capable of growth in human urine, where the main carbon sources are amino acids and peptides (6). While P. mirabilis dppA and cysJ mutants are attenuated in vivo, it was surprising that an oppB mutant had no defect in vivo. This result contrasts with a similar study of UPEC strain CFT073, in which both dppA and oppA mutants could not successfully compete with the wild-type parent during experimental infection (5). A possible answer may lie in the organization of the opp operons in these two species. The UPEC operon consists of the genes oppABCDF, while the P. mirabilis opp locus has two copies of oppA, designated oppA and oppA2 (which are 57% identical to each other, with 77% similar amino acid sequences). Perhaps the P. mirabilis OppA2 protein is able to import oligopeptides through another channel. Alternatively, other peptide import systems in P. mirabilis that are not present in UPEC may compensate for the oppB mutation.
The use of microarrays to study swarming has raised new questions about this long-studied phenomenon. Why are nutrient uptake pathways upregulated during consolidation? What is the role played by genes that were unexpected to vary in expression during swarming, such as the tellurite resistance (ter) genes or phage? Can any of the genes in this study be harnessed to control or prevent swarming on urinary catheters? How many swarming-regulated genes are also differentially regulated during infection? Perhaps this aptly named pathogen will now be less able to avoid questioning even as it changes forms.
Supplementary Material
Acknowledgments
This work was funded by Public Health Service grant AI059722 from the National Institutes of Health (H.L.T.M.). Analysis in the laboratory of D.A.R. is supported by University of Maryland School of Medicine internal funds. M.M.P. was supported in part by National Research Service Award F32 AI068324.
Editor: S. M. Payne
Footnotes
Published ahead of print on 5 April 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Allison, C., N. Coleman, P. L. Jones, and C. Hughes. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, C., and C. Hughes. 1991. Closely linked genetic loci required for swarm cell differentiation and multicellular migration by Proteus mirabilis. Mol. Microbiol. 5:1975-1982. [DOI] [PubMed] [Google Scholar]
- 3.Allison, C., H. C. Lai, D. Gygi, and C. Hughes. 1993. Cell differentiation of Proteus mirabilis is initiated by glutamine, a specific chemoattractant for swarming cells. Mol. Microbiol. 8:53-60. [DOI] [PubMed] [Google Scholar]
- 4.Allison, C., H. C. Lai, and C. Hughes. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583-1591. [DOI] [PubMed] [Google Scholar]
- 5.Alteri, C. J., S. N. Smith, and H. L. Mobley. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altman, P. L. 1961. Physical properties and chemical composition of urine: mammals. Part 1: man, p. 363-369. In D. L. Dittmer (ed.), Blood and other bodily fluids. Federation of American Societies for Experimental Biology, Washington, DC.
- 7.Armitage, J. P. 1981. Changes in metabolic activity of Proteus mirabilis during swarming. J. Gen. Microbiol. 125:445-450. [DOI] [PubMed] [Google Scholar]
- 8.Belas, R., D. Erskine, and D. Flaherty. 1991. Proteus mirabilis mutants defective in swarmer cell differentiation and multicellular behavior. J. Bacteriol. 173:6279-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belas, R., M. Goldman, and K. Ashliman. 1995. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J. Bacteriol. 177:823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belas, R., R. Schneider, and M. Melch. 1998. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J. Bacteriol. 180:6126-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burall, L. S., J. M. Harro, X. Li, C. V. Lockatell, S. D. Himpsl, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2004. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72:2922-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claret, L., and C. Hughes. 2000. Rapid turnover of FlhD and FlhC, the flagellar regulon transcriptional activator proteins, during Proteus swarming. J. Bacteriol. 182:833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemmer, K. M., and P. N. Rather. 2008. The Lon protease regulates swarming motility and virulence gene expression in Proteus mirabilis. J. Med. Microbiol. 57:931-937. [DOI] [PubMed] [Google Scholar]
- 15.Clemmer, K. M., and P. N. Rather. 2007. Regulation of flhDC expression in Proteus mirabilis. Res. Microbiol. 158:295-302. [DOI] [PubMed] [Google Scholar]
- 16.Coker, C., C. A. Poore, X. Li, and H. L. Mobley. 2000. Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect. 2:1497-1505. [DOI] [PubMed] [Google Scholar]
- 17.D'Agata, E. M. 2004. Rapidly rising prevalence of nosocomial multidrug-resistant, Gram-negative bacilli: a 9-year surveillance study. Infect. Control Hosp. Epidemiol. 25:842-846. [DOI] [PubMed] [Google Scholar]
- 18.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 19.Derouaux, A., B. Wolf, C. Fraipont, E. Breukink, M. Nguyen-Disteche, and M. Terrak. 2008. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J. Bacteriol. 190:1831-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufour, A., R. B. Furness, and C. Hughes. 1998. Novel genes that upregulate the Proteus mirabilis flhDC master operon controlling flagellar biogenesis and swarming. Mol. Microbiol. 29:741-751. [DOI] [PubMed] [Google Scholar]
- 21.Falkinham, J. O., III, and P. S. Hoffman. 1984. Unique developmental characteristics of the swarm and short cells of Proteus vulgaris and Proteus mirabilis. J. Bacteriol. 158:1037-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 23.Furness, R. B., G. M. Fraser, N. A. Hay, and C. Hughes. 1997. Negative feedback from a Proteus class II flagellum export defect to the flhDC master operon controlling cell division and flagellum assembly. J. Bacteriol. 179:5585-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghelardi, E., F. Celandroni, S. Salvetti, M. Ceragioli, D. J. Beecher, S. Senesi, and A. C. Wong. 2007. Swarming behavior of and hemolysin BL secretion by Bacillus cereus. Appl. Environ. Microbiol. 73:4089-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith, D. P., D. M. Musher, and C. Itin. 1976. Urease. The primary cause of infection-induced urinary stones. Invest. Urol. 13:346-350. [PubMed] [Google Scholar]
- 26.Gygi, D., M. M. Rahman, H. C. Lai, R. Carlson, J. Guard-Petter, and C. Hughes. 1995. A cell-surface polysaccharide that facilitates rapid population migration by differentiated swarm cells of Proteus mirabilis. Mol. Microbiol. 17:1167-1175. [DOI] [PubMed] [Google Scholar]
- 27.Hagberg, L., I. Engberg, R. Freter, J. Lam, S. Olling, and C. Svanborg Eden. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 29.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. U. S. A. 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatt, J. K., and P. N. Rather. 2008. Characterization of a novel gene, wosA, regulating FlhDC expression in Proteus mirabilis. J. Bacteriol. 190:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hay, N. A., D. J. Tipper, D. Gygi, and C. Hughes. 1999. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J. Bacteriol. 181:2008-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue, T., R. Shingaki, S. Hirose, K. Waki, H. Mori, and K. Fukui. 2007. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J. Bacteriol. 189:950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansen, A. M., C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2003. Visualization of Proteus mirabilis morphotypes in the urinary tract: the elongated swarmer cell is rarely observed in ascending urinary tract infection. Infect. Immun. 71:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, D. E., C. V. Lockatell, M. Hall-Craigs, H. L. Mobley, and J. W. Warren. 1987. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. J. Urol. 138:632-635. [DOI] [PubMed] [Google Scholar]
- 35.Jones, B. V., R. Young, E. Mahenthiralingam, and D. J. Stickler. 2004. Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect. Immun. 72:3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, H. E., and R. W. Park. 1967. The short forms and long forms of Proteus. J. Gen. Microbiol. 47:359-367. [DOI] [PubMed] [Google Scholar]
- 37.Kim, W., and M. G. Surette. 2004. Metabolic differentiation in actively swarming Salmonella. Mol. Microbiol. 54:702-714. [DOI] [PubMed] [Google Scholar]
- 38.Li, X., D. E. Johnson, and H. L. Mobley. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67:2822-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, X., H. Zhao, C. V. Lockatell, C. B. Drachenberg, D. E. Johnson, and H. L. Mobley. 2002. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect. Immun. 70:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liaw, S. J., H. C. Lai, and W. B. Wang. 2004. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect. Immun. 72:6836-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, Q., M. Z. Li, D. Leibham, D. Cortez, and S. J. Elledge. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8:1300-1309. [DOI] [PubMed] [Google Scholar]
- 42.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402. [DOI] [PubMed] [Google Scholar]
- 43.Marr, A. K., J. Overhage, M. Bains, and R. E. Hancock. 2007. The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153:474-482. [DOI] [PubMed] [Google Scholar]
- 44.Matsuyama, T., Y. Takagi, Y. Nakagawa, H. Itoh, J. Wakita, and M. Matsushita. 2000. Dynamic aspects of the structured cell population in a swarming colony of Proteus mirabilis. J. Bacteriol. 182:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mobley, H. L., and J. W. Warren. 1987. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 25:2216-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niki, H., A. Jaffe, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10:183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Fallon, E., S. Gautam, and E. M. D'Agata. 2009. Colonization with multidrug-resistant gram-negative bacteria: prolonged duration and frequent cocolonization. Clin. Infect. Dis. 48:1375-1381. [DOI] [PubMed] [Google Scholar]
- 48.Osborn, M. J., and L. Rothfield. 2007. Cell shape determination in Escherichia coli. Curr. Opin. Microbiol. 10:606-610. [DOI] [PubMed] [Google Scholar]
- 49.Overhage, J., M. Bains, M. D. Brazas, and R. E. Hancock. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearson, M. M., and H. L. Mobley. 2008. Repression of motility during fimbrial expression: identification of 14 mrpJ gene paralogues in Proteus mirabilis. Mol. Microbiol. 69:548-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson, M. M., and H. L. Mobley. 2007. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. J. Med. Microbiol. 56:1277-1283. [DOI] [PubMed] [Google Scholar]
- 52.Pearson, M. M., M. Sebaihia, C. Churcher, M. A. Quail, A. S. Seshasayee, N. M. Luscombe, Z. Abdellah, C. Arrosmith, B. Atkin, T. Chillingworth, H. Hauser, K. Jagels, S. Moule, K. Mungall, H. Norbertczak, E. Rabbinowitsch, D. Walker, S. Whithead, N. R. Thomson, P. N. Rather, J. Parkhill, and H. L. Mobley. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rauprich, O., M. Matsushita, C. J. Weijer, F. Siegert, S. E. Esipov, and J. A. Shapiro. 1996. Periodic phenomena in Proteus mirabilis swarm colony development. J. Bacteriol. 178:6525-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabbuba, N., G. Hughes, and D. J. Stickler. 2002. The migration of Proteus mirabilis and other urinary tract pathogens over Foley catheters. BJU Int. 89:55-60. [DOI] [PubMed] [Google Scholar]
- 55.Schild, S., R. Tamayo, E. J. Nelson, F. Qadri, S. B. Calderwood, and A. Camilli. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider, R., C. V. Lockatell, D. Johnson, and R. Belas. 2002. Detection and mutation of a luxS-encoded autoinducer in Proteus mirabilis. Microbiology 148:773-782. [DOI] [PubMed] [Google Scholar]
- 57.Stahl, S. J., K. R. Stewart, and F. D. Williams. 1983. Extracellular slime associated with Proteus mirabilis during swarming. J. Bacteriol. 154:930-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevenson, L. G., and P. N. Rather. 2006. A novel gene involved in regulating the flagellar gene cascade in Proteus mirabilis. J. Bacteriol. 188:7830-7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437-446. [DOI] [PubMed] [Google Scholar]
- 60.Turnbull, A. L., and M. G. Surette. 2008. d-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology 154:3410-3419. [DOI] [PubMed] [Google Scholar]
- 61.Verstraeten, N., K. Braeken, B. Debkumari, M. Fauvart, J. Fransaer, J. Vermant, and J. Michiels. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496-506. [DOI] [PubMed] [Google Scholar]
- 62.Walker, K. E., S. Moghaddame-Jafari, C. V. Lockatell, D. Johnson, and R. Belas. 1999. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 32:825-836. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 64.Williams, F. D., D. M. Anderson, P. S. Hoffman, R. H. Schwarzhoff, and S. Leonard. 1976. Evidence against the involvement of chemotaxis in swarming of Proteus mirabilis. J. Bacteriol. 127:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams, F. D., and R. H. Schwarzhoff. 1978. Nature of the swarming phenomenon in Proteus. Annu. Rev. Microbiol. 32:101-122. [DOI] [PubMed] [Google Scholar]
- 66.Yamanaka, K., T. Ogura, H. Niki, and S. Hiraga. 1996. Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250:241-251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.