Abstract
Yersinia enterocolitica is an important human pathogen. Y. enterocolitica must adapt to the host environment, and temperature is an important cue regulating the expression of most Yersinia virulence factors. Here, we report that Y. enterocolitica 8081 serotype O:8 synthesized tetra-acylated lipid A at 37°C but that hexa-acylated lipid A predominated at 21°C. By mass spectrometry and genetic methods, we have shown that the Y. enterocolitica msbB, htrB, and lpxP homologues encode the acyltransferases responsible for the addition of C12, C14 and C16:1, respectively, to lipid A. The expression levels of the acyltransferases were temperature regulated. Levels of expression of msbB and lpxP were higher at 21°C than at 37°C, whereas the level of expression of htrB was higher at 37°C. At 21°C, an lpxP mutant was the strain most susceptible to polymyxin B, whereas at 37°C, an htrB mutant was the most susceptible. We present evidence that the lipid A acylation status affects the expression of Yersinia virulence factors. Thus, expression of flhDC, the flagellar master regulatory operon, was downregulated in msbB and lpxP mutants, with a concomitant decrease in motility. Expression of the phospholipase yplA was also downregulated in both mutants. inv expression was downregulated in msbB and htrB mutants, and consistent with this finding, invasion of HeLa cells was diminished. However, the expression of rovA, the positive regulator of inv, was not affected in the mutants. The levels of pYV-encoded virulence factors Yops and YadA in the acyltransferase mutants were not affected. Finally, we show that only the htrB mutant was attenuated in vivo.
Lipopolysaccharide (LPS) is located in the outer leaflet of the outer membrane (OM) of Gram-negative bacteria. The molecular structure of LPS is rather unique: it is an amphiphilic compound with a hydrophobic region, lipid A, adjacent to a dense, negatively charged polysaccharide. Divalent cations bridge these negatively charged regions, causing the exclusion of phospholipids from the outer leaflet of the OM. This feature, together with the tight packing of lipid A hydrocarbon chains, creates a permeability barrier against toxic compounds (for a review, see references 49 and 50).
In Escherichia coli and Salmonella enterica serovar Typhimurium, lipid A is formed by a β(1′-6)-linked disaccharide of glucosamine phosphorylated at the 1 and 4′ positions, with positions 2, 3, 2′, and 3′ acylated with R-3-hydroxymyristoyl groups, the so-called lipid IVA, whose can be 2′- and 3′-R-3-hydroxymyristoyl groups further acylated with laureate (C12) and myristate (C14), respectively, by the actions of the so-called late acyltransferases HtrB (LpxL) and MsbB (LpxM), respectively (57). When E. coli is grown at 12°C, LpxP, the cold-temperature-specific late acyltransferase, acts instead of HtrB (LpxL), adding palmitoleate (C16:1) (57). Lipid A secondary acyltransferases have been shown to utilize primarily acyl-acyl carrier proteins (acyl-ACPs) as their acyl chain donors.
Yersinia enterocolitica is a human pathogen that causes a broad range of gastrointestinal syndromes (12). This organism is found widely in nature, in both aquatic and animal reservoirs. Infections are usually acquired by the ingestion of food or contaminated water, after which bacteria migrate through the intestinal tract to the terminal ileum (12). To infect humans, Y. enterocolitica must adapt to the host environment, and it possesses a number of virulence factors that help it to colonize the intestinal tract and to resist host defense mechanisms (44, 68). Temperature regulates most, if not all, virulence factors of yersiniae (44, 68). One example of a temperature-dependent trait is the expression of LPS O antigen. Optimum expression occurs when bacteria are grown at room temperature (RT; 22 to 25°C) (8, 11). In contrast, when they are grown at 37°C, the host temperature, only trace amounts of O antigen are produced (8, 11). This is because the two transcriptional units of the O-antigen gene cluster are repressed at 37°C (8, 11).
Several studies have shown that Gram-negative bacteria can modify lipid A in response to environmental conditions. For example, Salmonella and Pseudomonas modify the numbers and lengths of hydrocarbon chains attached to lipid A and add aminoarabinose and phosphoethanolamine to lipid A (26, 27, 31, 33, 34, 40). The available evidence shows that these lipid A-regulated changes are important for virulence (31, 33, 45). Various reports suggest that yersiniae modify lipid A in response to the growth temperature (6, 9, 38, 58, 59), and this pattern is particularly clear for Y. pestis. Thus, at 37°C, Y. pestis synthesizes a tetra-acyl lipid A lacking any secondary acylation (38, 58). However, at 21°C, lipid A is mainly hexa-acylated (38, 58). In this case, the 2′- and 3′-R-3-hydroxymyristoyl groups are acylated with laureate (C12) and palmitoleate (C16:1), with this form of lipid A resembling the lipid A produced by E. coli grown at 12°C (38, 58, 72). Recently, Rebeil and coworkers (59) have presented evidence explaining these temperature-dependent variations. The Y. pestis genome does not contain an htrB (lpxL) homologue, and the levels of expression of the late acyltransferase genes msbB (lpxM) and lpxP are higher at 21°C than at 37°C.
We and others have observed that Y. enterocolitica modifies lipid A in response to temperature, producing less acylated lipid A at 37°C than at 21°C (9, 52, 58). These data led us to hypothesize that Y. enterocolitica late acyltransferases could be temperature controlled. In this work, we first identified Y. enterocolitica homologues of msbB (lpxM), htrB (lpxL), and lpxP which are responsible for lipid A acylation. We then studied the roles of the gene products in temperature-dependent variations of lipid A. Finally, we examined the possible connection between the lipid A acylation status and the expression of other virulence factors of Y. enterocolitica and the roles of these acyltransferases in virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise indicated, Yersinia strains were grown in Luria-Bertani (LB) medium at either 21°C (RT) or 37°C. When appropriate, antibiotics were added to the growth medium at the following concentrations: ampicillin (Amp), 100 μg/ml for Y. enterocolitica and 50 μg/ml for E. coli; kanamycin (Km), 100 μg/ml in agar plates for Y. enterocolitica, 50 μg/ml in agar plates for E. coli, and 20 μg/ml in broth; chloramphenicol (Cm), 25 μg/ml; tetracycline (Tet), 12.5 μg/ml; trimethoprim (Tp), 100 μg/ml; and streptomycin (Str), 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype and/or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| C600 | thi thr leuB tonA lacY supE | 4 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)](Con) | |
| S17-1λpir | recA thi pro hsd(r− m+) RP4::2-Tc::Mu::Km Tn7 λ pir | |
| CC118-1λpir | Δ(ara-leu)7697 araD139 ΔlacX74 galE galK ΔphoA20 thi-1 rpsE rpoB argE(Am) recA1 | |
| Y. enterocolitica strains | ||
| 8081-R−M+ (YeO8) | R− M+ derivative of wild-type strain 8081; pYV+ | 80 |
| YeO8-ΔmsbBGB2 | YeO8 ΔmsbB::Km-GenBlock Kmr pYV+; msbB gene is inactivated | This study |
| YeO8-ΔhtrBGB | YeO8 ΔhtrB::Km-GenBlock Kmr pYV+; htrB gene is inactivated | This study |
| YeO8-ΔlpxPGB | YeO8 ΔlpxP::Km-GenBlock Kmr pYV+; lpxP gene is inactivated | This study |
| YeO8-ΔlpxPGB::pKNOCKYeinthtrB | YeO8-ΔlpxPGB htrB::pKNOCKYeinthtrBp Cmr pYV+; htrB and lpxP genes are inactivated | This study |
| YeO8::pGPLYePmsbB | YeO8 reporter strain with pGPLYePmsbB integrated into the msbB locus by single crossover; Ampr | This study |
| YeO8::pGPLYePhtrBTp | YeO8 reporter strain with pGPLYePhtrBTp integrated into the htrB locus by single crossover; Tpr | This study |
| YeO8::pGPLYePlpxP | YeO8 reporter strain with pGPLYePlpxP integrated into the lpxP locus by single crossover; Ampr | This study |
| YeO8-flhDC::lucFF | YeO8 reporter strain with pSRFlhDCO8 integrated into the flhDC locus by single crossover; Cmr | This study |
| YeO8-ΔmsbBGB2-flhDC::lucFF | YeO8-ΔmsbBGB2 reporter strain with pSRFlhDCO8 integrated into the flhDC locus by single crossover; Cmr | This study |
| YeO8-ΔhtrBGB-flhDC::lucFF | YeO8-ΔhtrBGB reporter strain with pSRFlhDCO8 integrated into the flhDC locus by single crossover; Cmr | This study |
| YeO8-ΔlpxPGB-flhDC::lucFF | YeO8-ΔlpxPGB reporter strain with pSRFlhDCO8 integrated into the flhDC locus by single crossover; Cmr | This study |
| YeO8-inv::phoA | YeO8 reporter strain with pINP41 integrated into the inv locus by single crossover; Cmr | This study |
| YeO8-ΔmsbBGB2-inv::phoA | YeO8-ΔmsbBGB2 reporter strain with pINP41 integrated into the inv locus by single crossover; Cmr | This study |
| YeO8-ΔhtrBGB-inv::phoA | YeO8-ΔhtrBGB reporter strain with pINP41 integrated into the inv locus by single crossover; Cmr | This study |
| YeO8-ΔlpxPGB-inv::phoA | YeO8-ΔlpxPGB reporter strain with pINP41 integrated into the inv locus by single crossover; Cmr | This study |
| YeO8-yplA::lacZYA | YeO8 reporter strain with pDHS45 integrated into the yplA locus by single crossover; Cmr | This study |
| YeO8-ΔmsbBGB2-yplA::lacZYA | YeO8-ΔmsbBGB2 reporter strain with pDHS45 integrated into the yplA locus by single crossover; Cmr | This study |
| YeO8-ΔhtrBGB-yplA::lacZYA | YeO8-ΔhtrBGB reporter strain with pDHS45 integrated into the yplA locus by single crossover; Cmr | This study |
| YeO8-ΔlpxPGB-yplA::lacZYA | YeO8-ΔlpxPGB reporter strain with pDHS45 integrated into the yplA locus by single crossover; Cmr | This study |
| Plasmids | ||
| pKNG101 | oriR6K Mob+sacB Strr | 37 |
| pGEM-T Easy | Cloning plasmid; Ampr | Promega |
| pKNOCK-Cm | oriR6K Mob+ Cmr | 3 |
| pTM100 | Mob+; derivative of pACYC184; Cmr Tetr | 46 |
| pUC4K | Source of GenBlock cassette; Ampr Kmr | Pharmacia |
| pCR-Blunt II-TOPO | Cloning plasmid; Ampr | Invitrogen |
| pGPL01 | Firefly luciferase transcriptional fusion suicide vector; Ampr | 32 |
| pRV34 | lucFF cloned into EcoRV site of pTM100; Cmr | M. Skurnik, unpublished data |
| pRV1 | Suicide vector; derivative of pJM703.1; Cmr | 65 |
| pGEMTΔmsbBGB | pGEM-T Easy containing ΔmsbB::Km-GenBlock; Ampr Kmr | This study |
| pGEMTΔhtrBGB | pGEM-T Easy containing ΔhtrB::Km-GenBlock; Ampr Kmr | This study |
| pGEMTΔlpxPGB | pGEM-T Easy containing ΔlpxP::Km-GenBlock; Ampr Kmr | This study |
| pKNGΔmsbBGB | pKNG101 containing ΔmsbB::Km-GenBlock; Strr Kmr | This study |
| pKNGΔhtrBGB | pKNG101 containing ΔhtrB::Km-GenBlock; Strr Kmr | This study |
| pKNGΔlpxPGB | pKNG101 containing ΔlpxP::Km-GenBlock; Strr Kmr | This study |
| pKNOCKYeinthtrB | pKNOCK-Cm containing htrB internal fragment; Cmr | This study |
| pTMYemsbB | 2.1-kb wild-type msbB locus cloned into pTM100; Tetr | This study |
| pTMYehtrB | 2.1-kb wild-type htrB locus cloned into pTM100; Tetr | This study |
| pTMYelpxP | 2-kb wild-type lpxP locus cloned into pTM100; Tetr | This study |
| pINP41 | pEP184 containing invΔ412::phoA; Cmr | 53 |
| pDHS45 | pFUSE containing yplXA′::lacZYA; Cmr | 63 |
| pSRFlhDCO8 | pRV1 containing flhDC::lucFF; Cmr | 10 |
| pRVProrovAlucFF | pRV1 containig rovA::lucFF; Cmr | This study |
| pGPLYePmsbB | pGPL01 containing a 980-bp DNA fragment corresponding to the msbB promoter region; Ampr | This study |
| pGPLYePlpxP | pGPL01 containing a 1.1-kb DNA fragment corresponding to the lpxP promoter region; Ampr | This study |
| pGPLYePhtrB | pGPL01 containing a 1-kb DNA fragment corresponding to the htrB promoter region; Ampr | This study |
| pGPLYePhtrBTp | Trimethoprim resistance cassette cloned into PstI site of pGPLYePhtrB; Tpr | This study |
Construction of msbB, htrB, and lpxP mutant strains and complementation plasmids.
Predicted Y. enterocolitica 8081 homologues of the E. coli msbB, htrB, and lpxP acyltransferase genes were identified in the Y. enterocolitica genome sequence from the Wellcome Trust Sanger Institute (EMBL accession number AM286415) (70) as locus tags YE2386, YE1612, and YE3829, respectively. DNA fragments of 2.1 kb for msbB and htrB and 2 kb for lpxP were PCR amplified (primers used are listed in Table 2), gel purified, and cloned into pGEM-T Easy (Promega) to obtain pGEMTtmsbB, pGEMThtrB, and pGEMTlpxP, respectively. These plasmids were amplified by inverse PCR using the method described by Byrappa et al. (14) (primers used are listed in Table 2) to delete internal fragments of 150, 100, and 56 bp in the coding regions of msbB, htrB, and lpxP, respectively. A kanamycin resistance cassette, obtained as a 1.4-kb PstI blunt-ended fragment from pUC4K (Pharmacia), was cloned into the plasmids obtained by inverse PCR to generate pGEMTΔmsbBGB, pGEMTΔhtrBGB, and pGEMTΔlpxPGB. Alleles containing the GenBlock cassette fragment from pUC4K, designated ΔmsbB::GenBlock and ΔlpxP::GenBlock, were gel purified after PvuII digestion of pGEMTΔmsbBGB and pGEMTΔmsbBGB, respectively, and cloned into SmaI-digested pKNG101, whereas the ΔhtrB::GenBlock allele was gel purified after SalI-XbaI digestion of pGEMTΔhtrBGB and cloned into similarly digested pKNG101 (Table 1). pKNG101 is a suicide vector that carries the defective pir-negative origin of replication of R6K, the RK2 origin of transfer, and an Str resistance marker (37). It also carries the sacBR genes that mediate sucrose sensitivity as a positive selection marker for the excision of the vector after double crossover (37). These three suicide plasmids were introduced by electroporation into E. coli S17-1λpir, from which the plasmids were mobilized into Y. enterocolitica strain YeO8. Strr Kmr transconjugates were selected after growth on Yersinia selective agar medium plates (Oxoid) supplemented with Km and Str. Bacteria from 10 individual colonies were pooled and allowed to grow in LB medium without any antibiotic overnight at RT. Bacterial cultures were serially diluted in LB medium lacking NaCl and containing 10% sucrose, and plates were incubated at RT. The recombinants that survived exposure to 10% sucrose were checked for their resistance to Str, and Kmr Strs recombinants were selected and named YeO8-ΔmsbBGB2, YeO8-ΔhtrBGB, and YeO8-ΔlpxPGB. The appropriate replacement of the wild-type alleles by the mutant ones was confirmed by PCR and Southern blotting (data not shown). To construct a double mutant lacking htrB and lpxP, an internal fragment of the htrB gene (386 bp) was amplified by PCR using Vent polymerase. The fragment was gel purified, digested with BamHI, and cloned into BamHI-EcoRV-digested pKNOCK-Cm (3) to obtain pKNOCKYeinthtrB. This plasmid was mobilized into YeO8-ΔlpxPGB, and Kmr Cmr transconjugates were selected after growth on Yersinia selective agar medium plates (Oxoid) supplemented with Km and Cm. Integration of the suicide vector into the htrB locus by homologous recombination was confirmed by Southern hybridization analysis (data not shown).
TABLE 2.
Primersa used in this study
| Target gene | Use | Sequence (5′ to 3′) |
|---|---|---|
| msbB | Mutagenesis | TAACCAACCCAATGCCAACC |
| GCGATGCCAGTTGGCACA | ||
| Inverse PCR | GAGTTTGTTGATTTCTTCGCGAC | |
| CAAACCGCAAGCGTGCGCTG | ||
| Complementation | TAACCAACCCAATGCCAACC | |
| GCGATGCCAGTTGGCACA | ||
| Amplification of promoter region | GCAAACTTTCCGGCAACACG | |
| GGCCGGTTTGACTGGCGCA | ||
| htrB | Mutagenesis | AGCGACAAAACCGGGAATGC |
| CAATGGCCATATAGGGCTGC | ||
| Inverse PCR | TGCAGACTTGGGGCCGGATG | |
| GCGCACCCAGTTCCAGCGTC | ||
| Complementation | AGCGACAAAACCGGGAATGC | |
| CAATGGCCATATAGGGCTGC | ||
| Amplification of promoter region | AGCGACAAAACCGGGAATGC | |
| CAATGGCCATATAGGGCTGC | ||
| Amplification of internal fragment | CGGATCCGTCAACGGCAAACAGCGGCAC | |
| GGGTTATTTGAAACCGGGATG | ||
| lpxP | Mutagenesis | TTCAGTAACTTGTACATGGCGACC |
| TAACAGCGAAGTCGTTGCCTG | ||
| Inverse PCR | TCAGCGCCTGCACCATGCCAC | |
| CAGATCAGGACTACGGCCCAC | ||
| Complementation | TTCAGTAACTTGTACATGGCGACC | |
| TAACAGCGAAGTCGTTGCCTG | ||
| Amplification of promoter region | CAAGCTTGGACTCTGTGGGTATGCGGT | |
| GCTTTGTCCATATCAGGGAAGC | ||
| rovA | Amplification of promoter region | ACAAATGTATATATACCGTCGATGC |
| CGCTTTGATCATAAATGGCTCG | ||
| lucFF | Amplification of coding region | GAGGAGAAATTAACTATGAGGGG |
| TTACAATTTGGACTTTCCGCC |
To complement the mutants, DNA fragments of 2.1 kb for msbB and htrB and 2 kb for lpxP were PCR amplified using Vent polymerase (Table 2 lists the primers used). The fragments, which contained the putative promoter and coding region of each acyltransferase gene, were gel purified, phosphorylated, and cloned into the ScaI site of the medium-copy-number plasmid pTM100 to obtain pTMYemsbB, pTMYehtrB, and pTMYelpxP. These plasmids were introduced into E. coli C600 and then mobilized into Y. enterocolitica strains by triparental conjugation using the helper strain E. coli HB101/pRK2013.
Isolation and analysis of lipid A.
Lipid As were extracted using an ammonium hydroxide-isobutyric acid method and subjected to negative-ion matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry analysis (23). Briefly, lyophilized crude cells (10 mg) were resuspended in 400 μl isobutyric acid-1 M ammonium hydroxide (5:3, vol/vol) and were incubated in a screw-cap test tube at 100°C for 2 h with occasional subjection to a vortex. Samples were cooled in ice water and centrifuged (at 2,000 × g for 15 min). The supernatant was transferred into a new tube, diluted with an equal volume of water, and lyophilized. The sample was then washed twice with 400 μl methanol and centrifuged (at 2,000 × g for 15 min). The insoluble lipid A was solubilized in 100 to 200 μl chloroform-methanol-water (3:1.5:0.25, vol/vol/vol). Analyses were performed with a Bruker Autoflex II MALDI-TOF mass spectrometer (Bruker Daltonics, Inc.) in negative reflective mode with delayed extraction. Each spectrum was an average of 300 shots. The ion-accelerating voltage was set at 20 kV. Dihydroxybenzoic acid (Sigma Chemical Co., St. Louis, MO) was used as a matrix. A sample of a few microliters of a lipid A suspension (1 mg/ml) was desalted with a few grains of ion-exchange resin (Dowex 50W-X8; H+) in an Eppendorf tube. A 1-μl aliquot of the suspension (50 to 100 μl) was deposited onto the target and covered with the same amount of the matrix suspended at 10 mg/ml in a 0.1 M solution of citric acid. Different ratios between the samples and dihydroxybenzoic acid were used when necessary. A peptide calibration standard (Bruker Daltonics) was used to calibrate the MALDI-TOF mass spectrometer. Further calibration for lipid A analysis was performed externally using lipid A extracted from E. coli strain MG1655 grown in LB medium at 37°C. Interpretation of the negative-ion spectra is based on data from earlier studies showing that ions with m/z values higher than 1,000 gave signals proportional to the corresponding lipid A species present in the preparation (6, 42, 52, 58). Important theoretical m/z values for the interpretation of peaks found in this study are as follows: lipid IVA, 1,405; C12, 182, C14, 210; C16:1, 236.2; aminoarabinose (AraNH), 131.1; and C16, 239 (Table 3).
TABLE 3.
Structural interpretations of lipid A species detected by mass spectrometry in this study
| m/z | Compositiona of lipid IVA | Lipid A |
||
|---|---|---|---|---|
| 3′ secondary substitution | 2′ secondary substitution | Modification | ||
| 1,178 | 2 × GlcN, 2 × HPO3, 3 × C14:0 (3-OH) | |||
| 1,388 | 2 × GlcN, 2 × HPO3, 3 × C14:0 (3-OH) | C14 | ||
| 1,405 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | |||
| 1,414 | 2 × GlcN, 2 × HPO3, 3 × C14:0 (3-OH) | C16:1 | ||
| 1,588 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | ||
| 1,614 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C14 | ||
| 1,642 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C16:1 | ||
| 1,773 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C16:1 | AraNH | |
| 1,797 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | C14 | |
| 1,880 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C16:1 | C16 | |
| 1,824 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | C16:1 | |
| 1,954 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | C16:1 | AraNH |
| 2,036 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | C14 | C16 |
| 2,063 | 2 × GlcN, 2 × HPO3, 4 × C14:0 (3-OH) | C12 | C16:1 | C16 |
Numbers with multiplication signs indicate numbers of units of GlcN, HPO3, and C14:0.
Construction of msbB::lucFF, htrB::lucFF, and lpxP::lucFF reporter fusions.
DNA fragments of 0.98, 1, and 1.1 kb containing the promoter regions of the msbB, htrB, and lpxP genes, respectively, were amplified by PCR using Vent polymerase, gel purified, and cloned into pCR-Blunt II-TOPO (Invitrogen). The cloned fragments were sequenced to ensure that no mistakes were introduced during amplification. Promoter regions were obtained as EcoRI fragments which were gel purified and cloned into the EcoRI site of the suicide vector pGPL01 (32). This vector contains an R6K origin of replication and a firefly luciferase gene (lucFF) without a promoter. Plasmids in which the luciferase gene was under the control of the msbB, htrB, and lpxP promoters were identified by restriction digestion analysis and named pGPLYePmsbB, pGPLYePhtrB, and pGPLYePlpxP, respectively. A trimethoprim resistance cassette, obtained as a PstI fragment from p34S-Tp (20), was cloned into PstI-digested pGPLYePhtrB to obtain pGPLYePhtrBTp. This plasmid, pGPLYePmsbB, and pGPLYePlpxP were introduced into YeO8 by electroporation, and strains in which the suicide vectors were integrated into the genome by homologous recombination were selected. Integration was confirmed by Southern blotting (data not shown).
Construction of a rovA::lucFF reporter fusion.
A DNA fragment of 1.1 kb containing the promoter region of rovA was amplified by PCR using Vent polymerase, gel purified, and cloned into HincII-digested pUC18 (Sigma) to obtain pUCProvA. The cloned fragment was sequenced to ensure that no mistakes were introduced during amplification. The firefly luciferase gene (lucFF) coding region was amplified by PCR using pRV34 as a template. The PCR fragment (1.4 kb) was gel purified and cloned into KpnI-digested blunt-ended pUCProvA to obtain pUCProvAlucFF, which contained the promoterless lucFF gene under the control of the rovA promoter region. This fragment (2.5 kb) was amplified by PCR using Vent polymerase, gel purified, and cloned into the EcoRV site of the suicide vector pRV1 to create pRVProrovAlucFF. This plasmid was introduced into Yersinia strains by electroporation, and the strains in which the suicide vector was integrated into the genome by homologous recombination were selected. Integration was confirmed by Southern blotting (data not shown).
Luciferase activity.
Reporter strains were grown on an orbital incubator shaker (at 250 rpm) until they reached late log phase. A 100-μl aliquot of the bacterial suspension was transferred into an Eppendorf tube and mixed with 100 μl of luciferase assay reagent (1 mM d-luciferin [Synchem] in 100 mM citrate buffer, pH 5). Luminescence was immediately measured with a luminometer (Hidex) and expressed as relative light units (RLU) per unit of optical density at 540 nm (OD540). All measurements were carried out in quadruplicate on at least three separate occasions.
Assessment of permeability to hydrophobic compounds.
Sensitivities to novobiocin (50 μg), crystal violet (40 μg), sodium dodecyl sulfate (SDS; 100 μg), and deoxycholate (DOC; 100 μg) on LB agar plates were assessed using the disk diffusion test. The antibiotic disks (concentration disks [diameter, 6.5 mm; Difco Laboratories]) were prepared by being loaded with the compounds dissolved in 20 μl of distilled water. The disks were dried overnight at 37°C and kept at 4°C until needed. Fresh exponentially growing bacteria were resuspended in phosphate-buffered saline (PBS) to a final concentration of 108 CFU/ml, and a lawn was prepared on each agar plate with a sterile swab. The loaded disks were placed onto the lawns, and the plates were incubated for 18 h at the growth temperature for the cultures. The diameters of the inhibition halos were measured, and after subtraction of the disk diameter, they were expressed in inhibition units (10 inhibition units is equal to 1 mm). All experiments were performed with duplicate samples from two independently grown cultures of bacteria.
Antimicrobial radial diffusion assay.
The antimicrobial activity of polymyxin B (purchased from Sigma) was assayed using the radial diffusion method described previously (41). Briefly, bacteria were grown in 5 ml of LB medium in a 15-ml Falcon tube, collected in the exponential phase of growth, and resuspended in PBS. An underlay gel that contained 1% (wt/vol) agarose (SeaKem LE agarose; FMC, Rockland, ME), 2 mM HEPES (pH 7.2), and 0.3 mg of tryptic soy broth (TSB) powder per ml was equilibrated at 50°C and inoculated with the different bacteria to a final concentration of 6.1 × 105 CFU per ml of molten gel. This gel was poured into standard petri dishes, and after solidification of the gel, small wells with capacities of 10 μl were carved. Aliquots of 5 μl of polymyxin B were added and allowed to diffuse for 3 h at 37°C. After that, a 10-ml overlay gel composed of 1% agarose and 6% TSB powder in water was poured on top of the previous gel and the plates were incubated overnight at 37°C. The next day, the diameters of the inhibition halos were measured to the nearest 1 mm and, after subtraction of the diameter of the well, were expressed in inhibition units (10 inhibition units = 1 mm). The minimal bactericidal concentration (MBC) was estimated by performing linear regression analysis (with the number of units versus the log10 concentration) and determining the x axis intercepts. All the experiments were run in quadruplicate on three independent occasions.
Analysis of motility and flhDC expression.
Phenotypic assays for swimming motility were initiated by stabbing 2 μl of an overnight culture at the center of an agar plate containing 0.3% agar and 1% tryptone (10, 79). Plates were analyzed after 24 h of incubation at RT, and the diameters of the halos indicating migration from the inoculation points were compared. Experiments were run in quadruplicate on three independent occasions.
To measure flhDC expression, plasmid pSRFlhDCO8 (10) carrying the transcriptional fusion flhDC::lucFF was integrated into the genomes of the strains by homologous recombination. Integration was confirmed by Southern blotting (data not shown). Reporter strains were grown on 1% tryptone on an orbital incubator shaker (at 250 rpm) until they reached late log phase, and luminescence was determined as described previously.
β-Galactosidase and AP activities.
β-Galactosidase activity was determined as described previously with bacteria grown in 1% tryptone at RT (47). Alkaline phosphatase (AP) activity in permeabilized cells was determined, and the results are expressed in enzyme units per unit of OD600 as described previously (43). Experiments were run in duplicate on three independent occasions.
Invasion assay.
Strains were grown aerobically for 4 h at RT, pelleted, and resuspended in PBS to an OD540 of 0.3. Bacterial suspensions were added to subconfluent HeLa cells at a multiplicity of infection of ∼100:1. After 60 min of infection, monolayers were washed twice with PBS and then incubated for an additional 90 min in medium containing gentamicin (100 μg/ml) to kill extracellular bacteria. This treatment was long enough to kill all extracellular bacteria. After this period, cells were washed three times with PBS and lysed with 1% Triton X-100 and bacteria were plated. Experiments were carried out in triplicate on at least two independent occasions. Invasiveness was calculated as follows: percent invasiveness = 100 × (number of bacteria resistant to gentamicin/initial number of bacteria added).
Analysis of Yop secretion.
Overnight cultures of Y. enterocolitica strains were diluted 1:50 in 25 ml of TSB supplemented with 20 mM MgCl2 and 20 mM sodium oxalate in 100-ml flasks. Cultures were incubated with aeration at 21°C for 2.5 h and then transferred to 37°C for 3 h. The OD540 of the culture was measured, and the bacterial cells were collected by centrifugation at 1,500 × g for 30 min. Ammonium sulfate (final concentration, 47.5% [wt/vol]) was used to precipitate proteins from 20 ml of the supernatant. After overnight incubation at 4°C, proteins were collected by centrifugation (at 3,000 × g for 30 min at 4°C) and washed twice with 1.5 ml of water. Dried protein pellets were resuspended in 50 to 80 μl of sample buffer and normalized according to the cell count. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) using 12% polyacrylamide gels, and proteins were visualized by Coomassie brilliant blue staining.
Analysis of OM proteins.
OM proteins were extracted from bacteria grown in 5-ml LB cultures either at 37°C or at RT as described previously (22), with modifications. Briefly, cells from 1.5-ml samples of the overnight cultures were recovered by centrifugation (at 3,500 × g for 3 min). Numbers of CFU were determined by plating serial dilutions. Cells were washed once with 1 ml TE buffer (10 mM Tris-HCl [pH 8]-1 mM EDTA) and finally resuspended in 800 μl TE buffer. Cells were broken by sonication with a Branson digital Sonifier (microtip diameter, 1/8 in.; amplitude, 10%) for three 30-s cycles: each cycle comprised three 10-s sonication steps separated by 1 s of no sonication, with 30 s of no sonication between the cycles. Unbroken cells were eliminated by centrifugation (at 3,500 × g for 2 min), and cell envelopes were recovered at 16,000 × g for 30 min. The inner membranes were solubilized in freshly prepared 2% sodium lauroyl sarcosinate (wt/vol) for 30 min at RT, and the insoluble OM proteins were recovered by centrifugation (at 16,000 × g for 30 min at RT). A second solubilization step for the inner membranes was carried out. Finally, the OM proteins were resuspended in 70 to 80 μl of sample buffer and normalized according to the cell count. Samples were analyzed by SDS-PAGE using 12% polyacrylamide gels, and proteins were visualized by Coomassie brilliant blue staining.
Animal experiments.
Six- to 7-week-old specific-virus-free male BALB/c mice were inoculated orally with 100 μl of a bacterial mixture containing 109 bacteria of the wild-type strain or an acyltransferase mutant strain. The mixture was serially diluted, and appropriate dilutions were plated to determine exact bacterial counts. At different time points (3 and 7 days) after infection, five animals per bacterial strain were euthanized by cervical dislocation and their spleens, livers, and Peyer's patches were aseptically removed, weighed, and homogenized in 2, 5, and 1 ml, respectively, of PBS. The bacterial loads recovered from the infected organs were determined by plating homogenates and serial dilutions onto Yersinia selective agar medium plates (Oxoid) to select YeO8 and onto Km-containing LB plates to select YeO8-ΔmsbBGB2, YeO8-ΔhtrBGB, and YeO8-ΔlpxPGB. Results were reported as log numbers of CFU per gram of tissue. Mice were treated in accordance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (directive 86/609/EEC) and in agreement with the guidelines of the Bioethical Committee of the University of the Balearic Islands.
Statistical analysis.
The results were analyzed by analysis of variance (ANOVA) or the one-sample t test using GraphPad Prism software (GraphPad Software Inc.). Results are given as means ± standard deviations (SD). A P value of <0.05 was considered to be statistically significant and was denoted in the figures with an asterisk.
RESULTS
Temperature-dependent changes in Y. enterocolitica lipid A structure.
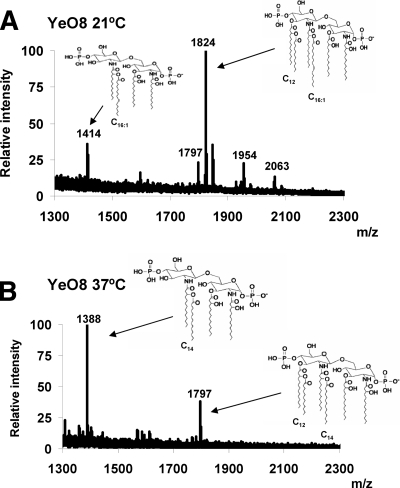
Lipid A species synthesized by Y. enterocolitica 8081 serotype O:8 (strain YeO8) grown either at 21 or 37°C were characterized by MALDI-TOF mass spectrometry (Fig. 1). Lipid A extracts from YeO8 grown at 21°C contained predominantly hexa-acylated species (m/z 1,824) (Fig. 1A) corresponding to two glucosamine residues, two phosphate groups, four 3-OH-C14 units, one C12 unit, and one C16:1 unit. Another peak (m/z 1,797) may represent a hexa-acylated form of lipid A containing four 3-OH-C14 units, one C12 unit, and one C14 unit. The species with m/z 1,414 has also been found in Y. pseudotuberculosis grown at RT (58) and may represent a tetra-acylated lipid A containing three 3-OH-C14 units and one C16:1 unit. Other species detected were consistent with the addition of aminoarabinose to the hexa-acylated form (m/z 1,954) and with the addition of C16 to the hexa-acylated form, producing hepta-acylated lipid A (m/z 2,063) (Fig. 1A).
FIG. 1.
Negative-ion MALDI-TOF mass spectrometry spectra for lipid A isolated from Y. enterocolitica serotype O:8 grown at 21°C (A) or at 37°C (B). Proposed structures corresponding to major peaks and acyl group positions follow previously reported structures for Yersinia and other Gram-negative bacteria. The results in both panels are representative of three independent lipid A extractions.
Lipid A extracts from cells grown at 37°C appeared to be identical to those described by Rebeil et al. and Oertelt et al. (52, 58). The main species were a tetra-acylated form (m/z 1,388), containing two glucosamine residues, two phosphate groups, three 3-OH-C14 units, and one C14 unit, and a hexa-acylated form (m/z 1,797) (Fig. 1B).
Roles of Y. enterocolitica lipid A late acyltransferases in temperature-dependent lipid A structure.
Lipid A acylation is dependent on the activities of the late acyltransferases HtrB (LpxL) and MsbB (LpxM) (57). A third acyltransferase, LpxP, acts instead of HtrB (LpxL) when bacteria are grown at 12°C (57). Our data were consistent with the idea that YeO8 homologues of MsbB, HtrB, and LpxP could be responsible for the addition of C12, C14, and C16:1, respectively, to lipid A. Three homologues of lipid A acyltransferase genes (corresponding to locus tags YE2386, YE1612, and YE3829, respectively) were identified in the YeO8 genome (EMBL accession number AM286415) (70). In silico analysis suggests that the three genes do not form part of any operon. Predicted MsbB, HtrB, and LpxP homologues have 65, 69, and 80% amino acid identities to E. coli MsbB (LpxM), HtrB (LpxL), and LpxP proteins, respectively. Furthermore, YeO8 MsbB has 84 and 94% amino acid identities to Y. pestis CO92 (YPO2063) and Y. pseudotuberculosis IP32953 (YPTB2046) MsbB homologues, whereas YeO8 HtrB has 91% amino acid identity to the Y. pseudotuberculosis IP32953 (YPTB2490) HtrB homologue. The Y. pestis genome does not contain an htrB homologue (59). Finally, YeO8 LpxP has 91 and 92% amino acid identities to Y. pestis CO92 (YPO3632) and Y. pseudotuberculosis IP32953 (YPTB3597) LpxP homologues. Each acyltransferase gene was mutated to determine whether these genes were responsible for the temperature-dependent lipid A variations. Growth rates were determined by measuring the OD540 at different time points until the cultures entered the stationary phase, and bacteria were enumerated by plating. No significant growth differences among strains at either RT or 37°C were found.
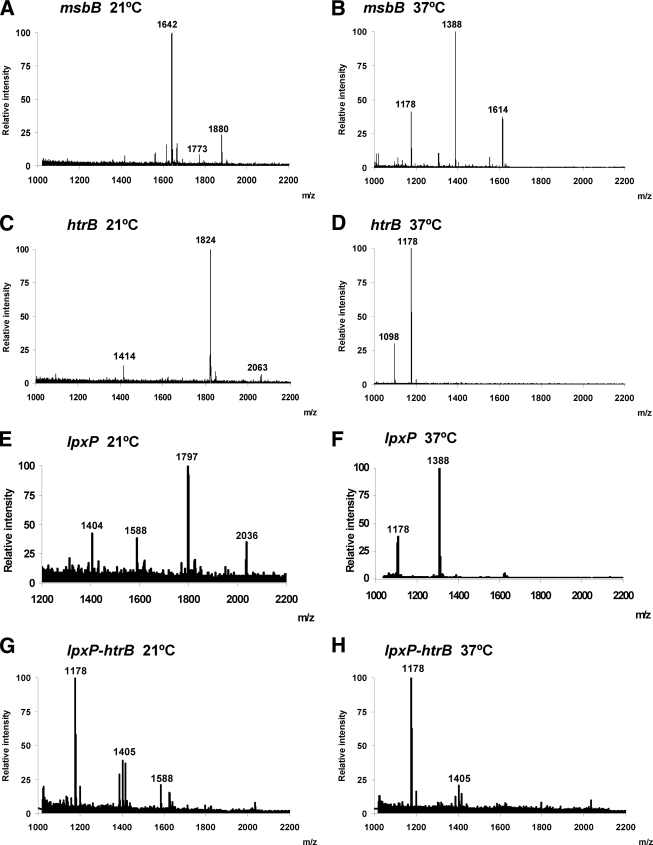
At 21°C, the msbB mutant (YeO8-ΔmsbBGB2) produced a lipid A form similar to that produced by the wild-type strain but lacking C12 (m/z 1,642). Other peaks may correspond to further additions of aminoarabinose (m/z 1,773) or C16 (m/z 1,880) (Fig. 2A). Lipid A extract from the mutant grown at 37°C lacked C12 and contained three major forms: a tetra-acylated form (m/z 1,388) also present in the sample from the wild-type strain, a penta-acylated form (m/z 1,614) containing four 3-OH-C14 units and one C14 unit, and a triacyl form of lipid A (m/z 1,178) (Fig. 2B). Lipid A from the htrB mutant (YeO8-ΔhtrBGB) lacked C14 (Fig. 2C and D). Lipid A species from the htrB mutant grown at 21°C were similar to those from the wild-type strain (compare Fig. 1A and Fig. 2C). In the sample from the htrB mutant grown at 37°C, the predominant form was triacyl lipid A (m/z 1,178). The lipid A sample from the lpxP mutant (YeO8-ΔlpxPGB) grown at 21°C lacked C16:1 and presented a major hexa-acylated form containing four 3-OH-C14 units, one C12 unit, and one C14 unit (m/z 1,797) (Fig. 2E). This molecular species could be substituted by C16 (m/z 2,036). Other species detected were consistent with penta-acylated lipid A lacking C12 (m/z 1,588) and tetra-acylated lipid A (m/z 1,405) lacking C12 and C14. These two molecular species have been found previously in yersiniae (58). Lipid A from the lpxP mutant grown at 37°C was tetra-acylated (m/z 1,388) (Fig. 2F). These results suggest that in cells grown at 21°C, HtrB acts in the absence of LpxP, adding C14 to lipid A. To further substantiate this finding, a double mutant lacking lpxP and htrB was constructed and its lipid A was analyzed. The lipid A sample from this strain grown at 21°C contained a molecular species with m/z 1,588, consistent with the absence of C14 in the main molecular species, with m/z 1,797, from the lpxP mutant. Other species detected were consistent with tetra- and triacylated lipid A forms (m/z 1,405 and 1,178, respectively). These species also lacked the 2′ secondary substitution. At 37°C, the predominant form was also triacyl lipid A (m/z 1,178), which is found in the htrB mutant at the same growth temperature.
FIG. 2.
Negative-ion MALDI-TOF mass spectrometry spectra for lipid A isolated from Y. enterocolitica serotype O:8 acyltransferase mutants. msbB, YeO8-ΔmsbBGB2 grown at 21°C (A) or at 37°C (B); htrB, YeO8-ΔhtrBGB grown at 21°C (C) or at 37°C (D); lpxP, YeO8-ΔlpxPGB grown at 21°C (E) or at 37°C (F); lpxP-htrB, YeO8-ΔlpxPGB::pKNOCKYeinthtrB grown at 21°C (G) or at 37°C (H). The results in all panels are representative of three independent lipid A extractions.
In summary, our results confirm the predicted functions of YeO8 msbB, htrB, and lpxP homologues, the products of which add C12, C14, and C16:1, respectively, to lipid A. Furthermore, our data suggest that MsbB- and LpxP-dependent acylations of lipid A occur mainly at 21°C whereas HtrB-dependent acylation is more apparent at 37°C in the wild-type strain.
Transcriptional regulation of msbB, htrB, and lpxP.
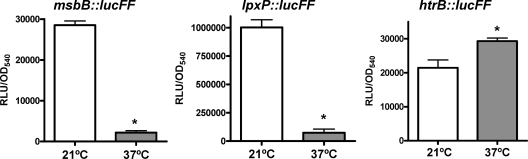
The results described above suggested that the expression and/or function of the three acyltransferases in YeO8 might be temperature controlled. To monitor transcription of the three late acyltransferase genes quantitatively, three transcriptional fusions in which a promoterless lucFF gene was under the control of the acyltransferase promoters were constructed. These fusions were introduced into YeO8, and the amount of light was determined. The expression of msbB::lucFF was 12-fold higher at 21°C than at 37°C (Fig. 3). lpxP::lucFF expression was also higher at 21°C than at 37°C (Fig. 3). In contrast, htrB::lucFF expression was higher at 37°C than at 21°C (Fig. 3). These results gave experimental support to our hypothesis that the expression of the three late acyltransferases is temperature regulated.
FIG. 3.
The expression levels of Y. enterocolitica serotype O:8 msbB, htrB, and lpxP acyltransferases are temperature regulated. The levels of acyltransferase expression by Y. enterocolitica serotype O:8 strains carrying the fusion msbB::lucFF, lpxP::lucFF, or htrB::lucFF and grown at 21 or 37°C were analyzed. Data are presented as means ± SD (n = 3). *, results are significantly different (P < 0.05; one-tailed t test) from the results for 21°C.
We asked whether signals other than temperature may alter the temperature regulation of the three acyltransferases. However, neither pH (ranging from 5.5 to 7) nor divalent-cation concentrations (low and high Mg2+ and Ca2+ levels) affected the temperature-dependent expression pattern of the three acyltransferases (data not shown).
Sensitivities of lipid A mutants to chemicals.
The OM functions as an efficient permeability barrier against many classes of molecules, including harmful agents such as detergents and antibiotics. Mutations in LPS are known to alter the OM permeability barrier against toxic compounds (49, 50). Therefore, we asked whether the OM barriers of the lipid A mutants were compromised. The sensitivities of the mutants to a panel of hydrophobic agents were tested by the disk diffusion method. No differences between the wild-type strain and the mutants in susceptibilities to crystal violet, DOC, and SDS were found, regardless of the growth temperature of the strains (data not shown). However, at 21°C the msbB, htrB, and lpxP mutants were more susceptible than the wild type to novobiocin (diameters of the halos of inhibition for each strain, 4.1 ± 0.7, 3.9 ± 0.6, 4.2 ± 0.6, and 2.7 ± 0.2 cm, respectively; P < 0.05 for each comparison between results for a mutant and the wild type). At 37°C, all strains were equally sensitive to novobiocin (data not shown).
In many pathogens, lipid A acylation is linked to resistance to antimicrobial peptides (51, 56). Therefore, we evaluated the resistance of the three mutants to the antimicrobial peptide polymyxin B. A wealth of data indicates that resistance to this peptide reflects well the resistance to other mammalian peptides (29, 35, 51, 75). When the strains were grown at 21°C, the MIC of polymyxin B for the lpxP mutant (0.32 ± 0.04 U/ml) was significantly lower (P < 0.05) than those for the wild-type and msbB and htrB mutant strains (0.48 ± 0.02, 0.42 ± 0.02, and 0.42 ± 0.03 U/ml, respectively). msbB and htrB mutants were as resistant as the wild-type strain (P > 0.05). At 37°C, the htrB mutant was more susceptible to polymyxin B (MIC, 0.17 ± 0.02 U/ml) than the wild-type and msbB and lpxP mutant strains (MICs for these strains, 0.38 ± 0.02, 0.32 ± 0.02, and 0.39 ± 0.05 U/ml, respectively; P < 0.05). Except for the lpxP mutant, the strains were more susceptible to polymyxin B when grown at 37°C than when grown at 21°C (P < 0.05 for the comparison of the MICs between temperatures for a given strain).
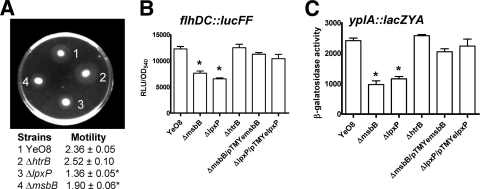
Motility and analysis of the flagellar regulon.
Y. enterocolitica is motile when grown at 21°C but not when grown at 37°C (79). Taking into account that 21°C is also the temperature at which lipid A is more acylated, we asked whether lipid A acylation plays any role in Yersinia motility. We quantified the migration of the wild-type and mutant strains in motility medium (1% tryptone-0.3% agar plates). Figure 4A shows that msbB and lpxP mutants were less motile than the wild type and the htrB mutant. Yersinia motility is related to the levels of flagellins, which in turn are regulated by the expression of flhDC, the flagellum master regulatory operon (10, 79). Therefore, we hypothesized that expression of flhDC could be lower in msbB and lpxP mutants than in the wild type and htrB mutant. To address this possibility, an flhDC::lucFF transcriptional fusion (10) was introduced into the chromosomes of the strains and the amount of light was determined. At 21°C, the amounts of light in msbB and lpxP mutants were smaller than those in the wild type and htrB mutant (Fig. 4B). Complementation of msbB and lpxP mutants with pTMYemsbB and pTMYelpxP, respectively, restored flhDC::lucFF expression to wild-type levels (Fig. 4B). When the strains were grown at 37°C, all strains produced the same amount of light (data not shown).
FIG. 4.
Motility and flhDC and yplA expression patterns are altered in Y. enterocolitica serotype O:8 acyltransferase mutants. (A) Results from a motility assay on a semisolid agar plate. Shown are the motilities after 24 h of incubation at RT. Data are presented as means ± SD (n = 3). Strains are listed according to the relevant genotype. (B) Analysis of flhDC expression by strains carrying the transcriptional fusion flhDC::lucFF. Results are given in RLU per OD540. (C) β-Galactosidase activity production by yplA′::lacZYA present in different strains (β-galactosidase values are given in Miller units and are means ± SD [n = 3]). *, results are significantly different (P < 0.05; one-tailed t test) from the results for YeO8 (wild-type strain).
The gene for YeO8 phospholipase A (YplA) belongs to the flagellar regulon and hence its expression is regulated by flhDC (62, 77, 78). Furthermore, YplA is secreted by the flagellar type III secretion system (62, 77, 78). Considering that flhDC expression was downregulated in msbB and lpxP mutants, we speculated that yplA expression could be affected in both mutants. The transcriptional fusion yplA::lacZYA (63) was introduced into the chromosomes of the wild type and the lipid A mutants and the β-galactosidase activities were measured. Indeed, the β-galactosidase activities were lower in msbB and lpxP mutants than in the wild type and the htrB mutant (Fig. 4C). Plasmids pTMYemsbB and pTMYelpxP complemented the msbB and lpxP mutants, respectively (Fig. 4C).
In summary, these results indicate that the flagellar regulon is downregulated in msbB and lpxP mutants, with a concomitant decrease in motility and downregulation of yplA expression.
Invasin in lipid A mutants.
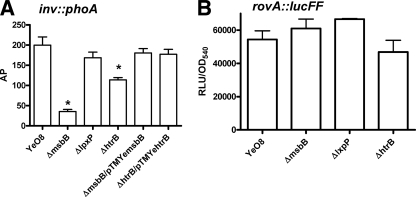
Inv is an OM protein of Y. enterocolitica responsible for invasion of the host (48, 54). Badger and Miller (7) have shown that motility and Inv are coordinately regulated. The reduced motility shown by msbB and lpxP mutants prompted us to determine whether inv expression was altered in the lipid A mutants. An inv::phoA translational fusion (53) was introduced into the genomes of the strains, and inv expression was monitored as AP activity (Fig. 5A). Levels of AP activity in the msbB and htrB mutants were significantly lower than those in the wild type and the lpxP mutant. The lowest AP activity was displayed by the msbB mutant. Plasmids pTMYemsbB and pTMYehtrB restored AP activity to wild-type levels. These differences in inv expression prompted us to study the abilities of the lipid A mutants to invade epithelial cells by using a gentamicin protection assay. The percentages of invasiveness of the msbB and htrB mutants for HeLa cells were significantly lower (13% ± 3% and 18% ± 4%, respectively) than that of the wild-type strain (26% ± 4%; P < 0.05). Plasmids pTMYemsbB and pTMYehtrB restored the invasiveness of the msbB and htrB mutants to wild-type levels (21% ± 3% and 27% ± 2%, respectively). The percentage of invasiveness of the lpxP mutant (25% ± 4%) was not significantly different from that of the wild type (26% ± 4%; P > 0.05).
FIG. 5.
inv expression is altered in Y. enterocolitica serotype O:8 acyltransferase mutants. (A) AP activities exhibited by wild-type (YeO8), YeO8-ΔmsbBGB2 (ΔmsbB), YeO8-ΔlpxPGB (ΔlpxP), YeO8-ΔhtrBGB (ΔhtrB), YeO8-ΔmsbBGB2/pTMYemsbB (ΔmsbB/pTMYemsbB), and YeO8-ΔhtrBGB/pTMYehtrB (ΔhtrB/pTMYehtrB) strains carrying an inv::phoA translational fusion (AP activity is expressed in enzyme units per OD600 unit, and data are means ± SD [n = 3]). (B) Analysis of rovA expression by strains carrying the transcriptional fusion rovA::lucFF. Results are given in RLU per OD540. *, results are significantly different (P < 0.05; one-tailed t test) from the results for YeO8 (wild-type strain).
RovA, a member of the Sly/Hor transcriptional activator family, is required for inv expression in Y. enterocolitica (60). Therefore, the low levels of inv expression in the msbB and htrB mutants could be caused by the downregulation of rovA expression, among other possibilities. To address this issue, a rovA::lucFF transcriptional fusion was introduced into the genomes of the wild type and the lipid A mutants and the amount of light was determined. Results shown in Fig. 5B demonstrate that rovA expression was not altered in the mutants.
Together, our data show that inv expression is downregulated in msbB and htrB mutants, although this effect is not caused by downregulation of the expression of rovA, the positive transcriptional regulator of inv.
Impact of lipid A acylation on production of YadA and Yops.
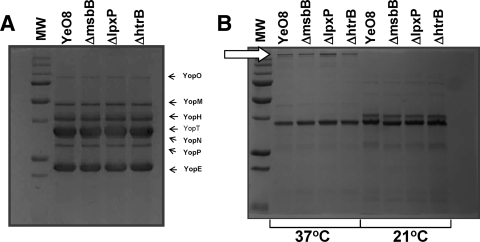
In several pathogens, the LPS status affects the expression of the type III secretion systems (5, 55, 74). In addition, a Salmonella msbB mutant is impaired in the secretion of proteins belonging to the Salmonella pathogenicity island 1 (SPI-1) type III secretion system (73). Therefore, we asked whether the production of the Yersinia pYV-encoded type III secretion system is altered in the lipid A mutants. At 37°C in the presence of low calcium concentrations, this system secretes a set of proteins called Yops that enable the pathogen to multiply extracellularly in lymphoid tissues (for a review, see references 17 and 18). Analysis of Yop secretion revealed that the three lipid A mutants secreted levels of Yops similar to those secreted by the wild-type strain (Fig. 6A).
FIG. 6.
The levels of expression of Yops and YadA are not affected in Y. enterocolitica serotype O:8 acyltransferase mutants. (A) SDS-PAGE analysis (the acrylamide concentration was 4% in the stacking gel and 12% in the separation one) and Coomassie brilliant blue staining of proteins from the supernatants of Ca2+-deprived cultures. (B) SDS-PAGE analysis (the acrylamide concentration was 4% in the stacking gel and 12% in the separation one) and Coomassie brilliant blue staining of OM proteins purified from strains grown in LB medium either at 21°C or at 37°C. The white arrow marks YadA protein. MW, molecular weight marker; YeO8, wild type; ΔmsbB, YeO8-ΔmsbBGB2; ΔlpxP, YeO8-ΔlpxPGB; ΔhtrB, YeO8-ΔhtrBGB. The results in both panels are representative of three independent experiments.
Another virulence gene carried by pYV is yadA, whose expression is induced only at 37°C (64). YadA is an OM protein mediating bacterial adhesion, bacterial binding to proteins of the extracellular matrix, and complement resistance (for a review, see reference 25). OM proteins were purified from bacteria grown at 37°C, and YadA expression was analyzed by SDS-PAGE (Fig. 6B). All strains produced similar amounts of YadA. The levels of other OM proteins were similar in all strains regardless of the growth temperature (Fig. 6B).
Taken together, these results suggest that the production of the pYV-encoded virulence factors Yops and YadA is not altered in the lipid A mutants.
Virulence of lipid A mutants.
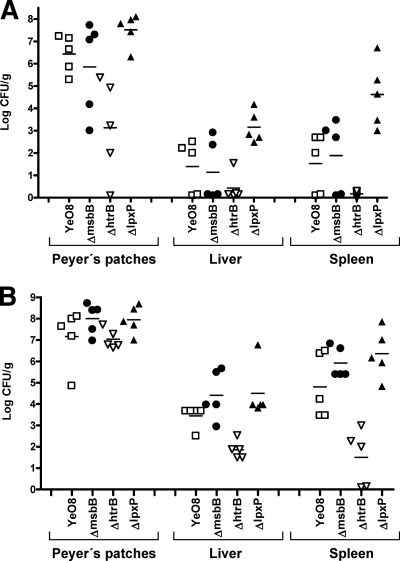
BALB/c mice were infected orogastrically, and 3 and 7 days postinfection, mice were dissected. The numbers of bacteria present in the Peyer's patches, spleens, and livers were determined by plating. Bacterial loads were recorded as log numbers of CFU per gram of tissue (Fig. 7). At 3 days postinfection, all strains colonized Peyer's patches (Fig. 7A). The msbB mutant colonized the organ as efficiently as the wild-type strain, whereas the load of lpxP mutant bacteria was higher than that of wild-type bacteria (P < 0.05; one-way ANOVA) (Fig. 7A). In contrast, the htrB mutant colonized the Peyer's patches less efficiently than the wild type (P < 0.05; one-way ANOVA) (Fig. 7A). Similar profiles for the spleen and liver were obtained (Fig. 7A). The htrB mutant colonized both organs less efficiently than the wild type (P < 0.05; one-way ANOVA); no differences between the msbB mutant and the wild type were found, whereas loads of lpxP mutant bacteria were higher than those of wild-type bacteria (P < 0.05; one-way ANOVA). At 7 days postinfection, increased loads of bacteria of all strains in all tissues were found (compare Fig. 7A and B). In Peyer's patches, the loads of bacteria of the three lipid A mutant strains and the wild-type strain were not significantly different (Fig. 7B). However, in spleen and liver tissues, the loads of htrB mutant bacteria were significantly lower than those of wild-type bacteria (P < 0.05; one-way ANOVA) (Fig. 7B). Loads of msbB and lpxP mutant bacteria were similar to those of wild-type bacteria (Fig. 7B).
FIG. 7.
Bacterial counts for mouse organs at 3 days postinfection (A) or 7 days postinfection (B). Mice were infected orally with 100 μl of a bacterial mixture containing 109 bacteria of the wild type (YeO8; □) or the acyltransferase mutant YeO8-ΔmsbBGB2 (ΔmsbB; •), YeO8-ΔhtrBGB (ΔhtrB; ▿), or YeO8-ΔlpxPGB (ΔlpxP; ▴). Results were reported as log numbers of CFU per gram of tissue. Symbols indicate results for individual mice, and horizontal lines indicate means for groups.
DISCUSSION
In contrast to other Enterobacteriaceae, pathogenic yersiniae show temperature-dependent variations in lipid A acylation (this work and references 6, 9, 38, 58, and 59). At 21°C, Y. enterocolitica synthesizes hexa-acylated lipid A containing four 3-OH-C14 units, one C12 unit, and either one C16:1 or one C14 unit. At 37°C, Y. enterocolitica produces mainly tetra-acylated lipid A and smaller amounts of hexa-acylated lipid A containing C14. The first aim of our study was to characterize the acyltransferases of Y. enterocolitica responsible for lipid A acylation and to determine whether their expression is temperature regulated.
By constructing mutants and analyzing their lipids As, we showed that msbB, htrB, and lpxP encode the acyltransferases responsible for the addition of C12, C14, and C16:1, respectively, to lipid A. Furthermore, Y. enterocolitica enzymes acylate the same lipid A positions as the E. coli ones. However, despite the similarity to the E. coli acyltransferases, Y. enterocolitica MsbB and HtrB add C12 and C14, respectively, whereas E. coli MsbB and HtrB transfer C14 and C12, respectively (this work and reference 57). Acyltransferases discriminate among acyl groups with different chain lengths by using the so-called hydrocarbon rulers (2, 76), but the specificities of homologous acyltransferases from different bacteria for a preferred chain length may result from a single amino acid change (76). Transcriptional analysis revealed that the expression of msbB was higher at RT than at 37°C, which was consistent with the reduced levels of lipid A molecules containing C12 (m/z 1,797) in the wild-type strain at 37°C. lpxP transcription was also higher at RT than at 37°C, and lipid A molecules from YeO8 grown at 37°C did not contain C16:1. There is a correlation between the expression patterns of msbB and lpxP and the detection of lipid A molecules containing C12 and C16.1. Nevertheless, we cannot rigorously rule out that posttranscriptional effects and/or enzyme catalytic activity sensitive to temperature may also be responsible for lack of acylation. The expression of htrB was higher at 37°C than at RT, although the temperature-dependent regulation was not as dramatic as that of the other two acyltransferase genes, thereby suggesting that htrB may also be expressed at 21°C. Indeed, HtrB-dependent lipid A acylation, i.e., addition of C14, could be detected at both temperatures. In addition, HtrB transfers C14 to the 2′-3-OH-C14 position of lipid A disaccharide in the lpxP mutant grown at 21°C, further indicating that htrB is expressed at 21°C. Similar findings for E. coli have been reported by Vorachek-Warren et al. (72). These authors have conclusively demonstrated that LpxP- and HtrB-dependent enzymatic activities are simultaneously present in E. coli membranes from bacteria grown at 12°C, a temperature at which LpxP activity is displayed by E. coli. Moreover, in an lpxP mutant grown at 12°C, HtrB is responsible for 2′-position acylation, hence indicating that HtrB activity is not compromised at 12°C. Why then does LpxP act instead of HtrB even though both enzymes are expressed at low temperatures (21°C for YeO8 and 12°C for E. coli)? It is reasonable to postulate that there must be competition between LpxP and HtrB activities during growth at low temperatures. In addition to other possible explanations, the composition of the acyl-ACP pool definitely influences the ability of the acyltransferases to catalyze acylation reactions. In this regard, it is known that the activity of the β-ketoacyl-ACP synthase is various magnitudes of order higher at low temperatures than at 37°C (28), which may result indirectly in diminished availability of C14 (C12 in E. coli)-ACP relative to C16:1-ACP. Another nonexclusive explanation is that the specific activity of LpxP may be higher than that of HtrB at low temperatures. However, to date the optimal conditions for assaying these enzymes have not been studied in depth.
A remaining issue is to explain at the molecular level the unique tetra-acyl lipid A (m/z 1,388) found in the wild type grown at 37°C. This species has been described previously for Y. enterocolitica (6, 52, 58) and is consistent with 3′ O deacylation of lipid A. Similarly deacylated lipid As have been found in S. enterica serovar Typhimurium and Helicobacter pylori (61, 67). For both pathogens, the hydrolase, named LpxR, removing the 3′-acyloxyacyl residue of lipid A has been identified recently (61, 67). In silico analysis of the genome of Y. enterocolitica revealed that this pathogen may encode an orthologue of this enzyme. Studies to examine whether Y. enterocolitica LpxR is indeed responsible for lipid A deacylation at 37°C are ongoing.
A second aim of this study was to test the hypothesis that the lipid A acylation status is important for Y. enterocolitica virulence. First, we assessed whether the permeability barrier of the OM was compromised in the acyltransferase mutants. Of the hydrophobic agents tested, only novobiocin displayed increased bactericidal activity against the three mutants, and this was true only when bacteria were grown at 21°C. These results suggest that the OM is not grossly altered in the mutants. In addition, the expression patterns of OM proteins in all strains were similar, further supporting the idea that the OM is not altered in the acyltransferase mutants. Second, we tested the susceptibilities of the mutants to polymyxin B, taken as a model antimicrobial peptide. At 21°C, the lpxP mutant was the strain most susceptible to polymyxin B, whereas at 37°C, the htrB mutant was the most susceptible strain. These results point out that in Y. enterocolitica, in contrast to other bacteria (71), the substitution of the 2′-R-3-hydroxymyristoyl group, and not that of the 3′ group, is required for polymyxin B resistance. Furthermore, the type of fatty acid also seems to be important because the lpxP mutant is susceptible to polymyxin B even though the 2′-R-3-hydroxymyristoyl group is substituted by C14.
A striking finding in our work is that the levels of expression of flhDC, yplA, and inv in the lipid A mutants were affected. Expression of flhDC was lower in the msbB and lpxP mutants than in the wild type, which was consistent with the reduced motility of these mutants. Our results are in good agreement with those previously reported showing that the levels of flhDC determine the levels of flagellin and motility (10, 79). Taking into consideration previous studies demonstrating that the expression levels of flhDC correlate with yplA expression (10, 79), we hypothesized that yplA expression was downregulated in msbB and lpxP mutants. Indeed, our results confirmed that this was the case. On the other hand, Badger and Miller (7) have shown that motility and Inv are coordinately regulated. Therefore, we speculated that inv expression could be altered in msbB and lpxP lipid A mutants. In partial agreement, we found that inv expression was downregulated in msbB and htrB mutants but not in the lpxP mutant. Of note, Inv levels correlated well with the abilities of the strains to invade HeLa cells. Collectively, our data support the notion that changes in lipid A acylation may act as a regulatory signal by acting on a transduction pathway(s). There are several mechanistic possibilities to explain the altered flhDC and inv expression patterns in the lipid A mutants. One could postulate that the expression of regulators for flhDC and inv could be altered in the mutants. So far at least three proteins, RovA, OmpR, and H-NS, have been identified as factors in the regulatory network connecting flhDC and inv expression levels in Yersinia (13, 24, 55, 60; C. M. Llompart and J. A. Bengoechea, unpublished data). Our data already showed that low inv expression found in the mutants was not caused by downregulation of rovA expression. On the other hand, considering that the factors studied either are expressed on the OM, as in the case of Inv and LPS, or require a membrane-associated system, as in the case of flhDC-dependent flagellum synthesis, we are keen to investigate whether changes in lipid A acylation could be detected by the so-called extracytoplasmic stress-responsive pathways, which in turn control gene expression patterns. We and others have shown that the Cpx system plays a role in LPS O antigen and inv regulation (11, 15) and that lipid A deacylation induces σE responses in E. coli (69). Experiments to test whether a Cpx- and/or σE-dependent pathway(s) underlies the regulatory connection between the lipid A status and the expression of other virulence factors are under way.
Ultimately, we examined whether msbB, lpxP, and htrB acyltransferases play a role in Y. enterocolitica virulence. In sharp contrast to other enterobacteria (16, 21, 39, 66), the msbB mutant was as virulent as the wild type. This could simply reflect the fact that msbB-dependent lipid A acylation was reduced at 37°C, the host temperature. Intriguingly, at 3 days postinfection, lpxP mutant bacteria were recovered from the organs of mice in higher numbers than wild-type bacteria. At present, we can only speculate on the underlying explanation. A tantalizing hypothesis is that substitution of C16:1 by C14 in lipid A somehow facilitates the survival of Y. enterocolitica during the initial stages of infection. Nevertheless, at 7 days postinfection, loads of lpxP mutant bacteria and wild-type bacteria were not significantly different, and this result is consistent with the lack of LpxP-dependent acylation at 37°C. Virulence experiments revealed that the htrB mutant was less virulent than the wild type. htrB mutants of other species also show defects in host-pathogen interactions (1, 19, 36). At 3 days postinfection, the htrB mutant was not able to colonize Peyer's patches as efficiently as the wild type and it did not disseminate into deeper organs. At least two nonexclusive explanations may account for the low-level colonization of Peyer's patches: (i) the level of penetration of the intestinal epithelium by the mutant is low and (ii) the mutant is cleared by a host defense mechanism present in the tissue. In support of the former, inv expression was lower in the mutant than in the wild type and the mutant was impaired in its ability to invade epithelial cells. However, the msbB mutant also expressed less inv in vitro than the wild type but it colonized Peyer's patches as efficiently as the wild type. Supporting the latter explanation, the htrB mutant was more susceptible than the wild type to polymyxin B, taken as a model of host antimicrobial peptides. This possibility alone cannot explain the mutant attenuation because the load of mutant bacteria increased at 7 days postinfection compared to that at 3 days postinfection. Another alternative may be that bacterial systems necessary to dismantle host defense mechanisms, like the pYV-encoded Yop arsenal of antihost proteins, are not fully functional in the mutant. However, Yop secretion in the htrB mutant was not affected. On the other hand, it is known that for efficient Yop translocation to the eukaryotic cell cytosol, intimate bacterium-cell interaction, which is thought to be mediated by Inv and YadA, is required (30). Although YadA expression was not affected, Inv-mediated adhesion was compromised in the mutant, thereby suggesting that Yop translocation by the mutant could be not as efficient as that by the wild type. Studies to test this possibility are ongoing.
Acknowledgments
We thank members of the Bengoechea lab for helpful suggestions and discussions. We also thank Martine Caroff for helpful advice on lipid A purification.
Fellowship support to C. Pérez-Gutiérrez, C. M. Llompart, and M. Reinés from the Spanish Ministry of Education, Govern de les Illes Balears, and Consejo Superior de Investigaciones Científicas (through the JAE-DOC program), respectively, is gratefully acknowledged. This work has been funded by grants from the Fondo de Investigación Sanitaria (no. PI03/0881) and the Consejo Superior de Investigaciones Científicas (Proyecto Intramural no. 200820I174) to J. A. Bengoechea.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 12 April 2010.
REFERENCES
- 1.Adin, D. M., N. J. Phillips, B. W. Gibson, M. A. Apicella, E. G. Ruby, M. J. Fall-Ngai, D. B. Hall, and E. V. Stabb. 2008. Characterization of htrB and msbB mutants of the light organ symbiont Vibrio fischeri. Appl. Environ. Microbiol. 74:633-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, V. E., E. I. Lo, C. K. Engel, L. Chen, P. M. Hwang, L. E. Kay, R. E. Bishop, and G. G. Prive. 2004. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 23:2931-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 4.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Augustin, D. K., Y. Song, M. S. Baek, Y. Sawa, G. Singh, B. Taylor, A. Rubio-Mills, J. L. Flanagan, J. P. Wiener-Kronish, and S. V. Lynch. 2007. Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J. Bacteriol. 189:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aussel, L., H. Thérisod, D. Karibian, M. B. Perry, M. Bruneteau, and M. Caroff. 2000. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 465:87-92. [DOI] [PubMed] [Google Scholar]
- 7.Badger, J. L., and V. L. Miller. 1998. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica 34. J. Bacteriol. 180:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bengoechea, J. A. 2003. Regulation of O-antigen biosynthesis in Yersinia enterocolitica. Adv. Exp. Med. Biol. 529:267-274. [DOI] [PubMed] [Google Scholar]
- 9.Bengoechea, J. A., K. Brandenburg, M. D. Arraiza, U. Seydel, M. Skurnik, and I. Moriyon. 2003. Pathogenic Yersinia enterocolitica strains increase the outer membrane permeability in response to environmental stimuli by modulating lipopolysaccharide fluidity and lipid A structure. Infect. Immun. 71:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 11.Bengoechea, J. A., L. Zhang, P. Toivanen, and M. Skurnik. 2002. Regulatory network of lipopolysaccharide O-antigen biosynthesis in Yersinia enterocolitica includes cell envelope-dependent signals. Mol. Microbiol. 44:1045-1062. [DOI] [PubMed] [Google Scholar]
- 12.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brzostek, K., M. Brzostkowska, I. Bukowska, E. Karwicka, and A. Raczkowska. 2007. OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 153:2416-2425. [DOI] [PubMed] [Google Scholar]
- 14.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson, K. E., J. Liu, P. J. Edqvist, and M. S. Francis. 2007. Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect. Immun. 75:4386-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clements, A., D. Tull, A. W. Jenney, J. L. Farn, S. H. Kim, R. E. Bishop, J. B. McPhee, R. E. Hancock, E. L. Hartland, M. J. Pearse, O. L. Wijburg, D. C. Jackson, M. J. McConville, and R. A. Strugnell. 2007. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J. Biol. Chem. 282:15569-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornelis, G. R. 2002. The Yersinia Ysc-Yop virulence apparatus. Int. J. Med. Microbiol. 291:455-462. [DOI] [PubMed] [Google Scholar]
- 18.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 19.DeMaria, T. F., M. A. Apicella, W. A. Nichols, and E. R. Leake. 1997. Evaluation of the virulence of nontypeable Haemophilus influenzae lipooligosaccharide htrB and rfaD mutants in the chinchilla model of otitis media. Infect. Immun. 65:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.d'Hauteville, H., S. Khan, D. J. Maskell, A. Kussak, A. Weintraub, J. Mathison, R. J. Ulevitch, N. Wuscher, C. Parsot, and P. J. Sansonetti. 2002. Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J. Immunol. 168:5240-5251. [DOI] [PubMed] [Google Scholar]
- 22.Domenech-Sanchez, A., L. Martinez-Martinez, S. Hernandez-Alles, C. M. del Carmen, A. Pascual, J. M. Tomas, S. Alberti, and V. J. Benedi. 2003. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob. Agents Chemother. 47:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hamidi, A., A. Tirsoaga, A. Novikov, A. Hussein, and M. Caroff. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46:1773-1778. [DOI] [PubMed] [Google Scholar]
- 24.Ellison, D. W., and V. L. Miller. 2006. H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188:5101-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 26.Ernst, R. K., S. M. Moskowitz, J. C. Emerson, G. M. Kraig, K. N. Adams, M. D. Harvey, B. Ramsey, D. P. Speert, J. L. Burns, and S. I. Miller. 2007. Unique lipid A modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. J. Infect. Dis. 196:1088-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 28.Garwin, J. L., A. L. Klages, and J. E. Cronan, Jr. 1980. Beta-ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem. 255:3263-3265. [PubMed] [Google Scholar]
- 29.Groisman, E. A. 1994. How bacteria resist killing by host-defence peptides. Trends Microbiol. 2:444-449. [DOI] [PubMed] [Google Scholar]
- 30.Grosdent, N., I. Maridonneau-Parini, M. P. Sory, and G. R. Cornelis. 2002. Role of Yops and adhesins in resistance of Yersinia enterocolitica to phagocytosis. Infect. Immun. 70:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 32.Gunn, J. S., and S. I. Miller. 1996. Pho-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darveau, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 34.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 35.Gutsmann, T., S. O. Hagge, A. David, S. Roes, A. Bohling, M. U. Hammer, and U. Seydel. 2005. Lipid-mediated resistance of Gram-negative bacteria against various pore-forming antimicrobial peptides. J. Endotoxin Res. 11:167-173. [DOI] [PubMed] [Google Scholar]
- 36.Jones, B. D., W. A. Nichols, B. W. Gibson, M. G. Sunshine, and M. A. Apicella. 1997. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect. Immun. 65:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria—inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 38.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan, S. A., P. Everest, S. Servos, N. Foxwell, U. Zahringer, H. Brade, E. T. Rietschel, G. Dougan, I. G. Charles, and D. J. Maskell. 1998. A lethal role for lipid A in Salmonella infections. Mol. Microbiol. 29:571-579. [DOI] [PubMed] [Google Scholar]
- 40.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehrer, R. I., M. Rosenman, S. S. Harwig, R. Jackson, and P. Eisenhauer. 1991. Ultrasensitive assays for endogenous antimicrobial polypeptides. J. Immunol. Methods 137:167-173. [DOI] [PubMed] [Google Scholar]
- 42.Lindner, B. 2000. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of lipopolysaccharides. Methods Mol. Biol. 145:311-325. [DOI] [PubMed] [Google Scholar]
- 43.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon for protein export signals. Proc. Natl. Acad. Sci. U. S. A. 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marceau, M. 2005. Transcriptional regulation in Yersinia: an update. Curr. Issues Mol. Biol. 7:151-177. [PubMed] [Google Scholar]
- 45.Merighi, M., C. D. Ellermeier, J. M. Slauch, and J. S. Gunn. 2005. Resolvase-in vivo expression technology analysis of the Salmonella enterica serovar Typhimurium PhoP and PmrA regulons in BALB/c mice. J. Bacteriol. 187:7407-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michiels, T., P. Wattiau, R. Brasseur, J.-M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by Yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 56:1242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 50.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nizet, V. 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 8:11-26. [PubMed] [Google Scholar]
- 52.Oertelt, C., B. Lindner, M. Skurnik, and O. Holst. 2001. Isolation and structural characterization of an R-form lipopolysaccharide from Yersinia enterocolitica serotype O:8. Eur. J. Biochem. 268:554-564. [DOI] [PubMed] [Google Scholar]
- 53.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 54.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez-Gutierrez, C., C. M. Llompart, M. Skurnik, and J. A. Bengoechea. 2007. Expression of the Yersinia enterocolitica pYV-encoded type III secretion system is modulated by lipopolysaccharide O-antigen status. Infect. Immun. 75:1512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 57.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 59.Rebeil, R., R. K. Ernst, C. O. Jarrett, K. N. Adams, S. I. Miller, and B. J. Hinnebusch. 2006. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J. Bacteriol. 188:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Revell, P. A., and V. L. Miller. 2000. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol. Microbiol. 35:677-685. [DOI] [PubMed] [Google Scholar]
- 61.Reynolds, C. M., A. A. Ribeiro, S. C. McGrath, R. J. Cotter, C. R. Raetz, and M. S. Trent. 2006. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 281:21974-21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmiel, D. H., E. Wagar, L. Karamanou, D. Weeks, and V. L. Miller. 1998. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect. Immun. 66:3941-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skurnik, M., and P. Toivanen. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174:2047-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skurnik, M., R. Venho, P. Toivanen, and A. Al-Hendy. 1995. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 17:575-594. [DOI] [PubMed] [Google Scholar]
- 66.Somerville, J. E., Jr., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stead, C. M., A. Beasley, R. J. Cotter, and M. S. Trent. 2008. Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. J. Bacteriol. 190:7012-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 69.Tam, C., and D. Missiakas. 2005. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli. Mol. Microbiol. 55:1403-1412. [DOI] [PubMed] [Google Scholar]
- 70.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tran, A. X., M. E. Lester, C. M. Stead, C. R. Raetz, D. J. Maskell, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186-28194. [DOI] [PubMed] [Google Scholar]
- 72.Vorachek-Warren, M. K., S. M. Carty, S. Lin, R. J. Cotter, and C. R. Raetz. 2002. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degrees C. J. Biol. Chem. 277:14186-14193. [DOI] [PubMed] [Google Scholar]
- 73.Watson, P. R., A. Benmore, S. A. Khan, P. W. Jones, D. J. Maskell, and T. S. Wallis. 2000. Mutation of waaN reduces Salmonella enterica serovar Typhimurium-induced enteritis and net secretion of type III secretion system 1-dependent proteins. Infect. Immun. 68:3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 75.Wiese, A., T. Gutsmann, and U. Seydel. 2003. Towards antibacterial strategies: studies on the mechanisms of interaction between antibacterial peptides and model membranes. J. Endotoxin Res. 9:67-84. [DOI] [PubMed] [Google Scholar]
- 76.Wyckoff, T. J., S. Lin, R. J. Cotter, G. D. Dotson, and C. R. Raetz. 1998. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 273:32369-32372. [DOI] [PubMed] [Google Scholar]
- 77.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U. S. A. 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, L., J. Radziejewska-Lebrecht, D. Krajewska-Pietrasik, P. Toivanen, and M. Skurnik. 1997. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 23:63-76. [DOI] [PubMed] [Google Scholar]