Abstract
Porphyromonas gingivalis is one of the most etiologically important microorganisms in periodontal disease. We found in a previous study that PG1385 (TprA) protein, a tetratricopeptide repeat (TPR) protein, was upregulated in P. gingivalis wild-type cells placed in a mouse subcutaneous chamber and that a tprA mutant was clearly less virulent in the mouse subcutaneous abscess model (M. Yoshimura et al., Oral Microbiol. Immunol. 23:413-418, 2008). In the present study, we investigated the gene expression profile of tprA mutant cells placed in a mouse subcutaneous chamber and found that 9 genes, including PG2102 (tapA), PG2101 (tapB), and PG2100 (tapC) genes, were downregulated in the tprA mutant compared with those in the wild type. Expression of a cluster of tapA, tapB, and tapC genes of the mutant was also downregulated in an in vitro culture with enriched brain heart infusion medium. The TprA protein has three TPR motifs known as a protein-protein interaction module. Yeast two-hybrid system analysis and in vitro protein binding assays with immunoprecipitation and surface plasmon resonance detection revealed that the TprA protein could bind to TapA and TapB proteins. TprA and TapB proteins were located in the periplasmic space, whereas TapA, which appeared to be one of the C-terminal domain family proteins, was located at the outer membrane. We constructed tapA, tapB, and tapC single mutants and a tapA-tapB-tapC deletion mutant. In the mouse subcutaneous infection experiment, all of the mutants were less virulent than the wild type. These results suggest that TprA, TapA, TapB, and TapC are cooperatively involved in P. gingivalis virulence.
Periodontal disease, the major cause of tooth loss in the general population of industrial nations (21, 37), is a chronic inflammatory disease of the periodontium that leads to erosion of the attachment apparatus and supporting bone for the teeth (1) and is one of the most common infectious diseases of humans (36). The obligately anaerobic Gram-negative bacterium Porphyromonas gingivalis has become recognized as a major pathogen for chronic periodontitis (7). P. gingivalis has been found to express numerous potential virulence factors, such as fimbriae, hemagglutinins, lipopolysaccharides, and various proteases that are capable of hydrolyzing collagen, immunoglobulins, iron-binding proteins, and complement factors (16, 17). Expression of these virulence factors is thought to be tightly regulated in response to environmental cues. In recent years, the search for virulence factors has been greatly facilitated by molecular genetics (27). Although a number of studies have shown gene expression of P. gingivalis being regulated by environmental stresses (13, 19, 35, 38, 41, 46, 55), gene expression of P. gingivalis cells in in vivo lesions is not completely understood. Our previous study (54) using a subcutaneous chamber model showed that 10 P. gingivalis proteins were upregulated in host tissues whereas four proteins were downregulated. Among the upregulated proteins, PG1089 (DNA-binding response regulator RprY), PG1385 (TPR domain protein), and PG2102 (immunoreactive 61-kDa PG91 antigen) were chosen for further analysis. Mouse abscess model experiments revealed that a mutant strain defective in PG1385 was clearly less virulent and a mutant defective in PG2102 also had a tendency to be less virulent than the wild-type parent strain. These results suggest that PG1385 and PG2102 proteins are involved in the virulence of P. gingivalis.
The PG1385 protein has three tetratricopeptide repeat (TPR) motifs. The TPR motif is a protein-protein interaction module found in multiple copies in a number of functionally different proteins that facilitate specific interactions with a partner protein(s) (3, 10).
In this study, we found that the PG1385 protein bound to each of the PG2101 and PG2102 proteins and that these mutant strains were less virulent, suggesting that PG1385, PG2101, and PG2102 proteins are cooperatively involved in P. gingivalis virulence.
MATERIALS AND METHODS
Strains and culture conditions.
All P. gingivalis strains and plasmids used in the study are shown in Table 1. P. gingivalis cells were grown anaerobically (10% CO2, 10% H2, 80% N2) in enriched brain heart infusion (BHI) medium and on enriched tryptic soy (TS) agar. For selection and maintenance of the antibiotic-resistant strains, the antibiotics erythromycin (Em) and tetracycline (Tc) were added to the medium at concentrations of 10 μg/ml and 0.5 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. gingivalis | ||
| W83 | Wild type | |
| KDP159 | tprA::[ermF ermAM]; Emr | 54 |
| KDP382 | ΔtapB::ermF; Emr | This study |
| KDP383 | ΔtapA::ermF; Emr | This study |
| KDP384 | Δ[tapA-tapB-tapC]::ermF; Emr | This study |
| KDP385 | ΔporT::[ermF-ermAM]; Emr | This study |
| KDP386 | ΔtapC::ermF; Emr | This study |
| E. coli | ||
| XL1-Blue | General-purpose host strain for cloning | Stratagene |
| BL21(DE3) | Host strain for expression vector pET-32a | Nippongene |
| S. cerevisiae | ||
| AH109 | Host strain for yeast two-hybrid system | Clontech |
| Plasmids | ||
| pBluescript II SK(−) | Apr; cloning vector (pBSSK) | Stratagene |
| pET-32a | Apr; expression vector | Novagen |
| pGEX-6P-1 | Apr; expression vector | GE Healthcare |
| pKD357 | Apr Emr; pBSSK containing ermF ermAM cassette located between upstream and downstream sequences of the porT gene | 43 |
| pKD1008 | Apr; pBSSK containing 0.3-kb tapC upstream and 0.5-kb tapC downstream regions | This study |
| pKD1009 | Apr Emr; ermF cassette was inserted into BamHI site of pKD1008 | This study |
| pKD1010 | Apr; pBSSK containing 0.3-kb tapB upstream and 0.5-kb tapB downstream regions | This study |
| pKD1011 | Apr Emr; ermF cassette was inserted into BamHI site of pKD1010 | This study |
| pKD1012 | Apr; pBSSK contains 0.3-kb tapA upstream and 0.5-kb tapA downstream regions | This study |
| pKD1013 | Apr Emr; ermF cassette was inserted into BamHI site of pKD1012 | This study |
| pKD1014 | Apr; pBSSK contains 0.3-kb tapA upstream and 0.5-kb tapC downstream regions | This study |
| pKD1015 | Apr Emr; ermF cassette was inserted into BamHI site of pKD1014 | This study |
| pKD1016 | Apr; pET-32a containing P. gingivalis tapB gene | This study |
| pKD1017 | Apr; pET-32a containing P. gingivalis tapA gene | This study |
| pKD1018 | Apr; pGEX-6P-1 containing P. gingivalis tprA gene | This study |
| pKD1019 | pGBKT7 containing P. gingivalis tprA gene | This study |
r, resistance; Ap, ampicillin.
Subcutaneous chamber model experiment.
A subcutaneous chamber model experiment was performed according to the method of Yoshimura et al. (54). Bacterial cells were grown at 37°C until an optical density at 550 nm (OD550) of 1.0 was reached. Cultures were then concentrated by centrifugation at 10,000 × g for 10 min, and cells were collected and resuspended in 1/30 of the original volume in fresh enriched BHI broth. Female BALB/c mice 8 to 10 weeks of age were used. Coil-shaped subcutaneous chambers were prepared and surgically implanted as previously described by Genco et al. (15). One week after implantation, the chambers were inoculated with 0.4 ml of a concentrated suspension of P. gingivalis in enriched BHI broth. Ninety minutes after inoculation, chamber fluid containing bacterial cells was aseptically removed from each implanted chamber by the use of a 25-gauge hypodermic needle and a syringe. Chamber fluid harvested from three mice was mixed and subjected to isolation of RNA for microarray analysis and real-time quantitative PCR (qPCR).
Microarray and data analyses.
Bacterial cells were lysed in TRIzol reagent (Invitrogen). RNA was isolated by TRIzol extraction followed by RNeasy column purification with genomic DNA digestion (DNase I) (Qiagen). Subsequently, synthesis of cDNA, target hybridization, washing, and scanning were carried out according to the Affymetrix protocol. Gene chips for P. gingivalis W83 (TI242619 60mer), in which numbers of probes per target gene, replicates, and total probes per chip were 19, 5, and 192,000, respectively, were purchased from Roche NimbleGen Inc. The gene chips were scanned, and the resulting image files were used to calculate and normalize the hybridization intensity data utilizing GeneChip operating software (Affymetrix). Single-microarray analysis measures a relative level of expression of a transcript (signal) and determines whether a transcript is present (P) or absent (A). Absolute analysis of each microarray was followed by comparison analysis using GeneSpringGX7 software (Silicon Genetics). The comparison estimates the magnitude of the change (i.e., the fold change of the normalized data) and the direction of the change (increase, decrease, or no change) of a transcript across the two arrays. Each experiment was performed twice, and only results for transcripts showing P/P were included here. Mean data for two sets of replicate samples were used for the comparison analysis. For most data sets, the results are shown as average values for severalfold change from the comparisons. A given transcript was designated “upregulated” when the average severalfold change value determined in a comparison of two sets of replicate samples represented an increase of at least 1.5-fold in the expression level. A given transcript was designated “downregulated” when the average severalfold change value determined in a comparison of two sets of replicate samples represented a decrease of at least 1.5-fold in the expression level.
Real-time qPCR.
Total RNA was reverse transcribed into cDNA with a SuperScript III first-strand synthesis system (Invitrogen). cDNA was used in real-time qPCR experiments performed in triplicate by using Brilliant II Fast SYBR green QPCR master mix (Stratagene) with an Mx3005P real-time PCR system (Stratagene) according to the manufacturer's instructions. The primers for the real-time analysis (see Table S1 in the supplemental material) were designed using Primer3 software (http://primer3.sourceforge.net/). Real-time qPCR conditions were as follows: 1 cycle at 95°C for 2 min and 35 cycles of 95°C for 5 s and 60°C for 20 s. At each cycle, the accumulation of PCR products was detected by the reporter dye from the double-stranded DNA (dsDNA)-binding SYBR green. To confirm that a single PCR product was amplified, a dissociation curve (melting curve) was constructed in the range of 55°C to 95°C after the PCR. All data were analyzed using Mx3005P software. The expression level of each targeted gene was normalized to that of gyrA (PG1386 encoding a DNA gyrase A subunit) (31). All PCRs were carried out in triplicate. The efficiency of primer binding was determined by linear regression by plotting the cycle threshold (CT) value versus the log of the cDNA dilution. Relative quantification of the transcript was determined using the comparative CT method (2−ΔΔCT) calibrated to gyrA. qPCR experiments were independently performed three times with comparable results.
Subcellular fractionation.
Subcellular fractionation of P. gingivalis cells was essentially performed according to the method of Murakami et al. (26). P. gingivalis cells were harvested by centrifugation at 10,000 × g for 30 min at 4°C and resuspended with 20 ml of phosphate-buffered saline (PBS) containing 0.1 mM N-p-tosyl-l-lysine chloromethyl ketone (TLCK), 0.1 mM leupeptin, and 0.5 mM EDTA. The cells were disrupted by two passes in a French pressure cell at 100 MPa. The remaining intact bacterial cells were removed by centrifugation at 3,000 × g for 10 min, and the supernatant was subjected to ultracentrifugation at 100,000 × g for 60 min. The cells were pelleted, and the supernatant was retained as the periplasmic-cytoplasmic fraction. The pellets were treated with 1% Triton X-100-PBS-20 mM MgCl2 for 30 min at 20°C. The outer membrane fraction was recovered as a precipitate by ultracentrifugation at 100,000 × g for 60 min at 4°C. The supernatant was obtained as the inner membrane fraction.
Spheroplast formation and proteinase treatment.
Spheroplast formation and proteinase treatment of P. gingivalis cells was essentially performed by a method previously described (12). After being suspended in 50 mM Tris acetate buffer (pH 7.8) containing 0.75 M sucrose, P. gingivalis cells were treated with lysozyme (final concentration, 0.1 mg/ml) on ice for 2 min. Conversion to spheroplasts was performed by slowly diluting the cell suspension over a period of 10 min with 2 volumes of cold 1.5 mM EDTA. After centrifugation at 10,000 × g for 10 min, the resulting precipitates were gently resuspended in 50 mM Tris acetate buffer (pH 7.8) containing 0.25 M sucrose and 10 mM MgSO4 (spheroplasts). The supernatants were used as the periplasm fraction, and the proteins in this fraction were precipitated with trichloroacetic acid and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis. Spheroplasts were treated on ice with proteinase K (final concentration, 1 mg/ml) in the presence or absence of 2% Triton X-100 for 1 h. After the proteinase K was quenched using phenylmethylsulfonyl fluoride (final concentration, 5 mM) for 5 min, the whole volume of the sample was mixed with 4 volumes of Laemmli sample buffer and subjected to SDS-PAGE and immunoblot analysis.
Protein electrophoresis and immunoblot analysis.
SDS-PAGE was performed by using the method of Laemmli (23). The gels were stained with 0.1% Coomassie brilliant blue (CBB) R-250. For immunoblot analysis, proteins on SDS-PAGE gels were electroblotted onto a polyvinylidene difluoride (PVDF) membrane. The blots were blocked with 5% skim milk for 1 h at room temperature, probed with anti-PG1385, anti-PG2101, anti-PG2102, or 1B5 monoclonal antibody (MAb) overnight at 4°C, washed, incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies, and finally detected with enhanced chemiluminescence (ECL) (GE Healthcare).
Protein purification and preparation of antisera.
P. gingivalis W83 genome sequence data were obtained from the TIGR website (http://www.tigr.org/). A genomic region that included the PG1385 gene was amplified by PCR from the chromosomal DNA of P. gingivalis W83 with the primer pair 5-rPG1385/BamHI and 3-rPG1385/EcoRI by the use of a PCR kit (Advantage-HF 2 PCR kit; Clontech). The amplified DNA fragment was cloned into a T-vector (pGEM-T Easy; Promega) and digested with BamHI and EcoRI. The resulting fragment was then inserted into the BamHI-EcoRI region of pGEX-6P-1 (GE Healthcare), and the recombinant expression plasmid was then transformed into Escherichia coli BL21(DE3). E. coli BL21(DE3) harboring the recombinant plasmid was inoculated into LB broth for large-scale culture experiments. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at a concentration of 0.1 mM, and this was followed by incubation for 3 h to overproduce the recombinant protein. The recombinant protein was purified with glutathione Sepharose beads (GE Healthcare). For removal of the glutathione S-transferase (GST) tag, the purified recombinant protein was incubated with PreScission proteases (GE Healthcare) at 4°C for 24 h. The protein was then further purified and concentrated by using Amicon Ultra filters (Millipore).
Genomic regions that included the PG2101 and PG2102 genes were amplified by PCR from the chromosomal DNA of P. gingivalis W83 by using the primer pair 5-rPG2101/KpnI and 3-rPG2101/NotI for the PG2101 gene and with the primer pair 5-rPG2102/KpnI and 3-rPG2102/NotI for the PG2102 gene and a PCR kit. The amplified DNA fragments were cloned into the T-vector and digested with KpnI and NotI. The resulting fragments were then inserted into the KpnI-NotI region of pET32a (Novagen), and the recombinant expression plasmids were transformed into E. coli BL21(DE3). E. coli BL21(DE3) harboring the recombinant plasmids was inoculated into LB broth for large-scale culture experiments. IPTG was added to the culture at a concentration of 0.1 mM, and this was followed by incubation for 3 h to overproduce the recombinant proteins. The recombinant proteins were purified with a Talon purification kit (Takara Bio). The proteins were then further purified and concentrated by using Amicon Ultra filtering (Millipore).
Recombinant PG2101-His (rPG2101-His) protein was mixed with TiterMax Gold (TiterMax), and the mixtures were injected into mice (BALB/c) subcutaneously, resulting in anti-2101 antiserum. Anti-PG2101 IgG was purified from the antiserum obtained from mice by the use of nProtein A Sepharose (GE Healthcare). Polyclonal rabbit antisera against recombinant PG1385 (rPG1385) and PG2102-His (rPG2102-His) proteins were from Sigma Genosys. 1B5 monoclonal antibody (MAb) (9) was kindly provided by Michael A. Curtis.
Yeast two-hybrid system.
A yeast two-hybrid 3 system was purchased from Clontech. Sau3AI-digested genomic DNA of P. gingivalis W83 was inserted into the BamHI site of pGADT7 to yield a genomic plasmid library. A genomic region that included the PG1385 gene was amplified by PCR from the chromosomal DNA of P. gingivalis W83 by the use of the primer pair Y2H-PG1385-Fw and Y2H-PG1385-Rv and a PCR kit. The amplified DNA fragment was cloned into the T-vector and digested using NcoI and BamHI. The resulting fragment was then inserted into the NcoI-BamHI region of pGBKT7 to yield pKD1019. Competent yeast cells (strain AH109) containing pKD1019 were transformed with 15 μg of genomic plasmid library and were plated onto minimal synthetic dropout (SD) agar lacking tryptophan, leucine, and histidine. The plates were incubated at 30°C for 7 days, and then transformants were streaked onto fresh SD agar and tested further for their ability to hydrolyze X-α-Gal (5-bromo-4-chloro-3-indolyl-d-α-galactoside).
Assays analyzing binding between rPG1385 and rPG2101-His and between rPG1385 and rPG2102-His.
rPG1385 (0 to 1.0 μg) was dissolved in PBS and added to 500 ng of rPG2101-His or rPG2102-His in a final volume of 200 μl and then incubated at 4°C for 2 h. Following incubation, Ni2+-chelate resin (Clontech) was added to the reaction mixture and incubated at 4°C for 2 h. Resin beads were recovered by centrifugation and washed three times with 500 μl of PBS. Binding between the proteins was also analyzed as follows. rPG1385 (500 ng) was dissolved in PBS and added to 0 to 1.0 μg of rPG2101-His or rPG2102-His in a final volume of 200 μl and then incubated at 4°C for 2 h. The resulting complexes were immunoprecipitated using anti-PG1385 antibody coupled with nProtein A Sepharose (GE Healthcare) at 4°C for 2 h. These precipitates were suspended in SDS-PAGE sample buffer and denatured by heating. Proteins on SDS-PAGE gels were blotted onto PVDF membranes and then subjected to immunoblotting using anti-PG1385, anti-PG2101, or anti-PG2102 antibody.
Surface plasmon resonance (BIAcore).
The interaction of rPG1385 with rPG2101-His or rPG2102-His was determined using surface plasmon resonance detection (BIAcore X100 system; GE Healthcare). rPG1385 (20 μg/ml) in 10 mM sodium acetate (pH 5.5) was immobilized on a CM5 carboxymethyl-dextran sensor chip using the aminecoupling method. rPG2101-His or rPG2102-His (0.016 μM to 10 μM) in HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] surfactant P 20) was passed over the surface of the sensor chip at a flow rate of 30 μl/min. The interaction was monitored by determining changes in surface plasmon resonance response at 25°C. After 2 min of monitoring, the same buffer was introduced onto the sensor chip in place of the rPG1385 solution to start the dissociation. Both the association rate constant (Ka) and the dissociation rate constant (Kd) were calculated by using BIAevaluation software (GE Healthcare) and1:1 (Langmuir) binding model software. The dissociation constant (Kd) was determined from Kd/Ka.
Construction of mutant strains.
The P. gingivalis PG2100 gene deletion mutant was constructed as follows. PG2100 upstream and PG2100 downstream DNA regions were amplified from P. gingivalis W83 chromosomal DNA by PCR using the primer pair 5-PG2100up/NotI and 3-PG2100up/BamHI for the PG2100 upstream region and the primer pair 5-PG2100dn/BamHI and 3-PG2100dn/KpnI for the PG2100 downstream region. The primers for the construction of mutant strains are listed in Table S1 in the supplemental material. The amplified DNAs were cloned into the T-vector and digested with NotI and BamHI for the PG2100 upstream DNA and with BamHI and KpnI for the PG2100 downstream DNA. The resulting fragments were inserted into the NotI-KpnI region of pBluescript II SK(−) to yield pKD1008. The 1.5-kb BamHI ermF DNA cartridge was inserted into the BamHI site of pKD1008, resulting in pKD1009 (ΔPG2100::ermF). P. gingivalis W83 was then transformed with BssHII-linearized pKD1009 DNA to yield strain KDP386.
P. gingivalis PG2101, PG2102, and PG2102-PG2100 gene deletion mutants were constructed essentially as described for the PG2100 gene deletion mutant. The primer pair 5-PG2101up/NotI and 3-PG2101up/BamHI and the primer pair 5-PG2101dn/BamHI and 3-PG2101dn/KpnI were used for the PG2101 upstream region and the PG2101 downstream region in construction of the PG2101 gene deletion mutant, respectively. The primer pair 5-PG2102up/NotI and 3-PG2102up/BamHI and the primer pair 5-PG2102dn/BamHI and 3-PG2102dn/KpnI were used for the PG2102 upstream region and the PG2102 downstream region in construction of the PG2102 gene deletion mutant, respectively. The primer pair 5-PG2102up/NotI and 3-PG2102up/BamHI and the primer pair 5-PG2100dn/BamHI and 3-PG2100dn/KpnI were used for the PG2102 upstream region and the PG2100 downstream region in construction of the PG2102-PG2100 gene deletion mutant, respectively.
The P. gingivalis porT deletion mutant was constructed as follows. P. gingivalis W83 was transformed with the BssHII-linearized pKD357 (43) DNA to yield strain KDP385.
Immunoprecipitation.
P. gingivalis cells were harvested and then dissolved with radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and 50 mM Tris-HCl; pH 8.0) and immunoprecipitated by the use of nProtein G agarose beads with 5 μg of anti-PG2102 polyclonal antibody. The resulting precipitate was dissolved with the same volume of the sample buffer and loaded on an SDS (10%) gel. Immunoblot analysis was performed with anti-P. gingivalis anionic surface polysaccharide 1B5 MAb (9).
Mouse virulence assay and statistical analysis.
Levels of virulence of the P. gingivalis W83 and mutant strains were determined by mouse subcutaneous infection experiments (28, 49, 53). Bacterial cells were grown at 37°C until an OD550 of 1.0 was reached. The cells were harvested and then resuspended and adjusted to a concentration of approximately 1 × 1012 CFU/ml in the enriched BHI broth. Female BALB/c mice (8 to 10 weeks of age) were challenged with subcutaneous injections of 0.1 ml of bacterial suspension at two sites on the depilated dorsal surface (0.2 ml per mouse). Injected mice were examined daily for survival. Three sets of experiments were carried out.
For data analysis, Kaplan-Meier plots were constructed and the log-rank test was used to evaluate the differences in mean survival rates in three experiments between mice infected with W83 parent strain and those infected with the mutant strains.
RESULTS
Gene expression profiling of the PG1385 mutant (tprA) inoculated into a mouse subcutaneous chamber.
We previously found that a strain defective in the PG1385 gene was clearly less virulent than the wild-type strain, indicating that PG1385 protein is involved in P. gingivalis virulence (54). The PG1385 gene encodes a TPR protein and is designated tprA. To investigate whether loss of the tprA gene product (TprA) influences in vivo expression of P. gingivalis genes, we determined the in vivo gene expression profile of the tprA mutant that was inoculated in a mouse subcutaneous chamber by using microarray analysis. Coil-shaped subcutaneous chambers were surgically implanted as previously described by Genco et al. (15). One week after implantation, P. gingivalis cells were inoculated into mouse subcutaneous chambers. Chamber fluid containing bacterial cells was aseptically removed from each implanted chamber 90 min after inoculation and subjected to isolation of total RNA for microarray analysis.
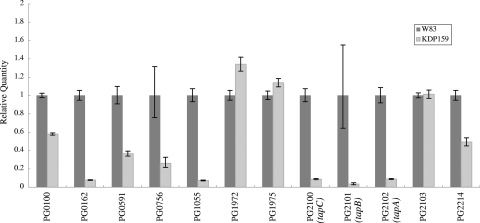
Genes of the tprA mutant with an average expression level that differed more than 1.5-fold (up or down) from those of the wild-type parent were identified, since the threshold was previously reported to be biologically significant (20, 24, 48). In the tprA mutant, 11 genes were upregulated, while 12 genes were downregulated (Table 2). We found that the upregulated genes were related to protein synthesis and to a hypothetical protein, as grouped by functional role categories defined by The Institute for Genomic Research (TIGR). Interestingly, PG1055 (thiol protease) and PG2102 (immunoreactive 61-kDa PG91 antigen), which were previously reported to have been upregulated in P. gingivalis wild-type cells placed in a mouse subcutaneous chamber (54), were found to be downregulated in the tprA mutant. To confirm the results obtained by microarray analysis, the RNA samples were subjected to real-time qPCR analysis. Expression of gyrA (a gene encoding a putative DNA gyrase A subunit, PG1386) was used for normalization. The 12 genes that were downregulated in the microarray analysis were examined using real-time qPCR, which showed that 9 of the 12 genes were downregulated (Fig. 1). Genes PG0162 (RNA polymerase sigma-70 factor, extracytoplasmic function [ECF] subfamily), PG1055, PG2100 (immunoreactive 63-kDa PG102 antigen), PG2101 (hypothetical protein), and PG2102 were markedly downregulated. Downregulation of genes for PG2100, PG2101, and PG2102 in the tprA mutant was also observed in cells incubated in enriched BHI medium (see Fig. S1 in the supplemental material).
TABLE 2.
P. gingivalis genes differentially expressed in the tprA mutant cells placed in a mouse subcutaneous chamber compared to those expressed in the wild-type cellsa
| TIGR no. | Identification | Avg. fold difference |
|---|---|---|
| Downregulated genes | ||
| PG0100 | Hypothetical protein | 0.59 |
| PG0162 | RNA polymerase sigma-70 factor, ECF subfamily | 0.57 |
| PG0591 | ISPg5, transposase Orf2 | 0.63 |
| PG0756 | Hypothetical protein | 0.47 |
| PG1055 | Thiol protease | 0.36 |
| PG1972 | Hemagglutinin protein HagB | 0.63 |
| PG1975 | Hemagglutinin protein HagC | 0.64 |
| PG2100 (tapC) | Immunoreactive 63-kDa PG102 antigen | 0.34 |
| PG2101 (tapB) | Hypothetical protein | 0.25 |
| PG2102 (tapA) | Immunoreactive 61-kDa PG91 antigen | 0.51 |
| PG2103 | Hypothetical protein | 0.24 |
| PG2214 | Hypothetical protein | 0.57 |
| Upregulated genes | ||
| PG0327 | Hypothetical protein | 1.92 |
| PG0373 | Hypothetical protein | 1.72 |
| PG0546 | Hypothetical protein | 1.83 |
| PG0635 | Ribosomal protein L11 methyltransferase | 1.69 |
| PG0656 | Ribosomal protein L34 | 2.02 |
| PG0722 | Hypothetical protein | 2.93 |
| PG0969 | S-Adenosylmethionine:tRNA ribosyltransferase-isomerase (putative protein) | 1.98 |
| PG1532 | Hypothetical protein | 1.92 |
| PG1866 | Hypothetical protein | 1.93 |
| PG2006 | Hypothetical protein | 1.69 |
| PG2225 | Hypothetical protein | 1.63 |
Mouse subcutaneous chamber experiments were performed as described in Materials and Methods. Total RNA was extracted from strains W83 (wild type) and KDP159 (tprA::erm). The extracted total RNA was analyzed by using a microarray as described in Materials and Methods. Numbers in the “Avg fold difference” column indicate the levels of gene expression in the tprA mutant versus the wild-type strain. The cutoff ratio for the levels of difference was 1.5.
FIG. 1.
Real-time qPCR analysis of gene expression in the tprA mutant inoculated into a mouse subcutaneous chamber. Mouse subcutaneous chamber experiments were performed as described in Materials and Methods. Total RNA was extracted from strains W83 (wild type) and KDP159 (tprA::ermF). The downregulated genes revealed by microarray analysis were selected for analysis by real-time qPCR. All PCRs were carried out in triplicate.
To determine whether the PG2102-, PG2101-, and PG2100-encoding genes, which are designated tapA (TprA-associated protein A gene), tapB, and tapC, respectively, are polycistronically transcribed, total RNA of the wild-type strain was subjected to PCR analysis. Total RNA was isolated from the wild-type strain; then, cDNA was synthesized and gene-specific primers were used for amplifying junction DNA among these genes. The junction DNA between the tapA and tapB genes and between the tapB and tapC genes was amplified, suggesting that these genes make up an operon (see Fig. S2 in the supplemental material).
Yeast two-hybrid analysis.
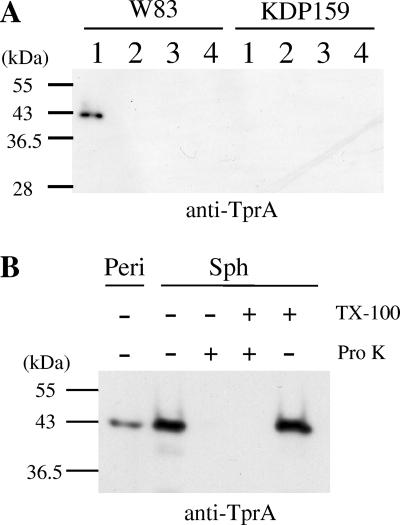
As another approach for clarifying the molecular mechanism responsible for the contribution of TprA protein to P. gingivalis virulence, we attempted to find P. gingivalis proteins that interacted with TprA protein, since it is likely that TprA protein, which is one of the TPR domain-containing proteins (3, 10), is associated with other proteins. First, we investigated the localization of TprA protein. The tprA mutant and its wild-type parent were fractionated into cytoplasm-periplasm, inner membrane, and outer membrane fractions and then subjected to SDS-PAGE and immunoblot analysis with anti-TprA antiserum (Fig. 2A). An anti-TprA-immunoreactive protein was found at a molecular mass of 43 kDa in the cytoplasm-periplasm fraction. For further analysis, experiments involving spheroplast formation and proteinase K treatment were performed (Fig. 2B). The 43-kDa anti-TprA immunoreactive protein band was observed in the periplasmic fraction. The 43-kDa protein, which was degraded by treatment with proteinase K, was also detected in the spheroplast fraction. In addition, the TprA protein possessed a signal sequence, as revealed by in silico analysis with SignalP V3.0 software (http://www.cbs.dtu.dk/services/SignalP/) (2). These results clearly indicate that the TprA protein is located in the periplasmic space, which is consistent with results of a previous study (45).
FIG. 2.
Subcellular localization of TprA protein. (A) Cytoplasm-periplasm, total membrane, inner membrane, and outer membrane fractions. Cells of strains W83 (wild type) and KDP159 (tprA::ermF) were fractionated and subjected to SDS-PAGE and immunoblot analysis using anti-TprA antiserum. Lanes: 1, cytoplasm-periplasm fraction; 2, total membrane fraction; 3, inner membrane fraction; 4, outer membrane fraction. (B) Spheroplast and periplasm fractions. Preparation of spheroplasts of P. gingivalis cells was performed as described in Materials and Methods, and the spheroplasts were subjected to proteinase K treatment (Pro K) in the presence or absence of 2% Triton X-100 (TX-100). Samples were subjected to SDS-PAGE followed by immunoblot analysis with anti-TprA antiserum. Sph, spheroplasts; Peri, periplasm fraction.
Yeast two-hybrid analysis was then performed using the tprA gene as bait. A number of P. gingivalis protein candidates interacting with TprA protein were found in the analysis (Table 3). They included PG0415 (peptidyl-prolyl cis-trans isomerase C [PPIC] type), PG0497 (5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase), PG1334 (band 7/Mec-2 family protein), PG2101 (TapB), and PG2200 (TPR domain protein) as a putative periplasmic protein.
TABLE 3.
TprA-associated proteins predicted from yeast two-hybrid system
| Protein category and TIGR no. | Descriptiona | Gene | TIGR functional role category |
|---|---|---|---|
| Putative periplasmic proteins | |||
| PG0415 | Peptidyl-prolyl cis-trans isomerase, PPIC type | Protein fate: protein folding and stabilization | |
| PG0497 | 5′-Methylthioadenosine/S-adenosylhomocysteine nucleosidase | mtn | Central intermediary metabolism: Other |
| PG1334 | Band 7/Mec-2 family protein | Unknown function: general | |
| PG2101 | Hypothetical protein | tapB | |
| PG2200 | TPR domain protein | Unknown function: general | |
| Putative inner membrane proteins | |||
| PG0081 | Hypothetical protein | ||
| PG0093 | HlyD family secretion protein | Transport and binding proteins: other | |
| PG0255 | Translation initiation factor IF-2 | infB | Protein synthesis: translation factors |
| PG0512 | Guanylate kinase | gmk | Purines, pyrimidines, nucleosides, and nucleotides: nucleotide and nucleoside interconversions |
| PG1068 | Conserved hypothetical protein | Hypothetical proteins: conserved | |
| PG1271 | Acetylornithine aminotransferase, putative | Amino acid biosynthesis: glutamate family | |
| PG1490 | TraG family protein | Mobile and extrachromosomal element functions: plasmid functions | |
| PG1768 | Magnesium chelatase | Transport and binding proteins: cations and iron-carrying compounds | |
| PG1824 | Enolase | eno | Energy metabolism: glycolysis-gluconeogenesis |
| PG2055 | Dihydroorotate dehydrogenase family protein | Purines, pyrimidines, nucleosides, and nucleotides: pyrimidine ribonucleotide biosynthesis | |
| PG1592 | HDIG domain protein | Unknown function: general | |
| PG1773 | PAP2 superfamily protein | Unknown function: general | |
| Putative outer membrane proteins | |||
| PG0026 | Hypothetical protein | ||
| PG0421 | Hypothetical protein | ||
| PG0809 | Hypothetical protein | ||
| Putative cytoplasmic proteins | |||
| PG0065 | Efflux transporter, RND family, MFP subunit | Transport and binding proteins: unknown substrate | |
| PG0137 | Aminoacyl-histidine dipeptidase | pepD-1 | Protein fate: degradation of proteins, peptides, and glycopeptides |
| PG0153 | Aspartyl-tRNA synthetase | aspS | Protein synthesis: tRNA aminoacylation |
| PG0384 | MutS2 family protein | DNA metabolism: other | |
| PG0394 | DNA-directed RNA polymerase, beta subunit | rpoB | Transcription: DNA-dependent RNA polymerase |
| PG0395 | DNA-directed RNA polymerase, beta′ subunit | rpoC | Transcription: DNA-dependent RNA polymerase |
| PG0445 | Peptidase T | pepT | Protein fate: degradation of proteins, peptides, and glycopeptides |
| PG0504 | Lipoate synthase | lipA | Biosynthesis of cofactors, prosthetic groups, and carriers: lipoate |
| PG0520 | Chaperonin, 60 kDa | groEL | Protein fate: protein folding and stabilization |
| PG0553 | Extracellular protease, putative | Protein fate: degradation of proteins, peptides, and glycopeptides | |
| PG0629 | ATP-NAD kinase | ppnK | Biosynthesis of cofactors, prosthetic groups, and carriers: pyridine nucleotides |
| PG0728 | Conserved hypothetical protein | Hypothetical proteins: conserved | |
| PG1195 | 8-Amino-7-oxononanoate synthase | bioF-2 | Biosynthesis of cofactors, prosthetic groups, and carriers: biotin |
| PG1242 | Replicative DNA helicase | dnaB | DNA metabolism: DNA replication, recombination, and repair |
| PG1622 | DNA topoisomerase IV, A subunit, putative | DNA metabolism: DNA replication, recombination, and repair | |
| PG1849 | DNA repair protein RecN | recN | DNA metabolism: DNA replication, recombination, and repair |
| PG1875 | Hemolysin | Cellular processes: pathogenesis | |
| PG1926 | Ribosomal protein L5 | rplE | Protein synthesis: ribosomal proteins (synthesis and modification) |
| Not determined | |||
| PG0232 | Zinc carboxypeptidase, putative | Protein fate: degradation of proteins, peptides, and glycopeptides | |
| PG1084 | Thioredoxin family protein | Energy metabolism: electron transport |
RND, resistance nodulation cell division; MFP, membrane fusion protein; HDIG, His-Asp-Ile-Gly.
Specific binding of TprA protein with TapA and TapB proteins.
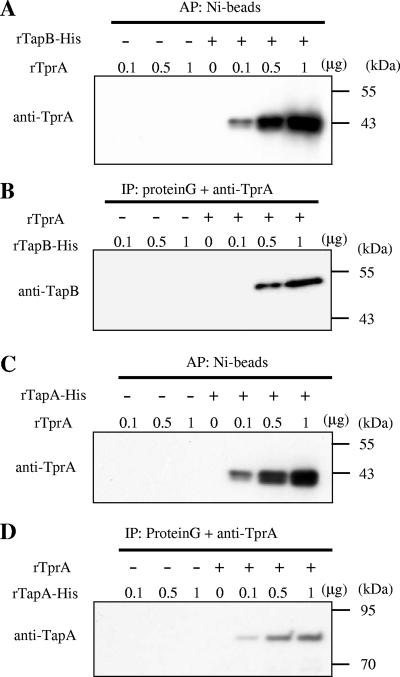
We examined the interaction of TprA protein with TapA, TapB, and TapC proteins in vitro using protein binding assays. We determined the binding activity of TprA protein with TapA and TapB proteins, since we could not obtain a TapC-soluble recombinant protein (Fig. 3). Polyhistidine-tagged recombinant TapA (rTapA-His) or TapB (rTapB-His) was incubated with Ni beads and recombinant TprA (rTprA). rTprA itself did not bind to Ni beads, as revealed by immunoblot analysis using anti-TprA antibody, whereas rTprA could bind to Ni beads in the presence of rTapA-His or rTapB-His, and the reaction was observed to occur in a concentration-dependent manner (Fig. 3A and C). Next, rTprA was mixed with rTapA-His or rTapB-His, immunoprecipitated with anti-TprA antibody, and subjected to immunoblot analysis with anti-TapA or anti-TapB antibody. The results were consistent with the results described above (Fig. 3B and D).
FIG. 3.
Binding of TprA protein to TapA and TapB proteins. (A and B) Binding of TprA and TapB. (A) rTprA (0 to 1.0 μg) was mixed with rTapB-His (0.5 μg) and was subjected to affinity purification (AP) by the use of Ni beads. The resulting samples were subjected to SDS-PAGE and immunoblot analysis with anti-TprA. (B) rTapB-His (0 to 1.0 μg) was mixed with rTprA (0.5 μg) and was immunopurified (IP) using protein G agarose with anti-TprA. The resulting samples were subjected to SDS-PAGE and immunoblot analysis with anti-TapB. (C and D) Binding of TprA and TapA. (C) rTprA (0 to 1.0 μg) was mixed with rTapA-His (0.5 μg) and was subjected to affinity purification using Ni beads. The resulting samples were subjected to SDS-PAGE and immunoblot analysis with anti-TprA. (D) rTapA-His (0 to 1.0 μg) was mixed with rTprA (0.5 μg) and was immunopurified using protein G agarose with anti-TprA. The resulting samples were subjected to SDS-PAGE and immunoblot analysis with anti-TapA.
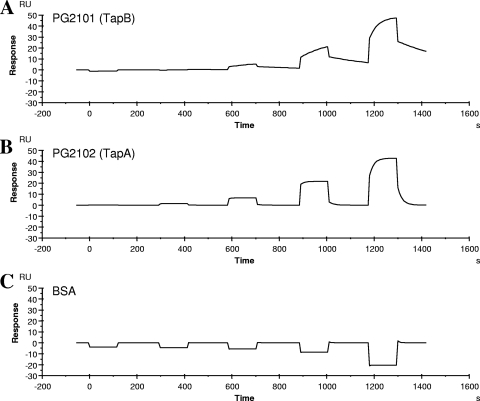
These specific interactions were further confirmed by surface plasmon resonance detection using the BIAcore system. The Kd values determined for interactions of rTprA with rTapA-His and rTapB-His were 1.89 × 10−4 M and 1.42 × 10−6 M, respectively (Fig. 4).
FIG. 4.
Surface plasmon resonance detection. Parameters of association and dissociation for rTapA-His and rTapB-His with rTprA were determined by surface plasmon resonance analysis (Biacore). Sensorgrams for the binding of rTapA-His or rTapB-His to rTprA immobilized on a CM5 sensor chip were overlaid at various concentrations of rTapA-His or rTapB-His. rTapA-His, rTapB-His, or bovine serum albumin (BSA) was injected at concentrations of 0.016, 0.08, 0.4, 2, and 10 mM. The Kd values of binding of rTprA to rTapA-His and rTapB-His were 1.89 × 10−4 M and 1.42 × 10−6 M, respectively. RU, resonance units.
Localization of TapA and TapB proteins.
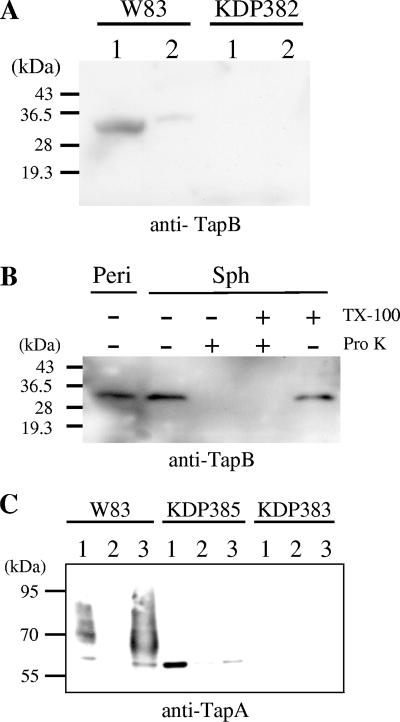
Cells of the wild-type and tapB mutant strains were fractionated into cytoplasm-periplasm, inner membrane, and outer membrane fractions and then subjected to SDS-PAGE and immunoblot analysis with anti-TapB antiserum (Fig. 5A). A 33-kDa anti-TapB-immunoreactive protein was found in the cytoplasm-periplasm fraction. To determine whether the 33-kDa anti-TapB-immunoreactive protein was located in the cytoplasm or periplasm, cells of the wild-type strain were subjected to spheroplast formation and proteinase K treatment followed by immunoblot analysis with anti-TapB (Fig. 5B). The 33-kDa anti-TapB-immunoreactive protein was found in the periplasm fraction. The 33-kDa protein was also found in the spheroplast fraction and disappeared after the proteinase K treatment. TapB protein possessed a signal sequence, as revealed by in silico analysis with SignalP V3.0 software. These results indicated periplasmic localization of TapB.
FIG. 5.
Localization of TapA and TapB proteins. (A and B) Subcellular localization of TapB protein. (A) Fractions of cytoplasm-periplasm (lanes 1) and total membrane (lanes 2) of W83 (wild type) and KDP382 (ΔtapB::ermF) were subjected to SDS-PAGE and immunoblot analysis using anti-TapB. (B) Spheroplast and periplasm fractions of P. gingivalis cells were separated as described in Materials and Methods. Spheroplasts were subjected to the proteinase K treatment (Pro K) in the presence or absence of 2% Triton X-100 (TX-100). Samples were subjected to SDS-PAGE followed by immunoblot analysis with anti-TapB. Sph, spheroplasts; Peri, periplasm fraction. (C) Subcellular localization of the TapA protein. Cytoplasm-periplasm, inner membrane, and outer membrane fractions of W83 (wild type), KDP385 (ΔporT::erm), and KDP383 (ΔtapA::ermF) were subjected to SDS-PAGE and immunoblot analysis using anti-TapA. Lanes: 1, cytoplasm-periplasm fraction; 2, inner membrane fraction; 3, outer membrane fraction.
To investigate the localization of TapA protein, the wild-type and tapA mutant cells were fractionated into cytoplasm-periplasm, inner membrane, and outer membrane fractions and then subjected to SDS-PAGE and immunoblot analysis with anti-TapA (Fig. 5C). A discrete protein band with a molecular mass of 60 kDa and diffuse protein bands with molecular masses of 65 to 95 kDa were detected in the outer membrane fraction of the wild-type strain but not in that of the tapA mutant. These protein bands were also observed in the cytoplasm-periplasm fraction, although with a much weaker reaction. These results indicated that TapA protein was located at the outer membrane.
TapA is one of the CTD proteins.
P. gingivalis has been shown to possess a novel family of outer membrane proteins that have a conserved RgpB C-terminal domain (CTD) (45, 51). The CTD has been proposed to play roles in the secretion of the proteins across the outer membrane and their attachment to the cell surface, probably via glycosylation (30, 43, 45). Very recently, we found a novel secretion system, the Por secretion system (PorSS), by which the CTD proteins may be secreted (42).
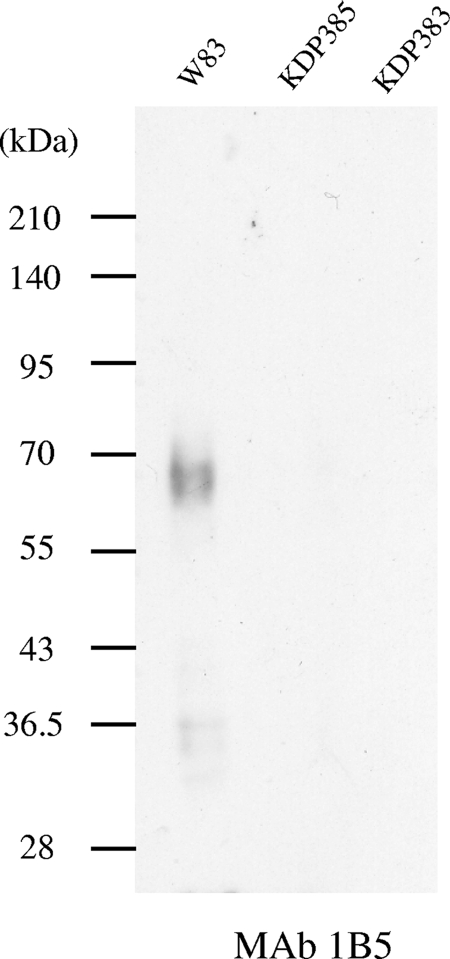
TapA (PG2102) was reported to belong to the CTD family of proteins and seemed to be secreted across the outer membrane and attach to the cell surface via glycosylation. TapA protein was immunoprecipitated from the wild-type and porT and tapA mutant strains by the use of anti-TapA. The immunoprecipitates were subjected to SDS-PAGE and immunoblot analysis using MAb 1B5, a monoclonal antibody that recognizes P. gingivalis anionic surface polysaccharides. MAb 1B5-immunoreactive diffuse protein bands with molecular masses of 65 to 95 kDa were found in the wild-type strain but not in the porT or tapA mutant strain (Fig. 6). The results suggested that the MAb 1B5-immunoreactive diffuse protein bands with molecular masses of 65 to 95 kDa were glycosylated forms of the TapA protein. In the porT mutant, the anti-TapA-immunoreactive protein with a molecular mass of 60 kDa was mainly found in the cytoplasm-periplasm fraction, suggesting that the TapA protein is one of the outer membrane proteins secreted via PorSS.
FIG. 6.
Glycosylation of TapA protein. TapA protein was immunoprecipitated from lysates of W83, KDP385 (ΔporT::erm) and KDP383 (ΔtapA::ermF) cells by the use of anti-TapA. The resulting immunoprecipitates were subjected to SDS-PAGE and immunoblot analysis with 1B5 MAb.
Contribution of the tapA-tapB-tapC operon to virulence of P. gingivalis.
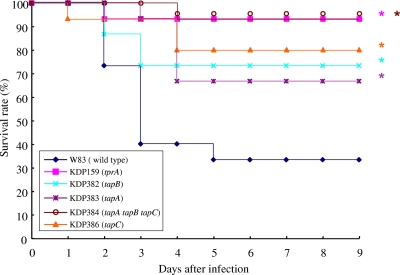
BALB/c mice were challenged with subcutaneous injections of bacterial suspension (2 × 1011 CFU per animal) (28, 49, 53), and their survival was monitored for 9 days. About 66.7% of the mice challenged with W83 died at the end of the experiment (9 days). In contrast, the survival rates of mice inoculated with the tprA, tapA, tapB, and tapC single mutants and the tapA-tapB-tapC deletion mutant were significantly higher (P < 0.05 [log-rank test]) than those of mice inoculated with the wild-type strain (Fig. 7). These results suggested that the tprA, tapA, tapB, and tapC genes were involved in P. gingivalis virulence.
FIG. 7.
Survival rates of mice challenged with the wild-type and mutant strains of P. gingivalis. Construction of the tapA, tapB, tapC, and tapA-tapB-tapC mutants is described in Materials and Methods. Genetic manipulation did not affect the expression of the downstream or upstream gene (see Fig. S3 in the supplemental material). Female BALB/c mice were intradermally inoculated with P. gingivalis cells into the back (approximately 2 × 1011 CFU), and then their survival was monitored daily for up to 9 days. The animal experiment, in which five mice were used for each bacterial strain, was performed three times. *, P < 0.05 versus the corresponding values for the wild-type littermates, as determined with the log-rank test.
DISCUSSION
The TPR motif was originally reported for cell division cycle proteins of Saccharomyces cerevisiae (18, 47). This motif is now known to be ubiquitous in nature, as it is found within functionally unrelated proteins from all genera. A TPR is defined as a degenerate 34-residue motif with a consensus amino acid arrangement of alternate large and small residues and highly conserved amino acids observed specifically at positions 8, 20, and 27 (47). These conserved residues allow the TPR to create a pair of antiparallel alpha helices. Multiple motifs, ranging in number from 3 to 16 among TPR proteins, lead to the formation of an alpha superhelical structure (11). This complex and unique structure gives rise to distinct substrate grooves that facilitate specific protein-protein interactions. The ability of TPR-containing proteins to interact with other proteins enables them to play a vital role in eukaryotic cell processes, such as mitosis, transcription repression, and protein import (14, 22, 50). Bacteria also utilize TPR proteins for a range of functions, including gene regulation, flagellar motor function, chaperone activity, gliding motility, and virulence (4, 5, 8, 29, 34, 44, 52). For example, FrzF from Myxococcus xanthus is a methyltransferase containing three TPR domains and it regulates the Frz chemosensory system, which controls cell reversals and gliding motility. In particular, TPRs in FrzF are involved in site-specific methylation of FrzCD, a metlyl-accepting chemotaxis protein (44). In addition, several chaperones required for the type III secretion system, including PcrH from Pseudomonas aeruginosa, LcrH from Yersinia species, and CesD from enteropathogenic E. coli, contain a TPR domain (4, 5, 52).
P. gingivalis W83 has at least 10 TPR proteins. However, none of them have been investigated so far. Structures of the TPR proteins are depicted with their putative subcellular location in Fig. S4 in the supplemental material. TprA protein contains three TPR domains. Okano et al. (35) reported that 19 proteins of P. gingivalis W83 are upregulated by aeration and that the 19 proteins include PG1385 (TprA), PG0045 (HtpG), PG0520 (60-kDa chaperonin), PG1208 (DnaK), PG0762 (trigger factor), PG0185 (RagA), and PG0618 (AhpC). Masuda et al. (25) reported that 14 proteins that include TprA were upregulated in P. gingivalis ATCC 33277 cells cultivated in a nutrient-poor medium. We have previously reported that TprA protein was upregulated in P. gingivalis W83 cells placed in a mouse subcutaneous chamber (54). Mouse subcutaneous infection experiments revealed that the tprA mutant was clearly less virulent. These findings suggest that expression of tprA is influenced by various environmental stresses and that TprA is involved in virulence of P. gingivalis.
TprA protein is located at the periplasm (45), which implies that TprA binds to other proteins at the periplasm or protruding from the inner or outer membrane. We performed a yeast two-hybrid assay and found TapB (a hypothetical protein) as a TprA-associated protein. Taking this result together with the finding that expression of TapA (as well as TprA) was induced in P. gingivalis cells placed in a mouse chamber, we assumed that TprA was related to products of a cluster of tapA, tapB, and tapC genes. Protein binding assays with immunoprecipitation and plasmon resonance detection revealed the interaction of TprA protein with TapA and TapB proteins. We could not examine the interaction between TprA protein and TapC protein in this study; however, TapC protein may interact with TprA protein, since TapC protein has 45% identity in amino acid sequence with TapA protein. Mouse subcutaneous infection experiments in this study revealed that the survival rate of mice with the tprA mutant was equivalent to that of mice with the tapA-tapB-tapC deletion mutant, suggesting that TprA protein plays a role in virulence of P. gingivalis in cooperation with TapA, TapB, and TapC proteins.
We also used a microarray to investigate the gene expression profile of the tprA mutant by inoculation of the mutant strain into a mouse subcutaneous chamber and comparison of the results to those obtained after inoculation with the wild-type strain. The results of microarray analysis revealed that 12 genes were upregulated and 11 genes were downregulated in the tprA mutant. Interestingly, a cluster of tapA, tapB, and tapC genes was found to be downregulated in the tprA mutant strain. Downregulation of the tapA, tapB, and tapC genes was confirmed by real-time qPCR analysis. At present, the mechanism of downregulation remains to be clarified.
TapA and TapC proteins belong to the C-terminal domain (CTD) family of proteins (45). The CTD has been proposed to play roles in the secretion of proteins across the outer membrane and their attachment to the surface of the cell, probably via glycosylation (30, 43, 45). Nguyen et al. reported that CTD is specifically conserved in proteins of distinct members of the phylum Bacteroidetes and suggested that CTD proteins are secreted by a novel secretion system (30). Very recently, we have found a novel secretion system by which CTD proteins are secreted and named it the Por secretion system (PorSS) (42). We found in the present study that TapA protein, which is an outer membrane protein, was modified by polysaccharides immunoreactive to the 1B5 MAb, resulting in production of diffuse protein bands with molecular masses of 65 to 95 kDa that were lacking in the porT mutant experiments. These results suggest that TapA protein is secreted by PorSS.
As revealed by the mouse subcutaneous infection experiments, the tapA and tapC mutants were less virulent than the wild-type parent strain, suggesting that the TapA and TapC CTD proteins are related to the virulence of P. gingivalis. Besides TapA and TapC proteins, P. gingivalis has a number of CTD proteins: gingipain proteinases (RgpA, RgpB, and Kgp), CPG70 carboxypeptidase (6), PrtT thiol proteinase, HagA hemagglutinin, Streptococcus gordonii binding protein (55), putative hemagglutinin, putative thiol reductase, putative fibronectin binding protein, putative Lys-specific proteinase, and putative von Willebrand factor domain protein. RgpA and Kgp contain C-terminal adhesins that are secreted and processed to form noncovalent complexes on the cell surface and are considered to be the major virulence factors of this bacterium (32, 33, 40). Gingipains have been linked directly to disease pathogenesis due to their ability to degrade host structural and defense proteins and the inability of mutants lacking functional Kgp or RgpA/B to cause alveolar bone loss in murine periodontitis models (39). The majority of these proteins are likely to play important roles in virulence of the bacterium, since they are involved in extracellular proteolytic activity, aggregation, heme and iron capture and storage, biofilm formation, and resistance to oxidative stress.
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid (18018032 and 20249073 to K.N.) for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Global COE Program at Nagasaki University (K.N.).
Editor: A. Camilli
Footnotes
Published ahead of print on 29 March 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Armitage, G. C. 1996. Periodontal diseases: diagnosis. Ann. Periodontol. 1:37-215. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 4.Bröms, J. E., P. J. Edqvist, A. Forsberg, and M. S. Francis. 2006. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol. Lett. 256:57-66. [DOI] [PubMed] [Google Scholar]
- 5.Bröms, J. E., A. L. Forslund, A. Forsberg, and M. S. Francis. 2003. PcrH of Pseudomonas aeruginosa is essential for secretion and assembly of the type III translocon. J. Infect. Dis. 188:1909-1921. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y. Y., K. J. Cross, R. A. Paolini, J. E. Fielding, N. Slakeski, and E. C. Reynolds. 2002. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J. Biol. Chem. 277:23433-23440. [DOI] [PubMed] [Google Scholar]
- 7.Christersson, L. A., C. L. Fransson, R. G. Dunford, and J. J. Zambon. 1992. Subgingival distribution of periodontal pathogenic microorganisms in adult periodontitis. J. Periodontol. 63:418-425. [DOI] [PubMed] [Google Scholar]
- 8.Core, L., and M. Perego. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509-1522. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 11.Das, A. K., P. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado-Partin, V. M., and R. E. Dalbey. 1998. The proton motive force, acting on acidic residues, promotes translocation of amino-terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J. Biol. Chem. 273:9927-9934. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, P. I., N. Slakeski, E. C. Reynolds, R. Morona, A. H. Rogers, and P. E. Kolenbrander. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 188:2454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodt, G., and S. J. Gould. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 5:78-111. [DOI] [PubMed] [Google Scholar]
- 18.Hirano, T., N. Kinoshita, K. Morikawa, and M. Yanagida. 1990. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell 60:319-328. [DOI] [PubMed] [Google Scholar]
- 19.Hosogi, Y., and M. J. Duncan. 2005. Gene expression in Porphyromonas gingivalis after contact with human epithelial cells. Infect. Immun. 73:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 21.Irfan, U. M., D. V. Dawson, and N. F. Bissada. 2001. Epidemiology of periodontal disease: a review and clinical perspectives. J. Int. Acad. Periodontol. 3:14-21. [PubMed] [Google Scholar]
- 22.King, R. W., J. M. Peters, S. Tugendreich, M. Rolfe, P. Hieter, and M. W. Kirschner. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279-288. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lo, A. W., C. A. Seers, J. D. Boyce, S. G. Dashper, N. Slakeski, J. P. Lissel, and E. C. Reynolds. 2009. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, T., Y. Murakami, T. Noguchi, and F. Yoshimura. 2006. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 72:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami, Y., M. Imai, H. Nakamura, and F. Yoshimura. 2002. Separation of the outer membrane and identification of major outer membrane proteins from Porphyromonas gingivalis. Eur. J. Oral Sci. 110:157-162. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama, K. 2003. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr. Protein Pept. Sci. 4:389-395. [DOI] [PubMed] [Google Scholar]
- 28.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 29.Newton, H. J., F. M. Sansom, J. Dao, A. D. McAlister, J. Sloan, N. P. Cianciotto, and E. L. Hartland. 2007. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect. Immun. 75:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, K. A., J. Travis, and J. Potempa. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J. Bacteriol. 189:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, K. A., J. Zylicz, P. Szczesny, A. Sroka, N. Hunter, and J. Potempa. 2009. Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien-Simpson, N. M., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409-426. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okabe, M., T. Yakushi, and M. Homma. 2005. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J. Biol. Chem. 280:25659-25664. [DOI] [PubMed] [Google Scholar]
- 35.Okano, S., Y. Shibata, T. Shiroza, and Y. Abiko. 2006. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics 6:251-258. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, R. C., L. J. Brown, and H. Loe. 1998. Periodontal diseases in the United States population. J. Periodontol. 69:269-278. [DOI] [PubMed] [Google Scholar]
- 37.Papapanou, P. N. 1999. Epidemiology of periodontal diseases: an update. J. Int. Acad. Periodontol. 1:110-116. [PubMed] [Google Scholar]
- 38.Park, Y., O. Yilmaz, I. Y. Jung, and R. J. Lamont. 2004. Identification of Porphyromonas gingivalis genes specifically expressed in human gingival epithelial cells by using differential display reverse transcription-PCR. Infect. Immun. 72:3752-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathirana, R. D., N. M. O'Brien-Simpson, G. C. Brammar, N. Slakeski, and E. C. Reynolds. 2007. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect. Immun. 75:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues, P. H., and A. Progulske-Fox. 2005. Gene expression profile analysis of Porphyromonas gingivalis during invasion of human coronary artery endothelial cells. Infect. Immun. 73:6169-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato, K., M. Naito, H. Yukitake, H. Hirakawa, M. Shoji, M. J. McBride, R. G. Rhodes, and K. Nakayama. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato, K., E. Sakai, P. D. Veith, M. Shoji, Y. Kikuchi, H. Yukitake, N. Ohara, M. Naito, K. Okamoto, E. C. Reynolds, and K. Nakayama. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280:8668-8677. [DOI] [PubMed] [Google Scholar]
- 44.Scott, A. E., E. Simon, S. K. Park, P. Andrews, and D. R. Zusman. 2008. Site-specific receptor methylation of FrzCD in Myxococcus xanthus is controlled by a tetra-trico peptide repeat (TPR) containing regulatory domain of the FrzF methyltransferase. Mol. Microbiol. 69:724-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seers, C. A., N. Slakeski, P. D. Veith, T. Nikolof, Y. Y. Chen, S. G. Dashper, and E. C. Reynolds. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelburne, C. E., R. M. Gleason, G. R. Germaine, L. F. Wolff, B. H. Mullally, W. A. Coulter, and D. E. Lopatin. 2002. Quantitative reverse transcription polymerase chain reaction analysis of Porphyromonas gingivalis gene expression in vivo. J. Microbiol. Methods 49:147-156. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 48.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachibana-Ono, M., A. Yoshida, S. Kataoka, T. Ansai, Y. Shintani, Y. Takahashi, K. Toyoshima, and T. Takehara. 2008. Identification of the genes associated with a virulent strain of Porphyromonas gingivalis using the subtractive hybridization technique. Oral Microbiol. Immunol. 23:84-87. [DOI] [PubMed] [Google Scholar]
- 50.Tzamarias, D., and K. Struhl. 1994. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369:758-761. [DOI] [PubMed] [Google Scholar]
- 51.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wattiau, P., B. Bernier, P. Deslee, T. Michiels, and G. R. Cornelis. 1994. Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. U. S. A. 91:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimura, M., Y. Nakano, Y. Yamashita, T. Oho, T. Saito, and T. Koga. 2000. Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect. Immun. 68:6912-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshimura, M., N. Ohara, Y. Kondo, M. Shoji, S. Okano, Y. Nakano, Y. Abiko, and K. Nakayama. 2008. Proteome analysis of Porphyromonas gingivalis cells placed in a subcutaneous chamber of mice. Oral Microbiol. Immunol. 23:413-418. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y., T. Wang, W. Chen, O. Yilmaz, Y. Park, I. Y. Jung, M. Hackett, and R. J. Lamont. 2005. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics 5:198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.