Abstract
The HT29MTXE12 (E12) cell line harbors an adherent mucus layer, providing a novel technique to model mucosal infection in vitro. In this study, we have characterized the interaction of Campylobacter jejuni with the E12 cell line and exploited its unique mucus layer to examine the potential efficacy of probiotic treatment to attenuate C. jejuni virulence properties. C. jejuni 81-176 colonized and reproduced in E12 mucus. Adhesion to and internalization of C. jejuni were enhanced in E12 cells harboring mucus compared to parental cells without mucus. Translocation of C. jejuni occurred at early time points following infection. C. jejuni aligned with tight junctions and colocalized with the tight junction protein occludin, suggesting a paracellular route of translocation. Probiotic strains Lactobacillus rhamnosus R0011, Lactobacillus helveticus R0052, Lactobacillus salivarius AH102, Bifidobacterium longum AH1205, a commercial combination of L. rhamnosus R0011 and L. helveticus R0052 (Lacidofil), and a cocktail consisting of L. rhamnosus, L. helveticus, and L. salivarius (RhHeSa) colonized E12 mucus and bound to underlying cells. Probiotics attenuated C. jejuni association with and internalization into E12 cells and translocation to the basolateral medium of transwells. Live bacteria and prolonged precolonization of E12 cells with probiotics were necessary for probiotic action. These results demonstrate the potential for E12 cells as a model of mucosal pathogenesis and provide a rationale for the further investigation of probiotics as prophylaxis against human campylobacteriosis.
The food-borne pathogen Campylobacter jejuni is among the most common bacterial causes of gastroenteritis (43). Serious postinfectious sequelae, such as reactive arthritis (35), irritable bowel syndrome (37), and the paralytic neuropathy Guillain-Barre syndrome (30), also have been associated with antecedent C. jejuni infections. Treatment of Campylobacter infections is usually supportive, while antibiotics are reserved for invasive disease (43). Currently there is no vaccine against Campylobacter, and preventive measures aimed at reducing environmental contamination are largely ineffective. Thus, there is a need for alternative strategies that would complement currently employed methods aimed at reducing C. jejuni-induced disease burden in humans.
Probiotics, “live microorganisms which when administered in adequate amounts confer a health benefit on the host” (44) or “commensal microorganisms that can be harnessed for health benefits” (34), are an attractive alternative intervention strategy both for poultry and for humans for the interruption of the C. jejuni infection cycle and/or treatment of active Campylobacter-related disease. Probiotics have been shown to attenuate the virulence of several enteropathogens in vitro (9, 36). Preliminary data from coculture (7) and animal studies (29, 41, 45) point to the potential effectiveness of probiotics in inhibiting C. jejuni infection. In a recent paper, Wine et al. (46) provide evidence for strain- and cell-type-specific reduction in C. jejuni invasiveness following coculture with probiotics. However, in general, the effect of probiotics on the pathogenicity of C. jejuni has received little attention.
Campylobacter research has been hampered by the absence of a simple and reliable animal model that mimics human campylobacteriosis (17) and by concern for the development of serious sequelae following experimental human challenge with C. jejuni. For these reasons, in vitro cell culture methods have been used extensively to study the virulence of this organism. However, a key disadvantage in the use of conventional cell culture methods is the lack of an associated mucus gel layer. The HT29MTXE12 (E12) cell line, a goblet cell-like subclone of the human colon carcinoma HT29 cell line, uniquely secretes an adherent mucus layer and forms tight junctions when polarized on transwell inserts (3). This novel model provides an opportunity to study in vitro the role of mucus and the relationship of mucus-associated factors, such as endogenous flora and probiotic organisms, on the colonization and pathogenic behavior of enteric pathogens.
In this study, we have characterized and exploited this novel model in order to examine the effectiveness of probiotics in protection against C. jejuni. We demonstrate that the presence of an associated mucus layer enhanced C. jejuni pathogenicity, and we provide evidence for bacterial translocation through the paracellular route. We show that some probiotic strains attenuate the virulence of C. jejuni. Further, we show that precolonization of E12 cells with probiotics is necessary for probiotic efficacy. These data provide evidence for the potential use of certain probiotics in intervention strategies against C. jejuni. In particular, these results suggest that the effectiveness of probiotics against human C. jejuni infection may require anticipatory use of this therapy to prevent infection in those at high risk rather than as active treatment for infected individuals.
MATERIALS AND METHODS
Infection assays. (i) Bacterial strains, routine maintenance, and culture.
C. jejuni 81-176 is a well-characterized and invasive strain that has been used in several studies (14). It was originally isolated in a milk-borne outbreak (20). C. jejuni 81-176 was routinely cultured on Mueller Hinton agar (Oxoid) and supplemented with Campylobacter selective supplement (Skirrow) at 37°C under microaerophilic conditions using CampyGen gas packs (Oxoid). Lactobacillus salivarius AH102 (9) and Bifidobacterium longum AH1205 (9) have previously been characterized at the Pharmabiotic Centre, Cork, Ireland. Lactobacillus helveticus R0052 (16, 36), Lactobacillus rhamnosus R0011 (36), and Lacidofil (5:95 mixture of L. helveticus and L. rhamnosus) were obtained as lyophilized products from the Institut Rosell-Lallemand, Montreal, Canada, and stored at 4°C. L. salivarius, L. rhamnosus, and L. helveticus were routinely grown in de Man, Rogosa, and Sharpe (MRS) broth. RhHeSa was a combination of L. rhamnosus, L. helveticus, and L. salivarius prepared by mixing equal volumes and optical densities at 600 nm (OD600) of these bacteria in tissue culture medium. B. longum was grown in MRS broth supplemented with 0.05% cysteine hydrochloride. All probiotic strains were grown at 37°C under microaerophilic conditions.
(ii) Cell lines.
The human colon adenocarcinoma cell line HT29 (22) was purchased from the American Type Culture Collection (ATCC HTB-38). HT29MTXE12 (E12) cells, which are methotrexate (MTX)-adapted HT29 cells (HT29MTX) (22) grown on transwell inserts and selected on the basis of the formation of an adherent mucus layer, tight junctions, and a confluent monolayer, were a kind gift from Per Artursson of the University of Uppsala, Sweden (3).
(iii) Cell culture.
E12 cells were routinely grown in Dulbecco's modified Eagle's medium (DMEM; BioWhittaker) supplemented with 10% fetal bovine serum (FBS) (Sigma), 2 mM l-glutamine (Gibco), and 1% nonessential amino acids (BioWhittaker). HT29 cells were grown in McCoy's 5A modified medium (Sigma) supplemented with 10% FBS. All cells were routinely maintained in 75-cm2 tissue culture flasks and incubated at 37°C in 5% CO2 in a humidified atmosphere. Cells were passaged when the confluence of the flasks was about 80%. Cells were trypsinized and seeded onto 24-well polyvinylidene difluoride (PVDF) plates (Corning). In order to polarize cells, they were seeded on transwell inserts (0.4-μm pore size, 12-mm diameter) (Corning), in which they formed a polarized monolayer and tight junctions. Cells were seeded at a density of 1 × 105 cells/well. Cells were grown for 48 h on 24-well plates prior to use and up to 21 days on transwell inserts. Penicillin (10 U/ml), 10 μg/ml streptomycin, and 0.25 μg/ml amphotericin B were added to the medium prior to seeding on transwells. Antibiotics were removed from the cells at least 24 h before infection assays.
(iv) Culture of C. jejuni for infection assays.
C. jejuni 81-176 was cultured from frozen stocks on Mueller Hinton agar for 24 h. One loop full of bacteria was then transferred to biphasic medium in 25-cm2 tissue culture flasks (Corning) consisting of Mueller Hinton agar and 6 ml of DMEM prepared as described above but supplemented with 2% FBS and without antibiotics, for 18 to 21 h. All bacterial incubations were under microaerophilic conditions generated by CampyGen gas packs. Bacteria were harvested from the biphasic medium, washed once in DMEM, and then diluted to an OD600 of ∼0.2 or the required OD, where indicated.
(v) Total association and gentamicin protection assay.
E12 and HT29 cells grown in 24-well plates (nonpolarized and no mucus) or polarized HT29 (21 days) and polarized E12 cells (7, 14, and 21 days) seeded on 24-well plates or 0.4-μm-pore-size transwells (Corning), respectively, were infected with C. jejuni harvested from Mueller Hinton agar/DMEM biphasic medium at a multiplicity of infection (MOI) of 500. Infected cells were incubated at 37°C under microaerophilic conditions using CampyGen gas packs (Oxoid) for 3, 24, and 48 h. Infection of E12 cells for 24 and 48 h with C. jejuni did not affect the viability of cells, as shown by trypan blue exclusion assays. One set of wells was washed six times with PBS, and the cells were lysed with 100 μl of 0.1% Triton X-100 in PBS for 15 min, after which serial dilutions of the cell lysate in PBS were plated on Mueller Hinton agar. To determine the amount of internalized bacteria, a second set of wells was washed with tissue culture medium and 400 μg/ml gentamicin sulfate in DMEM was added on both chambers of the transwells for 2 h. Cells were washed in PBS and lysed with 100 μl of 0.1% Triton X-100 in PBS, and serial dilutions of the lysate plated on Mueller Hinton agar plates. Mueller Hinton agar plates were incubated at 37°C under microaerophilic conditions for 48 to 72 h, after which bacterial CFU were enumerated.
To quantify bacteria that bound to underlying cells after penetrating the E12 mucus layer, 10 mM N-acetylcysteine (NAC; Sigma) in PBS containing 0.2 μM calcium chloride, 0.5 mM magnesium chloride, and 15 mM glucose was used, for 1 h with agitation at 70 rpm, to remove the mucus layer after the C. jejuni infection period. Cells were washed four times in PBS to remove NAC and lysed with 100 μl of 0.1% Triton X-100 in PBS for 15 min, and serial dilutions of the cell lysates were made in PBS.
(vi) Reproduction in E12 mucus.
E12 cells were infected with C. jejuni as described above (3 sets of wells). After 3 h, all wells were washed and a total association assay was performed on one set of wells. The remaining two sets of wells were further incubated with bacterium-free medium for 24 and 48 h, after which a total association assay was again performed. All wells were washed six times in tissue culture medium prior to total association assays. As a control, DMEM alone (supplemented with 2% FBS) was investigated for bacterial replication after 24 h. C. jejuni reproduction was also investigated directly in mucus harvested from E12 cell culture supernatants. Mucus was pooled from confluent E12 cell culture supernatants by centrifugation at 1,500 rpm for 5 min. Pelleted mucus was coincubated with C. jejuni in DMEM supplemented with 2% FBS (OD600 of ∼0.15) over a 24-h period. As a control, C. jejuni was incubated in DMEM alone without mucus over a 24-h period. Bacterial CFU were enumerated in mucus and medium alone (controls) at 0, 5, and 24 h as described above.
(vii) Translocation and TEER measurements.
The amount of bacteria that translocated to the basolateral medium of the transwells after apical infection with C. jejuni was determined by making serial dilutions of the basolateral medium in PBS, which was then plated on Mueller Hinton agar and incubated at 37°C under microaerophilic conditions. The basolateral medium was plated 5 and 24 h postinfection. To investigate the effect of C. jejuni on the barrier properties of E12 cells, following the infection of E12 cells with C. jejuni, transepithelial electrical resistance (TEER) was measured at 0, 5, 24, and 48 h postinfection using an EVOM X meter (World Precision Instruments) coupled to an Endohm chamber (World Precision Instruments), and the infected cells compared to noninfected cells whose TEER was measured at the same time points. The effect on E12 cell TEER of coincubation of probiotics and C. jejuni was also determined. E12 cells were precolonized with L. rhamnosus for 15 h prior to infection with C. jejuni for a further 48 h. TEER of E12 cells was measured at 0, 15, 24, and 48 h postinfection in coincubated cells, in cells infected with L. rhamnosus or C. jejuni alone, and in uninfected cells.
(viii) Infection studies with probiotics and C. jejuni.
Probiotics were grown in MRS broth for 6 to 9 h under microaerophilic conditions. Probiotics were pelleted by centrifugation at 3,000 rpm for 5 min and resuspended in prepared tissue culture medium (DMEM or RPMI) at an OD600 of ∼0.3 to 0.4. A total of 300 μl of probiotics in tissue culture medium was used to infect cells for an initial period of either 4 h or 15 h. The medium on the cells was then replaced with 300 μl of C. jejuni culture (OD600 of ∼0.15) harvested from biphasic medium. Infected cells were further incubated for 24 h. To minimize changes in the pH due to the probiotics (probiotics reduce the pH of medium), medium on cells was replaced with fresh medium at the 6th and 12th hours following infection. Medium replenishment was done carefully and did not disturb the mucous layer with its associated colonizing organisms. Medium replenishment did not affect probiotic colonization levels, as shown by total association assays. Typically, pH levels were maintained above 5.5. Coincubation with probiotics did not affect the viability of E12 cells, as shown by trypan blue exclusion assays. After the incubation period, a total association and invasion assay was performed to quantify bound and internalized C. jejuni and probiotic CFU. As a control, heat-treated L. rhamnosus (100°C for 10 min) was also used to infect E12 cells. Heat treatment resulted in dead but physically intact bacteria, as verified by Gram staining. Translocation of the bacteria from the apical to the basolateral aspect of cells was determined by making serial dilutions of the basolateral medium in PBS and plating on Mueller Hinton agar to quantify bacterial CFU in the basolateral medium. Mueller Hinton agar supplemented with Campylobacter selective supplement (Skirrows) was used to select for C. jejuni CFU, while MRS agar (MRS broth supplemented with 1% agar) was used to select for probiotics. For selection of C. jejuni when coincubated with L. rhamnosus, Campylobacter blood-free selective agar base (modified cefoperazone charcoal deoxycholate agar [CCDA]; Oxoid) supplemented with Campylobacter selective supplement was used.
(ix) Statistical analysis.
Experiments were conducted on at least three separate occasions in triplicates. Results are presented as the means ± standard deviations (error bars) of replicate experiments. TEER experiments were conducted with a sample size (n) of at least 5. Graphs were drawn using Microsoft Excel, and the unpaired Student t test was used to estimate statistical significance. A P value of <0.05 was considered significant.
Microscopy. (i) Embedding polarized E12 cells in liver.
The HT29MTXE12 cell line is a subclone of the parent HT29 cell line, which secretes an adherent mucus gel when it becomes polarized on transwell inserts for 21 days (3). The mucus layer typically increased in thickness with time. Conventionally, E12 cells were studied and used at 7, 14, and 21 days postseeding on transwells. After cells were grown on transwells for the required time, the transwell inserts were washed with PBS, excised, embedded between two flat pieces of chicken liver tissue to protect the mucus layer (18), frozen in isopentane cooled by liquid nitrogen, and placed in Cryomold intermediates (Tissue-Tek) containing OCT compound (Santa Cruz). The embedded transwell inserts were further frozen in isopentane cooled by liquid nitrogen, prior to storage at −20°C. Sections were made using a cryostat and fixed onto poly-l-lysine precoated microscope slides (VWR International) using 4% formaldehyde in PBS (formalin). Slides were examined by fluorescence microscopy (see “Imaging” below).
(ii) Periodic acid-Schiff (PAS) staining for mucus.
Slides were immersed in periodic acid solution (Sigma) for 5 min and Schiff's reagent (Sigma) for 15 min and counterstained with hematoxylin solution (Sigma) for 90 s. Slides were rinsed with six changes of distilled water between reagents, left to dry, and mounted in xylene dibutylphthalate (DPX; BDH) prior to fluorescence microscopy.
(iii) TAMRA staining of C. jejuni and probiotics.
Bacteria were fluorescently labeled with carboxytetramethylrhodamine (TAMRA; Molecular Probes) (6, 28) and used in infection assays. TAMRA stains the cell cytoplasm by reacting with amines to form stable amide bonds and is conventionally used in our laboratory for vital staining (6). Bacteria were incubated in 10 μg/ml of TAMRA in PBS for 30 min at 37°C in the dark and washed four times in PBS (6, 28).
(iv) Antibody detection of C. jejuni colonization of E12 mucus.
E12 cells were infected with C. jejuni, and the transwells containing cells were embedded in chicken liver as described above. Slides were fixed for 15 min with 4% formaldehyde in PBS, blocked with 1% bovine serum albumin (BSA) in PBS for 1 h, and then incubated with a 1:200 dilution of rabbit C. jejuni antiserum (28) in 1% BSA for 1 h. Cells were incubated with a 1:500 dilution of goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Invitrogen) for 1 h followed by a 1:5,000 dilution of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma) in PBS for 30 min in the dark, to stain cell nuclei. Slides were washed in PBS, air dried, and then mounted in Dako fluorescent mounting medium. All washes between treatments were carried out six times in PBS. All antibody dilutions were in 1% BSA in PBS. Slides were visualized by immunofluorescence microscopy.
(v) Colocalization of C. jejuni with occludin.
E12 cells were cultured for 14 days and infected with C. jejuni for 3 h or 24 h. For some experiments, TAMRA-stained bacteria were used to infect cells. Cells were washed six times in PBS, and the mucolytic agent NAC was incubated with cells for 2 or 3 h to remove the mucus layer. Following the removal of mucus, transwells were washed in PBS and fixed with 4% formalin for 1 h. In the case in which TAMRA (red)-stained bacteria were used to infect E12 cells, the following procedure was used to stain the tight junction protein occludin after fixing in formalin: cells were blocked with 20% rabbit serum (Dako) in 1% BSA for 1 h and incubated for 1 h with a 1:100 dilution of goat anti-occludin antibody (Santa Cruz) followed by a 1:500 dilution of rabbit anti-goat IgG conjugated to Alexa Fluor 488 (Invitrogen). Thus, C. jejuni was stained red, and occludin was stained green. All washes between treatments were carried out six times with PBS. Transwells were allowed to dry, and the inserts were excised from the transwell and mounted onto a microscope slide using Dako fluorescent mountant. Slides were visualized by both immunofluorescence and confocal microscopy (see “Imaging” below).
(vi) Differential staining.
This protocol was used to discriminate between adherent and internalized bacteria (6). E12 cells were cultured for 14 days and infected with TAMRA-stained C. jejuni for 24 h or 48 h, the mucus layer was removed, and the cells were fixed as described above. Cells were blocked with 20% goat serum in 1% BSA in PBS. To stain adherent bacteria, cells were incubated with a 1:200 dilution of rabbit anti-C. jejuni followed by a 1:500 dilution of goat anti-rabbit IgG conjugated to Alexa Fluor 488. Adherent bacteria stained yellow (green and red), while internalized bacteria stained red (TAMRA only). All washes between treatments were carried out six times in PBS. Transwells were allowed to dry, and inserts excised and mounted onto a microscope slide using Dako fluorescent mountant. This procedure was repeated after C. jejuni was used to infect E12 cells for 48 h to reveal bacteria that remained adherent and internalized 48 h postinfection.
(vii) Differential staining of probiotics and C. jejuni.
L. rhamnosus was stained with TAMRA as described above for C. jejuni. TAMRA (red)-stained L. rhamnosus was used to infect E12 cells for an initial period of 4 h, after which the medium on the cells was replaced with 300 μl of C. jejuni at an OD600 of ∼0.15. Probiotics and C. jejuni were then further incubated for 24 h, after which the filter was excised from the transwell and embedded in chicken liver and sections were made in a cryostat and fixed onto a microscope slide as described above. Medium on the infected cells was replaced with fresh medium at the 6th and 12th hours postinfection to minimize changes in pH. Sections were then stained with a 1:200 dilution of rabbit-raised anti-C. jejuni antibody and revealed with a 1:400 dilution of donkey anti-rabbit IgG conjugated to Alexa Fluor 488 (Molecular Probes). All antibodies were diluted in 1% BSA in PBS, and incubations were for 1 h at room temperature. DAPI (1:4,000) was added to the secondary antibody solution to detect cell nuclei. Red-stained L. rhamnosus and green-stained C. jejuni cells could be observed in mucus and bound to cells by both immunofluorescence and laser scanning confocal microscopy (LSCM).
To detect both C. jejuni and L. rhamnosus bound to cells, 10 mM NAC was used to remove the mucus layer as described above, immediately after the incubation period. Cells were then fixed in 4% formaldehyde in PBS for 30 min, after which rabbit-raised anti-C. jejuni and donkey-raised anti-rabbit IgG conjugated to Alexa Fluor 488 were used to detect C. jejuni bound to cells as described above. Red-stained probiotics and green stained-C. jejuni bound to cells were observed by both immunofluorescence and confocal microscopy.
(viii) Imaging.
Immunofluorescence microscopy was carried out using an Olympus BX 51 light microscope and a JFX U-RFL-T fluorescent unit. Confocal microscopy was carried out using an LSM 510 laser scanning confocal microscope.
(ix) SEM.
Scanning electron microscopy (SEM) was carried out according to a protocol described by Sherman et al. (36). E12 cells were cultured for 14 days and infected with C. jejuni for 24 h as described above. Uninfected cells were used as controls. Cells were washed in PBS and fixed with 2.5% glutaraldehyde in Sorenson's phosphate buffer, pH 7.4, overnight. The fixing solution was replaced with Sorenson's phosphate buffer for 10 min, after which cells were incubated in 1% osmium tetroxide for 1 h at room temperature. Cells were then dehydrated in increasing grades of ethanol (from 70% to 100%). Cells were critically point dried, sputter coated with gold, and visualized using a scanning electron microscope (JSM 5410 LV).
RESULTS
The interaction of C. jejuni with mucus-adherent E12 cells. (i) Growth, polarization, and C. jejuni colonization of E12 mucus.
The thickness of the mucus layer and the transepithelial resistance of E12 cells typically increased from 7 through 14 to 21 days postseeding (Fig. 1Ai and ii). Using immunofluorescence microscopy, C. jejuni could be seen abundantly colonizing E12 mucus by 24 h postinfection (Fig. 1Bii). The organisms penetrated the mucus layer and could be found attached to and beneath the monolayer by 24 h postinfection (Fig. 1Biii). In addition, there was a modest increase in C. jejuni numbers due to reproduction in the monolayer between 3 h (6.9 × 106 CFU/well) and 24 h (1.9 × 107 CFU/well) and a further increase at 48 h following infection (3.6 × 107 CFU/well) (Fig. 1C). Furthermore, direct incubation of C. jejuni in mucus harvested from E12 cell culture supernatants led to a 10-fold increase in the number of recovered bacteria after 24-h coincubation compared with the number of bacteria recovered from tissue culture medium alone (data not shown). This indicates that E12 cells can sustain C. jejuni viability and E12 mucus encourages reproduction of the organism.
FIG. 1.
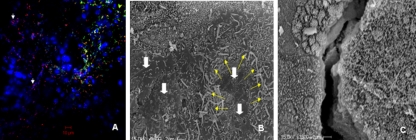
Interaction of C. jejuni with E12 cells. (A) Change in E12 cell mucus layer thickness and transepithelial resistance over time. The deep pink Schiff's reagent-stained mucus layer clearly increases in thickness between 7, 14, and 21 days. (i) Cells are seen adherent to the transwell filter. Remnants of liver tissue cells used for sample preparation can be visualized both above and below the filter. (ii) There is a marked increase in TEER compared to that for 21-day-cultured HT29 cells (21D HT29). Data are presented as means ± standard deviations (error bars). n = 10 for each group. Magnification, ×400. Scale bars, 150 μm. (B) Immunofluorescent micrograph of C. jejuni colonizing 21-day-cultured E12 cells. (i) Cell nuclei are visible following staining with DAPI on the transwell filter. (ii) Same field as that shown in panel Bi, showing extensive colonization by C. jejuni stained green within the mucus layer and predominantly located at the periphery of underlying cells. (iii) Some organisms (arrows) can be seen below the cell layer and above the transwell filter (translocation). Bringing organisms into focus results in some lack of definition of the filter and mucus in panels Bii and iii. Magnification, ×400. Scale bars, 10 μm. (C) C. jejuni reproduction in association with E12 cells. Three sets of 21-day-cultured E12 cells were infected with C. jejuni for 3 h. Medium was replaced with bacterium-free medium, and two sets of wells were further incubated for 24 and 48 h. A total association assay (n = 7) was performed after washes at each time point. * denotes significant difference (P < 0.05) between 24 and 48 h compared to 3 h. Error bars indicate standard deviations.
(ii) C. jejuni colonized mucus and bound to and invaded E12 cells.
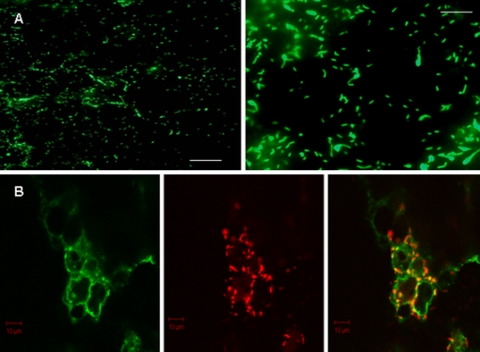
C. jejuni adhesion and internalization into underlying E12 cells were investigated by differential staining and LSCM. Differential staining with TAMRA (internal and external bacteria stained red) and Alexa Fluor 488 (external bacteria stained yellow/green) permitted the discrimination and visualization of bound and internalized bacteria after the mucus layer was removed. Using this approach, C. jejuni was visualized both adhering to and internalized into E12 cells (Fig. 2A). In agreement with previous studies (42), internalized C. jejuni bacteria were visualized in close proximity to and around cell nuclei (Fig. 2A). In addition, C. jejuni remained bound and internalized 48 h postinfection without affecting the viability of E12 cells, as shown by trypan blue exclusion assays (data not shown), indicating that the E12 cell line tolerates prolonged host-pathogen interactions.
FIG. 2.
Confocal and electron micrographic images of C. jejuni on E12 cells. (A) Differential immunofluorescence distinguishes internalized (red-stained) and external adherent (yellow-green) C. jejuni organisms following infection of E12 cells. Internalized organisms commonly coalesce around cell nuclei (arrows), which are stained blue with DAPI. Magnification, ×400. Scale bar, 10 μm. Typically, some areas of the monolayer harbored mainly internalized bacteria (as seen to left of image), while in other areas both internalized and externally adherent bacteria could be visualized (right side of micrograph). (B) SEM of C. jejuni adhering to E12 cells. Organisms typically coalesced around cell-cell interfaces (yellow arrows). White arrows indicate relative effacement of microvilli on cells that are heavily colonized with C. jejuni compared to those in the upper part of the micrograph with intact microvilli and few organisms. Magnification, ×3,500. Scale bar, 2 μm. (C) SEM of two adjacent uninfected cells (slightly separated at the cell-cell junction during processing) with intact microvilli. Magnification, ×3,500. Scale bar, 2 μm.
The interaction of C. jejuni with E12 cells was also visualized by SEM. Organisms were not evenly distributed on the surface of E12 cells but appeared to cluster around cell-cell junctions (Fig. 2B). In keeping with a recent report (32), microvilli appeared effaced on cells heavily colonized with bacteria compared to areas on the surface of cells lacking bacteria (Fig. 2B and C).
(iii) C. jejuni translocation and effect on monolayer resistance.
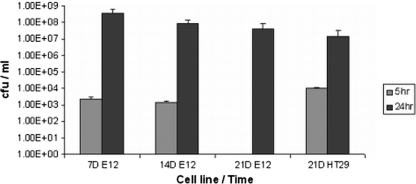
As observed in Fig. 1Biii, C. jejuni 81-176 translocated to the basolateral compartment following apical infection of E12 cells. Enumeration of bacteria in the basolateral compartment using plate assays revealed that modest numbers had translocated by 5 h postinfection (with the exception of 21-day E12 cell cultures) (Fig. 3). The reason for the lack of translocation at 5 h in day 21 E12 cells compared with day 21 HT29 cells is not clear but may reflect the particularly high TEER of E12 monolayers at 21 days (Fig. 1Aii). By 24 h, as many as 108 organisms were present in the basolateral compartment. As C. jejuni did not reproduce when introduced directly into the basolateral compartment of control wells (with overlying E12 monolayers) and given a lack of any translocation by the probiotic L. helveticus (data not shown), these results suggested a specific translocation mechanism for C. jejuni. Very high translocation levels at later time points indicate either a very efficient translocation mechanism or the possibility that some disruption of the monolayer has started to occur. It is also noteworthy that there were similar levels of C. jejuni translocation in E12 and HT29 cells.
FIG. 3.
Recovery of C. jejuni translocated to the basolateral medium after 5 h and 24 h of apical infection using 7-, 14-, and 21-day-cultured E12 cells compared to 21-day-cultured HT29 polarized cells (n = 8). Error bars indicate standard deviations. Results are presented as CFU/ml.
There was a discernible ovoid or circular pattern of C. jejuni adherence to E12 cells, particularly at lower magnification (Fig. 4A). The ovoid or circular shapes rendered suggested that the organisms were preferentially adherent to cell-cell junctions. On this basis and in the light of previous studies (27, 40), we hypothesized that bacteria were preferentially attaching to cell-cell junction structures in order to translocate the monolayer paracellularly. To investigate this further, we examined if C. jejuni colocalized with the tight junction protein occludin. Following infection and removal of the mucus layer, C. jejuni could be found aligning with the tight junctions of E12 cells and colocalizing with occludin as early as 3 h postinfection (Fig. 4B).
FIG. 4.
Pattern of C. jejuni adhesion to E12 cells. (A) Two different magnifications are shown, indicating a circular or ovoid pattern of distribution of C. jejuni as it clusters toward cell-cell junctions. Scale bars, 10 μm (left) and 5 μm (right). Magnification, ×200 (left) and ×400 (right). (B) Confocal microscopy of the tight junction protein occludin and C. jejuni. There is clear pericellular distribution of C. jejuni organisms (stained red) and colocalization with green-stained occludin protein (yellow spots). Magnification, ×630. Scale bars, 10 μm.
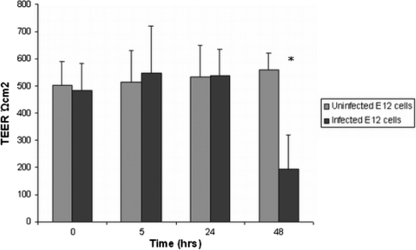
To investigate the effect of C. jejuni on the barrier properties of E12, 21-day cultured cells were infected with bacteria and the TEER in infected cells was measured over a period of 48 h in comparison to uninfected cells. By 24 h postinfection, the TEER had not fallen, despite translocation of large numbers of organisms to the basolateral compartment (Fig. 5). Only by 48 h postinfection was there a significant reduction in the TEER values of infected cells, indicating that tight junctions were disrupted. These results indicate that early translocation of C. jejuni occurs without disrupting the tight junctions of E12 cells, and the microscopy results support the contention that the organisms translocate via a paracellular route.
FIG. 5.
Transepithelial resistance values of 21-day E12 cells following C. jejuni infection remain unaffected despite very high C. jejuni translocation efficiency at 24 h. There is no evidence of loss of TEER until later time points (48 h). * denotes significant difference (P < 0.00005) compared to 48 h uninfected E12 cells (n = 12). Error bars indicate standard deviations.
(iv) Comparison of C. jejuni pathogenicity in E12 cells with that in non-mucus-adherent cells.
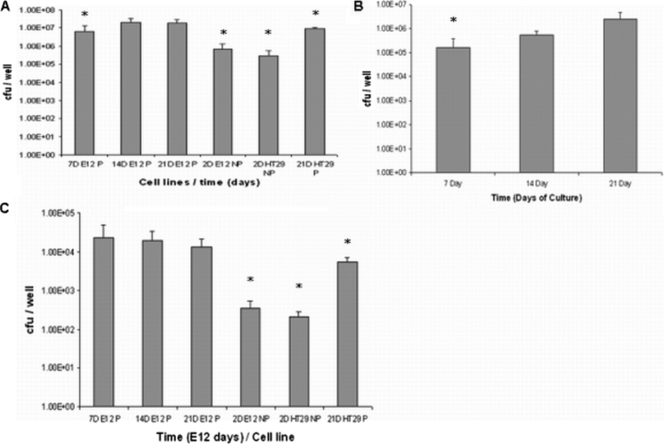
We exploited the presence of an adherent mucus layer on E12 cells to determine the effect of mucus on the virulence of C. jejuni using total association assays and the gentamicin protection assay. Polarized and nonpolarized HT29 parent cell lines, which do not harbor or secrete mucus, and nonpolarized E12 cells which secrete mucus into the culture supernatant but do not harbor an adherent mucus layer were used as controls. Not surprisingly (given the thick mucus layer), there was an increase in C. jejuni total association (colonized, adherent, and internalized bacteria) to E12 cells by more than 100-fold in the presence of a mucus layer (7-, 14-, and 21-day-cultured E12 cells) compared to nonpolarized E12 and HT29 cells and by 10-fold in the presence of mucus compared to 21-day-cultured polarized HT29 cells (Fig. 6A).
FIG. 6.
Colonization, adhesion, and invasion of E12 cells. (A) C. jejuni total association (mucus-colonized, adherent, and invaded organisms) to polarized E12 cells compared to parental cell lines. Day 7-, 14-, and 21-day-cultured E12 cells are indicated as 7DE12P, 14DE12P, and 21DE12P, respectively. 2DE12NP, 2-day-cultured nonpolarized E12 cells; 2DHT29NP, 2-day-cultured nonpolarized HT29 cells; 21D HT29P, 21-day-cultured polarized HT29 cells (no mucus layer). * denotes significant difference compared to 21DE12P. 7DE12P, P < 0.005; 2DE12NP, P < 0.005; 2DHT29NP, P < 0.0005; 21DHT29P, P < 0.05 (n = 11). Error bars indicate standard deviations. (B) Adhesion of C. jejuni to E12 cells after removal of the mucus layer. * denotes significant difference compared to 14- and 21-day-cultured E12 cells. 14-day E12, P < 0.05; 21 -day E12, P < 0.05 (n = 8). Error bars indicate standard deviations. (C) C. jejuni internalization into polarized E12 cells compared to parental cell lines. * denotes significant difference compared to 21DE12P. 2DE12NP, P < 0.005; 2DHT29 NP, P < 0.05; 21DHT29P, P < 0.05 (n = 8). Error bars indicate standard deviations.
C. jejuni binding to underlying E12 cells after penetrating the mucus layer was also investigated using 7-, 14-, and 21-day-cultured E12 cells. Cells were infected with C. jejuni, and the mucolytic agent N-acetylcysteine was used to remove the E12 mucus layer, after which the CFU of adherent and internalized bacteria were quantified (see Materials and Methods). C. jejuni binding to E12 cells increased from 7-day-cultured (1.6 × 105 CFU/well) through 14-day-cultured (5.6 × 105 CFU/well) to 21-day-cultured E12 cells (2.5 × 106 CFU/well) (Fig. 6B). In 7-, 14-, and 21-day-cultured E12 cells, the relative proportion of mucus-colonizing bacteria that had adhered to E12 cells increased from 2.6% to 2.8% to 15.2%, respectively.
Internalization also was enhanced in mucus-adherent cells (E12) compared to non-mucus-adherent parental HT29 cells (Fig. 6C). Invasion was approximately 100-fold higher in polarized E12 cells (with mucus) than in nonpolarized E12 and HT29 cells (without a mucus layer) and 10-fold higher than in polarized HT29 cells (21 days in culture). As a control, the probiotic bacterium L. helveticus R0052 did not invade E12 cells (data not shown). Thus, C. jejuni total association and internalization increased when E12 and HT29 cells became polarized and further increased in the presence of mucus. The ratio of internalized to adherent bacteria was approximately a log-fold higher in E12 than that in HT29 cells, i.e., between 1:27 and 1:200 in E12 cells compared with approximately 1:2,000 in HT29 cells. Taken together, these results suggest that both mucus and cell polarity promote the efficiency of C. jejuni total association and internalization.
The effect of probiotics on the pathogenicity of C. jejuni. (i) Probiotics colonized E12 mucus and adhered to underlying cells.
Having established E12 cells as a suitable model of Campylobacter infection, we used it to examine the effect of probiotics on the pathogenicity of C. jejuni. Immunofluorescence and confocal microscopy were used to demonstrate probiotic colonization of E12 mucus and binding to underlying cells. Briefly, E12 cells were precolonized with TAMRA (red)-stained L. rhamnosus for 4 h and infected with C. jejuni for a further 24 h. After the incubation period, C. jejuni was labeled green with an in-house, whole-cell anti-C. jejuni antibody (see Materials and Methods). To detect probiotic organisms (L. rhamnosus) and C. jejuni organisms that adhered to underlying E12 cells, the infection process was repeated as described above, except that the mucus layer was removed (see Materials and Methods) prior to labeling adherent C. jejuni green with antibody. L. rhamnosus and C. jejuni were visualized both colonizing E12 mucus and bound to underlying cells by confocal microscopy (data not shown). Colocalization of C. jejuni and L. rhamnosus also could be observed with mucus on merged confocal images. Colocalization of at least some L. rhamnosus and C. jejuni organisms raises the possibility of either a direct interaction between these organisms or a shared affinity for a common receptor in intestinal mucus. In contrast, there was little evidence of colocalization of organisms when adherent to cells, and the circular adhesion pattern seen for C. jejuni (Fig. 4A) was not evident for either organism when used to coinfect E12 cells (data not shown). Total association assays indicated that approximately 107 organisms were present in mucus and adherent to cells after 24 h for each of the probiotic strains tested.
(ii) Probiotics attenuated C. jejuni total association to E12 cells.
Individual probiotic isolates and combinations thereof were investigated for their effect on the pathogenicity of C. jejuni using E12 cells. Three strains of Lactobacillus (L. rhamnosus R0011, L. helveticus R0052, and L. salivarius AH102) and B. longum AH1205 were used in addition to a commercial combination of L. rhamnosus and L. helveticus (Lacidofil) and an in-house combination comprising L. rhamnosus, L. helveticus, and L. salivarius (RhHeSa) mixed equally (∼109 CFU/well each) (see Materials and Methods). These probiotics were used to initially colonize 14-day-old E12 cells for 4 h prior to C. jejuni infection. Some of the isolates and combinations reduced total association of C. jejuni to E12 cells (Fig. 7A). However, other preparations had no effect (e.g., L. helveticus), and in general, the relative reduction was modest (Fig. 7A).
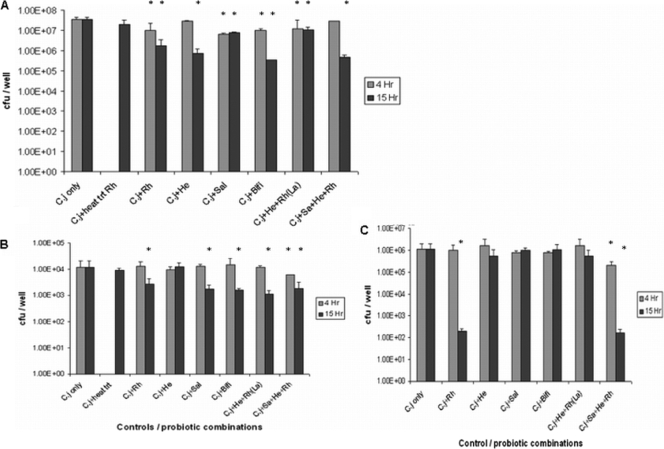
FIG. 7.
Effect of probiotics and probiotic cocktails on C. jejuni total association to (A), invasion of (B), and translocation across (C) 14-day-cultured E12 cells. (A) E12 cells were precolonized with probiotics and probiotic combinations for 4 and 15 h prior to infection with C. jejuni (C.j) for a further 24 h. Rh, L. rhamnosus; heat trt Rh, heat-treated Rh; He, L. helveticus; Sal, L. salivarius; Bifi, B. longum; La, L. rhamnosus and L. helveticus. Error bars indicate standard deviations. * denotes significant difference compared to C. jejuni only (n = 9). P < 0.005 for all groups. (B) E12 cells were precolonized with probiotics for 4 and 15 h prior to infection with C. jejuni for a further 24 h. The gentamicin protection assay was used to quantify internalized C. jejuni CFU. * denotes statistical difference compared to C. jejuni only (n = 8). P < 0.05 for all groups. Error bars indicate standard deviations. (C) E12 cells cultured for 14 days were precolonized with probiotics for 4 and 15 h prior to infection with C. jejuni for a further 24 h (n = 8). Serial dilutions of the basolateral medium were made in PBS and plated onto solid medium. Error bars indicate standard deviations. * denotes statistical difference compared to C. jejuni only. Rh, P < 0.05; 4 h Sa+He+Rh, P < 0.05; 15 h Sa+He+Rh, P < 0.05.
However, when probiotic strains were allowed to “precolonize” monolayers for 15 h, the effect on C. jejuni pathogenicity was considerably enhanced (Fig. 7A). Probiotic colonization numbers did not increase significantly at 15 h compared with 4 h (data not shown). Only L. salivarius and Lacidofil did not show more marked inhibition of total association of C. jejuni after 15 h compared with 4-h precolonization. Many of the preparations reduced total C. jejuni association 1- to 1.5-log-fold, with the most effective reduction induced by B. longum, by which C. jejuni recovery was reduced from 9.2 × 106 to 3.3 × 105. Given the accentuation of the effect following prolonged precolonization, further experiments were performed at both 4 and 15 h after colonization with probiotic isolates.
(iii) Precolonization of E12 cells with probiotics inhibited C. jejuni internalization.
Only the RhHeSa probiotic combination attenuated C. jejuni internalization after 4-h precolonization (Fig. 7A). However, with the exception of L. helveticus, precolonization of E12 cells with probiotic preparations for 15 h prior to infection with C. jejuni significantly attenuated C. jejuni invasion (Fig. 7B). Precolonization of E12 cells with heat-treated L. rhamnosus for 15 h had no effect on C. jejuni invasion. The preparations combining more than one isolate tended to have the greatest effect on attenuation of invasion, with an approximately 90% reduction in recovered C. jejuni compared to the baseline (Fig. 7B).
(iv) Precolonization of E12 cells with probiotics attenuated C. jejuni translocation to the basolateral medium.
With the exception of RhHeSa, no probiotic strain or combination of strains affected C. jejuni translocation after a 4-h preincubation period with E12 cells (Fig. 7C). However, after 15-h precolonization, L. rhamnosus on its own or combined with L. helveticus and L. salivarius markedly attenuated C. jejuni translocation (Fig. 7C). L. helveticus and Lacidofil reduced C. jejuni translocation after preincubation for 15 h, but this difference was not statistically significant. L. salivarius and B. longum did not affect C. jejuni translocation, despite their attenuation of C. jejuni total association and internalization. In addition, following precolonization of E12 cells with L. rhamnosus for 15 h and infection with C. jejuni for a further 48 h, L. rhamnosus did not prevent the previously described C. jejuni-mediated disruption of tight junctions (data not shown).
DISCUSSION
Campylobacter research has been hampered by the lack of an easily accessible animal model that mimics human Campylobacter infection. Therefore, conventional tissue culture has been of major importance in exploring C. jejuni pathogenicity (14, 21, 27, 42). The E12 cell line provides an important additional tool for Campylobacter research by mimicking in vitro a fundamentally important feature of mucosal infection, the overlying mucus layer. In this study, the interaction of C. jejuni with E12 cells was characterized in depth. We believe that E12 cells provide an important novel technique to complement recent seminal studies looking at the effect of specific mucin molecules on C. jejuni pathogenicity (26, 39). C. jejuni colonization and reproduction in mucus, binding to and invasion of underlying cells, and translocation to the basolateral aspects of cells were demonstrated, validating the E12 model as suitable for the study of Campylobacter-host interactions.
The route of C. jejuni translocation to the basolateral aspects of cells is a subject of debate. While some studies have described C. jejuni transcellular transcytosis (5, 15), others have implicated the paracellular route of translocation (4, 27). Disruption of tight junctions by C. jejuni also has been shown in other studies (8, 24). In this study, C. jejuni aligned with tight junctions and colocalized with occludin as early as 3 h postinfection, suggesting paracellular passage of organisms. Although many bacterial pathogens affect tight junctions and the related pathogen Helicobacter pylori attaches near intercellular boundaries (12), this appears to be the first report providing evidence of physical alignment of a bacterial species with tight junctions and colocalization with occludin. Other important nonbacterial human pathogens, including Toxoplasma gondii (2) and hepatitis C virus (23), are known to interact directly with tight junction proteins. Indeed, passage of C. jejuni paracellularly without affecting the integrity of tight junctions is reminiscent of T. gondii. Further experiments examining the nature of the interaction of C. jejuni with occludin and other cell junction components are required.
In previous studies, C. jejuni motility has been shown to increase in viscous solutions (11) and in media mimicking the viscosity of mucus (38). In addition, in vitro studies using conventional cell lines have demonstrated increased C. jejuni binding and internalization into intestinal cells following preincubation in media of increased viscosity (38), crude human mucus (6), and purified mucins (10). In this study we confirm that the overlying mucus layer supports C. jejuni reproduction and enhances adhesion to and invasion of the underlying epithelium. C. jejuni replication in mucus may ensure bacterial persistence in the host since it is rich in nutrients and is essentially microaerophilic. Whether the increased pathogenicity in the presence of mucus is caused by increased viscosity or other factors is not clear. However, it is unlikely that the observed increase in C. jejuni total association and invasion into E12 cells was due to increased bacterial numbers secondary to reproduction in mucus alone, because increasing the inoculum did not result in an increase in C. jejuni total association and invasion in other experiments (data not shown).
In vivo, commensal bacteria establish themselves in the gut shortly after birth and subsequently maintain a population in dynamic equilibrium with the intestinal mucosa (13, 31). Animal and human studies of the effects of probiotics on the gut involve at least a period of persistent colonization for the effects to be observed (33). Despite this, most in vitro studies of probiotic action typically involve short exposure of model epithelia to the probiotic strains used (1, 19, 25, 46). It is possible that such brief pretreatment of cells with probiotics fails to reproduce at least some of the effects that would be seen in vivo. The E12 model is suited to the study of more persistent colonization and, in this manner, more accurately reflects conditions in vivo. We observed a striking difference in the effectiveness of nearly all the probiotic strains used when cells were precolonized for 15 h compared to 4 h. In nearly all cases, prolonged precolonization allowed for much more effective attenuation of C. jejuni virulence. In future studies, it will be useful to explore even longer time periods (e.g., 24 or even 48 h) to see if further reduction in C. jejuni pathogenicity occurs.
One of the potential mechanisms by which probiotics work is to reduce luminal pH (33, 36). All the organisms used in this study are lactic acid producing. C. jejuni is inactivated at low pH (pH 5) (38), and during coculture with L. salivarius we observed that C. jejuni lost viability at a pH of 4.7 (unpublished results). It is possible that probiotic-induced acidity contributes to the effects on C. jejuni virulence in our study. However, in order to avoid a drastic reduction in pH that would affect cell and organism viability, we repeatedly replenished the E12 cell culture medium during coculture of probiotics with C. jejuni. In addition, complex effects of different probiotic strains on various aspects of C. jejuni pathogenicity (invasion, adhesion, and translocation) are unlikely to be explained by the effects of changes in pH alone.
Mechanisms of probiotic-induced alteration in pathogenicity may be generalized and nonspecific (e.g., consumption of local nutrients and interference with host binding sites). Alternatively, a wide variety of more-specific mechanisms of action have been ascribed to probiotics during protection of the mucosa from infecting organisms in vitro and in animal models (31, 33, 36). It is not clear what mechanisms of action were involved in probiotic-induced attenuation of C. jejuni pathogenicity in our experiments. In order to avoid a nonspecific effect of low pH on C. jejuni viability (38), we repeatedly replenished the E12 cell culture medium during coculture. In addition, we feel that probiotic-induced changes in epithelial barrier function are unlikely to have caused reduced translocation of C. jejuni, as there was no concomitant effect of these commensals on TEER (data not shown). However, a noteworthy feature of our experiments was the heterogeneity in the ability of probiotics to attenuate different aspects of C. jejuni pathogenicity. Probiotic strain-specific variation in attenuation of C. jejuni invasiveness, using conventional cell lines, also was recently reported (46). These findings suggest different mechanisms of action for the different probiotic strains and cocktails used. These data also suggest that the mechanisms by which an organism such as C. jejuni colonizes mucus, invades epithelial cells, and translocates to the basolateral aspects of cells are, at least to some extent, mutually exclusive and independent of one another.
These studies point to the potential for a number of well-characterized probiotic isolates to prevent and/or attenuate C. jejuni disease in humans. The possibility that such an approach could also be used to reduce chicken broiler colonization is also attractive as an effective means to reduce Campylobacter infections in humans (29). Our studies provide a rational basis for the further investigation of specific probiotic strains for future in vitro and clinical studies (for example, the prevention of C. jejuni infection in travelers). Our data point to the likelihood that probiotic treatment may have a greater potential as a method of prophylaxis against C. jejuni infection rather than as a method of treatment of an acute illness.
In summary, we describe a novel model of Campylobacter infection. E12 cells are unique in harboring an adherent, overlying mucus layer, providing an opportunity to study the interaction of enteric pathogens with mucus, mucin, endogenous flora, and related luminal factors in a manner that more accurately represents conditions in vivo compared to conventional cell culture methods. This model confirms the contribution of mucus to the enhancement of C. jejuni pathogenicity, provides strong supportive evidence for paracellular passage of C. jejuni through interaction with tight junction proteins, and establishes the potential efficacy of probiotic treatment for the prevention and attenuation of C. jejuni-related disease in humans.
Acknowledgments
This work was supported by a principal investigator grant from Science Foundation Ireland (SFI). We also acknowledge funding support from the Children's Research Centre, Our Lady's Hospital for Sick Children, Crumlin, Dublin 12, Ireland.
We thank Brendan Dolan, Anne Cullen, and Cormac O'Connell for guidance in fluorescence, confocal, and electron microscopy, respectively.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Altenhoefer, A., S. Oswald, U. Sonnenborn, C. Enders, J. Schulze, J. Hacker, and T. A. Oelschlaeger. 2004. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 40:223-229. [DOI] [PubMed] [Google Scholar]
- 2.Barragan, A., F. Brossier, and L. D. Sibley. 2005. Transepithelial migration of Toxoplasma gondii involves an interaction of intercellular adhesion molecule 1 (ICAM-1) with the parasite adhesin MIC2. Cell Microbiol. 7:561-568. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, I., P. Stenberg, P. Artursson, and T. Kissel. 2001. Transport of lipophilic drug molecules in a new mucus-secreting cell culture model based on HT29-MTX cells. Pharm. Res. 18:1138-1145. [DOI] [PubMed] [Google Scholar]
- 4.Beltinger, J., J. del Buono, M. M. Skelly, J. Thornley, R. C. Spiller, W. A. Stack, and C. J. Hawkey. 2008. Disruption of colonic barrier function and induction of mediator release by strains of Campylobacter jejuni that invade epithelial cells. World J. Gastroenterol. 14:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bras, A. M., and J. M. Ketley. 1999. Transcellular translocation of Campylobacter jejuni across human polarised epithelial monolayers. FEMS Microbiol. Lett. 179:209-215. [DOI] [PubMed] [Google Scholar]
- 6.Byrne, C. M., M. Clyne, and B. Bourke. 2007. Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 153:561-569. [DOI] [PubMed] [Google Scholar]
- 7.Chaveerach, P., L. J. Lipman, and F. van Knapen. 2004. Antagonistic activities of several bacteria on in vitro growth of 10 strains of Campylobacter jejuni/coli. Int. J. Food Microbiol. 90:43-50. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M. L., Z. Ge, J. G. Fox, and D. B. Schauer. 2006. Disruption of tight junctions and induction of proinflammatory cytokine responses in colonic epithelial cells by Campylobacter jejuni. Infect. Immun. 74:6581-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corry, J. E., D. E. Post, P. Colin, and M. J. Laisney. 1995. Culture media for the isolation of campylobacters. Int. J. Food Microbiol. 26:43-76. [DOI] [PubMed] [Google Scholar]
- 10.de Melo, M. A., and J. C. Pechere. 1988. Effect of mucin on Campylobacter jejuni association and invasion on HEp-2 cells. Microb. Pathog. 5:71-76. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero, R. L., and A. Lee. 1988. Motility of Campylobacter jejuni in a viscous environment: comparison with conventional rod-shaped bacteria. J. Gen. Microbiol. 134:53-59. [DOI] [PubMed] [Google Scholar]
- 12.Guttman, J. A., and B. B. Finlay. 2009. Tight junctions as targets of infectious agents. Biochim. Biophys. Acta 1788:832-841. [DOI] [PubMed] [Google Scholar]
- 13.Holzapfel, W. H., P. Haberer, J. Snel, U. Schillinger, and J. H. Huis in't Veld. 1998. Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85-101. [DOI] [PubMed] [Google Scholar]
- 14.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, L., B. D. Tall, S. K. Curtis, and D. J. Kopecko. 2008. Enhanced microscopic definition of Campylobacter jejuni 81-176 adherence to, invasion of, translocation across, and exocytosis from polarized human intestinal Caco-2 cells. Infect. Immun. 76:5294-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jandu, N., Z. J. Zeng, K. C. Johnson-Henry, and P. M. Sherman. 2009. Probiotics prevent enterohaemorrhagic Escherichia coli O157:H7-mediated inhibition of interferon-gamma-induced tyrosine phosphorylation of STAT-1. Microbiology 155:531-540. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, R., K. A. Krogfelt, S. A. Cawthraw, W. van Pelt, J. A. Wagenaar, and R. J. Owen. 2008. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev. 21:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keely, S., A. Rullay, C. Wilson, A. Carmichael, S. Carrington, A. Corfield, D. M. Haddleton, and D. J. Brayden. 2005. In vitro and ex vivo intestinal tissue models to measure mucoadhesion of poly (methacrylate) and N-trimethylated chitosan polymers. Pharm. Res. 22:38-49. [DOI] [PubMed] [Google Scholar]
- 19.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32:691-701. [DOI] [PubMed] [Google Scholar]
- 20.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 21.Krause-Gruszczynska, M., M. Rohde, R. Hartig, H. Genth, G. Schmidt, T. Keo, W. Konig, W. G. Miller, M. E. Konkel, and S. Backert. 2007. Role of the small Rho GTPases Rac1 and Cdc42 in host cell invasion of Campylobacter jejuni. Cell Microbiol. 9:2431-2444. [DOI] [PubMed] [Google Scholar]
- 22.Lesuffleur, T., A. Barbat, E. Dussaulx, and A. Zweibaum. 1990. Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Cancer Res. 50:6334-6343. [PubMed] [Google Scholar]
- 23.Liu, S., W. Yang, L. Shen, J. R. Turner, C. B. Coyne, and T. Wang. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacCallum, A., S. P. Hardy, and P. H. Everest. 2005. Campylobacter jejuni inhibits the absorptive transport functions of Caco-2 cells and disrupts cellular tight junctions. Microbiology 151:2451-2458. [DOI] [PubMed] [Google Scholar]
- 25.Mack, D. R., S. Ahrne, L. Hyde, S. Wei, and M. A. Hollingsworth. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAuley, J. L., S. K. Linden, C. W. Png, R. M. King, H. L. Pennington, S. J. Gendler, T. H. Florin, G. R. Hill, V. Korolik, and M. A. McGuckin. 2007. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J. Clin. Invest. 117:2313-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteville, M. R., and M. E. Konkel. 2002. Fibronectin-facilitated invasion of T84 eukaryotic cells by Campylobacter jejuni occurs preferentially at the basolateral cell surface. Infect. Immun. 70:6665-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mooney, A., C. Byrne, M. Clyne, K. Johnson-Henry, P. Sherman, and B. Bourke. 2003. Invasion of human epithelial cells by Campylobacter upsaliensis. Cell Microbiol. 5:835-847. [DOI] [PubMed] [Google Scholar]
- 29.Morishita, T. Y., P. P. Aye, B. S. Harr, C. W. Cobb, and J. R. Clifford. 1997. Evaluation of an avian-specific probiotic to reduce the colonization and shedding of Campylobacter jejuni in broilers. Avian Dis. 41:850-855. [PubMed] [Google Scholar]
- 30.Nachamkin, I., J. Engberg, M. Gutacker, R. J. Meinersman, C. Y. Li, P. Arzate, E. Teeple, V. Fussing, T. W. Ho, A. K. Asbury, J. W. Griffin, G. M. McKhann, and J. C. Piffaretti. 2001. Molecular population genetic analysis of Campylobacter jejuni HS:19 associated with Guillain-Barre syndrome and gastroenteritis. J. Infect. Dis. 184:221-226. [DOI] [PubMed] [Google Scholar]
- 31.Neish, A. S. 2009. Microbes in gastrointestinal health and disease. Gastroenterology 136:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemelka, K. W., A. W. Brown, S. M. Wallace, E. Jones, L. V. Asher, D. Pattarini, L. Applebee, T. C. Gilliland, Jr., P. Guerry, and S. Baqar. 2009. Immune response to and histopathology of Campylobacter jejuni infection in ferrets (Mustela putorius furo). Comp. Med. 59:363-371. [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, S. C., A. L. Hart, M. A. Kamm, A. J. Stagg, and S. C. Knight. 2009. Mechanisms of action of probiotics: recent advances. Inflamm. Bowel Dis. 15:300-310. [DOI] [PubMed] [Google Scholar]
- 34.O'Hara, A. M., and F. Shanahan. 2007. Mechanisms of action of probiotics in intestinal diseases. ScientificWorldJournal 7:31-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pope, J. E., A. Krizova, A. X. Garg, H. Thiessen-Philbrook, and J. M. Ouimet. 2007. Campylobacter reactive arthritis: a systematic review. Semin. Arthritis Rheum. 37:48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, P. M., K. C. Johnson-Henry, H. P. Yeung, P. S. Ngo, J. Goulet, and T. A. Tompkins. 2005. Probiotics reduce enterohemorrhagic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 73:5183-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, J. L., and D. Bayles. 2007. Postinfectious irritable bowel syndrome: a long-term consequence of bacterial gastroenteritis. J. Food Prot. 70:1762-1769. [DOI] [PubMed] [Google Scholar]
- 38.Szymanski, C. M., M. King, M. Haardt, and G. D. Armstrong. 1995. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect. Immun. 63:4295-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu, Q. V., M. A. McGuckin, and G. L. Mendz. 2008. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J. Med. Microbiol. 57:795-802. [DOI] [PubMed] [Google Scholar]
- 40.van Alphen, L. B., N. M. Bleumink-Pluym, K. D. Rochat, B. W. van Balkom, M. M. Wosten, and J. P. van Putten. 2008. Active migration into the subcellular space precedes Campylobacter jejuni invasion of epithelial cells. Cell Microbiol. 10:53-66. [DOI] [PubMed] [Google Scholar]
- 41.Wagner, R. D., S. J. Johnson, and D. Kurniasih Rubin. 2009. Probiotic bacteria are antagonistic to Salmonella enterica and Campylobacter jejuni and influence host lymphocyte responses in human microbiota-associated immunodeficient and immunocompetent mice. Mol. Nutr. Food Res. 53:377-388. [DOI] [PubMed] [Google Scholar]
- 42.Watson, R. O., and J. E. Galan. 2008. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO. 2000. Campylobacter. Fact Sheet No 255. WHO, Geneva, Switzerland.
- 44.WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. WHO, Geneva, Switzerland. http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf.
- 45.Willis, W. L., and L. Reid. 2008. Investigating the effects of dietary probiotic feeding regimens on broiler chicken production and Campylobacter jejuni presence. Poult. Sci. 87:606-611. [DOI] [PubMed] [Google Scholar]
- 46.Wine, E., M. G. Gareau, K. Johnson-Henry, and P. M. Sherman. 2009. Strain-specific probiotic (Lactobacillus helveticus) inhibition of Campylobacter jejuni invasion of human intestinal epithelial cells. FEMS Microbiol. Lett. 300:146-152. [DOI] [PubMed] [Google Scholar]