Abstract
Our previous structural studies on intact, infectious murine norovirus 1 (MNV-1) virions demonstrated that the receptor binding protruding (P) domains are lifted off the inner shell of the virus. Here, the three-dimensional (3D) reconstructions of recombinant rabbit hemorrhagic disease virus (rRHDV) virus-like particles (VLPs) and intact MNV-1 were determined to ∼8-Å resolution. rRHDV also has a raised P domain, and therefore, this conformation is independent of infectivity and genus. The atomic structure of the MNV-1 P domain was used to interpret the MNV-1 reconstruction. Connections between the P and shell domains and between the floating P domains were modeled. This observed P-domain flexibility likely facilitates virus-host receptor interactions.
Murine norovirus 1 (MNV-1) (3, 14, 15) and rabbit hemorrhagic disease virus (RHDV) are members of the genera Norovirus and Lagovirus of the family Caliciviridae that offer a comparison to recombinant human norovirus (rNV) virus-like particles (VLPs) for assessing the structures and roles of domains within the capsid proteins of this family of viruses. Calicivirus particles contain 180 copies of the 56- to 76-kDa major capsid protein (Orf2), which is comprised of the internal/buried N terminus (N), shell (S), and protruding (P) domains (9, 10). The S domain, an eight-stranded β-barrel, forms an ∼300-Å contiguous shell around the RNA genome. A flexible hinge connects the shell to a “protruding” (P) domain at the C-terminal half of the capsid protein, which can be further divided into a globular head region (P2) and a stem region (P1) that connects the shell domain to P2. The accompanying article (13) describes the determination of the structure of the P domain of MNV-1 to a resolution of 2.0 Å.
We recently determined the cryo-transmission electron microscopy (TEM) structure of MNV-1 to ∼12-Å resolution (4) and found that, compared to rNV VLPs (10) and San Miguel sea lion virus (SMSV) (1, 2), the protruding domains are rotated by ∼40° in a clockwise fashion and lifted up by ∼16 Å. To better understand the unusual conformation of MNV-1 and whether it is unique to this particular member of the calicivirus family, the ∼8-Å cryo-TEM structures of infectious MNV-1 and the VLPs of RHDV were determined.
MNV-1 was produced as previously described (4). Three liters of cell culture yielded 0.5 to 1.0 mg of purified virus with a particle/PFU ratio of less than 100. Baculovirus expression and purification of recombinant RHDV (rRHDV) VLPs were performed as previously described (8). Cryo-electron microscopy (EM) data were collected at the National Resource for Automated Molecular Microscopy (NRAMM) facility in San Diego, CA (4). Images were collected at a nominal magnification of ×50,000 at a pixel size of 0.1547 nm at the specimen level using Leginon software (12) and processed with Appion software (5). The contrast transfer function for each set of particles from each image was estimated and corrected using ACE2 (a variation of ACE [7]). Particle images were automatically selected (11). The final stacks of particle images contained 20,425 MNV virions and 7,856 rRHDV VLPs, and EMAN 3D (6) was used for the reconstructions. Resolutions were estimated by Fourier shell correlations (FSC) of the three-dimensional (3D) reconstructions and application of a cutoff of 0.5. An amplitude correction of the final electron density was performed using GroEL small-angle X-ray scattering (SAXS) data.
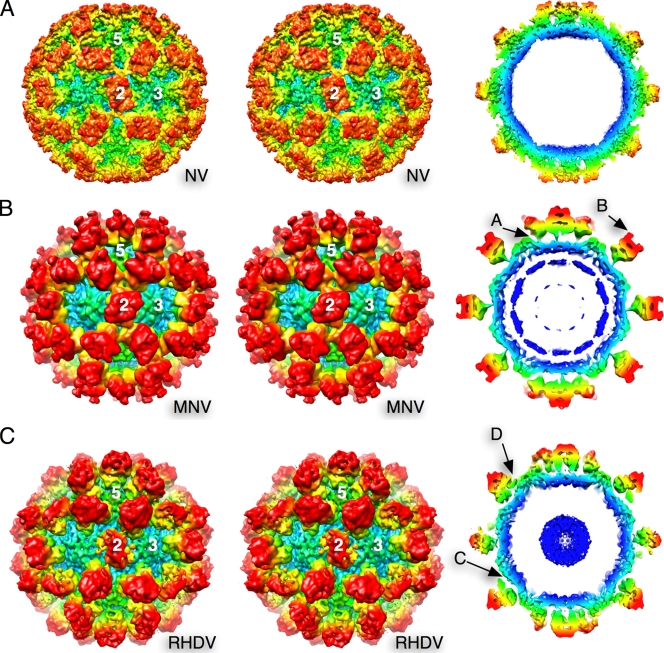
3D reconstructions of MNV-1 and rRHDV were calculated to resolutions of 8 Å and 8.1 Å, respectively (Fig. 1). The P domains of rNV VLPs rest directly on top of the shell domain (10) (Fig. 1A). In contrast, the P domains of MNV-1 are lifted and rotated above the shell of the capsid (4) (Fig. 1B). At this higher resolution, there was a clear connection between the P1 domain and the shell domain in all three capsid subunits (Fig. 1B, arrow A). Unlike the smooth protruding domains of rNV, MNV-1 has two clear “horns” (arrow B), not dissimilar to those observed for the sapoviruses (1, 2). There also are islands of density in the interior of the shell, directly beneath the 5-fold axes, that may represent ordered regions of RNA.
FIG. 1.
Stereo diagrams (left) and thin sections (right), with radius coloring, of rNV (A), MNV-1 (B), and an rRHDV VLP (C). For rNV, the atomic coordinates (10) were used. In MNV, arrow A indicates the thin connector between the P1 and S domains. Arrow B denotes the horns found at the tips of the P2 domains. Arrow C denotes the large gap between the P1 and S domains in the rRHDV VLP. Arrow D denotes the false connectivity in rRHDV VLPs between the P1 domain and the S domain near the 5-fold axes.
As with MNV-1, there is a marked gap between the P and S domains in the rRHDV VLP (Fig. 1C, arrow C). This gap is not as pronounced as in MNV-1 because the P domains are not rotated as in MNV-1. In this electron density map, the A/B dimers appear to be touching the shell domain near the 5-fold axes. This contact difference between the A/B dimers and the C/C dimers could be the reason why the tops of the C/C dimers appear to be markedly disordered compared to the A/B dimers in rRHDV and the C/C dimers in MNV-1.
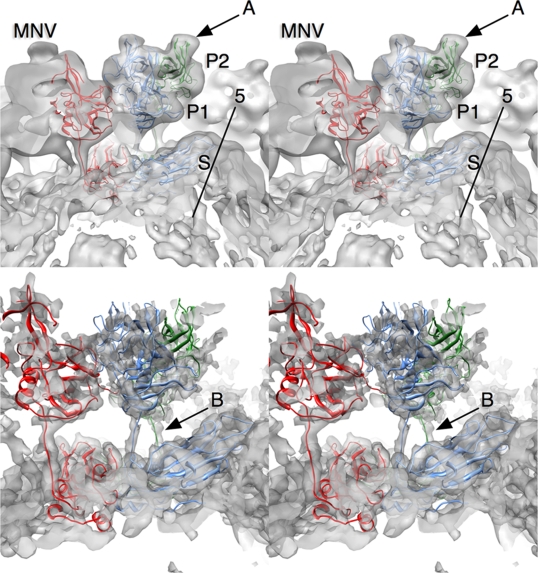
Shown in Fig. 2 is the fitting of the atomic structures of the MNV-1 P domain (13) and the rNV S domains into the MNV-1 3D reconstruction electron density. The horns (arrow A, loops A′-B′ and E′-F′) observed at the tips of the P domain match exceedingly well with the electron density. As discussed in the accompanying publication (13), the A′-B′ and E′-F′ loops displayed two discrete conformations, a closed structure, where the two loops were tightly associated, and an open structure, where the loops were splayed apart. The horns of the closed conformation fit better into the reconstruction, as the E′-F′ loop in the open form jutted out of the density at the base of the horns. The unmodified density in the lower panel of Fig. 2 shows fine features in the shell domain and a very clear connection between the shell and P1 domains. The connections between the P1 and S domains were of sufficient quality to build a basic backbone model by uncoiling the linker region (arrow B). The P domain in the unfiltered 3D reconstruction was far less ordered than the S domain (Fig. 2). This was likely due to movement of the entire P domain with respect to the shell.
FIG. 2.
Fitting of the MNV-1 P domain and the rNV shell domain into the MNV-1 electron density. A, B, and C subunits are represented by blue, green, and red, respectively. The electron density is shown in transparent gray. The top panel is the 8.0-Å-resolution 3D reconstruction modified using a low-pass filter. The bottom panel is the reconstruction without modification. The horns on the tops of the P domains are denoted by arrow A. Arrow B denotes the connection between the S and P domains.
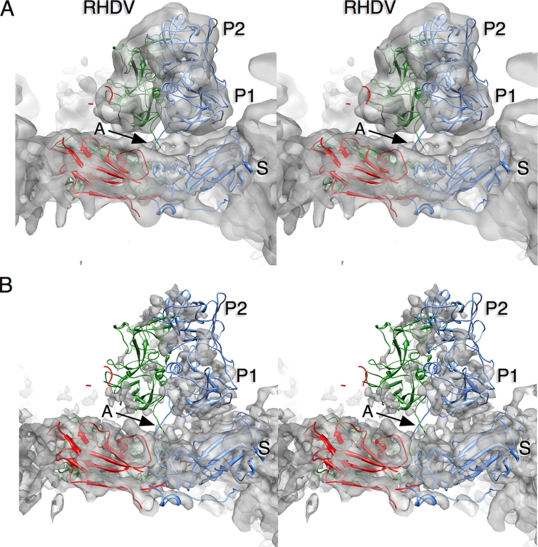
Using the structure of rNV VLP P domains for modeling, the rRHDV P domains are lifted off the surface of the shell, but not rotated as with MNV-1. This places the bottom edge of the A subunit P1 domain near the S domain at the 5-fold axes. The P-domain dimers of rNV and rRHDV have a more “arch-like” shape than MNV-1. Unlike in MNV-1, the electron densities of the C/C dimers in rRHDV are far more diffuse than those of the A/B dimers (Fig. 3B) and the connector between the S and P1 domains is not clear. During fitting, the connector region was not as extended as with MNV-1. This may afford greater flexibility, leading to more diffuse electron density.
FIG. 3.
Fitting of the rNV atomic structure into the rRHDV VLP electron density. The upper stereo image shows the 8.1-Å-resolution 3D reconstruction after modification by a low-pass filter. Below is the same reconstruction prior to density modification.
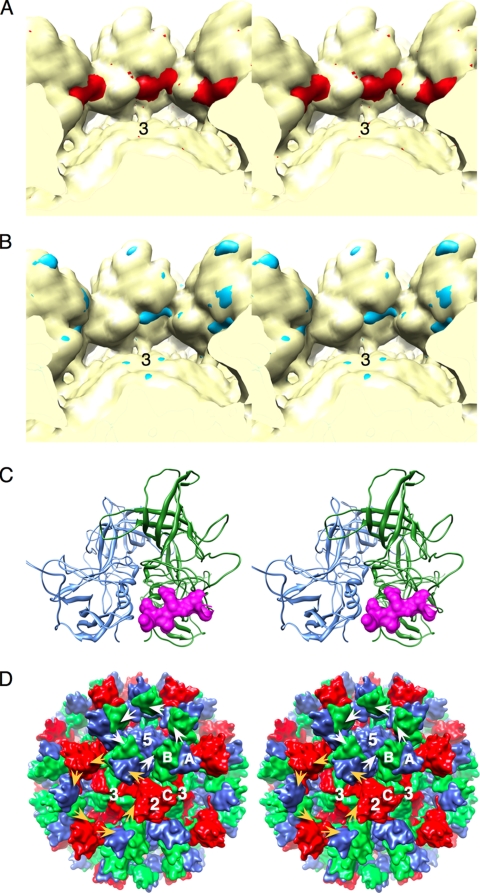
When the atomic models for the MNV-1 P domains (13) were placed into the cryo-TEM electron density (Fig. 4), the C termini extended deep into the cores of adjacent P domains. Possible connections not accounted for by the P-domain structures were also observed in the electron density between the P domains. A bulge between the P1 and P2 domains in the 3D reconstruction indicated a possible interaction between the C termini and the adjacent P domains. These same interactions were observed in the crystal lattice. This highly mobile C terminus may be a flexible tether between the P domains in the intact virion.
FIG. 4.
Possible carboxyl-terminus interactions between the P domains of MNV-1. (A) Stereo image of MNV-1 calculated to 12-Å resolution with (red) and without (yellow) the last 10 residues of the P domain. (B) The calculated MNV-1 density with the carboxyl terminus removed (yellow) overlaid onto the 3D reconstruction of MNV-1 (blue). Note the strands of difference density that roughly correspond to the C terminus in panel A. (C) The C-terminus interactions observed in the structure of the MNV-1 P domains. Shown in blue and green are ribbon diagrams of an A/B P-domain dimer. In mauve is a surface rendering of the C terminus from a crystallographically related dimer. (D) Surface rendering of the final MNV-1 model with possible interactions between the P domains in MNV-1. The carboxyl termini of the A subunits (blue) interact with the counterclockwise-related B subunits around the 5-fold axes (white arrows). Around the 3-fold (quasi-6-fold) axes, the C subunits interact with the A subunits and the B subunits interact with the C subunits (orange arrows).
It is absolutely clear that the hinge region between the S and P domains affords a remarkable degree of flexibility in the P domains that is not genus specific or related to differences between rVLPs and authentic virions. The simplest explanation for the role of this transition is that it gives the P domains flexibility that may be used to optimize interactions with cell receptors during attachment and entry. In this way, the P domains can increase their avidity for the cell surface by being more facile in adapting to the presentation of cellular recognition motifs.
Acknowledgments
This work was supported by startup funds from the Donald Danforth Plant Science Center to T.J.S. and the University of Michigan to C.E.W. and from the National Institutes of Health (AI054483-07 and AI065982-04 to H.W.V.; AI080611 and 1-U54-AI-057153 subcontract to C.E.W.) and by the University of Otago and New Zealand Health Research Council (07/050B to V.L.Y. and V.K.W.). Cryo-TEM data were collected at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the National Center for Research Resources' P41 program (RR17573). Work in the laboratory of J.S. was funded in part by the University of Michigan Center for Structural Biology.
We thank the staff of NRAMM for all of their help.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Chen, R., J. D. Neill, M. K. Estes, and B. V. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, R., J. D. Neill, and B. V. V. Prasad. 2002. Crystallization and preliminary crystallographic analysis of San Miguel sea lion virus: an animal calicivirus. J. Struct. Biol. 141:143-148. [DOI] [PubMed] [Google Scholar]
- 3.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. I. Virgin. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 4.Katpally, U., C. E. Wobus, K. Dryden, H. W. I. Virgin, and T. J. Smith. 2007. Unexpected structural differences between authentic norovirus and virus like particles. J. Virol. 82:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander, G. C., S. M. Stagg, N. R. Voss, A. Cheng, D. Fellmann, J. Pulokas, C. Yoshioka, C. Irving, A. Mulder, P. W. Lau, D. Lyumkis, C. S. Potter, and B. Carragher. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 7.Mallick, S. P., B. Carragher, C. S. Potter, and D. J. Kriegman. 2005. ACE: automated CTF estimation. Ultramicroscopy 104:8-29. [DOI] [PubMed] [Google Scholar]
- 8.Peacey, M., S. Wilson, M. A. Baird, and V. K. Ward. 2007. Versatile RHDV virus-like particles: incorporation of antigens by genetic modification and chemical conjugation. Biotechnol. Bioeng. 98:968-977. [DOI] [PubMed] [Google Scholar]
- 9.Prasad, B. V., D. O. Matson, and A. W. Smith. 1994. Three-dimensional structure of calicivirus. J. Mol. Biol. 240:256-264. [DOI] [PubMed] [Google Scholar]
- 10.Prasad, B. V. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 11.Roseman, A. M. 2003. Particle finding in electron micrographs using a fast local correlation algorithm. Ultramicroscopy 94:225-236. [DOI] [PubMed] [Google Scholar]
- 12.Suloway, C., J. Pulokas, D. Fellmann, A. Cheng, F. Guerra, J. Quispe, S. Stagg, C. S. Potter, and B. Carragher. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41-60. [DOI] [PubMed] [Google Scholar]
- 13.Taube, S., J. R. Rubin, U. Katpally, T. J. Smith, A. Kendall, J. A. Stuckey, and C. E. Wobus. 2010. High-resolution X-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J. Virol. 84:5695-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wobus, C. E., S. M. Karst, L. B. Thackray, K.-O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackensie, K. Y. Green, and H. W. I. Virgin. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wobus, C. E., L. B. Thackray, and H. W. I. Virgin. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]