Abstract
Murine noroviruses (MNV) are closely related to the human noroviruses (HuNoV), which cause the majority of nonbacterial gastroenteritis. Unlike HuNoV, MNV grow in culture and in a small-animal model that represents a tractable model to study norovirus biology. To begin a detailed investigation of molecular events that occur during norovirus binding to cells, the crystallographic structure of the murine norovirus 1 (MNV-1) capsid protein protruding (P) domain has been determined. Crystallization of the bacterially expressed protein yielded two different crystal forms (Protein Data Bank identifiers [PDB ID], 3LQ6 and 3LQE). Comparison of the structures indicated a large degree of structural mobility in loops on the surface of the P2 subdomain. Specifically, the A′-B′ and E′-F′ loops were found in open and closed conformations. These regions of high mobility include the known escape mutation site for the neutralizing antibody A6.2 and an attenuation mutation site, which arose after serial passaging in culture and led to a loss in lethality in STAT1−/− mice, respectively. Modeling of a Fab fragment and crystal structures of the P dimer into the cryoelectron microscopy three-dimensional (3D) image reconstruction of the A6.2/MNV-1 complex indicated that the closed conformation is most likely bound to the Fab fragment and that the antibody contact is localized to the A′-B′ and E′-F′ loops. Therefore, we hypothesize that these loop regions and the flexibility of the P domains play important roles during MNV-1 binding to the cell surface.
Murine noroviruses (MNV) are members of the family Caliciviridae, which contains small icosahedral viruses with positive-sense, single-stranded RNA genomes (18). MNV is related to human noroviruses (HuNoV), which cause most of the sporadic cases and outbreaks of infectious nonbacterial gastroenteritis worldwide in people of all ages (4, 15, 28, 36, 38, 64). However, noroviruses are an understudied group of viruses due to the previous lack of a tissue culture system and small-animal model. Since its discovery in 2003 (23), MNV has become an increasingly important model to study norovirus biology (66). The availability of a small-animal model, cell culture, and reverse-genetics system, combined with many shared characteristics of human and murine noroviruses, allows detailed studies of norovirus biology (7, 23, 63, 65, 66).
The norovirus genome is organized into 3 major open reading frames (ORFs), which encode the nonstructural polyprotein (∼200 kDa) and the major (VP1; ∼58-kDa) and minor (VP2; ∼20-kDa) capsid proteins (18). Recently, a putative ORF-4 was identified in MNV, but the existence of that product and its function remain unknown (60). Norovirus capsids are formed from 180 copies of VP1 arranged with T=3 icosahedral symmetry (9, 25, 46-48). Each capsid protein is divided into an N-terminal arm (N), a shell (S), and a C-terminal protruding (P) domain, with the last two domains connected by a short hinge. VP1 self-assembles into virus-like particles (VLPs) in baculovirus, mammalian, and plant expression systems (21, 22, 50, 57, 67). The S domain forms a smooth shell around the viral genome but is unable to bind to receptors (3, 55). The P domain dimerizes, forming arch-like structures on the capsid surface, and is subdivided into P1 (the stem of the arch) and P2 (the top of the arch) subdomains. The sequence of the P2 subdomain is the least conserved, followed by the P1 and S domains with the highest degree of conservation. While the S domain of Norwalk virus (NV) is required in order to form VLPs in a baculovirus expression system, the P domains contribute to stability by intermolecular interactions (3, 24). The homodimeric interactions of the HuNoV P domain, observed by crystallographic studies of VLPs, is retained when the protein region is expressed in a bacterial expression system (55). In addition, the norovirus P domain, specifically the P2 subdomain, contains the sites for antigenicity, immune-driven evolution, and cell binding (13a, 20, 25, 32, 41, 51, 56). For MNV-1, the Fab fragment of the neutralizing antibody A6.2 binds to the outermost tip of the P2 subdomain and is thought to prevent infection by blocking capsid-receptor interaction (25).
Early steps in the norovirus life cycle are determinants of norovirus tropism (19) and thereby determine the outcome of a viral infection. While the tropism of HuNoV remains unknown, MNV-1 has a tropism for murine macrophages and dendritic cells in vitro and in vivo (62, 65). Recent studies from our laboratory demonstrated that MNV-1 binds to sialic acid on murine macrophages, in particular on the ganglioside GD1a (58). It subsequently enters murine macrophages and dendritic cells in a pH-independent manner (43). To better understand MNV-cell surface binding, we expressed, purified, and determined the high-resolution structure of the MNV-1 P domain at 2.0-Å resolution. Here, we show that, similar to HuNoV P domains (10, 55), recombinant MNV-1 P domains can be expressed and fold in a biologically correct manner. This was shown by the ability of the recombinant MNV-1 P domain to bind murine macrophages, to competitively inhibit MNV-1 infection, and to be recognized by the neutralizing antibody A6.2, which interferes with macrophage binding. Expressed P domain yielded different crystal forms with significant structural differences in the outermost loops of the P2 subdomains. Overall, the MNV-1 P-domain crystal structures show tertiary structures similar to those of HuNoV P domains, with the greatest structural variation in the polypeptide loops on the outer surface of the P domain corresponding to the mobile regions among the various crystal forms. In particular, one of these loops, E′-F′, was observed in “open” and “closed” conformations. Modeling of a Fab fragment and the crystal structures of the P domain into the cryoelectron microscopy three-dimensional (3D) reconstruction of the Fab/MNV-1 complex indicated that the “closed” conformation is the form likely being bound by the neutralizing antibody A6.2. Two sequences located in the A′-B′ and E′-F′ loops were identified as epitopes for A6.2. Biological support for the in silico modeling data comes from a recombinant MNV-1 in which amino acids of the Norwalk virus E′-F′ loop replaced those of MNV-1 and that was no longer neutralized by A6.2. We hypothesize that flexibility in the E′-F′ loop is important for virus-cell interaction and that A6.2 might sterically block viral binding to the cell surface and/or prevent structural changes in the viral capsid required during receptor interaction. In addition, a channel at the interphase of the P dimer was identified that is stabilized by an “ionic lock” (i.e., a bridge formed by two sets of opposing arginine and glutamic acid residues). We hypothesize that the ionic lock may act as a trigger for structural changes important during infection, possibly at the level of host cell entry. Together, these data identify several potential movements within the MNV-1 P domain, which points to the flexibility of the MNV-1 capsid.
MATERIALS AND METHODS
Cell culture and virus stocks.
RAW 264.7 (murine macrophages) and 293T cells were purchased from the ATCC (Manassas, VA) and maintained as described previously (65). A plaque-purified MNV-1 clone (GV/MNV1/2002/USA), MNV-1.CW1 (65), was used at passage 6 for all infections.
Expression and purification of the MNV-1 P domain.
The P domain of MNV-1 (residues 225 to 541) was cloned into a pMCSG7 expression vector with a tobacco etch virus (TEV)-cleavable NH2-terminal 6-histidine tag (C. Brown and J. Delproposto, unpublished data). The protein was expressed overnight at 20°C in Escherichia coli strain C41 cells. The cells were subsequently lysed by sonication in buffer containing 50 mM Tris, pH 7.5, 500 mM NaCl, and 10% glycerol. The supernatant was incubated on Ni-nitrilotriacetic acid (NTA) affinity agarose (Qiagen), and the protein eluted with buffer containing 300 mM imidazole. The protein, along with TEV protease, was dialyzed overnight at 4°C against 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol (DTT), and 10% glycerol to remove the His6 tag. The MNV-1 P domain was further purified by gel filtration on a Superdex 200 column (GE Healthcare) and eluted primarily as a monomer. The protein was then concentrated to 4.7 to 7.5 mg/ml for crystallization.
Binding competition assay.
RAW 264.7 cells were plated in 12-well dishes at 2 × 105 cells per well and pretreated on ice for 1 h with different concentrations of P domain, bovine serum albumin (BSA), or plain buffer (50 mM Tris, pH 7.5, 150 mM NaCl) in a volume of 0.5 ml per well. The cells were infected with MNV-1 at the indicated multiplicities of infection (MOI) in a total volume of 1 ml per well for 1 h on ice. The cells were washed three times with 2 ml phosphate-buffered saline (PBS), 0.5 ml cell culture medium was added, and the cells were incubated for 8 and 18 h at 37°C and 5% CO2. Virus titers were analyzed by plaque assay as previously described (65). Cell viability was monitored in parallel with WST-1 reagent (Roche, NJ), following the manufacturer's recommendations, and the absorbance was measured after 120 min.
Neutralization assay.
Different concentrations of purified monoclonal antibody (MAb) (A6.2, anti-MNV-1 capsid [65], and isotype control [mouse IgG1; ZyMed, CA]) or buffer (50 mM Tris, pH 7.5, 150 mM NaCl) were incubated with 0.1 mg P domain at 37°C in a volume of 0.25 ml. After 1 h, 2 × 106 RAW 264.7 cells (resuspended in 0.25 ml ice-cold hybridoma supernatant containing FcγR MAb 2.4G2 [30a]) were added to the P-domain-antibody complexes and placed on a rocker at 4°C for 1 h. The cells were washed four times with 1 ml buffer (50 mM Tris, pH 7.5, 150 mM NaCl) to remove unbound P domain. Equal numbers of RAW 264.7 cells were analyzed by SDS-PAGE using standard protocols (33). The presence of P domain in the various samples was detected using a polyclonal rabbit serum raised against purified MNV-1 (65) and Western blot analysis. Quantification of bound P domain was performed by measuring relative band intensities using Adobe Photoshop as described previously (37).
Crystallization and structure determination.
Monoclinic crystals of the murine P domain were obtained by the hanging-drop vapor diffusion method. Drops containing 4.7 mg/ml P domain, 10 mM Tris, pH 7.5, 10 mM NaCl, 0.5 mM DTT, 90 mM glycine, 0.2 M sodium acetate, and 10% (wt/vol) polyethylene glycol (PEG) 3350 were allowed to equilibrate at 20°C against a reservoir containing 0.4 M sodium acetate and 20% (wt/vol) PEG 3350. Block-shaped crystals grew after 2 months. X-ray diffraction data to 2.0 Å were collected at 100 K on crystals cryoprotected with reservoir buffer plus 30% glycerol at the Life Sciences Collaborative Access Team (LS-CAT) beamline (21-ID-F) at the Advanced Photon Source, Argonne IL. The data were processed and scaled using HKL2000 (42). The protein crystallized in space group P21 with 2 molecules of the P domain in the asymmetric unit, resulting in a solvent content of 56%. The structure was determined by molecular replacement (34) using the human P-domain structure (Protein Data Bank identifier [PDB ID], 2OBR) (6) as the search model. Fitting and building of the MNV-1 P-domain crystal structure into the electron density was performed using COOT (14) and refined to an R factor of 19.1% using Refmac (39) in the CCP4 program suite (2). The final structure was verified via omit maps and analyzed using a series of validation programs (Procheck, SFcheck, Molprobity, Wasp, Errat, map2model, and Prove) (11, 13, 27, 30, 40, 44, 61). All residues were in the allowed regions of the Ramachandran plot with the exception of residues 537 to 541, which were disordered in the structure.
An orthorhombic crystal form of the P domain was obtained by equilibrating hanging drops containing 7.3 mg/ml P-domain protein, 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.2 M lithium sulfate, and 30% (wt/vol) PEG 4000 against a reservoir containing 0.2 M lithium sulfate and 30% (wt/vol) PEG 4000 at 4°C. Square plate-shaped crystals formed within 3 weeks and continued to grow for up to 2 months. The growth conditions were sufficient for cryoprotection, and no further buffer exchanges were necessary. Data to 2.0-Å resolution were collected at 100 K on the LS-CAT beamline (21-ID-G) at the Advanced Photon Source. The crystals grew in the space group C2221 with a single P-domain monomer in the asymmetric unit. The data were processed using HKL2000 (42), and the structure was determined by molecular replacement (34) using our previously determined MNV-1 P-domain structure as the search model. The structure was built using the program Coot (14) and refined with Refmac (39) to a working R factor of 19%. The final structure was verified and analyzed in the same fashion as the monoclinic form. All residues were in the allowed regions of the Ramachandran plot. Crystal structures were confirmed, and potential model bias was removed from the molecular-replacement solutions by building models into a series of iterative build omit maps (59).
Cryoelectron microscopy of an MNV-Fab complex.
The MNV-1-Fab model was constructed using the pseudo-atomic model for MNV-1 described in the accompanying paper (24), the ∼22-Å-resolution 3D image reconstruction of the MNV-1-Fab complex previously described (25), the crystal structure of the MNV-1 P-domain dimer in the “closed” conformation, and the structure of Fab17 (31, 52, 54). The electron density from the previous 3D image reconstruction of the MNV-1-Fab complex was then overlaid onto this pseudo-atomic model of MNV-1, and the structure of the antibody, Fab17, was manually placed into the envelope corresponding to the bound Fab. The variable domains (VL and VH) were fitted separately from the constant domains (CL and CH1) by rotating the constant domains about the flexible elbow region, the connection between the variable and constant domains. The likely epitope region was further examined using the web-based tools at PISA (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; 29).
Mutagenesis of MNV-1.CW1*.
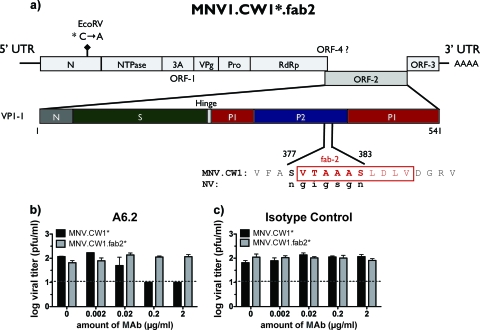
Mutagenesis of MNV-1.CW1* was based on the reverse-genetics system (63), using the pMNV* vector, containing cDNA of the entire genome of MNV-1.CW1. The asterisk refers to a silent mutation in the N terminus to distinguish wild-type and recombinant viruses (63). Site-directed mutagenesis of the genome was performed using the Quikchange XL kit (Stratagene, CA) according to the manufacturer's recommendations. Amino acids (aa) 377 to 383 (SVTAAAS) in the Fab2-binding epitope of MNV-1.CW1* were exchanged for the structurally corresponding amino acids 334 to 340 (NGIGSGN) from NV (PDB ID, 1IHM) (46) using a structural alignment performed with VAST (17). The following forward (F) and reverse (R) primers were used to generate recombinant virus, designated MNV-1.CW1*.fab2 (5′->3′): F, CCTACCAGGGCAGGGTGTTCGCCAGCGTCACTGCTGCGGCCTCTCTTGACTTGGTGGATGGCAGG; R, CCTGCCATCCACCAAGTCAAGAGAGGCCGCAGCAGTGACGCTGGCGAACACCCTGCCCTGGTAGG. Introduction of the desired mutations was confirmed by sequencing ORF-2.
Generation of recombinant MNV-1.
Recombinant MNV was generated using reverse genetics as previously described by Ward et al. (63). Briefly, a transfection mixture containing 1.5 μg of the MNV-1 cDNA plasmid (63) was prepared and incubated with FuGene HD (Roche) at a 1:2 ratio according to the manufacturer's recommendations. The mixture was transfected into a T75 flask with 1 × 106 293T cells in a volume of 10 ml and incubated for 72 h at 37°C and 5% CO2. The transfected 293T cells were lysed by freeze-thawing, and viral titers were analyzed by plaque assay (65). Individual plaques were plaque purified three times and amplified in RAW 264.7 cells. The final working stock was verified by sequencing the ORF-2 cDNA.
Plaque neutralization assay.
The plaque neutralization assay was performed as described previously (65) with the following modifications. Different concentrations of purified MAb (A6.2, anti-MNV-1 capsid) and isotype control (IgG1; Zymed, CA) were incubated with 666 PFU of either MNV-1.CW1* or MNV-1CW1*.fab2 for 30 min at 37°C prior to performing the plaque assay.
Protein structure accession numbers.
Coordinates and structure factors for both the monoclinic and orthorhombic crystal structures were deposited in the Protein Data Bank under PDB accession codes 3LQ6 and 3LQE, respectively.
RESULTS AND DISCUSSION
Bacterially expressed MNV-1 P domain competes for cell surface binding and inhibits infection.
Fast protein liquid chromatography (FPLC) analysis of the bacterially expressed MNV-1 P domain (residues 225 to 541 of VP1) lacking the 6×His tag showed that the P domain from MNV-1 exists in multiple oligomeric states (data not shown). Comparison with a protein standard indicated two main populations of proteins consistent with the molecular weight of monomers and dimers, in addition to the void fraction. Under these particular purification conditions, the predominant species was monomeric.
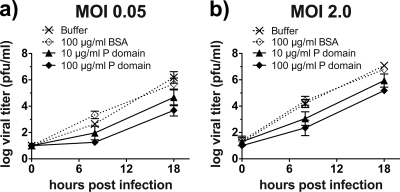
To determine whether bacterially expressed P domains folded correctly, the ability of FPLC-purified P domain (containing the His tag) to compete with MNV-1 for binding to the surfaces of murine macrophages was analyzed (Fig. 1). RAW 264.7 cells were pretreated with different concentrations of P domain, an irrelevant protein (BSA), or buffer and infected with MNV-1, and viral growth was measured by plaque assay at 0, 8, and 18 h postinfection. Compared to the buffer-treated infection, pretreatment with different concentrations of P domain significantly reduced MNV-1 infection in a dose-dependent manner irrespective of the MNV-1 infectious dose (Fig. 1). Pretreatment of cells with BSA did not affect MNV-1 infection. For example, after a single round of infection (8 h) at an MOI of 0.05, 0.1 mg/ml P domain inhibited infection 446-fold compared to BSA. This suggested that bacterially expressed P domain competed in vitro with MNV-1 for binding to the host cell surface, i.e., viral receptor(s).
FIG. 1.
MNV-1 infection of murine macrophages is impaired by recombinantly expressed P dimer. RAW 264.7 cells (2 × 105) were pretreated with the indicated concentrations of P dimer, BSA, or solvent control for 1 h on ice to prevent P-dimer uptake. The cells were infected with MNV-1 for 1 h at an MOI of 0.05 (a) or 2.0 (b), and unbound virus and P dimer were washed off. The infections were allowed to proceed at 37°C for 8 or 18 h. Viral titers were determined by plaque assay. Cell viability throughout the experiment was monitored using WST-1 reagent and remained above 90% (data not shown). Titers are presented as means ± standard errors (SE) from duplicate samples in three independent experiments.
Binding of the MNV-1 P domain to murine macrophages is inhibited by an MNV neutralizing antibody.
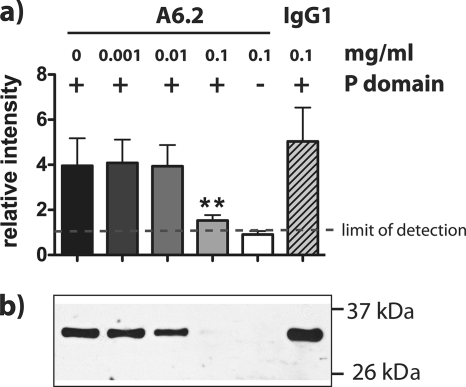
The MAb A6.2 recognizes a conformational epitope in the surface-exposed hypervariable P2 domain of VP1 and has been shown to neutralize MNV-1 infection (25, 65). To address whether antigenic properties of MNV-1 are preserved in bacterially expressed P domain, neutralization of P-domain binding to RAW 264.7 cells by MAb A6.2 was determined (Fig. 2). FPLC-purified P domain (containing the His tag) was pretreated with different concentrations of A6.2 or isotype control and incubated with RAW 264.7 cells. P-domain binding to RAW 264.7 cells was determined by Western blotting, and the band intensity was quantified (Fig. 2A). Probing with anti-MNV-1 serum revealed a band around 35 kDa, consistent with the calculated mass of the MNV-1 P domain (34.5 kDa) (Fig. 2B). A6.2, but not the isotype control, reduced binding of P domain to RAW 264.7 cells in a dose-dependent manner (Fig. 2B). This suggested that the antigenic properties in the A6.2 binding region were preserved between MNV-1 and the bacterially expressed P domain. Furthermore, these data are consistent with our hypothesis that A6.2 neutralizes MNV-1 infection by preventing virion attachment to the cell surface (25) either because of the sheer bulk of the antibody or by fixing the conformation of the P domain into the closed conformation (see below). Taken together, the infection competition (Fig. 1) and antibody binding experiments (Fig. 2) demonstrate that bacterially expressed MNV-1 P domain folds in a biologically relevant manner.
FIG. 2.
Binding of MNV-1 P dimer to murine macrophages is inhibited by the monoclonal antibody A6.2 in a dose-dependent manner. P dimer (+) or buffer (−) (100 μg) was incubated with the indicated concentrations of A6.2 or isotype control (IgG1) for 1 h at 37°C. RAW 264.7 cells (2 × 106), resuspended in host cell medium containing anti-FcγR MAb 2.4G2, were added and incubated for 1 h on ice. Unbound P dimer was washed off, and equal numbers of RAW 264.7 cells were analyzed by SDS-PAGE and Western blotting. (a) Quantitative analysis was performed, measuring the relative intensities of the P-domain-specific bands from three independent experiments. The limit of detection was set to background intensity levels. Statistical analysis was performed using the t test; **, P < 0.01. The error bars indicate standard errors. (b) The P-domain-specific band in a representative Western blot probed with a polyclonal antibody against MNV-1.
Crystal structures of MNV-1 P domain reveal regions of flexibility.
As stated above, purified MNV-1 P domain existed primarily in a monomeric state as determined by FPLC. The FPLC-purified monomeric MNV-1 P domain without the His tag was used for crystallization trials. However, only the dimeric structure was apparent in all crystal forms. Two crystal forms of the MNV-1 P domain were determined; a monoclinic form (space group P21) with a dimer in the asymmetric unit and an orthorhombic form (space group C2221), where the dimer formed across a crystallographic 2-fold axis. The structures were determined by molecular replacement and refined to a resolution of 2 Å (Table 1). The overall molecular topologies and dimer interfaces in both crystal structures were very similar (root mean square deviation [RMSD] = 0.45 Å2), and amino acids 228 to 540 were resolved in both structures.
TABLE 1.
Data collection and phasing and refinement statistics
| Data collection | Valuea |
|
|---|---|---|
| Monoclinic | Orthorhombic | |
| Space group | P21 | C2221 |
| Unit cell a, b, c (Å) | 57.75, 86.26, 83.51 | 86.26, 105.22, 84.05 |
| Unit cell α, β, γ (Å) | 90.0, 110.95, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.97872 | 0.97856 |
| Resolution (Å) | 2.0 (2.03-2.0) | 2.0 (2.07-2.00) |
| Rsym (%)b | 7.8 (37) | 7.1 (27) |
| <I/sI>c | 10 (2) | 10 (10) |
| Completeness (%)d | 99.4 (93.3) | 93.3 (68.1) |
| Redundancy | 3.7 (2.8) | 4.4 (3.5) |
| Refinement | ||
| Resolution (Å) | 2.0 | 2.0 |
| R factor (%)e | 17.87 | 19.06 |
| Rfree (%)f | 23.18 | 25.64 |
| No. of protein atoms | 5,307 | 2,676 |
| No. of water molecules | 650 | 304 |
| No. of unique reflections | 48,795 | 24,409 |
| Rmsdg | ||
| Bonds | 0.017 | 0.026 |
| Angles | 1.898 | 2.139 |
Statistics for the highest-resolution bin of reflections are in parentheses.
Rsym = ΣhΣj l Ihj − <Ih> l/ΣhΣjIhj, where Ihj is the intensity of observation j of reflection h and <Ih> is the mean intensity for multiply recorded reflections.
Intensity signal-to-noise ratio.
Completeness of the unique diffraction data.
R factor = Σh I IFoI − IFcI I/ΣhIFoI, where Fo and Fc are the observed and calculated structure factor amplitudes for reflection h.
Rfree was calculated against a 10% random sampling of the reflections that were removed before structure refinement.
RMSD of bond lengths and bond angles.
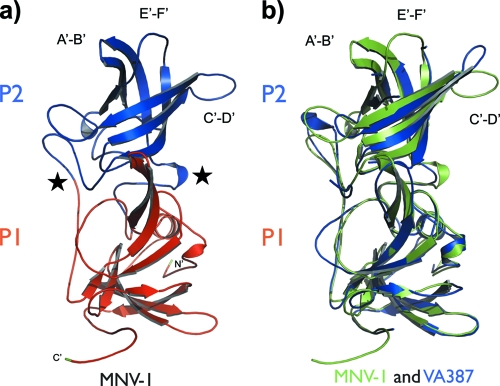
The observed topology of the MNV-1 monomer is unique to the P domains of noroviruses (5, 6, 10, 46) (Fig. 3). The structure can be divided into two subdomains, P1 and P2. The MNV-1 P1 subdomain consists of N-terminal residues 229 to 277 and C-terminal residues 416 to 541, which fold into two highly twisted antiparallel β-sheets and a single α-helix. The P2 subdomain is formed from an extensive insertion (aa 278 to 415) and folds into a six-strand antiparallel β-barrel. The interface between the two subdomains within a monomer consists of a randomly coiled polypeptide chain. The C terminus of the MNV-1 P domain is partially disordered and flexible. As this flexible tail stretches out into the neighboring P dimer, it may be involved in the polymerization of the subunits in the viral particle (see reference 24).
FIG. 3.
Structure comparison of the MNV-1 P domain with the VA387 P domain. (a) Cartoon of the MNV-1 P-domain structure with the P1 subdomain in red and the P2 subdomain in blue. Surface-exposed loops in the P2 subdomain are labeled A′-B′ (aa 299 to 300), C′-D′ (aa 342 to 350), and E′-F′ (aa 378 to 388). The N and C termini of the MNV-1 P domain are shown in green. The arrows indicate regions of β-strands. The coils represent helices. The stars designate the hinge regions between P1 and P2. (b) Superimposition of MNV-1 (green) and VA387 (blue) (PDB ID, 2OBR [6]) P-domain structures.
Structural comparisons of the two crystal forms of MNV-1 described here, as well as comparison with those of the closely related HuNoV capsid proteins, enabled a detailed analysis of the dynamics of the P domains. At one level, the molecular dynamics is manifested by rigid-body rotation of the P2 subdomain relative to the P1 subdomain. This motion would allow the outer P2 subdomain to flex relative to the rigid core of the viral capsid. Superimposition of the P1 subdomains of the P-domain structures of the murine capsid on that of the HuNoV capsid (46) suggested a concerted rotation of the P2 subdomain relative to the P1 subdomain (data not shown). This rotation produces a shift of up to 3 Å in the Cα positions of the residues distal to the hinge region. This is enabled by two flexible regions, or hinges, connecting the subdomains. The first hinge region is comprised of residues 283 to 288, and the second region is located in the interdomain loop residues 407 to 415 (Fig. 3A). A very similar interdomain rotation was evident when the murine structure was compared to the structure of the HuNoV VA387 strain (6), with maximum Cα shifts of 3 Å, and the same two hinge regions are implicated in the domain rotation (Fig. 3B).
Analysis of the MNV-1 P-domain dimers (Fig. 4) using AreaiMol (45) showed that the dimer interface in the P-domain structure is extensive and consists of 3,079 Å2 of buried surface area (data not shown). The interfaces in the two MNV-1 P-domain crystal structures are identical, suggesting that only the dimer form of the protein crystallizes. Most of the dimer contacts occur due to hydrogen bonding between residues in the P1 subdomain (data not shown). In particular, there are contacts between side chains of 310 helices (aa 283 to 285) and α-helices (aa 455 to 464) of both P1 subdomains. In the P2 subdomain, portions of the C′-D′ loop (aa 341 to 351) make extensive intermolecular contacts with portions of a β-sheet in a symmetry-related P1 subdomain.
FIG. 4.
Orthogonal views of the MNV-1 P-domain dimer. (a and b) Surface diagrams of the MNV-1 P-domain dimer. Individual monomers are surface colored in blue and green. The ribbon diagrams of the P1 domains are colored red, while the P2 domains are colored blue. (a) Side view of the dimer perpendicular to the 2-fold axis. (b) Top view looking down the 2-fold axis. A close-up of the ionic lock is shown in the inset. (c) Slice through the surface of the P-domain dimer interface showing the channel extending from one end of the dimer to the other along the 2-fold axis. (d) Slice through the surface view of the P domain, looking down into the channel (∼8 Å in diameter). This view is a 90° rotation from the orientation shown in panel c.
The interaction of the two P2 subdomains results in an approximately 20-Å-deep and 50-Å-wide canyon at the dimer interface (Fig. 4). A crater approximately 30 Å in diameter is located at the center of the canyon. A narrow (∼8-Å-wide) channel runs the entire length (40 Å) of the P-domain dimer (Fig. 4C and D). On top of this channel sits an ionic lock, which consists of two salt bridges formed between 2 residues, E338 and R396, from each P2 subdomain, which are absolutely conserved among all MNV sequences available to date (Fig. 4B).
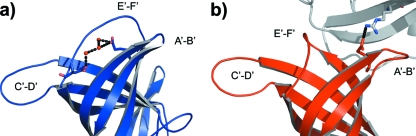
The existence of two crystal structures presented an opportunity to investigate alternative P-domain conformations. Superimposition of the two MNV-1 P-domain structures using the SSM superimpose routine in COOT (14) indicated a high degree of structural identity between the monomers within the two crystal structures (RMSD = 0.45 Å2) (Fig. 5 and data not shown). Most notable were changes in the variable loops on the outer surface of the P2 subdomain (Fig. 5). Two of the surface loops, E′-F′ (aa 379 to 388) and A′-B′ (aa 299 to 300), showed large displacements, with Ala 382 showing the largest displacement (Cα movement of 16 Å). Additional topological changes were also observed in the surface-exposed loop of the C-terminal P1 subdomain (aa 361 to 367). Not surprisingly, the loops with the largest deviations between the MNV-1 P-domain structures were also the most flexible regions of the structures, as indicated by the high thermal parameters (40 to 56 A2). A third loop, C′-D′ (aa 342 to 350), on the surface of the P2 subdomain is stabilized by contacts with an adjacent P1 subdomain in the dimer and therefore displays relatively small variations in position and lower thermal parameters (30 to 40 A2) compared to the A′-B′ and E′-F′ loops. The A′-B′ and E′-F′ loops displayed two unique conformations: a closed structure, where the two loops were tightly associated, and an open structure, where the loops were splayed apart (Fig. 5). Of note, the two loop conformations were observed within the two independent P-domain monomers in the monoclinic crystal structure, highlighting the large variation in the positions and conformations of the outer surface loops of the P2 subdomain. In molecule B, these loops are in a closed conformation, which is stabilized by a series of hydrogen bonds to water molecules (Fig. 6A). In molecule A, they are in an open conformation, which is the result of crystal packing (Fig. 6B). The open conformation is stabilized by hydrophilic contacts with an adjacent molecule in the crystal lattice. Specifically, a salt bridge was formed between E303 and R476 from a symmetry-related molecule (Fig. 6B). The closed conformation of the loops (Fig. 5 and 6A) is observed in both monomers in the orthorhombic structure of the symmetric P-domain dimer. Whether the two distinct conformations represent two naturally occurring static states, each with a different biological function; two of many possible structures in this dynamic portion of the P domain; or an artifact of crystal packing is unknown. Regardless, these significant variations in the conformations of several flexible loops on the surface of the P2 subdomain represent a second type of molecular flexibility, in addition to the rigid-body rotation of the P2 subdomain relative to the P1 subdomain described above.
FIG. 5.

Flexibility within the MNV-1 P2 subdomains. Shown is a superimposition of the P2 subdomains from the two crystal structures (blue, closed conformation; red, open conformation) showing variation in the E′-F′ and A′-B′ loop conformations.
FIG. 6.
Stabilization of the MNV-1 P2 subdomain flexible loops. (a) The closed loop conformation (blue) is stabilized by hydrogen bonds with a series of water molecules. (b) Interaction of the open loop structure (red) with the P1 subdomain of a symmetry-related P-domain molecule (gray) highlighting the salt bridge formed between E303 and R476.
The neutralizing monoclonal antibody A6.2 binds the outermost tip of the closed conformation of the MNV-1 P2 domain.
Our previous work showed that Fab fragments of the neutralizing antibody A6.2 bound to residues in the P2 subdomain located in the A′-B′ and E′-F′ loops (25). In that study, pseudo-atomic models had been created using cryoelectron microscopy image reconstructions of MNV-1 (∼12-Å resolution), MNV-1-FabA6.2 (∼22-Å resolution), and the atomic structure of recombinant Norwalk virus (rNV). With the atomic structure of the MNV-1 P domains presented here and the high-resolution (∼8-Å) image reconstruction of MNV-1 presented in the accompanying publication (24), the pseudo-atomic modeling of the MNV-1-FabA6.2 complex was revisited to better define this particular neutralizing epitope.
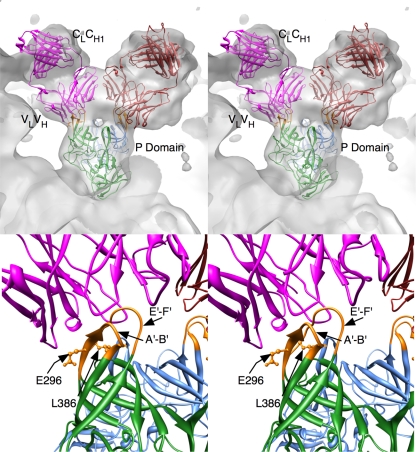
For this analysis, the MNV-1-Fab model was constructed using the pseudo-atomic model described in the accompanying paper (24), the ∼22-Å resolution 3D image reconstruction of the MNV-1-Fab A6.2 complex previously described (25), an MNV-1 P-domain dimer, and the structure of Fab17 (31, 52, 54). As detailed in the accompanying paper (24), the high-resolution image reconstruction of MNV-1 has marked “horns” (protrusions made by loops A′-B′ and E′-F′) at the top of the P domains that closely match the closed conformation. When the open conformation was placed into the 8-Å 3D image reconstruction, the E′-F′ loop protruded noticeably from the sides of the horns. Therefore, dimers of the closed conformation were used to create a pseudo-atomic model of MNV-1. The electron density from the previous 3D image reconstruction of the MNV-1-Fab A6.2 complex was then overlaid onto this model of MNV-1, and the structure of the antibody, Fab17, was placed into the envelope corresponding to the bound Fab. During the fitting process, it became apparent that the horns of the P domains needed to be placed into the hypervariable cleft between the heavy and light chains. However, this necessitated fitting the variable domains (VL and VH) separately from the constant domains (CL and CH1) by rotating the constant domains about the flexible elbow region, the connection between the variable and constant domains. This in effect changed the elbow angle to ∼180°. As shown in Fig. 7, these modifications yielded an excellent fit of the antibody into the 3D reconstruction envelope.
FIG. 7.
3D modeling of an antibody-P-domain complex. Shown are stereo diagrams of Fab17 and the MNV-1 P domains fitted into the 3D image reconstruction of the MNV-1-Fab complex, in gray. The blue and green ribbon figures represent the A and B subunits in the MNV-1 structure, respectively. The mauve and red ribbon figures represent the two copies of Fab17 bound to the P-domain dimer. At the bottom is shown a close up of the MNV-1-Fab interactions with the contact residues highlighted in orange in the P-domain dimer. Also noted are the locations of residues E296 and L386, the attenuation and escape mutation sites, respectively.
The likely epitope region was further examined using the web-based tools in PISA (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; 29). Contact was localized to a portion of the A′ β-strand and A′-B′ loop (aa 294 to 303), as well as the E′-F′ loop (aa 379 to 388) (Fig. 8), which form the horns on the P2 domain. The total contact was ∼860 Å2, which is typical for epitope-paratope interactions. In this model, the horns of the P2 domain fit snugly into the cleft between the heavy- and light-chain hypervariable regions and make contact with all six antigen binding loops (complementary determining regions). In our previous analysis using the structure of the rNV P domain (25), there was an additional possible contact with parts of the C′-D′ loop and the D′ β-strand (aa 345 to 358). However, the extension of the horn pushes the antibody away from making contacts with the flatter regions of the P2 head. Therefore, the epitope for this antibody is clearly limited to the outermost tips of the P2 domain formed by the A′-B′ (i.e., Fab1) and E′-F′ (i.e., Fab2) protrusions.
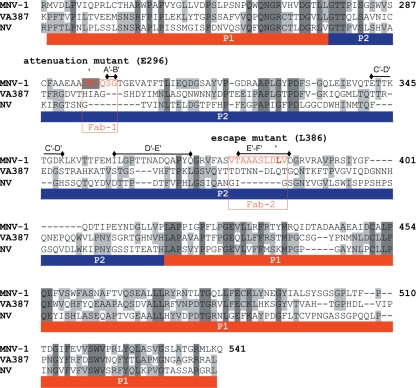
FIG. 8.
Structural alignment of the P domains from MNV-1 with VA387 and Norwalk virus. The P-domain structures of VA387 (PDB ID, 2OBR, [6]) and NV (PDB ID, 1IHM [46]) were aligned with the MNV-1 P-domain structure using VAST (17). The A6.2 Fab binding sites are highlighted in red. The red and blue bars beneath the sequences represent the P1 and P2 subdomains, respectively. The arrows indicate the locations of the known escape mutation (L386) and attenuation mutation (E296). The black bars above the alignment denote the surface-exposed loop regions in the P2 domain.
Modeling the MNV-1-Fab complex indicated that MNV-1 with the E′-F′ loop in the closed conformation represents an antigenically active structure. Further biological conformation for the involvement of the E′-F′ loop in the A6.2 epitope comes from two independent experiments. Previously, Lochridge and Hardy (32) identified a neutralization escape mutant that was localized to L386, located in the C-terminal half of the Fab2 site (Fig. 8). In addition, we took advantage of the reverse-genetics system to create the first chimeric virus of MNV-1 and NV. Specifically, a 7-amino-acid sequence of MNV-1 (aa 377 to 383), encompassing the N-terminal half of the predicted Fab2-binding site, was exchanged for the structurally corresponding region of NV (Fig. 9A). Recombinant MNV-1.CW1*.fab2 was successfully recovered and tested in a plaque neutralization assay (Fig. 9B and C). A6.2 was unable to neutralize MNV-1.CW1*.fab2 (Fig. 9B). This was in contrast to successful neutralization of the wild-type virus, MNV-1.CW1*, by A6.2 (Fig. 9B). Isotype control antibody was unable to neutralize either virus (Fig. 9C). Taken together, these data provide convincing evidence that the E′-F′ loop is an important part of the A6.2 binding site, and disruption of interaction with amino acids in either the N-terminal or C-terminal half of this Fab2-binding site leads to loss of neutralization.
FIG. 9.
Recombinant MNV-1 with mutations in the Fab2-binding site is no longer neutralized by A6.2. (a) Schematic of the MNV-1.CW1*.fab2 genome structure showing the ORF-1 polyprotein (N terminus [N], NTPase, 3A-like [3A], VPg, protease [Pro], and RNA-dependent RNA polymerase [RdRp]), ORF-2 encoding the major capsid protein (VP1), and ORF-3. Recombinant MNV-1.CW1* contains a silent mutation in the N-terminal protein compared to MNV-1.CW1 to introduce a unique EcoRV site (63). Shown below is a further close-up of the schematic of VP1 showing the N, S, and P domains. To mutate the Fab2-binding site (red), residues 377 to 383 (SVTAAAS) were replaced with the structurally corresponding residues 334 to 340 (NGIGSGN) in NV, generating the recombinant mouse-human chimera MNV-1.CW1*.fab2. (b) Neutralization of recombinant MNV-1.CW1*, but not MNV-1.CW1.fab2*, by A6.2 in a plaque neutralization assay. (c) Inability of the isotype control (IgG1) MAb to neutralize virus. The limit of detection is indicated by the dashed line. Titers are presented as means ± SE from duplicate samples in three independent experiments.
This limited epitope at the outermost tips of the MNV-1 capsid is significantly different than a number of other antibodies that contact a broader, flatter surface (52). It is interesting that the antibody contact is limited to these loops and that they have two distinct conformations in the crystal structures. While it is not unexpected that the antibody contact area includes the known escape mutation site for this antibody (L386) (32), it is interesting that it also overlaps the attenuation site, E296 (65) (Fig. 8). This is akin to studies of other viruses demonstrating that epitopes and regions of viral capsids that change during host adaptation can overlap (1, 53).
It should be noted that there are several limitations in analyzing the exact details of the antibody-virus interactions. The structure of Fab17 is only an approximation for that of the bound A6.2, and as was seen in the case of Fab17 (31, 52), flexibility of the hypervariable region may allow the antibody to mold itself to the epitope region. As was shown in our previous studies, where the structures of human rhinovirus 14, the Fab, and the Fab-virus complex were all known (52), pseudo-atomic models made from 3D reconstructions at this resolution can be off by several angstroms (8), and this can have a significant effect on predicted contact points. Nevertheless, while the exact contacts can only be approximated, confirmation comes from the fact that the structure of the P-domain horns and the 3D image reconstruction are of sufficient resolution to limit the epitope region to these outermost loops. In addition, two different mutant viruses, a neutralization escape mutant (32) and the chimeric MNV-1.CW1*.fab2 (Fig. 9), provide biological confirmation of the in silico modeling results of the A6.2 epitope.
Conformational changes in the MNV-1 capsid are hypothesized to play an important role during infection.
It is well known that for several viruses (e.g., poliovirus [35, 49] and reovirus [16]), binding to viral receptors leads to reversible and irreversible conformational changes in the virion. We hypothesize that conformational changes occur in the MNV-1 capsid that facilitate specific steps in the viral life cycle (e.g., endosomal escape or uncoating). One of those changes could occur via opening of the E′-F′ loop and/or via opening of the ionic lock and subsequently the channel between the two P domains. The latter is consistent with recent data demonstrating that removal of a calcium ion from a salt bridge between pentameric subunits of simian virus 40 (SV40) induces a conformational change and triggers disassembly (26). Additional experiments are needed to test this hypothesis.
Acknowledgments
Work in the laboratory of C.E.W. was funded by startup funds from the University of Michigan; a career development grant from the NIH/NIAID Regional Center of Excellence for Bio-defense and Emerging Infectious Diseases Research (RCE) Program, Region V Great Lakes RCE (NIH award 1-U54-AI-057153); and R01 AI080611. Work in the laboratory of J.A.S. was funded in part by the University of Michigan Center for Structural Biology. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (grant 085P1000817).
We thank Clay Brown and Jim Delproposto (LSI, University of Michigan, Ann Arbor, MI) for cloning of the MNV-1 P-domain expression plasmid and Jennifer Meagher (LSI, University of Michigan, Ann Arbor, MI) for advice on purification protocols. We are indebted to Nicole Koropatkin (University of Michigan, Ann Arbor, MI) for critical comments and helpful suggestions on the manuscript.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, S. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. [DOI] [PubMed] [Google Scholar]
- 5.Bu, W., A. Mamedova, M. Tan, M. Xia, X. Jiang, and R. S. Hegde. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, S., Z. Lou, M. Tan, Y. Chen, Y. Liu, Z. Zhang, X. C. Zhang, X. Jiang, X. Li, and Z. Rao. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhry, Y., M. A. Skinner, and I. G. Goodfellow. 2007. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 88:2091-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Che, Z., N. H. Olson, D. Leippe, W. M. Lee, A. G. Mosser, R. R. Rueckert, T. S. Baker, and T. J. Smith. 1998. Antibody-mediated neutralization of human rhinovirus 14 explored by means of cryoelectron microscopy and X-ray crystallography of virus-Fab complexes. J. Virol. 72:4610-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R., J. D. Neill, J. S. Noel, A. M. Hutson, R. I. Glass, M. K. Estes, and B. V. Prasad. 2004. Inter- and intragenus structural variations in caliciviruses and their functional implications. J. Virol. 78:6469-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, J. M., A. M. Hutson, M. K. Estes, and B. V. Prasad. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175-9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colovos, C., and T. O. Yeates. 1993. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2:1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Donaldson, E. F., L. C. Lindesmith, A. D. Lobue, and R. S. Baric. 2010. Viral shape-shifting: non virus invasion of the human immune system. Nat. Rev. Microbiol. 8:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes, J., D. Tang, G. Leone, and P. W. Lee. 1994. Binding of reovirus to receptor leads to conformational changes in viral capsid proteins that are reversible upon virus detachment. J. Biol. Chem. 269:17043-17047. [PubMed] [Google Scholar]
- 17.Gibrat, J. F., T. Madej, and S. H. Bryant. 1996. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6:377-385. [DOI] [PubMed] [Google Scholar]
- 18.Green, K. Y. 2007. Caliciviridae, p. 949-980. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 19.Guix, S., M. Asanaka, K. Katayama, S. E. Crawford, F. H. Neill, R. L. Atmar, and M. K. Estes. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81:12238-12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy, M. E., T. N. Tanaka, N. Kitamoto, L. J. White, J. M. Ball, X. Jiang, and M. K. Estes. 1996. Antigenic mapping of the recombinant Norwalk virus capsid protein using monoclonal antibodies. Virology 217:252-261. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 21a.Harrington, P. R., B. Yount, R. E. Johnston, N. Davis, C. Moe, and R. S. Baric. 2002. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 76:730-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 24.Katpally, U., N. R. Voss, T. Cavazza, S. Taube, J. R. Ruben, V. L. Young, J. Stuckey, V. K. Ward, H. W. Virgin IV, C. E. Wobus, and T. J. Smith. 2010. High-resolution cryo-electron microscopy structures of Murine Norovirus 1 and Rabbit Hemorrhagic Disease Virus reveals marked flexibility in the receptor binding domains. J. Virol. 84:5836-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katpally, U., C. E. Wobus, K. Dryden, H. W. Virgin IV, and T. J. Smith. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 82:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano, M. A., L. Xing, H. Tsukamoto, T. Inoue, H. Handa, and R. H. Cheng. 2009. Calcium bridge triggers capsid disassembly in the cell entry process of simian virus 40. J. Biol. Chem. 284:34703-34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirillova, O., and W. Minor. 2006. Map2mod—a server for evaluation of crystallographic models and their agreement with electron density maps. Bioinformatics 22:1660-1661. [DOI] [PubMed] [Google Scholar]
- 28.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262-S269. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 30a.Li, W., M. H. Sofi, S. Rietdijk, N. Wang, C. Terhorst, and C. H. Chang. 2007. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity 27:763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H., T. J. Smith, W. M. Lee, A. G. Mosser, R. R. Rueckert, N. H. Olson, R. H. Cheng, and T. S. Baker. 1994. Structure determination of an Fab fragment that neutralizes human rhinovirus 14 and analysis of the Fab-virus complex. J. Mol. Biol. 240:127-137. [DOI] [PubMed] [Google Scholar]
- 32.Lochridge, V. P., and M. E. Hardy. 2007. A single-amino-acid substitution in the P2 domain of VP1 of murine norovirus is sufficient for escape from antibody neutralization. J. Virol. 81:12316-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDermott, B. M., Jr., A. H. Rux, R. J. Eisenberg, G. H. Cohen, and V. R. Racaniello. 2000. Two distinct binding affinities of poliovirus for its cellular receptor. J. Biol. Chem. 275:23089-23096. [DOI] [PubMed] [Google Scholar]
- 36.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, R. K., H. Qadota, T. J. Stark, K. B. Mercer, T. S. Wortham, A. Anyanful, and G. M. Benian. 2009. CSN-5, a component of the COP9 signalosome complex, regulates the levels of UNC-96 and UNC-98, two components of M-lines in Caenorhabditis elegans muscle. Mol. Biol. Cell 20:3608-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno-Espinosa, S., T. Farkas, and X. Jiang. 2004. Human caliciviruses and pediatric gastroenteritis. Semin. Pediatr. Infect. Dis. 15:237-245. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 40.Nayal, M., and E. Di Cera. 1996. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 256:228- 234. [DOI] [PubMed] [Google Scholar]
- 41.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 43.Perry, J. W., S. Taube, and C. E. Wobus. 2009. Murine norovirus-1 entry into permissive macrophages and dendritic cells is pH-independent. Virus Res. 143:125-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pontius, J., J. Richelle, and S. J. Wodak. 1996. Deviations from standard atomic volumes as a quality measure for protein crystal structures. J. Mol. Biol. 264:121-136. [DOI] [PubMed] [Google Scholar]
- 45.Potterton, E., P. Briggs, M. Turkenburg, and E. Dodson. 2003. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59:1131-1137. [DOI] [PubMed] [Google Scholar]
- 46.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 47.Prasad, B. V., M. E. Hardy, X. Jiang, and M. K. Estes. 1996. Structure of Norwalk virus. Arch. Virol. Suppl. 12:237-242. [DOI] [PubMed] [Google Scholar]
- 48.Prasad, B. V., R. Rothnagel, X. Jiang, and M. K. Estes. 1994. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68:5117-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Racaniello, V. R. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. U. S. A. 93:11378-11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santi, L., L. Batchelor, Z. Huang, B. Hjelm, J. Kilbourne, C. J. Arntzen, Q. Chen, and H. S. Mason. 2008. An efficient plant viral expression system generating orally immunogenic Norwalk virus-like particles. Vaccine 26:1846-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siebenga, J. J., M. F. Beersma, H. Vennema, P. van Biezen, N. J. Hartwig, and M. Koopmans. 2008. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J. Infect. Dis. 198:994-1001. [DOI] [PubMed] [Google Scholar]
- 52.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, T. J., R. H. Cheng, N. H. Olson, P. Peterson, E. Chase, R. J. Kuhn, and T. S. Baker. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A. 92:10648-10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, T. J., N. H. Olson, R. H. Cheng, H. Liu, E. S. Chase, W. M. Lee, D. M. Leippe, A. G. Mosser, R. R. Rueckert, and T. S. Baker. 1993. Structure of human rhinovirus complexed with Fab fragments from a neutralizing antibody. J. Virol. 67:1148-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan, M., R. S. Hegde, and X. Jiang. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 78:6233-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taube, S., A. Kurth, and E. Schreier. 2005. Generation of recombinant norovirus-like particles (VLP) in the human endothelial kidney cell line 293T. Arch. Virol. 150:1425-1431. [DOI] [PubMed] [Google Scholar]
- 58.Taube, S., J. W. Perry, K. Yetming, S. P. Patel, H. Auble, L. Shu, H. F. Nawar, C. H. Lee, T. D. Connell, J. A. Shayman, and C. E. Wobus. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 83:4092-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terwilliger, T. C., R. W. Grosse-Kunstleve, P. V. Afonine, N. W. Moriarty, P. D. Adams, R. J. Read, P. H. Zwart, and L. W. Hung. 2008. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D Biol. Crystallogr. 64:515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thackray, L. B., C. E. Wobus, K. A. Chachu, B. Liu, E. R. Alegre, K. S. Henderson, S. T. Kelley, and H. W. Virgin IV. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81:10460-10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaguine, A. A., J. Richelle, and S. J. Wodak. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191-205. [DOI] [PubMed] [Google Scholar]
- 62.Ward, J. M., C. E. Wobus, L. B. Thackray, C. R. Erexson, L. J. Faucette, G. Belliot, E. L. Barron, S. V. Sosnovtsev, and K. Y. Green. 2006. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol. Pathol. 34:708-715. [DOI] [PubMed] [Google Scholar]
- 63.Ward, V. K., C. J. McCormick, I. N. Clarke, O. Salim, C. E. Wobus, L. B. Thackray, H. W. Virgin IV, and P. R. Lambden. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 104:11050-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Widdowson, M. A., S. S. Monroe, and R. I. Glass. 2005. Are noroviruses emerging? Emerg. Infect. Dis. 11:735-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wobus, C. E., L. B. Thackray, and H. W. Virgin IV. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, X., N. A. Buehner, A. M. Hutson, M. K. Estes, and H. S. Mason. 2006. Tomato is a highly effective vehicle for expression and oral immunization with Norwalk virus capsid protein. Plant Biotechnol. J. 4:419-432. [DOI] [PubMed] [Google Scholar]