Abstract
Bovine herpesvirus 1 (BoHV-1) and BoHV-5 are closely related pathogens of cattle, but only BoHV-5 is considered a neuropathogen. We engineered intertypic gD exchange mutants with BoHV-1 and BoHV-5 backbones in order to address their in vitro and in vivo host ranges, with particular interest in invasion of the brain. The new viruses replicated in cell culture with similar dynamics and to titers comparable to those of their wild-type parents. However, gD of BoHV-5 (gD5) was able to interact with a surprisingly broad range of nectins. In vivo, gD5 provided a virulent phenotype to BoHV-1 in AR129 mice, featuring a high incidence of neurological symptoms and early onset of disease. However, only virus with the BoHV-5 backbone, independent of the gD type, was detected in the brain by immunohistology. Thus, gD of BoHV-5 confers an extended cellular host range to BoHV-1 and may be considered a virulence factor but does not contribute to the invasion of the brain.
Bovine herpesvirus 1 (BoHV-1) and BoHV-5 belong to the subfamily Alphaherpesvirinae and are closely related pathogens of cattle (22). The protein repertoire of the two viruses averages 82% amino acid identity (20). Both viruses are neurotropic, but only BoHV-5 can significantly replicate in the central nervous system (CNS) to cause encephalitis of either naturally infected cattle or experimentally inoculated laboratory animals (2, 5, 6, 12, 40, 41, 44). Glycoprotein D (gD) is accepted as the critical and essential receptor-binding protein of many alphaherpesviruses (reviewed in references 8 and 48). The main gD receptors identified to date include members of the tumor necrosis factor (TNF) receptor family (HveA) and the poliovirus receptor family (HveB or nectin 2 and HveC or nectin 1) (28, 42, 51). Furthermore, a modified form of heparan sulfate, 3-O-sulfated heparan sulfate, can mediate herpesvirus entry (46). J1.1-2 cells (J cells) represent a subpopulation of thymidine kinase-negative baby hamster kidney (BHK) cells selected for their property of being resistant to infection with herpes simplex virus type 1 (HSV-1), HSV-2, and BoHV-1. The expression of nectin 1 in those cells rendered them susceptible to BoHV-1 infection and replication, which suggests that nectin 1 can serve as a receptor for BoHV-1 gD (gD1) (16, 18, 28). Interestingly, we observed that BoHV-5 was able to productively replicate in J cells without the nectin 1 receptor.
According to a previously reported sequence comparison of BoHV-1 and BoHV-5 (20), the highest divergence between the two viruses mapped to the latency-related region and the immediate-early proteins (less than 75% amino acid identity) BICP0, BICP4, and BICP22. Glycoprotein E (gE) was also listed in this category, with 74% amino acid identity between gE of BoHV-1 (gE1) and gE5. This fact also gave ample reason for attempts to map the neurovirulent phenotype of BoHV-5 to the gE5 molecule (3, 4, 13). In contrast, the highest sequence similarities between the two viruses were described for proteins involved in viral DNA replication and processing as well as certain virion proteins. Among others, the predicted amino acid sequences of gD1 and gD5 were listed as being 98% identical (20). However, our own analysis using the European Molecular Biology software suite (43) revealed only 79.9% amino acid identity. Obviously, the most extensive difference between gD1 and gD5 maps to a glycine-rich stretch located in the molecule's ectodomain, between amino acids (aa) 280 and 330 of gD5, in close vicinity to the transmembrane region.
Based on these considerations, we hypothesized that BoHV-5 was able to make use of a cellular receptor that is unavailable to BoHV-1. To test this hypothesis, the gD genes were removed from bacterial artificial chromosomes (BACs) harboring the genome of either BoHV-5 or BoHV-1 (27). In a second step, gD exchange viruses were created by the cotransfection of the gD-less BACs with appropriate plasmids carrying either the gD1 or gD5 gene and appropriate flanking sequences. The newly generated viruses included an intertypic BoHV-5 mutant carrying gD1 in the place of gD5 and a corresponding BoHV-1 carrying gD5. These mutants, together with appropriate revertant mutants, were then used to explore their ability to infect J cells in vitro and their ability to cause neurological disease and invade the brain in vivo, in a previously established mouse model (2). Our results indicate that gD5 confers an extended host range to BoHV-1 but is nonessential for the invasion of the brain.
MATERIALS AND METHODS
Viruses and cells.
Madin-Darby bovine kidney (MDBK) cells, simian virus 40 (SV40)-transformed African green monkey kidney (COS-1) cells, and J1.1-2 cells (J cells) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with a mixture of penicillin and streptomycin and 5% to 10% fetal calf serum (FCS). J cells are derived from BHKtk− cells and have been selected for resistance against HSV infection (16). Vero 2.2 cells were grown in DMEM supplemented with G418 (500 μg/ml), 10% FCS at 37°C, and 5% CO2 (47). Wild-type (wt) BoHV-5 (strain N569), wt BoHV-1 (strain Jura), recombinant BoHV-5 (rBoHV-5), rBoHV-5gD1HA, rBoHV-1, rBoHV- 1gD5V5, recombinant-revertant BoHV-5gD5V5 (rrBoHV-5gD5V5), and rrBoHV-1gD1YFP were propagated in MDBK cells as described previously (25, 40).
Mice.
All animals were cared for and used in accordance with Swiss laws for animal experimentation. Mouse strain AR129, which was used throughout this study, is derived from 129Sv/Ev (H-2b) mice. AR129 mice have genetically deleted type 1 interferon receptors combined with a RAG-2 knockout (30).
Plasmids.
More details of all constructions are provided at http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html. All primers are listed in Table 1.
TABLE 1.
Synthetic oligonucleotides used in this study
| Oligonucleotide | Sequencea | Amplification product |
|---|---|---|
| P1 | 5′-GATCGAGCTCGTTAACCGACTCGGATTTTGGGC-3′ | 5′ of gD5 homology arm |
| P2 | 5′-GATCGCGGCCGCAGTTGCTCGCTCGCAGCAAC-3′ | |
| P3 | 5′-GATCACCGGTGCGGCCCGGGCCCTCCCCCG-3′ | 3′ of gD5 homology arm |
| P4 | 5′-GATCGGTACCGTTAACGCGGTCGCCTGTAGCATGACGAAGC-3′ | |
| P5 | 5′-ATAAGAATGCGGCCGCATGAAGGGCCGACATTGGCCGTGc-3′ | gD1 ORF fused to HA |
| P6 | 5′-GATCGAATTCTCAAGCATAATCTGGAACATCATATGGATACCCGGGCAGCGCGCTGTAGt-3′b | |
| P7 | 5′-GATCGGATCCATGCGGAGGCTGGCGCTGCT-3′ | gD5 ORF fused to V5 |
| P8 | 5′-GATCAAGCTTTCACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCCCCGGGCAGCGCGCTGTAGT-3′c | |
| P9 | 5′-GATCGAGCTCGTTAACCTCCGACTACGCGCTCTACG-3′ | 5′ of gD1 homology arm |
| P10 | 5′-GATCGGATCCGTTCGCCCGCTCGCAGCA-3′ | |
| P11 | 5′-GATCAAGCTTGGGGCCTAGGCCCTCCCCC-3′ | 3′ of gD1 homology arm |
| P12 | 5′-GATCGGTACCGTTAACGCGCCGAGAGCACGGC-3′ | |
| P13 | 5′-GATCTCTAGAACCCGCATCCGCGGTGGCTTT-3′ | 5′ of gD1 flank and gD1 ORF without stop codon |
| P14 | 5′-GATCGGATCCCCCGGGCAGCGCGCTGTAGTT-3′ | |
| P15 | 5′-AGTCGGATCCTGAGCGGCCTAGGCCCTCCCCCGA-3′ | 3′ of gD1 flank |
| P16 | 5′-GCATGAATTCAGGATCGACGCCAGTTGGCGCCGGAA-3′ | |
| P17 | 5′-ACGTGGATCCCATGGTGAGCAAGGGCGAGGA-3′ | YFP ORF |
| P18 | 5′-GATCGGATCCCTTGTACAGCTCGTCCATGC-3′ | |
| P19 | 5′-GGGCGACTAGAGATACACTCGCCCCGCGCGGCTGCTGCGAGCGGGCGAACcctcgaggtcgacataactt-3′ | Kanamycin ORF flanked by 50-bp homology arms for gD1 locus |
| P20 | 5′-GGAGCCGGGGCTAGGAGCAAAGGGGGCGGTCGGGGGAGGGCCTAGGCCGCgagcccttaattaaccggtg-3′ | |
| gD5upper | 5′-CGGAGGCTGGCGCTGCTGT-3′ | gD5-DIG-labeled probe |
| gD5lower | 5′-ACAGCGTGCGCCCCACCTGC-3′ | |
| gD1upper | 5′-GACGACGAGCTGGGACTGATT-3′ | gD1-DIG-labeled probe |
| gD1lower | 5′-CGGGGGTCTGACTCTC-3′ |
Underlining indicates the restriction site used for cloning. The binding part of the primers is shown in lowercase type.
The letters in boldface type indicate an HA tag.
The letters in boldface type indicate a V5 tag.
p118 contains a gene expression cassette providing kanamycin resistance flanked by two FLP recombination target (FRT) sites (see also http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html).
p302ΔgD1HA contains a gene expression cassette providing kanamycin resistance, flanked by two FRT sites, and homology arms to target the glycoprotein D locus (gDS) of the BoHV-5 BAC (fBoHV-5) (27). Primers P1, P2, P3, and P4 were used for the amplification and cloning of the homology arms, i.e., 477 bp upstream and 421 bp downstream of gD5, respectively.
p302ΔkanR contains the same homology arms as p302ΔgD1HA. In this construct, the homology arms flank the gD1 open reading frame (ORF), which is fused to a hemagglutinin (HA) epitope coding sequence at its 3′ end. Primers P5 and P6 were used for the amplification and cloning of the gD1HA ORF.
p309 contains homology arms to target the gD5 locus that flank the gD5 ORF fused to a V5 epitope sequence at its 3′ end. Primers P7 and P8 were used for the amplification and cloning of the gD5V5 ORF.
p307 contains a gD5V5 ORF flanked with extended homology arms to target the gD1 locus. The upstream 1,134-bp homology arm was amplified with primers P9 and P10. The downstream 1,136-bp homology arm was amplified with primers P11 and P12.
pCS156 has a total size of 7,107 bp and contains a 4,182-bp fragment consisting of gD1 fused to the yellow florescent protein (YFP) coding sequences, flanked by homology arms to target the gD1 locus. The upstream homology arm, together with the gD1 coding sequence without the stop codon, was amplified with primers P13 and P14. The downstream homology arm was amplified with primers P15 and P16. The YFP sequence was amplified with primers P17 and P18.
Red recombinations for gD-deleted BoHV BACs. (i) fBoHV-5ΔgDkanR BAC.
Vector p302ΔgD1HA was used for the construction of fBoHV-5ΔgDkanR BAC by Red recombination (19). The 2,832-bp transfer fragment containing 477 bp upstream of the gD5 start codon, a kanamycin cassette flanked by FRT sites, and 421 bp downstream of the gD5 stop codon was excised with HpaI from p302ΔgD1HA, gel purified, and electroporated into arabinose-induced Escherichia coli strain DH10B cells harboring fBoHV5 BAC and pKD46 (19). Electroporated cells were selected on LB agar plates containing 25 μg/ml kanamycin. The resulting recombinant BoHV-5 BAC was designated fBoHV-5ΔgDkanR.
(ii) fBoHV-1ΔgDkanR BAC.
The 2,010-bp DNA transfer fragment was amplified by PCR using p118 as a template. It contains a kanamycin cassette for selection in E. coli, flanked by 50 bp of homology upstream of the start codon of the gD1 ORF and 50 bp of homology downstream of the stop codon of the gD1 ORF. Primers P19 and P20 were used for amplification are listed in Table 1. The resulting PCR product was DpnI digested in order to remove residual template DNA. Next, the transfer fragment was electroporated into arabinose-induced E. coli strain DH10B cells harboring fBoHV-1 BAC and pKD46 (19). Electroporated cells were selected on LB agar plates containing 25 μg/ml kanamycin. The resulting recombinant BoHV1 BAC was designated fBoHV1ΔgDkanR.
DNA preparation from virions.
Herpesviral DNAs were extracted as described previously (23).
Generation of recombinant viruses. (i) rBoHV-5gD1HA.
To generate the gD-exchanged recombinant, BoHV-5 expressing BoHV-1 gD, p302ΔkanR was digested with HpaI, gel purified, and cotransfected with fBoHV-5ΔgDkanR BAC DNA into Vero 2.2 cells by use of Lipofectamine reagent (Invitrogen) as described previously (45). fBoHV-5ΔgDkanR BAC DNA transfected alone did not lead to viral progeny. However, after cotransfection with the purified fragment from p302ΔkanR, rBoHV-5gD1HA emerged and was passaged three times in MDBK cells.
(ii) rrBoHV-5gD5V5.
To generate the BoHV-5 gD revertant, an HpaI-digested and gel-purified fragment derived from p309 was cotransfected with fBoHV-5ΔgDkanR BAC DNA into Vero 2.2 cells as a described above. The resulting recombinant virus progeny, rrBoHV-5gDV5, was passaged three times in MDBK cells.
(iii) rBoHV-1gD5V5.
To generate the gD-exchanged BoHV-1 recombinant expressing BoHV-5 gD (rBoHV-1gD5V5), an HpaI-digested and gel-purified fragment derived from vector p307 and fBoHV-1ΔgDkanR BAC DNA were cotransfected into COS-1 cells as described above. The resulting recombinant virus progeny, rBoHV-1gD5V5, was passaged three times in MDBK cells.
(iv) rrBoHV-1gD1YFP.
To generate the BoHV-1 gD revertant mutant, vector pCS156 was XbaI/EcoRI digested, gel purified, and cotransfected with fBoHV-1ΔgDkanR DNA into MDBK cells as described above. The resulting recombinant virus progeny, rrBoHV-1gD1YFP, was passaged three times in MDBK cells.
Cre-mediated excision of F plasmid in BAC-derived BoHV recombinants.
MBDK cells were cotransfected with the DNA of selected viral mutants and cre-expressing vector p116.006 (21). Three days posttransfection, five viral plaques were randomly collected and plaque purified three times. Finally, viral DNA from non-green fluorescent protein (GFP) fluorescent progeny was extracted and characterized by restriction enzyme analysis in order to verify the deletion of the BAC cassette.
For in vitro experiments, the BAC cassette from rrBoHV-1gD1YFP was deleted by cre-mediated recombination and the selection of GFP-negative, YFP-positive progeny virus. However, the BAC cassette in the other recombinant viruses used in this study was retained in order to have permanent in vitro tracers. For the in vivo experiments performed in this study, the BAC cassette in the rBoHV-1gD5V5 and rBoHV-5gD1HA viruses was deleted in order to obtain virulence close to those of the respective wild-type viruses (data not shown).
Indirect immunofluorescence.
MDBK cells or J1.1-2 cells were infected and incubated until discernible plaques had formed. The cells were then fixed in 3% paraformaldehyde and permeabilized with 0.2% Triton X-100 (Fluka Chemie). After blocking with 2% albumin from bovine serum albumin (BSA) (Sigma-Aldrich), the cells were incubated with mouse monoclonal IgG2a (1:500 in phosphate-buffered saline [PBS]) against either the HA epitope (Santa Cruz Biotechnology) or the V5 epitope (Invitrogen). For visualization, Cy3-conjugated AffiniPure goat anti-mouse IgG was used (Jackson ImmunoResearch Laboratories).
Southern blot hybridization.
Digested virus and BAC DNAs were separated on 0.7% agarose gels for 24 to 48 h at 30 V and then transferred onto a nylon membrane (Roche) overnight. Transferred DNA was then hybridized with gD5-digoxigenin (DIG)- or gD1-DIG-labeled probes overnight at 68°C. For detection by chemiluminescence, CDP-Star substrate (Roche) was used. The gD5-DIG- and gD1-DIG-labeled probes were amplified by PCR (PCR DIG probe synthesis kit; Roche). The primers used for amplification (gD5upper, gD5lower, gD1upper, and gD1lower) are listed in Table 1.
Virus growth analysis.
MDBK cells were infected with the respective viruses at either a multiplicity of infection (MOI) of 0.01 or an MOI of 5. After 2 h at 4°C, the temperature was shifted to 37°C for 1 h to allow virus penetration. The inoculum was removed before the cells were washed twice with PBS and overlaid with fresh DMEM. At 0, 24, 48, 72, and 96 h (MOI of 0.01) and at 0, 6, 12, 18, and 24 h (MOI of 5) following the temperature shift, the infected cells were scraped into the culture medium and clarified by centrifugation (311 × g for 10 min). The supernatant was then removed to be analyzed separately. The cell pellet was resuspended in fresh DMEM. Infectious virus was harvested following three cycles of freezing-thawing and low-speed centrifugation to remove cell debris. Infectivities of the supernatants and the pellets were titrated separately in 96-well plates. Each titration was performed in three independent assays. Viral titers were determined as 50% tissue culture infective doses (TCID50)/ml in MDBK cells.
Soluble receptors.
The following soluble receptors carrying either the entire ectodomain (VCC) or the single N-terminal V domain (V) of human nectins were used as previously described: human nectin 1 [N1(V)-Fc], human nectin 2 [N2(V)-Fc], human nectin 3 [N3(VCC)-Fc], and human nectin 4 [N4(VCC)-Fc] (14, 15, 24, 38). Briefly, the PCR amplification products were cloned into the COS Fc Link vector (SmithKline Beecham) and transfected into COS cells with FuGENE6 reagent according to the manufacturer's instructions. The severe acute respiratory syndrome (SARS) coronavirus S (SARS-S)-Fc construct was a gift from F. Neipel, Erlangen University. The ectodomain of SARS-S was cloned into vector pAB61 (7) and transfected into HEK 293T cells. Soluble proteins were affinity purified from the cell medium with Affigel-protein A or HiTrap-protein A, as detailed and referenced elsewhere previously (7, 14, 37).
Inhibition of infection.
Soluble receptor proteins were incubated with stocks of infectious virus in order to test for the inhibition of infection. Triplicate aliquots of rBoHV-5 and rBoHV-1gD5V5, corresponding to an MOI of 5 for 20,000 MDBK cells, were preincubated individually with each of the soluble receptors at a 200 nM concentration. A total of 200 nM SARS-S spike glycoprotein, 200 nM BSA, or DMEM without added proteins was used as a control. After 1 h of incubation at 37°C, the aliquots were transferred into J cell monolayers in 96-well plates. After 2 h, these inocula were removed, and the cells were supplemented with 100 μl fresh medium before incubation at 37°C for 48 h. The emergence of plaques was recorded, and GFP-positive cells were counted with a fluorescence microscope.
Design of the animal experiments.
Forty-three AR129 mice (8 weeks of age, all female) were divided into groups of 7 to 10 and used for intraperitoneal (i.p.) inoculation with 107 TCID50 of the viruses specified in Table 2. Four siblings of the same mice were used as mock-infected controls.
TABLE 2.
Infection, incubation times, and status at necropsy
| Infection | No. of animals/group | No. of animals (dpi) |
|
|---|---|---|---|
| Diseaseda | Healthyb | ||
| BoHV-1 | 8 | 5 (18, 20, 23)e | 3 (29) |
| rrBoHV-1 | 9 | 3 (21, 25, 29) | 6 (15, 18) |
| rBoHV-1gD5 | 9 | 8 (14, 15, 18, 19, 23) | 1 (26) |
| BoHV-5 | 10 | 10 (11, 12, 13, 14, 16, 19, 21, 28) | 0 |
| rBoHV-5gD1 | 7 | 7 (12, 17) | 0 |
| Mock | 4 | 0 | 4 (17, 29) |
Number of animals with disease signs at the time of necropsy. dpi equals the incubation time.
Number of animals without apparent disease signs before necropsy. The day(s) of necropsy is in parentheses.
Individual mice were euthanized as soon as they developed severe disease symptoms. Exceptions are listed in Table 2. At necropsy, the brain, kidney, liver, spleen, intestine, and lung were collected. Each organ was divided into two parts. One part was fixed with formalin solution in order to be used for hematoxylin and eosin (HE) staining or immunohistochemistry (IHC). The remaining part was shock-frozen in liquid nitrogen and conserved at −80°C for further analysis by real-time PCR.
DNA detection and quantification.
Primers and probes for quantitative real-time PCR (TaqMan) used to amplify sequences within the open reading frames of glycoprotein B of BoHV-1 and BoHV-5, PCR conditions, and usage of plasmid positive controls were previously described (2). For the internal control, the previously developed TaqMan assay reagent for the 18S rRNA kit was used (AB) according to the manufacturer's instructions. The data were analyzed with a 7900 HT Fast real-time system detector (AB). The absolute quantitation of virus copies per cell was done according to AB user bulletin 2 as previously described (2).
Extraction of viral DNA from mouse brains.
The tissue from one half-brain sagittally divided was homogenized by use of a TissueRuptor apparatus (Qiagen), and DNA from 50 mg of brain was extracted by using the QIAamp DNA minikit (Qiagen).
Immunohistology and antigen detection.
Immunohistology was performed with paraffin-embedded brain, kidney, liver, spleen, intestine, and lung tissue sections fixed in formalin solution (4%). Three- to four-micrometer-thick organ sections were mounted on positively charged slides and then deparaffinized in xylene and rehydrated by being dipped into a graded ethanol series (100, 96, and 70%). After counterstaining with hemalaun for 4 min and washing in water at an ambient temperature, the sections were digested with a proteinase K solution (Real proteinase K diluent [catalog number S2032] and 40× concentrate [catalog number S2019]; Dako) for 10 min at room temperature (RT). Afterwards, endogenous peroxidase activity was blocked by incubation in 3% H2O2 for 15 min, and the nonspecific binding of antibodies was eliminated by treatment with blocking solution (Cytomation protein block serum free, catalog number X0909; Dako) for 10 min at RT. The sections were then incubated overnight with monoclonal antibody (mcAb) 141 (specific for gCs of both BoHV-1 and BoHV-5 [35]) at a 1:20 dilution in PBS (pH 8) and were incubated for 30 min with an anti-mouse secondary antibody conjugate containing a dextran polymer labeled with horseradish peroxidase (EnVision K4001; Dako). Incubations were done at room temperature. Between every step, slides were rinsed with PBS (pH 8).
The development of the color reaction by use of the aminoethyl carbazole (AEC) (Red) substrate kit (catalog number 00-2007; Invitrogen), added after washings, was visualized with a microscope and stopped by rinsing with PBS. As a positive control, wt BoHV-1-infected cells were used.
RESULTS
Generation and genotypic characterization of recombinant BoHV-5 and BoHV-1 with intertypic gD.
A two-step strategy was used to exchange the gD-encoding sequences between BoHV-5 and BoHV-1. In a first step, previously described BACs (27) harboring the genomes of either BoHV-1 (fBoHV-1) or BoHV-5 (fBoHV-5) were used to replace the gD-coding sequences with a kanamycin resistance cassette (see http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html).
In a second series of experiments, infectious viruses were rescued by homologous recombination. Notably, gD-less BACs did not provide infectious progeny upon transfection. Therefore, gD-less BAC DNAs were cotransfected with plasmids containing gD sequences supplemented with marker tags and appropriate flanking sequences. As a result, the following four new viruses were generated and characterized by restriction enzyme analysis and Southern blotting as well as by the identification of their markers (see http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html): (i) rBoHV-1gD5V5, a BoHV-1 that encoded gD5 with a V5 tag at its C terminus; (ii) rrBoHV-1gD1YFP, a rescue virus that encoded gD1 with a yellow fluorescent protein fused to its C terminus; (iii) rBoHV-5gD1HA, a BoHV-5 mutant expressing gD1 with an HA tag fused to its C terminus; and (iv) rrBoHV-5gD5V5, a rescue mutant differing from its ancestor by a gD5 fused to a C-terminal V5 tag.
Marker tag analysis of the newly generated viruses.
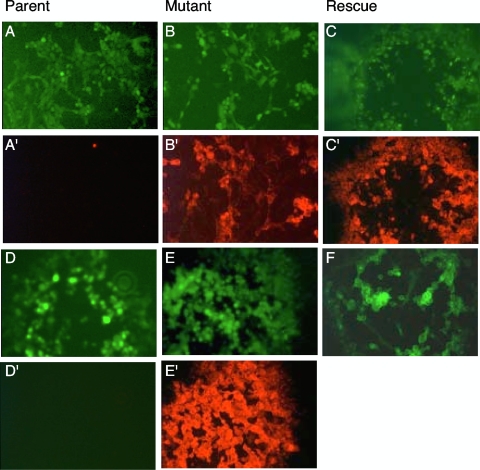
Three of the new viruses carried a gD fused to a C-terminal epitope, which was expected to be recognized by the appropriate monoclonal antibodies. The fourth virus carried a yellow fluorescent protein marker. To test for the presence and functionality of those markers, MDBK cells were infected with parent, mutant, and rescue viruses and analyzed for plaque development by fluorescent microscopy (Fig. 1). Emerging green fluorescence was due to the replication of the viruses and based on the expression of a GFP cassette embedded in the BAC sequences. The replication and plaque formation of those viruses provided evidence that they all contained a functional gD. To avoid interference between yellow and green fluorescence in rrBoHV-1gD1YFP, the GFP cassette was removed from this virus by Cre recombination, and this rescue virus was shown to provide yellow fluorescence in the absence of green fluorescence (Fig. 1F). After successful plaque development, the monolayers were fixed for immunolabeling with the appropriate monoclonal antibodies. As shown in Fig. 1A′ through E′, each virus with a predicted tag did react properly in this assay. Thus, each of the new viruses could be addressed by fluorescence and/or immunohistology (see also http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html).
FIG. 1.
Phenotyping of parent, mutant, and rescue viruses by fluorescence microscopy. MDBK cells were infected with various viruses to be analyzed at 72 hpi. The top two rows show plaques of BAC-derived rBoHV-5 and its mutants, whereas the bottom two rows show BAC-derived rBoHV-1 and its mutants. The top row shows green fluorescent plaques of rBoHV-5 (A), rBoHV-5gD1HA (B), and rrBoHV-5gD5V5 (C). In the second row, the same dishes were stained with mcAbs and a red fluorescent Cy3 conjugate (mcAb against the HA tag [A′ and B′] and mcAb against the V5 tag [C′]). In the third row, the following fluorescent plaques are shown: green fluorescent rBoHV-1 (D), rBoHV-1gD5V5 (E), and yellow fluorescent rrBoHV-1gD1EYFP (F). In the fourth row (D′ and E′), the same viruses as those in the third row were stained with mcAb against V5 and a red fluorescent Cy3 conjugate.
Viral growth analysis.
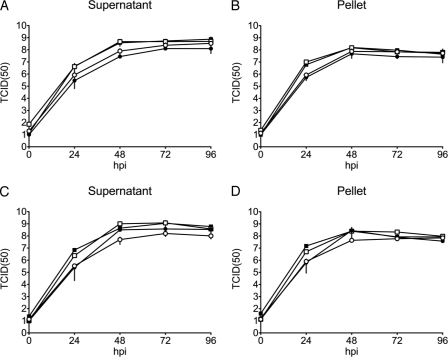
Both cell-free and cell-associated virus productions were determined by growth curve experiments. Each analysis was done in triplicate. The results shown in Fig. 2 indicated that all mutant and revertant viruses grew with similar kinetics and to similar titers as those of their wild-type ancestors. Thus, the growths of the different viruses in MDBK cells were not significantly affected either by the exchange of gD or from retaining the BAC cassettes within the viral genome (see also http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html).
FIG. 2.
Growth kinetics of recombinant BoHV-5 mutants and recombinant BoHV-1 mutants versus wt BoHV-5 and wt BoHV-1. MDBK cells were infected at an MOI of 0.01 with different viruses and harvested at various times postinoculation. The virus yields at each time point were determined by titration. (A and B) rBoHV-5 (full squares), rBoHV-5gD1 (open squares), rBoHV-1 (full circles), and rBoHV-1gD5 (open circles). (C and D) wt BoHV-5 (full squares), rrBoHV-5gD5 (open squares), wt BoHV-1 (full circles), and rrBoHV-1gD1 (open circles). Cell-free virus (supernatant) (A and C) and cell-associated virus (pellets) (B and D) were titrated separately. The x axis represents the time scale postinfection. Virus titers (y axis) are expressed as TCID50/ml. Mean values with standard deviations (error bars) are shown.
Type-specific gD determines the host range of BoHV-1 and -5 in vitro.
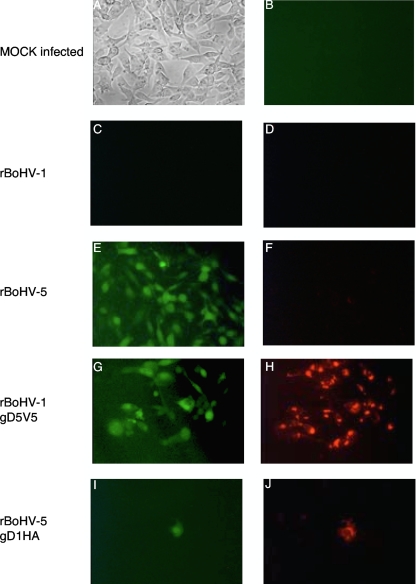
J1.1-2 cells (J cells) are known to be resistant to infection with HSV-1, HSV-2, and BoHV-1 (16). However, we had previously observed that these cells were susceptible to BoHV-5. To test whether this property could be mapped to gD5, we produced viral stocks in MDBK cells and inoculated J cells with the different viruses, using equal infectious doses (MOI of 5) as determined by titration with MDBK cells. The results at 72 hours postinfection (hpi) are shown in Fig. 3. Mock infection or infection with BoHV-1 did not lead to plaque formation at all (Fig. 3A, to C). In contrast, fluorescent plaques developed continuously over time upon the inoculation of J cells with either BoHV-5 or BoHV-1gD5 (Fig. 3E and G). Plaques due to infection with the latter virus could also be stained immunologically by using a monoclonal antibody against the V5 tag (Fig. 3H). Wild-type gD5 (Fig. 3F) did not react with monoclonal antibodies against the HA tag. However, inoculation with BoHV-5gD1 did occasionally lead to the detection of single green fluorescent J cells (Fig. 3I). As expected, such cells could also be stained with a monoclonal antibody against the HA tag (Fig. 3J). However, neither the spread of virus infection to neighboring cells nor plaque formation was observed.
FIG. 3.
Type-specific gD determines the host range of BoHV-1 and BoHV-5. J cells were mock infected (top row) or infected at an MOI of 5 with BoHV recombinants, all of which express the green fluorescent marker upon successful infection and replication. At 72 hpi, cells were fixed and stained with monoclonal antibodies and a secondary antibody carrying Cy3. (A and B) Mock-infected cells neither developed plaques (A) nor provided any green or red fluorescence (B). (C and D) rBoHV-1 neither gave rise to green fluorescence (C) nor provided red fluorescence (D) due to Cy3 conjugate binding to mcAb against V5. (E and F) rBoHV-5 caused the development of green fluorescent plaques (E) but did not bind mcAb against HA (F). (G and H) rBoHV-1gD5V5 caused green fluorescent plaque formation (G) and the binding of mcAb against V5 (H). (I and J) rBoHV-5gD1HA provided single green fluorescent cells (I), which also stained positive with mcAb against the HA tag (J).
In summary, these data strongly indicate that BoHV-5 can use host entry receptors, which are generally not accessible to BoHV-1, and that gD5 contributes a major part to the apparent host range divergence of the two viruses in vitro.
Inhibition of infection by preincubation with soluble nectins.
J cells lack nectin 1, nectin 2, and HevA but express the nectin 3 ortholog of human nectin 3 (15, 16). Since gD5 was able to mediate the infection of J cells with BoHV-5 and rBoHV-1gD5, a question arose regarding whether or not saturating gD5 with the ectodomain of human nectin 3 could block infectivity. Therefore, replicate aliquots of infectious virus stocks were preincubated before the inoculation of J cells with each one of the following soluble receptors: human nectin 1, human nectin 2, human nectin 3, and human nectin 4, respectively (for details see Materials and Methods) (14, 15, 24, 38). Preincubations with equal concentrations of BSA or SARS-S glycoprotein or DMEM without added protein were used as controls. Forty-eight hours after inoculation, the emergence of plaques was recorded, and GFP-positive cells were counted with a fluorescence microscope. The results are summarized in Fig. 4 (see also http://www.vetvir.uzh.ch/aboutus/publikationen/supplements/glycoprotein.html). Human nectin 3 reduced the number of infectious events with rBoHV-5 and rBoHV-1gD5V5 in J cells almost completely. Compared to human nectin 3, human nectin 1 exerted a somewhat lesser effect, whereas human nectins 2 and 4 provided an intermediate effect (80 to 90% reduction). As expected, BSA (in comparison to incubation without protein) affected infectivity only marginally. As a further negative control, we used a totally unrelated protein, the surface protein, S, of severe acute respiratory syndrome coronavirus (SARS-S). According to our experience, the addition of such negative controls may inhibit virus infection by about 25% (14). The addition of SARS-S-Fc reduced virus infectivity between 25% and 40% compared to that with BSA.
FIG. 4.
Inhibition of infection by preincubation with soluble nectins. Relative infectivity rates are shown. Viruses were incubated with soluble proteins as described in Materials and Methods before being inoculated onto monolayers of J cells. Fluorescent cells were counted at 48 hpi. Fluorescent cell numbers emerging from virus incubated with 200 nM BSA were set as 100% (bars measured on the y axis). The identities of the proteins used for coincubation with virus stocks are indicated beneath each pair of bars. Each experiment was done at least in triplicate (mean values are shown, and the error bars represent the standard errors of the means).
Using one-way analysis of variance and either Dunnet's or also Newman-Keuls multiple-comparison test, we found that the difference between BSA and SARS-S was not statistically significant. The inhibition of BoHV-5gD with nectin 1 was significant against BSA (P < 0.05) but not against SARS-S. All other competitors provided significant inhibition compared to either BSA or SARS-S. There was no significant difference between the effects of various nectins. The results indicate that gD5 was able to interact with a variety of soluble human nectins, even though their native counterparts were not expressed in J cells. The differences between nectin 3 on one side and nectins 1, 2, and 4 on the other side may reflect different affinities of gD5 for the various nectins. Altogether, these experiments suggested that gD5 carries binding sites for a number of receptors and their human orthologs. Thus, the gD5 receptor-binding domain(s) seems to act in a rather promiscuous manner, which is surprising compared to data for wt HSV-1 and unrestricted HSV-1 mutants (15).
Infection of AR129 mice.
Having established that rBoHV-1gD5 had an extended host range in vitro, it was of interest to know its properties in vivo. For this purpose, groups of 7 to 10 AR129 mice were inoculated intraperitoneally with 107 TCID50 of either BoHV-5, BoHV-1, rBoHV-1gD5, rBoHV-5gD1, or rrBoHV-1. For animal welfare reasons, all animals were checked twice daily for clinical symptoms and were euthanized upon the development of severe disease. The healthy animals at a given time point postinoculation were considered “survivors.” A chart showing the survival times of the animals in each group is presented in Fig. 5. Both clinically and in terms of survival, the mice could be divided into two different statistically significant groups. Mice inoculated with wt BoHV-5, BoHV-5gD1, and BoHV-1gD5 experienced an early onset of disease and had to be euthanized significantly earlier (mean, 12 to 15 days postinfection [dpi]) than the mice inoculated with either wt BoHV-1 or rrBoHV-1 (mean, 22 to 25 dpi). The calculated P values for significant differences varied between 0.001 and 0.005 (Gehan-Breslow-Wilcoxon test).
FIG. 5.
Survival times of AR129 mice upon infection with different BoHVs. Groups of 7 to 10 mice were inoculated intraperitoneally with 107 TCID50 of virus and checked twice daily for their health status. The mice were euthanized upon the development of severe disease symptoms. The x axis gives the survival times postinoculation. The y axis gives the percentage of mice in each group at any given time point. The symbols used for each virus are given at the right.
Erected fur was the single clinical symptom that was observed for all mice that succumbed to disease. Surprisingly, all diseased mice showed central nervous symptoms, although two different types could be distinguished and grouped with the virus used for inoculation. Diseased mice of three groups (BoHV-1, rrBoHV-1, and rBoHV-1gD5) showed ataxia, weakness in the rear limbs, and hunched back. The mice inoculated with either BoHV-5 or rBoHV-5gD1 showed even more severe neurological symptoms, i.e., stumbling, body tremor, ptosis, and distorted body shape. Only mice inoculated with BoHV-5 also showed body turnover and apparent pain during walking.
Taken together, two surprises came with these experiments: first was the fact that BoHV-1-infected mice developed disease with neurological symptoms, although with a significant delay, and second was the finding that the mice inoculated with BoHV-1gD5 grouped with BoHV-5 in terms of survival time and with BoHV-1 in terms of clinical disease signs.
We conclude from this experiment that BoHV-1gD5 was significantly more virulent in AR129 mice than either its parent, BoHV-1, or its rescue mutant rrBoHV-1gD1. Indeed, it seemed that the BoHV-1 mutant carrying gD5 was almost equally as virulent as the neurotropic virus BoHV-5. However, BoHV-5gD1 was indistinguishable from BoHV-5 in terms of both survival time and type of clinical disease.
Invasion of the brain.
To initially test whether or not the disease signs in the mice could be associated with the ability of the respective viruses to invade the brain, DNA was extracted from brain samples and subjected to real-time PCR analysis. Based on the results (Fig. 6), it was possible to divide the mice into two significantly different groups (P < 0.01 by unpaired t test), those with high yields of viral DNA in the brain (BoHV-5 and rBoHV-5gD1) and those with low or no yields of viral DNA in the brain (BoHV-1, rBoHV-1gD5, and rrBoHV-1gD1). As expected for this type of mouse, no histological signs of encephalitis could be detected in any of the brain samples upon HE staining (Fig. 7).
FIG. 6.
DNA of BoHV-5 and BoHV-5gD1 in brain tissue. DNA was extracted from the brains of each animal and subjected to real-time PCR and quantification of the viral DNA load per cell as described in Materials and Methods. The viruses used for inoculation are listed on the x axis. Box plots refer to the y axis and show mean values, the 75th percentile (box), as well as the maximal and minimal values (whiskers).
FIG. 7.
Histology (HE) and immunohistology of brain sections. (A) HE staining of a section of mouse brain from an animal inoculated with BoHV-1. This picture is representative of all sections analyzed independent of the inoculated virus. (B to D) Immunohistology using a monoclonal antibody recognizing gCs of both BoHV-1 and BoHV-5. (B) Negative brain section of a mouse inoculated with BoHV-1. This picture is representative of all brain sections from mice inoculated with either BoHV-1, BoHV-1gD5, or rrBoHV-1. (C) Representative picture of a brain section from a mouse inoculated with rBoHV-5gD1. This picture is also representative of animals inoculated with BoHV-5. Neurons in the brain stem area show positive intracytoplasmic staining (red cytoplasmic staining). (D) Cutout of the section outlined in C. Bar, 100 μm.
However, upon immunohistological staining using a monoclonal antibody recognizing gCs of both BoHV-1 and BoHV-5, viral antigen could be detected in neurons in 7 out of 10 brain stems of mice inoculated with BoHV-5 and all 7 brains of mice inoculated with BoHV-5gD1 (Fig. 7). In contrast, we did not detect any viral antigen in the brain sections from all the other mice.
We conclude from these experiments that invasion of the brain is a property of BoHV-5 but does not reside within gD5. Indeed, gD5 is apparently nonessential for invasion of the brain.
DISCUSSION
The present study focused on the contributions of the type-specific entry mediator-binding glycoprotein gD, specified by either BoHV-1 or BoHV-5, to the infectivity of cell cultures and virulence in vivo, which led to the following new insights. (i) gD was interchangeable between BoHV-1 and BoHV-5 without significantly affecting the replication properties of the respective viruses in conventional cell cultures. (ii) However, type-specific gD was able to determine the host range of both BoHV-1 and -5. Interestingly, gD1 was associated with a narrow host range, whereas gD5 provided the ability to infect cells that were not accessible through gD1. (iii) To explore this further, viruses carrying gD5 were tested for their abilities to be blocked by a range of soluble receptor molecules, i.e., nectins 1, 2, 3, and 4 or control proteins, such as SARS coronavirus glycoprotein S or BSA. Surprisingly, all four nectins were able to abolish the infectivity of viruses carrying gD5. In contrast, the control protein BSA had almost no effect. Interestingly, the SARS-S glycoprotein was able to interfere with infectivity, at least to a limited extent. Since the SARS-S glycoprotein targets angiotensin-converting enzyme 2 (ACE-2), which is expressed on the surface of kidney cells, we suggest that this interference may be attributed to steric hindrance (34). Thus, gD5 was blockable by an extraordinary broad range of nectins. (iv) In vivo, we found that a BoHV-1 mutant carrying gD5 was almost equally as virulent as the neurotropic BoHV-5. However, the replacement of gD5 by gD1 in BoHV-5 did not reduce the ability of this virus to invade the brain, while the replacement of gD1 with gD5 in BoHV-1 did not enhance the ability to infect secondary neurons.
Among all of the different herpesvirus entry mediators known to date, nectin 1 has been shown to serve as a receptor for BoHV-1 (9, 10, 16-18, 26, 28, 49). In contrast, the potential receptors for BoHV-5 have not yet been determined. This study can only partially help to shed light on this issue. However, the successful inhibition of infectivity with soluble nectins together with our host range studies in vitro suggest that gD5 may bind to and make use of an extremely broad range of receptors compared to its counterparts in other alphaherpesviruses, such as wild-type HSV-1 and unrestricted HSV-1 mutants (15).
The crystal structures of BoHV gDs have not been determined. However, on the basis of overall similarity and the conservation of cysteine residues throughout the molecules, which greatly determine the three-dimensional structure through disulfide bonds, one may assume that the general formation of the BoHV gDs should be similar to that of HSV gDs (10). The binding of HSV gDs to their entry mediators has been mapped predominantly to the N-terminal portion of the molecule, although more C-terminally located fragments also contribute to entry functions (10, 17, 26, 49). Therefore, it is interesting that only few differences between gD1 and gD5, i.e., 6 aa substitutions, map within the first 100 aa from the N terminus of the mature BoHV gD molecules (1; our unpublished data). However, gD5 differs from gD1 in an N-linked glycosylation site (aa 23 to 25 [NYT]). Threonine at position 25 in gD1 is replaced by isoleucine in gD5, which abrogates the glycosylation site (1, 50). We suggest that due to lesser glycosylation, gD5 might have a more flexible three-dimensional structure, which may facilitate binding to a broader range of entry mediators. However, glycosylation may not be the single determinant of the broader host range attributed to gD5. The highest divergence between gD1 and gD5 maps to a fragment between aa 280 and 330 of the molecules, which still belongs to the ectodomain but is located close to the transmembrane region (1). In HSV gD, this fragment (aa 260 to 310) has been referred to as the profusion domain (PFD), which is required for interactions with other viral glycoproteins, i.e., gB, gH, and gL, to proceed with the entry process (26, 29). Interestingly, aa 60 to 210 of HSV gD can be replaced, while the segment at aa 210 to 310 is essential (39). Having this in mind, it is interesting that despite the differences in their PFDs, gD1 and gD5 apparently interacted successfully with their heterologous interaction partners. However, the details of one gD type interacting with the other partners of the entry complex may influence the entry pathway as well as the entire dynamics of viral replication.
The in vivo studies revealed a number of interesting new findings. Based on our previously established model, we inoculated AR129 mice with our new viruses (2). Unexpectedly, almost all of them developed neurological symptoms and had to be euthanized before the planned end of our study. Thus, all of our BAC-derived viruses had retained a significant degree of neurovirulence. However, disease developed early or late, depending on the virus type used for inoculation. Wild-type BoHV-1 and the corresponding rescue mutant showed a delayed onset of disease, whereas wild-type BoHV-5 and BoHV-5gD1 as well as BoHV-1gD5 showed an early onset. Thus, gD5 appeared to increase the virulence of BoHV-1, whereas gD1 did not abrogate the virulence of BoHV-5. Clinically and virologically (PCR and immunohistology), the mice could also be separated into two groups. However, in this case BoHV-1gD5 did not group with BoHV-5 and BoHV-5gD1 but rather grouped with the viruses with BoHV-1 backbones. Only BoHV-5 viruses, regardless of their gD types, were able to invade and replicate in the brain. This observation is in agreement with data from previous reports, which attributed the phenotype of neuroinvasion to gE rather than gD (3, 4, 11-13, 36). However, in the case of BoHVs, the contribution of gD to neuroinvasiveness had not been addressed previously, and it was therefore important to look at it. Nevertheless, these results and considerations cannot explain the occurrence of neurological disease symptoms in mice inoculated with the BoHV-1 backbone. In this context, we accidentally observed that both BoHV-1- and BoHV-5-derived viruses were able to target the adrenal gland of the mice, causing severe necrosis (data not shown). The adrenal medulla consists of differentiated postganglionic cells, which may provide direct access to the CNS (33). Indeed, intraperitoneal inoculation of C3H/HeN mice with HSV-1 leads to severe adrenal necrosis due to viral replication (31, 32). The authors of that study reported that HSV-1 was able to advance from the medullar gland to the spinal cord and brain stem, thus causing neurological symptoms. Presently, we can only speculate that this pathway may also be open to BoHV-1 and BoHV-5, but this issue presents an interesting topic for further research. Future work may also focus on the fine-mapping of the receptor-binding fragment of either gD type and on the identification of the receptor used by gD5, for example, a bovine ortholog of human nectin 3.
Acknowledgments
We thank Eva Loepfe and Kati Zlinszky for excellent technical support and Monika Engels and Sarah Albini for enlightening discussions. We thank F. Neipel, Erlangen University, for the SARS-S-Fc construct.
This work was supported by grant 3100A0-12498 from the Swiss National Science foundation to M.A.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Abdelmagid, O. Y., H. C. Minocha, J. K. Collins, and S. I. Chowdhury. 1995. Fine mapping of bovine herpesvirus-1 (BHV-1) glycoprotein D (gD) neutralizing epitopes by type-specific monoclonal antibodies and sequence comparison with BHV-5 gD. Virology 206:242-253. [DOI] [PubMed] [Google Scholar]
- 2.Abril, C., M. Engels, A. Liman, M. Hilbe, S. Albini, M. Franchini, M. Suter, and M. Ackermann. 2004. Both viral and host factors contribute to neurovirulence of bovine herpesviruses 1 and 5 in interferon receptor-deficient mice. J. Virol. 78:3644-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Mubarak, A., and S. I. Chowdhury. 2004. In the absence of glycoprotein I (gI), gE determines bovine herpesvirus type 5 neuroinvasiveness and neurovirulence. J. Neurovirol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mubarak, A., Y. Zhou, and S. I. Chowdhury. 2004. A glycine-rich bovine herpesvirus 5 (BHV-5) gE-specific epitope within the ectodomain is important for BHV-5 neurovirulence. J. Virol. 78:4806-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashbaugh, S. E., K. E. Thompson, E. B. Belknap, P. C. Schultheiss, S. Chowdhury, and J. K. Collins. 1997. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn. Invest. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 6.Belknap, E. B., J. K. Collins, V. K. Ayers, and P. C. Schultheiss. 1994. Experimental infection of neonatal calves with neurovirulent bovine herpesvirus type 1.3. Vet. Pathol. 31:358-365. [DOI] [PubMed] [Google Scholar]
- 7.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume, G., M. Amasio, E. Avitabile, A. Cerretani, C. Forghieri, T. Gianni, and L. Menotti. 2007. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev. Med. Virol. 17:313-326. [DOI] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 10.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 11.Ch'ng, T. H., P. G. Spear, F. Struyf, and L. W. Enquist. 2007. Glycoprotein D-independent spread of pseudorabies virus infection in cultured peripheral nervous system neurons in a compartmented system. J. Virol. 81:10742-10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdhury, S. I., B. J. Lee, D. Mosier, J. H. Sur, F. A. Osorio, G. Kennedy, and M. L. Weiss. 1997. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. J. Comp. Pathol. 117:295-310. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury, S. I., B. J. Lee, A. Ozkul, and M. L. Weiss. 2000. Bovine herpesvirus 5 glycoprotein E is important for neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J. Virol. 74:2094-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cocchi, F., M. Lopez, L. Menotti, M. Aoubala, P. Dubreuil, and G. Campadelli-Fiume. 1998. The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl. Acad. Sci. U. S. A. 95:15700-15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchi, F., L. Menotti, V. Di Ninni, M. Lopez, and G. Campadelli-Fiume. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J. Virol. 78:4720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly, S. A., D. J. Landsburg, A. Carfi, J. C. Whitbeck, Y. Zuo, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 79:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly, S. A., J. J. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologs in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 19.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delhon, G., M. P. Moraes, Z. Lu, C. L. Afonso, E. F. Flores, R. Weiblen, G. F. Kutish, and D. L. Rock. 2003. Genome of bovine herpesvirus 5. J. Virol. 77:10339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Oliveira, A. P., D. L. Glauser, A. S. Laimbacher, R. Strasser, E. M. Schraner, P. Wild, U. Ziegler, X. O. Breakefield, M. Ackermann, and C. Fraefel. 2008. Live visualization of herpes simplex virus type 1 compartment dynamics. J. Virol. 82:4974-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engels, M., and M. Ackermann. 1996. Pathogenesis of ruminant herpesvirus infections. Vet. Microbiol. 53:3-15. [DOI] [PubMed] [Google Scholar]
- 23.Engels, M., C. Giuliani, P. Wild, T. M. Beck, E. Loepfe, and R. Wyler. 1986. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 6:57-73. [DOI] [PubMed] [Google Scholar]
- 24.Fabre, S., N. Reymond, F. Cocchi, L. Menotti, P. Dubreuil, G. Campadelli-Fiume, and M. Lopez. 2002. Prominent role of the Ig-like V domain in trans-interactions of nectins. Nectin3 and nectin4 bind to the predicted C-C′-C"-D beta-strands of the nectin1 V domain. J. Biol. Chem. 277:27006-27013. [DOI] [PubMed] [Google Scholar]
- 25.Fraefel, C., J. Zeng, Y. Choffat, M. Engels, M. Schwyzer, and M. Ackermann. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 68:3154-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. U. S. A. 102:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabev, E., C. Fraefel, M. Ackermann, and K. Tobler. 2009. Cloning of bovine herpesvirus type 1 and type 5 as infectious bacterial artificial chromosomes. BMC Res. Notes 2:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 29.Gianni, T., M. Amasio, and G. Campadelli-Fiume. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284:17370-17382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grob, P., V. E. Schijns, M. F. van den Broek, S. P. Cox, M. Ackermann, and M. Suter. 1999. Role of the individual interferon systems and specific immunity in mice in controlling systemic dissemination of attenuated pseudorabies virus infection. J. Virol. 73:4748-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irie, H., Y. Harada, E. Kurokawa, M. Saito, Y. Sugawara, H. Ohami, and W. Mori. 1987. Early adrenal infection by herpes simplex virus type-1 (Miyama + GC strain): special reference to inoculation dose and spread from the adrenal to the central nervous system. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 53:325-331. [DOI] [PubMed] [Google Scholar]
- 32.Irie, H., Y. Harada, H. Yoshihashi, T. Kimura, M. Kojima, M. Kataoka, M. Saito, Y. Sugawara, and W. Mori. 1989. Spread of herpes simplex virus type-1 (Miyama +GC strain) to the central nervous system after intraperitoneal inoculation: the role of the myenteric plexus of the gut. Arch. Virol. 105:247-257. [DOI] [PubMed] [Google Scholar]
- 33.Irie, H., W. Mori, Y. Harada, E. Kurokawa, M. Yamada, and S. Nii. 1986. Replication of herpes simples virus in the adrenal of mouse. Igakuno Ayumi 137:309-310. [Google Scholar]
- 34.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liman, A., M. Engels, G. Meyer, and M. Ackermann. 2000. Glycoprotein C of bovine herpesvirus 5 (BHV-5) confers a distinct heparin-binding phenotype to BHV-1. Arch. Virol. 145:2047-2059. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Z. F., M. C. Brum, A. Doster, C. Jones, and S. I. Chowdhury. 2008. A bovine herpesvirus type 1 mutant virus specifying a carboxyl-terminal truncation of glycoprotein E is defective in anterograde neuronal transport in rabbits and calves. J. Virol. 82:7432-7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez, M., M. Aoubala, F. Jordier, D. Isnardon, S. Gomez, and P. Dubreuil. 1998. The human poliovirus receptor related 2 protein is a new hematopoietic/endothelial homophilic adhesion molecule. Blood 92:4602-4611. [PubMed] [Google Scholar]
- 38.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2alpha (PRR2alpha or HveB) and nectin2delta are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menotti, L., G. Nicoletti, V. Gatta, S. Croci, L. Landuzzi, C. De Giovanni, P. Nanni, P. L. Lollini, and G. Campadelli-Fiume. 2009. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl. Acad. Sci. U. S. A. 106:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzler, A. E., A. A. Schudel, and M. Engels. 1986. Bovine herpesvirus 1: molecular and antigenic characteristics of variant viruses isolated from calves with neurological disease. Arch. Virol. 87:205-217. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, G., M. Lemaire, J. Lyaku, P. P. Pastoret, and E. Thiry. 1996. Establishment of a rabbit model for bovine herpesvirus type 5 neurological acute infection. Vet. Microbiol. 51:27-40. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 43.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 44.Rock, D. L., W. A. Hagemoser, F. A. Osorio, and D. E. Reed. 1986. Detection of bovine herpesvirus type 1 RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J. Gen. Virol. 67:2515-2520. [DOI] [PubMed] [Google Scholar]
- 45.Saeki, Y., C. Fraefel, T. Ichikawa, X. O. Breakefield, and E. A. Chiocca. 2001. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol. Ther. 3:591-601. [DOI] [PubMed] [Google Scholar]
- 46.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99: 13-22. [DOI] [PubMed] [Google Scholar]
- 47.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 186:74-86. [DOI] [PubMed] [Google Scholar]
- 48.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 49.Spear, P. G., S. Manoj, M. Yoon, C. R. Jogger, A. Zago, and D. Myscofski. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 344:17-24. [DOI] [PubMed] [Google Scholar]
- 50.Tikoo, S. K., M. D. Parker, J. V. van den Hurk, J. Kowalski, T. J. Zamb, and L. A. Babiuk. 1993. Role of N-linked glycans in antigenicity, processing, and cell surface expression of bovine herpesvirus 1 glycoprotein gIV. J. Virol. 67:726-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]