Abstract
The initial wave of swine-origin influenza A virus (pandemic H1N1/09) in the United States during the spring and summer of 2009 also resulted in an increased vigilance and sampling of seasonal influenza viruses (H1N1 and H3N2), even though they are normally characterized by very low incidence outside of the winter months. To explore the nature of virus evolution during this influenza “off-season,” we conducted a phylogenetic analysis of H1N1 and H3N2 sequences sampled during April to June 2009 in New York State. Our analysis revealed that multiple lineages of both viruses were introduced and cocirculated during this time, as is typical of influenza virus during the winter. Strikingly, however, we also found strong evidence for the presence of a large transmission chain of H3N2 viruses centered on the south-east of New York State and which continued until at least 1 June 2009. These results suggest that the unseasonal transmission of influenza A viruses may be more widespread than is usually supposed.
The recent emergence of swine-origin H1N1 influenza A virus (pandemic H1N1/09) in humans has heightened awareness of how the burden of morbidity and mortality due to influenza is associated with the appearance of new genetic variants (5) and of the genetic and epidemiological determinants of viral transmission (8). The emergence of pandemic H1N1/09 is also unprecedented in recorded history as it means that three antigenically distinct lineages of influenza A virus—pandemic H1N1/09 and the seasonal H1N1 and H3N2 viruses— currently cocirculate within human populations.
Although the presence of multiple subtypes of influenza A virus may place an additional burden on public health resources, it also provides a unique opportunity to compare the patterns and dynamics of evolution in these viruses on a similar time scale. Indeed, one of the most interesting secondary effects of the current H1N1/09 pandemic has been an increased vigilance for cases of influenza-like illness and hence an intensified sampling of seasonal H1N1 and H3N2 viruses during the typical influenza “off-season” (i.e., spring-summer) in the northern hemisphere. Because the influenza season in the northern hemisphere generally runs from November through March, with a usual peak in January or February, influenza viruses sampled outside of this period are of special interest.
The current model for the global spatiotemporal dynamics of influenza A virus is that the northern and southern hemispheres represent ecological “sinks” for this virus, with little ongoing viral transmission during the summer months (9). In contrast, more continual viral transmission occurs within the tropical “source” population (13) that is most likely centered on an intense transmission network in east and southeast Asia (10). However, the precise epidemiological and evolutionary reasons for this major geographic division, and for the seasonality of influenza A virus in general, remain uncertain (1, 4). Evidence for this “sink-source” ecological model is that viruses sampled from successive seasons in localities such as New York State do not usually form linked clusters on phylogenetic trees, indicating that they are not connected by direct transmission through the summer months (7). Similar conclusions can be drawn for the United States as a whole and point to multiple introductions of phylogenetically distinct lineages during the winter (6), followed by complex patterns of spatial diffusion (14). However, despite the growing epidemiological and phylogenetic data supporting this model, it is also evident that there is relatively little sequence data from seasonal influenza viruses that are sampled from April to October in the northern hemisphere. Hence, it is uncertain whether extended chains of transmission can occur during this time period, even though this may have an important bearing on our understanding of influenza seasonality.
To address these issues, we examined the evolutionary behavior of seasonal H1N1 and H3N2 viruses as they cocirculated during a single time period—(late) April to June 2009—within a single locality (New York State). Not only are levels of influenza virus transmission in the northern hemisphere usually very low during this time period, but in this particular season the human host population was also experiencing the emerging epidemic of pandemic H1N1/09.
MATERIALS AND METHODS
Sequence data.
The New York State sequence data for the seasonal specimens were identified by the influenza virus surveillance program and the genome amplified directly from the swab specimen (16) at the Wadsworth Center, New York State Department of Health. Genome sequence data was generated at the J. Craig Venter Institute for the Influenza Genome Sequencing Project and are freely available on GenBank from where they were downloaded. A number of procedures were used to minimize the possibility of contamination. Portions of primary clinical specimens for molecular analysis, including those for whole-genome sequencing, were divided into aliquots and lysed in an accessioning room located far from the culture facility, and forwarded directly to the extraction laboratory. Once there, they were handled in a separate biosafety cabinet from cultured isolates and extracted on automated nucleic acid extractors dedicated to primary samples that are never used to extract cultured isolates. In addition, negative extraction controls were included on every run to check for carryover of contaminating material, and the negative extraction controls contained carrier nucleic acid to ensure that even extremely low levels of contamination would be carried through and be detectable in the subsequent assay. The entire workflow is unidirectional and staff were not permitted to return to the accessioning or other ‘clean’ areas. For the multisegment reverse transcription-PCR amplification, the RNA was isolated in clean rooms and using equipment dedicated to preamplification procedures. No template controls were included in both the RNA extraction and genomic amplification procedures to ensure that amplification was specific to the respective specimens.
For H1N1 we obtained seven new sequences sampled between 27 April and 26 May 2009 from five different counties in New York State. For H3N2, a larger sample of 41 viruses was obtained between 27 April and 1 June 2009 and representing 20 different counties, although predominantly from the southeast of the state (Duchess, Nassau, Orange, Putnam, Ulster, and Westchester counties). These sequences were compared to all those global H1N1 and H3N2 sequences available on GenBank and covering the years 2008 and 2009 (although most of the “background” viruses are also sampled from the United States) and for which the exact day of sampling was available. Because of the very large number of background HA1 and NA gene sequences available for these viruses, which are therefore more representative, our analyses focused on these gene segments. This resulted in data sets of the following size: seasonal H1N1 HA1 = 652 sequences, 1,050 nucleotides (nt); NA = 365 sequences, 1,410 nt; H3N2 HA1 = 1,258 sequences, 1,040 nt; and NA = 375 sequences, 1,407 nt. Because of the lack of insertions and deletions all sequence alignments were undertaken by hand using the SE-AL program (kindly distributed by Andrew Rambaut).
Phylogenetic analysis.
Phylogenetic trees were first inferred using the maximum-likelihood (ML) method available in the PHYML package (2) and using SPR branch-swapping. In all cases, the GTR+Γ4 model of nucleotide substitution was utilized. We also performed PHYML analyses using the simpler HKY85+Γ4 substitution model and recovered tree topologies that contained the same major groupings with respect to the New York State viruses (which are the only phylogenetic patterns of interest in the present study). To further assess the robustness of our analyses, we undertook an additional phylogenetic analysis utilizing the ML approach available in the GARLI package (15), adjusting the run length parameter (to 100,000 steps) to improve the search quality. This analysis again made use of the GTR+Γ4 substitution model. Because of the similarity of the resulting phylogenies, only those generated by PHYML are shown here. Finally, for all data sets, a bootstrap resampling procedure was undertaken using 1,000 replicate neighbor-joining trees and the parameter settings for the GTR+Γ4 model of nucleotide substitution determined in PHYML. This analysis was undertaken using the PAUP* package (12). All relevant parameter values for the analyses described above are available from the authors on request.
RESULTS AND DISCUSSION
Seasonal H1N1.
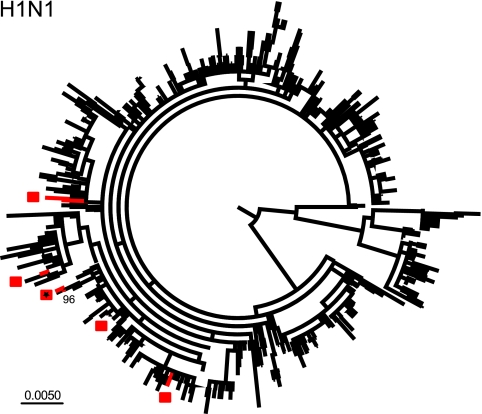
Because of the very small size of the seasonal H1N1 data set sampled from New York State (n = 7) from April to May 2009, few strong conclusions could be drawn on the overall molecular epidemiology of this virus in this population. However, it is clear that all but two of the HA1 sequences from New York State fall into disparate locations on the phylogenetic tree, suggesting that they represent independent entries into this population and for which there has likely been no significant onward transmission (Fig. 1). A similar pattern is observed in the NA phylogeny (see Fig. S1 in the supplemental material). The interesting exception are two viruses (A/New York/3768/2009 and A/New York/3467/2009) sampled on 13 and 26 May from the adjacent Steuben and Ontario counties, respectively, and which form a monophyletic group in the NA phylogeny and which are closely related in the HA1 tree (forming a clade with A/Massachusetts/4/2009 sampled on 27 January 2009; Fig. 1, asterisk). Such close clustering is compatible with the hypothesis that these viruses are linked by a direct chain of transmission. However, it is also noteworthy that these viruses are characterized by relatively long branch lengths (in marked contrast to the H3N2 transmission cluster described below), which may be indicative of multiple unsampled links within the New York State transmission chain, independent entries from other locations, or greater antigenic diversity which may in turn increase the possibility of transmission. Similarly, it is notable that all seven newly sampled New York State viruses are closely related to viruses sampled in the early months (January to March) of 2009 in the United States, most notably the isolate from Massachusetts. It is therefore possible that despite their diverse phylogenetic locations, these viruses have also been transmitted within the greater U.S. population to at least the end of April. Clearly, a greater sampling of viruses from multiple locations is needed to confirm this hypothesis.
FIG. 1.
ML tree (unrooted) of 652 HA1 sequences of seasonal H1N1 virus sampled globally during the period from 2008 to 2009. The seven viruses sampled from New York State during the period 27 April to 26 May are shaded red, with each likely introduction event marked by a square. A possible transmission cluster is denoted with an asterisk; the bootstrap value for this node is also shown.
Seasonal H3N2.
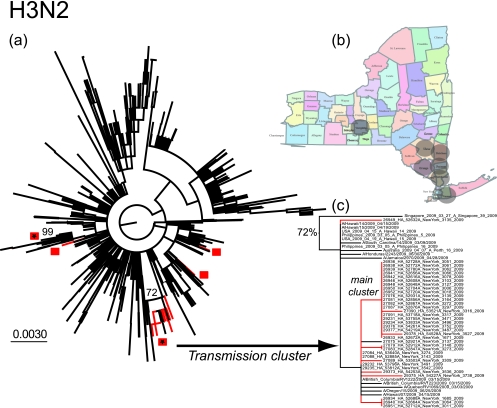
As with H1N1, our analysis of the H3N2 virus population from April to June 2009 reveals that there have again been multiple entries of the virus into New York State, reflected in the presence of distinct clusters of viruses on both the HA1 (Fig. 2) and NA (see Fig. S2 in the supplemental material) phylogenies. The most striking result, however, is the presence of a cluster of 21 very closely related viruses that was sampled over a time period of 5 weeks from 27 April to 1 June 2009 and characterized by very short branch lengths (including predominantly identical sequences). That all but one of these viruses was sampled from six adjacent counties in the most populous south-east region of New York State strongly suggests that they have been sampled from a direct chain of transmission (Fig. 2b and c). Importantly, this putative transmission cluster was observed in both the HA and the NA phylogenies. In addition, the strict quality controls in place at the Wadsworth Center and the fact that members of this cluster were extracted at different times makes cross-contamination highly unlikely. That a single member of this cluster was sampled from Tompkins county in central New York State suggests that this transmission chain may extend even further in time and space. Indeed, that an additional 13 viruses fall very close to the transmission cluster in all phylogenies, being separated from the main cluster by other U.S. viruses (Fig. 2), supports the notion that this transmission chain is widespread. A second possible transmission cluster concerns two viruses (A/New York/3104/2009 and A/New York/3280/2009) sampled on 29 April and 3 May in Wyoming and Orange counties, respectively. Although this cluster contains only two isolates, and from counties that do not share a border, the sequences in question are identical, which again suggests that they form part of a larger transmission chain.
FIG. 2.
(a) ML tree (unrooted) of 1,258 HA1 sequences of seasonal H3N2 sampled globally during the period from 2008 to 2009. The 41 viruses sampled from New York State during the period 27 April to 1 June are shaded red, with each likely introduction event marked by a square. Possible transmission clusters are noted by asterisks, and their associated bootstrap values are also shown. Although a bootstrap value <70% is observed for the main transmission cluster, this group of sequences is strongly supported (0.84) under the approximate likelihood ratio test available in PHYML. (b) Counties of New York State from where the putative H3N2 transmission cluster was sampled (shaded circles). (c) Magnification of the 21-sequence cluster that provides strong evidence for the unseasonal transmission of H3N2.
The intense interest in pandemic H1N1/09 has ignited an extensive program of surveillance for influenza-like illness and genome sequencing. This has enabled us to detect, for the first time, the extended transmission of seasonal influenza A virus during the influenza “off-season” in the northern hemisphere. Although most lineages of influenza virus undoubtedly die-off during the summer months, so that the influenza epidemics that occur during any specific season are most likely caused by the seeding, each winter, of viruses ultimately derived from a global reservoir population, our study clearly shows that the onset of spring in the northern hemisphere does not necessarily spell the immediate end of virus transmission: that at least 21 viruses (and likely many more) sampled across 5 weeks in seven counties form a tight monophyletic group in the H3N2 phylogenies represents strong evidence that influenza A virus has been transmitted in the population for at least this period. In addition, our phylogenetic analysis reveals that multiple lineages of H1N1 and H3N2 also circulate during the influenza off-season, again illustrating how influenza A virus is easily able to exploit human contact networks. The remaining question is whether such unseasonal transmission will produce a chain long enough that it enables the virus to persist from epidemic to epidemic; that is, to survive the entire summer period? From the large-scale phylogenetic data presented to date there is no strong evidence for such extended transmission, as lineages of viruses sampled from successive influenza seasons tend not to cluster together (7).
More generally, the observation of the unseasonal transmission of H3N2 (and perhaps H1N1) may have implications for our understanding of the causes of seasonality in influenza virus. In particular, that the virus is able to establish transmission networks in the warmer and more humid months of late April to June in New York State indicates that the virus is able to spread in what may be suboptimal climatic conditions (3, 11) if a sufficient number and density of susceptible hosts is available. Although the spring and summer of 2009 was highly unusual in that H1N1/09 as well as seasonal influenza viruses cocirculated in the U.S. population, summer waves of influenza virus transmission are often observed during pandemics. We therefore suggest that the more intensive sampling of influenza virus genetic diversity during the summer months in the northern hemisphere may represent a useful additional way to explore the issues that underpin the distinctive seasonality of influenza A virus.
Supplementary Material
Acknowledgments
This study was in part funded by NIH grant GM080533 and NIH contract HHSN272200900007C. D.E.W. is funded in part by NIH/NIAID P01 AI059576-05 and the New York State Department of Health. K.S.G. is supported in part by the New York State Department of Health.
Footnotes
Published ahead of print on 17 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Dowell, S. F. 2001. Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg. Infect. Dis. 7:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Mol. Biol. Evol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 3.Lowen, A. C., S. Mubareka, J. Steel, and P. Palese. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 3:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowen, A. C., and P. Palese. 2009. Transmission of influenza virus in temperate zones is predominantly by aerosol, in the tropics by contact: a hypothesis. PLoS Curr. Influenza 2009 Aug 17. RRN1002. [DOI] [PMC free article] [PubMed]
- 5.Morens, D. M., J. K. Taubenberger, and A. S. Fauci. 2009. The persistent legacy of the 1918 influenza virus. N. Engl. J. Med. 361:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson, M. I., L. Edelman, D. J. Spiro, A. R. Boyne, J. Bera, R. Halpin, E. Ghedin, M. A. Miller, L. Simonsen, C. Viboud, and E. C. Holmes. 2008. Molecular epidemiology of H3N2 and H1N1 influenza virus during a single epidemic season in the United States. PLoS Pathog. 4:e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson, M. I., L. Simonsen, C. Viboud, M. A. Miller, J. Taylor, K. St. George, S. B. Griesemer, E. Ghedin, N. A. Sengamalay, D. J. Spiro, I. Volkov, B. T. Grenfell, D. J. Lipman, J. K. Taubenberger, and E. C. Holmes. 2006. Stochastic processes are key determinants of the short-term evolution of influenza A virus. PLoS Pathog. 2:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann, G., T. Noda, and Y. Kawaoka. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambaut, A., O. G. Pybus, M. I. Nelson, C. Viboud, J. K. Taubenberger, and E. C. Holmes. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell, C. A., T. C. Jones, I. G. Barr, N. J. Cox, R. J. Garten, V. Gregory, I. D. Gust, A. W. Hampson, A. J. Hay, A. C. Hurt, J. C. de Jong, A. Kelso, A. I. Klimov, T. Kageyama, N. Komadina, A. S. Lapedes, Y. P. Lin, A. Mosterin, M. Obuchi, T. Odagiri, A. D. Osterhaus, G. F. Rimmelzwaan, M. W. Shaw, E. Skepner, K. Stohr, M. Tashiro, R. A. Fouchier, and D. J. Smith. 2008. The global circulation of seasonal influenza A (H3N2) viruses. Science 320:340-346. [DOI] [PubMed] [Google Scholar]
- 11.Shaman, J., V. Pitzer, C. Viboud, M. Lipsitch, and B. T. Grenfell. 2009. Absolute humidity and the seasonal onset of influenza in the continental US. PLoS Curr. Influenza. 2009 Dec 17. RRN1136. [DOI] [PMC free article] [PubMed]
- 12.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates. Sunderland, MA.
- 13.Viboud, C., W. J. Alonso, and L. Simonsen. 2006. Influenza in tropical regions. PLoS Med. 3:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viboud, C., O. N. Bjornstad, D. L. Smith, L. Simonsen, M. A. Miller, and B. T. Grenfell. 2006. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312:447-451. [DOI] [PubMed] [Google Scholar]
- 15.Zwickl, D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum-likelihood criterion. Ph.D. dissertation. University of Texas, Austin.
- 16.Zhou, B., M. E. Donnelly, D. T. Scholes, K. St. George, M. Hatta, Y. Kawaoka, and D. E. Wentworth. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza A viruses. J. Virol. 83:10309-10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.