Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is a B-lymphotropic herpesvirus strongly linked to both lymphoproliferative diseases and Kaposi's sarcoma. The viral latency program of KSHV is central to persistent infection and plays important roles in the pathogenesis of KSHV-related tumors. Up to six polypeptides and 18 microRNAs are known to be expressed in latency, but it is unclear if all major latency genes have been identified. Here, we have employed array-based transcript profiling and limiting-dilution reverse transcription-PCR (RT-PCR) methodologies to explore this issue in several KSHV-infected cell lines. Our results show that RNAs encoding the K1 protein are found at low levels in most latently infected cell lines. The gene encoding v-IL-6 is also expressed as a latent transcript in some contexts. Both genes encode powerful signaling molecules with particular relevance to B cell biology: K1 mimics signaling through the B cell receptor, and v-IL-6 promotes B cell survival. These data resolve earlier controversies about K1 and v-IL-6 expression and indicate that, in addition to core latency genes, some transcripts can be expressed in KSHV latency in a context-dependent manner.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8, is a B-lymphotropic herpesvirus that is strongly linked to several human tumors (see reference 25 for a review). These include Kaposi's sarcoma (KS), which is an endothelial neoplasm (9), and tumors associated with two B-lymphoproliferative diseases, primary effusion lymphoma (PEL) (7) and multicentric Castleman's disease (MCD) (57). Like all herpesviruses, KSHV has two distinct genetic programs, latency and lytic replication. In latency, viral gene expression is heavily restricted, with only a few KSHV genes being transcribed; the viral genome is maintained as a nuclear plasmid, no viral progeny are produced, and the host cell survives. In contrast, in lytic infection all the viral genes are expressed, in a temporally regulated cascade. Viral DNA is extensively replicated, and progeny virions are assembled and released, with concomitant death of the host cell. Because they retain the full complement of viral genes, cells that are latently infected can “switch” to the lytic program upon receipt of appropriate stimuli.
Both cycles are essential to viral propagation and maintenance in the population. Lytic replication, of course, generates the virus that is required for spread from cell to cell and person to person. Latency ensures viral persistence in infected hosts, providing a lifetime of opportunities for lytic reactivation and spread. In addition, latency is thought to play an important role in herpesviral tumorigenesis. In Epstein-Barr virus (EBV), the latency program is strongly immortalizing and plays a key role in triggering EBV-related lymphomas (33). In KSHV, although latency is not powerfully immortalizing in vitro, the latency program is on in all PEL cells and the vast majority of endothelial tumor cells (“spindle cells”) in advanced KS lesions (16, 54, 59). One latent viral protein, v-FLIP, is critical for the survival of PEL cells (28) and for the characteristic spindle-like morphology of KS tumor cells (27), via upregulation of NF-κB (12, 22). Other latent proteins include (i) LANA, known to be important for viral genomic persistence (1, 2, 14), for upregulation of ß-catenin (24), and for inhibition of p53 (23) and Rb (50), (ii) v-Cyc, a viral homolog of cyclin D (10, 41), and (iii) kaposins A, B, and C, a family of polypeptides that are active in signal transduction (43, 45, 52). In B cells (but not in endothelial cells), another protein, v-IRF3 (aka LANA-2), is also produced in latency and inhibits induction of type I interferons and the function of p53 (51). Twelve pre-microRNAs (pre-miRNAs) are also encoded in the latency program; these are processed to generate 18 mature miRNAs (5, 53, 61).
Although there is general agreement about the assignment of the above-mentioned genes to the latency program (17), there has been considerable debate about whether additional loci might also be expressed in latency. Particular controversy has surrounded 3 loci: K1, K2 (v-IL-6), and K15. K1 and K15 aroused early interest because both are transmembrane signaling molecules that are located at the left and right hand termini (respectively) of the linear viral genome (32). In both herpesvirus saimiri (HVS) and EBV, genes at these genomic positions encode important signaling molecules that have been shown to have a role in lymphoproliferation (15, 32), and in EBV, both of the syntenic loci (LMP1 and LMP2) are known latent genes (33). LMP2 and K1 both mimic aspects of signaling through the B cell antigen receptor (15, 36, 39), and LMP1 and K15 both bind TNF receptor-associated factors (TRAFs) and activate NF-κB (4, 64). However, in many KSHV-infected cell lines, K1 is expressed at very low levels and is also clearly induced during lytic replication (35). Since most latently infected populations harbor a variable subpopulation of lytically induced cells, it has been difficult to determine if the occasional detection of low levels of K1 mRNA in uninduced populations is due to authentic latent expression or to the subpopulation of lytically infected cells; in recent years, the consensus in the field has favored the latter interpretation. K15, once named LAMP (for latency-associated membrane protein), is also currently held to be a lytic gene for similar reasons. The KSHV K2 gene encodes v-IL-6, a viral homolog of interleukin-6 (IL-6), which is a major proinflammatory cytokine with additional important effects on B cell survival and growth (11). Like K15, the K2 gene is strongly induced during lytic KSHV replication and is present at low levels in uninduced cell lines (46). Some analyses (6, 30, 48) have favored classifying it as a lytic gene, while others (47) suggest that it can be expressed in some PEL cells that do not stain for lytic markers, though these cells represented only 5% of all LANA+ cells. Most recent reviews, however, have not included v-IL-6 on the list of known latent genes (19, 25, 29, 31).
Here, we have revisited the question of which viral genes are expressed in latency, using several different approaches. First, we generated latently infected cells by KSHV infection of several different cell lines, including cells of endothelial and mesenchymal lineages. Two of these lines displayed very low levels of spontaneous lytic reactivation; array-based transcript profiling in these cells revealed consistent expression of the K1 gene. One of two stably latent lines also expresses transcripts from both strands of the K2/v-IL-6 gene in uninduced cells. Expression of these genes in latency was confirmed by a limiting-dilution reverse transcription-PCR (RT-PCR) assay. These results resolve earlier controversies about the expression of the K1 gene and clearly indicate that v-IL-6 can also be expressed in latency in some contexts. Moreover, they affirm that the latency program, while containing a set of core genes whose expression is invariant, can also include other genes whose expression depends upon the cellular context.
MATERIALS AND METHODS
Tissue culture, virus preparation, and infection.
HFF (human foreskin fibroblast) cells were obtained from the ATCC (CRL-2522) and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). SLK cells were cultured in DMEM with 10% FBS. BJAB and BCBL-1 cells were cultured in RPMI medium supplemented with 10% FBS, 55uM beta-mercaptoethanol, and 2 mM glutamine. TIME cells were cultured as previously described (62). Stable SLK.219 and HFF.219 cells were constructed by infecting SLK cells with rKSHV.219 produced from Vero cells harboring the recombinant virus (courtesy of J. Vieira) and SLK.219 cells, respectively. SLK.219 cells were induced to enter lytic replication with 1.2 mM valproic acid (VA) and AdRTA (adenovirus harboring the ORF50 cDNA).
Flow cytometry.
SLK and TIME cells were subjected to flow cytometric analysis to detect expression of green fluorescent protein (GFP) and red fluorescent protein (RFP). Cells were analyzed using a FACSCalibur (BD Biosciences), and the resulting data were analyzed using FlowJo. Cells were prepared for flow cytometric analysis by trypsinization to harvest the cells, followed by a 5′ fixation period with 2% paraformaldehyde.
RNA preparation.
HFF and SLK cell samples were harvested in RLT buffer, and RNA was isolated using an RNeasy minikit according to the manufacturer's protocol (Qiagen). BJAB and BCBL-1 samples were harvested in RNA-Bee solution, and RNA was isolated according to the manufacturer's protocol (Tel-Test, Inc.). BJAB and BCBL-1 RNAs that were subject to microarray analysis were then repurified using the RNeasy minikit according to the manufacturer's protocol (Qiagen); RNAs that were subjected to Northern analysis were used without further purification. Poly(A)-enriched RNAs were made using an Oligotex mRNA midi kit according to the manufacturer's protocol (Qiagen).
KSHV tiling microarray construction, sample labeling, and microarray hybridization.
The KSHV genome sequence used for the microarray design was obtained from GenBank accession number U75698.1. Thirteen thousand seven hundred forty-six 60-mer KSHV-specific oligonucleotide probes were printed in duplicate using Agilent technology. Each probe overlapped a neighboring probe by 40 nucleotides (nt), resulting in a tiling design with probes offset by 20 nucleotides. Probes were designed from sequences of both strands of the genome. The custom tiling microarray also contained control probes (approximately 1,500) and probes detecting host transcripts (approximately 16,311). This custom KSHV tiling microarray can be obtained from Agilent using the custom design identification number 017577.
Total RNA from the experimental samples was purified as indicated above. The reference RNA was a mixture of RNAs of both infected and uninfected cells, including TIME, HUVEC, BJAB, BCBL-1, and SLK cells. The integrity of the RNAs was analyzed using a model 2100 Bioanalyzer (Agilent), or the RNAs were analyzed using conventional formaldehyde-agarose gel electrophoresis and staining. RNAs were quantified using an ND1000 spectrophotometer (Nanodrop). A Quick Amp labeling kit (Agilent) was used according to the manufacturer's protocol to generate labeled cRNA from 450 ng of total RNA. Experimental samples were labeled with Cy5, and reference samples were labeled with Cy3 and then competitively hybridized to the custom KSHV tiling microarray according to the manufacturer's protocol for the whole human genome oligonucleotide microarrays (Agilent). Hybridized microarrays were washed according to the manufacturer's protocol (Agilent) and scanned with a GenePix 4000B scanner (Axon Instruments) and all feature intensities collected using GenePix Pro 6.0 software. TIFF images of scanned slides were analyzed using Feature Extraction Software, version 9.5.3 (Agilent), using a custom grid file (017577_D_F_20070822). All reported raw microarray data are MIAME (minimum information about a microarray experiment) compliant and are stored in NCBI's Gene Expression Omnibus (GEO) (21), accessible through GEO accession number GSE20443 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20443). Processed data are also provided as information file S1 (best viewed with Java Treeview) in the supplemental material.
Cell dilution and qRT-PCR.
Duplicate 2-fold serial dilutions of SLK.219 cells were made, starting with approximately 1,500 cells in the 1st dilution and approximately 3 cells in the 10th dilution. The cell dilutions were deposited in individual wells of a 96-well tissue culture plate. Three hours after plating, cells were gently washed with phosphate-buffered saline (PBS) and then lysed with Cells-to-CT lysis buffer (AM1724; Ambion) according to the manufacturer's protocol. The lysis reactions were stopped by the addition of stop buffer containing a control RNA, XenoRNA (4386996; Ambion). RNA lysates from each dilution were subjected to gene-specific RT using a high-capacity cDNA reverse transcription kit (4374966; Applied Biosystems). cDNAs were used as templates for TaqMan quantitative PCR (qPCR) analysis. (The sequences of the gene-specific RT primers and the primer-probe sets [used for qPCR] are provided in information file S2 in the supplemental material.) If a threshold cycle (CT) value for a specific gene could not be determined for either of the duplicate qPCRs, the transcript was considered below the detection limit. All data are presented as 40 minus the average CT value.
Northern blotting.
Poly(A)-enriched RNAs (200 ng) were resolved by formaldehyde-agarose gel electrophoresis in MOPS (morpholinepropanesulfonic acid) buffer. Resolved RNAs were transferred to a Nytran SuPerCharge membrane according to the Turboblotter manufacturer's protocol (Whatman, Schleicher & Schuell). Membranes were UV cross-linked using a UV Stratalinker 2400 (Stratagene). Membranes were prehybridized using ULTRAhyb buffer (Ambion) at 65°C for more than 1 h. [32P]UTP-labeled riboprobes were generated by performing T3 or T7 in vitro transcription reactions (Ambion), using PCR templates. PCR products, using T3/T7-linked primers spanning specific regions of the KSHV genome (K1 primers [TAATACGACTCACTATAGGGAATGCATCCTTGCCAATATCC and AATTAACCCTCACTAAAGGGATTTCATTTCGTCCGTTTGGT] and v-IL-6 primers [TAATACGACTCACTATAGGGACCCGCAGTGATCAGTATCGT and AATTAACCCTCACTAAAGGGATAGTGTATGCCGCGTTAGCA]), served as templates for these transcription reactions. Labeled riboprobes were hybridized to the membranes in ULTRAhyb buffer at 65°C for approximately 16 h. Membranes were first washed two times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS at 65°C for 5 min. Then, the membranes were washed in a high-stringency buffer (0.2× SSC, 0.1% SDS) for various amounts of time at 65°C. The length of time was empirically determined for each probe such that the wash conditions resulted in minimal nonspecific hybridization. Blots were exposed to phosphor storage screens (GE Healthcare). Screens were analyzed using a Storm 860 scanner (Molecular Dynamics).
3′ RACE.
3′ rapid amplification of cDNA ends (3′ RACE) analysis for determination of the 3′ end of K1 was performed using a RLM-RACE kit according to the manufacturer's protocol (Ambion). 3′ RACE was performed on poly(A)-enriched RNA from BCBL-1, BCBL-1 induced with 600 μM valproic acid, and iSLK.219 induced with doxycycline for 48 h. The 5′ upstream-gene-specific primer used for PCR was SC-535 (TTGCCCATTGTCAAAAACAA). All the reactions revealed the same splice and 3′ end, indicated in Fig. 5; however, the genomic sequences for the K1-ORF4 region are different in KSHV of BCBL-1 and rKSHV.219 (a derivative of KSHV of JSC-1 cells). To simplify reporting of the newly identified splice site and 3′ end (which are identical in BCBL-1 cells and in iSLK.219 cells), these important sequence elements are transposed on the KSHV sequence (GenBank accession number U75698.1) and presented in Fig. 5.
FIG. 5.
K1 transcript structure. (A) The K1 transcript structure is based on RT-PCR and 3′ RACE analysis performed in this study and 5′ RACE analysis performed by Bowser et al. (3). (B) The nucleotide positions of important structural elements include the 5′ nucleotide of the transcript (nt 29), the splice donor site (nt 981), the splice acceptor site (nt 2625), the predicted poly(A) signal (nt 2937 to 2942), the site of polyadenylation (nt 2972), and the coding region (nt 105 to 971). All coordinates are based on the KSHV genome sequence (GenBank accession number U75698.1).
RESULTS
Generation of stably infected cells with low spontaneous-reactivation rate.
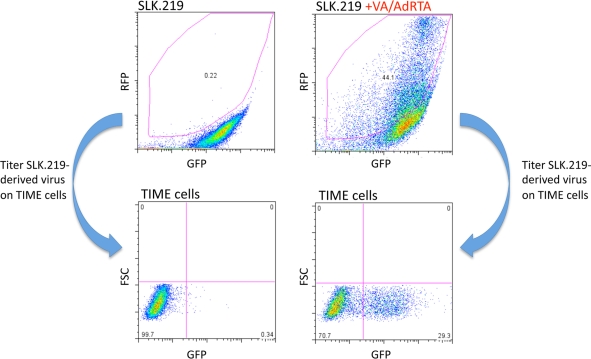
The central difficulty in determining the latency status of genes that are also induced during lytic replication is contamination of the pool of uninduced RNAs with transcripts emanating from the subpopulation of cells undergoing spontaneous lytic reactivation. To bypass this problem, we examined two cell lines that we empirically discovered to display very low levels of spontaneous lytic reactivation upon KSHV infection: human foreskin fibroblasts (HFF) and SLK endothelial cells. We derived stably latent cell lines from each cell type by infection with rKSHV.219, a recombinant KSHV-bearing a puromycin resistance locus, which allowed selection of puromycin-resistant mass cultures (63). In addition, rKSHV.219 harbors a constitutively expressed GFP gene as well as an RFP locus under the control of the lytic PAN promoter. Cells latently infected by rKSHV.219 are GFP positive but lack RFP expression; when lytic replication is induced, productively induced cells rapidly become RFP+.
Both lines display very low levels of RFP spontaneously; Fig. 1 shows the data for SLK.219 cells. As can be seen, only 0.22% of uninduced SLK.219 cells displayed RFP expression as determined by flow cytometry, and culture supernatants from these cells had virtually no infectivity (as revealed by the ability to transduce GFP expression to naïve TIME cells) (Fig. 1). Following lytic induction (here, by exposure to valproate and an adenovirus expressing the KSHV lytic switch protein, RTA), 44% of the cells expressed RFP and supernatants of these cells efficiently transduced the GFP marker into naïve TIME cells. HFF cells had similarly low levels of basal RFP expression but also generated infectious virus upon induction (data not shown). This indicates that the strict restriction on lytic gene expression in the ground state was not the result of any defect in these cells but a reflection of authentic, reversible latency.
FIG. 1.
SLK.219 cells produce infectious virus. SLK.219 cells were either mock treated or induced to enter lytic replication with 1.2 mM valproic acid (VA) and AdRTA (adenovirus harboring the ORF50 cDNA). Reactivated cells express RFP. Extent of reactivation was measured using fluorescence-activated cell sorting (FACS) analysis. Only VA/AdRTA-treated SLK.219 cells display substantial reactivation. Virus in the supernatants of the mock- and VA/AdRTA-treated SLK.219 cells was concentrated by centrifugation, resuspended in TIME cell medium, and then used to infect TIME cells. The rKSHV.219 virus harbors a constitutively expressed GFP gene. Infected TIME cells are detected by measuring GFP fluorescence after receiving SLK.219-derived virus.
Transcriptome analysis by tiling microarray.
To examine the patterns of KSHV gene expression in these latent populations, we constructed a custom DNA array that tiles across the KSHV genome using synthetic 60-mer oligonucleotides, each oligonucleotide offset from its neighbor by a 20-bp interval (S. Chandriani, Y. Xu, and D. Ganem, submitted for publication). The arrayed oligonucleotides span the entire 137.5 kb of unique genomic DNA but do not include the terminal repeats, which are 85% G+C and are therefore likely to hybridize nonspecifically to many sequences. Tiling oligonucleotides were derived from the sequences of both strands of the KSHV genome. Total RNA from each sample was labeled using the Eberwine labeling protocol, which results in Cy5 (or Cy3)-labeled, linearly amplified cRNA that is hybridized to the tiling microarray (20). Figure 2 shows a semiquantitative depiction of the resulting hybridization profiles. For reference, the hybridization pattern of uninduced BCBL-1 cells is shown (Fig. 2A). BCBL-1 is a latently infected B cell line derived from primary effusion lymphoma (PEL) cells; such PEL lines have been the backbone of most prior studies of latent KSHV gene expression. However, they have substantial rates of spontaneous lytic reactivation (2 to 4%), and the resulting lytic infection also releases substantially more virus than most other infected cell types. As shown in Fig. 2A, uninduced BCBL-1 cells display an extensive pattern of transcripts emanating from many regions of both strands of the viral genome. This pattern is derived from lytically infected cells contaminating the prep (Chandriani et al., submitted, and data not shown), illustrating how difficult it is to identify latent genes in such a context. In contrast, the hybridization pattern from SLK/rKSHV.219 (Fig. 2B) or HFF/rKSHV.219 (Fig. 2C) was much more restricted. In both rKSHV-infected lines, in addition to clear expression of the major latency locus (MLL; open reading frames [ORFs] 71 to 73), strong expression of the coding strand of ORF K1 and ORF4 is evident. Several additional active regions are evident in SLK/rKSHV.219 cells but not HFF/rKSHV.219 cells; these regions map to the sense and antisense strands of ORF K2 (the gene for v-IL-6). Conversely, hybridization to oligonucleotides from ORF75 was detected in latently infected HFF cells but not SLK/rKSHV.219 cells.
FIG. 2.
KSHV transcriptome analysis during latent infection. KSHV tiling microarray data (ordered by genome position) are displayed for 13,746 unique probes. In each group of arrays (A, B, C), the color bar indicates the fold change relative to the level for the appropriate mock-infected cells. (A) Viral gene expression analysis of uninfected BJAB and uninduced BCBL-1 cells. (B) Viral expression analysis of stably infected SLK.219 cells and the parental SLK cells. (C) Viral expression analysis of stably infected HFF.219 and the parental HFF cells. The asterisk indicates a series of neighboring probes that detect a transcript antisense to ORF K9 that is likely a spurious transcript (see text for details).
We did not detect a clear signal from ORF K15 in either uninduced line. However, we note an important caveat in interpreting this negative result: the tiling microarray employed in this study was generated using the M-type KSHV sequence, while the strains that were studied were P type. As there is extensive sequence divergence between the M-type and P-type strains in the region to the right of ORF75, which includes K15, the absence of signal here is not conclusive evidence of lack of K15 expression (26, 49). In fact, we observed poor hybridization to K15 oligonucleotides even after induction (not shown), reaffirming that this divergence compromises our ability to detect K15 transcripts.
Finally, both lines display strong expression of an RNA complementary to ORF K9, the coding region for v-IRF1, a lytic protein involved in the inhibition of interferon induction. This antisense hybridization signal is denoted by an asterisk in Fig. 2. However, this transcript was not seen in latency in any cell line (including SLK) infected with wild-type, PEL-derived KSHV stocks. On closer inspection, we noted that the K9 region is directly adjacent to the puromycin/GFP/RFP insertion in rKSHV.219; we infer that the K9 antisense transcript is most likely an artifact of this insertion. Accordingly, we have not characterized this transcript further.
It is noteworthy that SLK and HFF are not the only lines displaying ORF K1-ORF4 RNAs in the absence of induction. Such RNAs are also prominent in uninduced BCBL-1 cells (Fig. 2) as well as in HUVEC and TIME endothelial cells by 24 h after infection with KSHV (Chandriani et al., submitted). In all three cases, many other transcripts are also evident, reflective of ongoing lytic reactivation, but importantly, in no population of uninduced KSHV-infected cells are ORF K1-ORF4 RNAs absent.
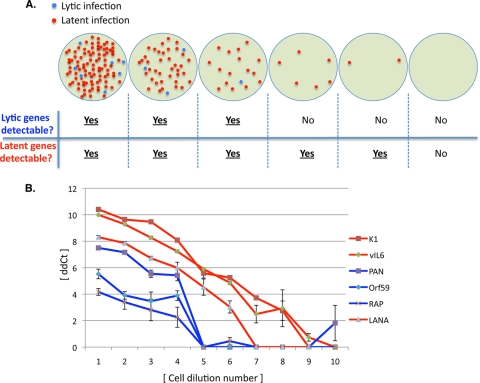
Limiting-dilution RT-PCR strategy for identifying latent versus lytic RNAs.
To confirm that the RNAs visualized in SLK cells in Fig. 2 are indeed latently expressed, we examined a subset of these RNAs in a different assay, based on limiting-dilution RT-PCR. The principle of this assay is outlined in Fig. 3A. This principle relies on the fact that in latently infected cells, only a small subpopulation of cells spontaneously enters the lytic cycle. Accordingly, when such cells are serially diluted, lytically infected cells are lost earlier in the dilution series than are latently infected cells. Therefore, if RNA is prepared from each dilution sample and examined by RT-PCR, signals from known lytic RNAs should disappear at earlier dilutions than signals from latent RNAs. Of course, such an assay can be affected by several variables, such as the abundance of the given RNA and the efficiency with which it can be amplified. To make certain that we accurately identify lytic RNAs in this analysis, we employed PCR primer sets for three different genes (PAN, ORF K8/RAP, and ORF59). PAN is a particularly useful marker of lytic cells because it is so abundantly expressed (∼105 copies/cell) (56); we also explicitly verified that the PAN primers we selected were as efficient for PCR amplification as those we chose for the LANA gene, the canonical latency marker (see Fig. S1 in the supplemental material). Each RT-PCR test was done using quantitative, real-time PCR, and CT values were obtained for each sample (see Materials and Methods).
FIG. 3.
Detection of latent transcripts by use of serial cell dilution analysis. (A) Schematic of the method for evaluating latent viral gene expression. Uninduced, KSHV-infected cultures often harbor a small subpopulation of spontaneously reactivated cells. When such cultures are serially diluted, large dilutions are likely to generate some purely latently infected group of cells. qRT-PCR can be performed with each dilution to detect lytic and latent genes. Therefore, the ability to detect lytically expressed genes in a dilution series will end before the ability to detect latently expressed genes. (B) The 2-fold cell dilution series was generated on uninduced SLK.219 cells, ranging from dilution 1 (with approximately 1,500 cells) to dilution 10 (with approximately 3 cells). Six viral RNAs were analyzed by qRT-PCR with each dilution, and the results of the analysis are presented as 40 minus the CT value. Three classic lytic genes are indicated by the blue curves. LANA, a known latently expressed gene, is indicated by the blue curve. The remaining two genes (K1 and v-IL-6) are likely expressed during latent infection, as they are detectable in cell dilutions in which lytic genes are not.
Figure 3B shows the results of this experiment, which was done with SLK/rKSHV.219 cells. As can be seen, quantitative RT-PCR (qRT-PCR) signals from the PAN gene decay more rapidly than those from LANA, disappearing by the 5th serial 2-fold dilution. The same is also true for the two other lytic genes selected for examination, K8/RAP and ORF59. In contrast, amplicons from the K1 and K2/v-IL-6 regions displayed the pattern expected for latent genes, similar to what was found for the LANA control. These data corroborate the array profiling data and reinforce the conclusion that, in these cells, both transcripts are behaving as latent gene products.
To further explore the expression of these RNAs, we examined polyadenylated RNA from BCBL1 and iSLK.219 cells, pre- and postinduction, by Northern blotting with strand-specific probes for ORF K1 (Fig. 4A) or ORF K2/v-IL-6 (Fig. 4B). BCBL1 cells were induced with valproic acid. iSLK.219 cells are SLK/rKSHV.219 cells that also harbor a doxycycline-inducible RTA gene. (RTA is the KSHV transcription factor that is responsible for the switch to lytic replication [42, 60].) As such, lytic replication in these cells can be induced with doxycycline. As can be seen, K1 transcripts were not clearly detectable prior to induction in iSLK.219 cells but accumulated steadily postinduction (Fig. 4A), similar to what we had previously observed in BCBL-1 cells (35). As expected from earlier expression studies (30, 46), the same was true for v-IL-6 transcripts in iSLK.219 (Fig. 4B) (and for RNAs antisense to v-IL-6) (Chandriani et al., submitted). In contrast, low levels of v-IL-6 mRNA were detectable in BCBL-1 cells preinduction, likely due to the higher frequency of spontaneous lytic reactivation in these cells. Thus, SLK/rKSHV.219 cells do not differ importantly from PEL cells or other latently infected cells in terms of the levels of K1 and v-IL-6 transcripts in latency. Rather, they differ primarily in their low background level of lytic reactivation, which allows the use of sensitive methods, like linear amplification/array hybridization, to detect them without the accompanying noise from lytic transcripts.
FIG. 4.
Northern analysis of K1 and v-IL-6. Poly(A)-enriched RNAs from indicated cells were resolved by formaldehyde-agarose gel electrophoresis, transferred to nylon membranes, and hybridized to radioactively labeled riboprobes that detect K1 (A) and v-IL-6 (B) transcripts.
The fine structure of latent K1 mRNA.
The 5′ end of K1 mRNAs has been mapped by Bowser and colleagues (3); however, the rest of the mRNA has not been characterized. Because of the potential of ORF K1 to influence signaling in latency, we considered it important to make certain that this ORF was not disrupted by RNA splicing or editing. Accordingly, we amplified cDNA from the coding region of ORF K1 from uninduced SLK/rKSHV.219 cells by RT-PCR with several primer sets and sequenced the corresponding cDNA fragments. This established that the entire coding region is unspliced and unedited. Because we could identify no canonical poly(A) addition signal in the immediate 3′ noncoding region of ORF K1, and because numerous oligonucleotides from the ORF4 locus were also recognized on the array (Fig. 2), we wondered if this transcript proceeded into the downstream ORF4 locus, which does harbor a poly(A) signal. To examine the 3′ portion of the transcript, we carried out a 3′ RACE analysis using an anchored oligo(dT) primer and an upstream primer from within the K1 coding region. Sequencing of the principal product of this reaction (Fig. 5B) revealed that this RNA is spliced, from a 5′ splice donor site just distal to the UGA stop codon of K1 to a 3′ splice acceptor in the distal ORF4 gene; the resulting intron is a canonical one with GT…AG termini. (This 3′ splice acceptor site is also used for intragenic splicing within ORF4 [58].) This experiment also mapped the poly(A) addition site to position 2972, some 30 nt 3′ to a noncanonical ACUAAA poly(A) addition signal. [According to prior mutational analyses, this variant poly(A) signal functions at ca. 10% efficiency relative to that of the canonical AAUAAA signal (55), which may provide another reason why this transcript is so nonabundant during infection.] The fine structure of K1 mRNA is schematized in Fig. 5A. The presence of some ORF4 sequences in this RNA likely accounts for some of the ORF4 hybridization signal observed in Fig. 2. However, we also note that in the K1 Northern blot shown in Fig. 4A, a second, larger transcript is also observed; this species could be a bicistronic RNA, bearing intact both K1 and ORF4 sequences, polyadenylated at this site. Certainly, such an RNA could contribute as well to the ORF4 signal seen in Fig. 2. However, because the ORF4 gene is 3′ to K1 in such an RNA, this is unlikely to generate ORF4 protein (a complement control protein [58]) in latency unless an internal ribosome entry site (IRES) is located in the intergenic region.
DISCUSSION
These results settle a longstanding controversy concerning whether the K1 and K2 (v-IL-6) genes can be expressed in latency. We find ORF K1 to be expressed at low levels in virtually all uninduced cell lines in culture. (It may be no accident that latent expression of the K1 gene is so low level, as overexpression of K1 has deleterious consequences for cell biology, often being linked to growth deregulation and tumor formation [18, 40].) Clearly, the viral IL-6 locus can also be expressed at low levels in latency, but its expression in this context is not invariant; for example, it is expressed in SLK but is not detectable in uninduced HFF cells. Transcripts from ORF75 display the converse pattern, on in HFF and off in SLK. Our ability to unambiguously detect these RNAs in latency is attributable to several factors. First, and most obvious, is the development of cell lines with extremely low background levels of spontaneous reactivation. Second, the use of linear amplification prior to labeling and hybridization doubtlessly increases the sensitivity of our microarray analysis over the sensitivities of earlier studies based primarily on Northern blotting of unamplified RNA. Finally, the limiting-dilution assay whose results are shown in Fig. 3 employs exponential amplification by RT-PCR, making it extremely sensitive for transcript detection; in this case, latent RNAs are distinguished from lytic ones by the removal of lytically infected cells from the tested cell population by dilution.
Although both K1 and v-IL-6 mRNAs in latency are extremely nonabundant, their products are powerful signaling molecules; even low-level expression is likely to be sufficient to influence cell physiology. Given that KSHV is a lymphotropic herpesvirus, it is interesting that these two genes both affect signaling pathways of great importance to B cell biology. For example, K1 protein bears a cytosolic ITAM motif and mimics signaling through the B-cell receptor (BCR), activating tyrosine kinases Lyn and Syk, as well as PLC-gamma2 and vav, but does not depend upon exogenous ligands to do so (36, 39). The consequences of this signaling include activation of cytosolic calcium fluxes, phosphorylation of Akt, activation of AP-1 and NFAT transcription factors, and upregulation of numerous cytokines, chemokines, and growth factors, including IL-1, IL-8, Rantes, and vascular endothelial growth factor (VEGF) (37). Such cytokines could contribute not only to the biology of lymphatic infection by KSHV but also to the development of Kaposi's sarcoma, which is also characterized by an inflammatory microenvironment (25). Moreover, the links between K1 expression, Akt activation, and VEGF production may have direct relevance for KS generation, since it has been shown that ectopic expression of K1 can substantially extend the survival of primary endothelial cells (65). In the same study, Wang et al. (65) detected K1 protein expression (by immunohistochemistry [IHC]) in some KS tumors that lacked late lytic transcripts for ORF26, which is consistent with our finding that K1 can be expressed in latency.
Our finding that K1 is expressed in latency may also help to explain an earlier, seemingly paradoxical result reported by Lee et al. (38). They observed that when K1-based constructs were exogenously expressed in naïve BJAB cells, strong signaling was observed but that when the same K1 constructs were transfected into PEL cells, signaling (as judged by Ca2+ transients or tyrosine phosphorylation) was greatly reduced. Since PEL cells are tonically exposed to latent K1 expression, it is likely that this result reflects desensitization of the signaling pathway by prior, chronic K1 signaling.
Our analysis cannot determine whether K15 transcripts are expressed in latency, because of the divergence between the M-type sequences on the array and the P-type sequences in our RNA samples. We note with interest that in the infected HFF cell line, ORF75 transcripts were detected in uninduced cells; since some K15 RNAs can read through into this region (26), it is conceivable that this signal reflects latent K15 transcription. However, definitive resolution of this question will require more experimentation with a redesigned array.
The expression of v-IL-6 in some latency contexts is also of considerable interest. This protein is homologous to host IL-6, a proinflammatory cytokine that is also a potent stimulator of B cell survival and differentiation (34), but the two proteins display some interesting differences. Human IL-6 acts via signaling through a heterodimer of gp130 (the signaling subunit) and p80, the high-affinity IL-6 binding subunit (34). In contrast, v-IL-6 does not require the high-affinity p80 subunit, being able to signal via interaction with gp130 alone (44). A second important difference is that v-IL-6 is very poorly secreted from cells, being mostly retained in the endoplasmic reticulum of expressing cells (13). So, unlike for cellular IL-6, which acts efficiently in a paracrine manner, much of v-IL-6's action appears to be autocrine. We speculate that latent expression of v-IL-6 in KSHV's B cell reservoir extends the survival of these cells and promotes viral persistence in vivo. Consistent with this, Chen et al. recently observed that PEL cells transduced with shRNAs directed against v-IL-6 displayed reduced growth rates and enhanced apoptosis (13). Such a result would not be anticipated if v-IL-6 were solely a lytic gene, unless it had residual paracrine activity. The findings of Chen et al. and our finding that many latent cells can express the protein, taken together, are most consistent with the idea that low-level v-IL-6 production in latency promotes cell-autonomous B cell survival in PEL cells.
Our finding that v-IL-6 transcripts are not invariably associated with latency but are on in only some cell lines suggests that in addition to a core latency program, KSHV may harbor a repertoire of viral genes that are expressed only for selected cell types or conditions. We are aware of two other observations with KSHV that are consistent with this idea. First, v-IRF3, also called LANA-2, is expressed in PEL cells but not latently infected KS spindle cells (51); we also failed to detect it in latently infected SLK and HFF cells (Fig. 2). Second, agonism of the Notch signaling pathway in latently infected PEL cells can induce expression of selected viral genes in the absence of lytic induction, effectively creating an expanded repertoire of latency-like transcripts (8). Moreover, in EBV infection, at least three different programs of latency are described, each of which is characterized by expression of a distinct, though overlapping, set of latent transcripts. The idea that latency has some plasticity, while not prominently emphasized at the textbook level, is entirely in keeping with what is known about mammalian gene expression. After all, in latency, the nuclear viral episome is transcribed primarily by the host transcriptional machinery, like any other piece of host chromatin. It would be surprising indeed if a 165-kb segment of chromatinized DNA could respond to different signals or environments in only one of two ways, i.e., by either expressing a fixed subset of latency genes or turning on the entire lytic program. Just how much plasticity the KSHV genome will support is unknown, but the fact that significant differences emerge even on analysis of a small set of cell lines suggests that the virus's capacity for variegation in latency may have been substantially underestimated.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute.
We thank Jinjong Myoung for supplying iSLK.293 cells.
Footnotes
Published ahead of print on 10 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowser, B. S., S. M. DeWire, and B. Damania. 2002. Transcriptional regulation of the K1 gene product of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:12574-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkmann, M. M., M. Pietrek, O. Dittrich-Breiholz, M. Kracht, and T. F. Schulz. 2007. Modulation of host gene expression by the K15 protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 81:42-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. S., J. Nicholas, J. M. Orenstein, R. B. Mann, P. G. Murray, P. J. Browning, J. A. DiGiuseppe, E. Cesarman, G. S. Hayward, and R. F. Ambinder. 1999. Heterogeneity of viral IL-6 expression in HHV-8-associated diseases. J. Infect. Dis. 180:824-828. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H., D. P. Dittmer, Y. C. Shin, Y. Hong, and J. U. Jung. 2005. Role of Notch signal transduction in Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 79:14371-14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhary, P. M., A. Jasmin, M. T. Eby, and L. Hood. 1999. Modulation of the NF-kappa B pathway by virally encoded death effector domains-containing proteins. Oncogene 18:5738-5746. [DOI] [PubMed] [Google Scholar]
- 13.Chen, D., G. Sandford, and J. Nicholas. 2009. Intracellular signaling mechanisms and activities of human herpesvirus 8 interleukin-6. J. Virol. 83:722-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 15.Damania, B., J. K. Choi, and J. U. Jung. 2000. Signaling activities of gammaherpesvirus membrane proteins. J. Virol. 74:1593-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmer, D. P. 2003. Transcription profile of Kaposi's sarcoma-associated herpesvirus in primary Kaposi's sarcoma lesions as determined by real-time PCR arrays. Cancer Res. 63:2010-2015. [PubMed] [Google Scholar]
- 18.Douglas, J., B. Dutia, S. Rhind, J. P. Stewart, and S. J. Talbot. 2004. Expression in a recombinant murid herpesvirus 4 reveals the in vivo transforming potential of the K1 open reading frame of Kaposi's sarcoma-associated herpesvirus. J. Virol. 78:8878-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberwine, J. 1996. Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. Biotechniques 20:584-591. [DOI] [PubMed] [Google Scholar]
- 21.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field, N., W. Low, M. Daniels, S. Howell, L. Daviet, C. Boshoff, and M. Collins. 2003. KSHV vFLIP binds to IKK-gamma to activate IKK. J. Cell Sci. 116:3721-3728. [DOI] [PubMed] [Google Scholar]
- 23.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 24.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 25.Ganem, D. 2007. Kaposi's sarcoma-associated herpesvirus, p. 2847-2888. In D. Knipe and P. Howley (ed.), Fields virology, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 26.Glenn, M., L. Rainbow, F. Aurade, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossmann, C., S. Podgrabinska, M. Skobe, and D. Ganem. 2006. Activation of NF-{kappa}B by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J. Virol. 80:7179-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199:993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Horenstein, M. G., N. J. Moontasri, and E. Cesarman. 2008. The pathobiology of Kaposi's sarcoma: advances since the onset of the AIDS epidemic. J. Cutan. Pathol. 35(Suppl. 2):40-44. [DOI] [PubMed] [Google Scholar]
- 30.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 32.Jung, J. U., J. K. Choi, A. Ensser, and B. Biesinger. 1999. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin. Cancer Biol. 9:231-239. [DOI] [PubMed] [Google Scholar]
- 33.Kieff, E., and A. Rickinson. 2007. EBV and its replication, p. 2603-2654. In D. Knipe and P. Howley (ed.), Fields virology, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA.
- 34.Kishimoto, T. 2006. Interleukin-6: discovery of a pleiotropic cytokine. Arthritis Res. Ther. 8(Suppl. 2):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagunoff, M., and D. Ganem. 1997. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology 236:147-154. [DOI] [PubMed] [Google Scholar]
- 36.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, B. S., S. H. Lee, P. Feng, H. Chang, N. H. Cho, and J. U. Jung. 2005. Characterization of the Kaposi's sarcoma-associated herpesvirus K1 signalosome. J. Virol. 79:12173-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, B. S., M. Paulose-Murphy, Y. H. Chung, M. Connlole, S. Zeichner, and J. U. Jung. 2002. Suppression of tetradecanoyl phorbol acetate-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus by K1 signal transduction. J. Virol. 76:12185-12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, H., J. Guo, M. Li, J. K. Choi, M. DeMaria, M. Rosenzweig, and J. U. Jung. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol. Cell. Biol. 18:5219-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, H., R. Veazey, K. Williams, M. Li, J. Guo, F. Neipel, B. Fleckenstein, A. Lackner, R. C. Desrosiers, and J. U. Jung. 1998. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat. Med. 4:435-440. [DOI] [PubMed] [Google Scholar]
- 41.Li, M., H. Lee, D. W. Yoon, J. C. Albrecht, B. Fleckenstein, F. Neipel, and J. U. Jung. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 71:1984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 43.McCormick, C., and D. Ganem. 2005. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 307:739-741. [DOI] [PubMed] [Google Scholar]
- 44.Molden, J., Y. Chang, Y. You, P. S. Moore, and M. A. Goldsmith. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625-19631. [DOI] [PubMed] [Google Scholar]
- 45.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H. G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 47.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 51.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheets, M. D., S. C. Ogg, and M. P. Wickens. 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18:5799-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 58.Spiller, O. B., M. Robinson, E. O'Donnell, S. Milligan, B. P. Morgan, A. J. Davison, and D. J. Blackbourn. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 77:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umbach, J. L., and B. R. Cullen. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venetsanakos, E., A. Mirza, C. Fanton, S. R. Romanov, T. Tlsty, and M. McMahon. 2002. Induction of tubulogenesis in telomerase-immortalized human microvascular endothelial cells by glioblastoma cells. Exp. Cell Res. 273:21-33. [DOI] [PubMed] [Google Scholar]
- 63.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225-240. [DOI] [PubMed] [Google Scholar]
- 64.Wang, L., M. M. Brinkmann, M. Pietrek, M. Ottinger, O. Dittrich-Breiholz, M. Kracht, and T. F. Schulz. 2007. Functional characterization of the M-type K15-encoded membrane protein of Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 88:1698-1707. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., D. P. Dittmer, C. C. Tomlinson, F. D. Fakhari, and B. Damania. 2006. Immortalization of primary endothelial cells by the K1 protein of Kaposi's sarcoma-associated herpesvirus. Cancer Res. 66:3658-3666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.