Abstract
HIV-1 escape mutants are well known to be selected by immune pressure via HIV-1-specific cytotoxic T lymphocytes (CTLs) and neutralizing antibodies. The ability of the CTLs to suppress HIV-1 replication is assumed to be associated with the selection of escape mutants from the CTLs. Therefore, we first investigated the correlation between the ability of HLA-A*1101-restricted CTLs recognizing immunodominant epitopes in vitro and the selection of escape mutants. The result showed that there was no correlation between the ability of these CTLs to suppress HIV-1 replication in vitro and the appearance of escape mutants. The CTLs that had a strong ability to suppress HIV-1 replication in vitro but failed to select escape mutants expressed a higher level of PD-1 in vivo, whereas those that had a strong ability to suppress HIV-1 replication in vitro and selected escape mutants expressed a low level of PD-1. Ex vivo analysis of these CTLs revealed that the latter CTLs had a significantly stronger ability to recognize the epitope than the former ones. These results suggest that escape mutations are selected by HIV-1-specific CTLs that have a stronger ability to recognize HIV-1 in vivo but not in vitro.
HIV-1-specific cytotoxic T lymphocytes (CTLs) have an important role in the control of HIV-1 replication during acute and chronic phases of an HIV-1 infection (5, 28, 33). On the other hand, HIV-1 can escape from the host immune system by various mechanisms. These may include the appearance of HIV-1 carrying escape mutations in its immunodominant CTL epitopes as well as Nef-mediated downregulation of HLA class I molecules. There is a growing body of evidence for the former mechanism, i.e., that CTLs targeting immunodominant HIV-1 epitopes select escape mutants in chronically HIV-1-infected individuals (18, 20, 36), whereas the latter mechanism was proved by demonstrating that HIV-1-specific CTLs fail to kill Nef-positive-HIV-1-infected CD4+ T cells but effectively kill Nef-defective-HIV-1-infected ones or that they suppress the replication of Nef-defective HIV-1 much more than that of Nef-positive HIV-1 (12, 13, 42, 45).
It is speculated that HIV-1 immunodominant epitope-specific CTLs have the ability to suppress HIV-1 replication and effectively select escape mutants. However, the correlation between this ability of the CTLs and the appearance of escape mutants is still unclear, because it is not easy to evaluate the ability of HIV-1-specific CTLs to exert a strong immune pressure in vivo. To examine this ability, most previous studies measured the number of HIV-1-specific CTLs or CD8+ T cells and the CTL activity against target cells prepulsed with the epitope peptide or those infected with HIV-1 recombinant vaccinia virus (6, 7, 23, 46). However, the results obtained from such experiments do not reflect the ability of the CTLs to exert immune pressure in vivo. We and other groups previously utilized an assay to directly evaluate the ability of the CTLs to suppress HIV-1 replication in vitro (1, 17, 18, 42, 43). This assay may be better for evaluation of immune pressure by HIV-1-specific CTLs than other assays, because the ability of the CTLs to suppress HIV-1 replication is directly measured in cultures of HIV-1-infected CD4+ T cells incubated with HIV-1-specific CTL clones. But it still remains unknown whether this assay reflects immune pressure in vivo.
In the present study, we investigated whether HIV-1-specific CTLs having a strong ability to suppress HIV-1 replication could positively select escape mutants. Since HLA-A*1101 is known to be an HLA allele relatively associated with a slow progression to AIDS (32), it is speculated that some HLA-A*1101-restricted CTLs would have a strong ability to suppress HIV-1 replication in vitro. Therefore, we first focused on 4 well-known HLA-A*1101-restricted CTL epitopes in the present study. We investigated the frequency of CTLs specific for these epitopes in chronically HIV-1-infected individuals, the ability of these CTLs to suppress HIV-1 replication in vitro, and whether the escape mutants were selected by the CTLs. Furthermore, we analyzed the expression of Programmed Death-1 (PD-1) on these CTLs ex vivo and antigen recognition of them.
MATERIALS AND METHODS
Patient samples.
Informed consent was obtained from all subjects according to the Declaration of Helsinki. For sequence analysis, blood specimens were collected in EDTA. Plasma and peripheral blood mononuclear cells (PBMCs) were separated from heparinized whole blood. Patient HLA type was determined by standard sequence-based genotyping.
Sequence of autologous virus.
Viral RNA was extracted from samples of plasma from HIV-1-infected patients by the use of a QIAamp MinElute virus spin kit (Qiagen), and cDNA was synthesized from the RNA with SuperScript RNase H-reverse transcriptase and random primers (Invitrogen). The Nef region and the Gag region were amplified by nested PCR using Taq DNA polymerase (Promega). The PCR products were then agarose gel purified and sequenced directly or cloned by use of a TOPO TA cloning kit (Invitrogen). All DNA sequencing was performed by using a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) and an ABI Prism 310 genetic analyzer. The regions of Gag349, Nef73, and Nef84 epitopes were sequenced directly in 124, 121, and 122 individuals, respectively, while those of Nef73 and Nef84 epitopes were sequenced for cloned samples from 10 and 11 individuals, respectively.
Cells.
C1R cells expressing HLA-A*1101 (C1R-A*1101) and transporter associated with antigen processing (TAP)-defective RMA-S cells expressing HLA-A*1101 (RMA-S-A*1101) were previously generated and were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and 0.15 mg/ml hygromycin B.
Generation of CTL clones.
Peptide-specific CTL clones were generated from an established peptide-specific bulk CTL culture by seeding 0.8 cell/well into U-bottomed 96-well microtiter plates (Nunc, Roskilde, Denmark) together with 200 μl of cloning mixture (RPMI 1640 medium supplemented with 10% FCS and 200 U/ml human recombinant interleukin-2, 5 × 105 irradiated allogeneic PBMC from a healthy individual, and 1 × 105 irradiated C1R-A*1101 cells prepulsed with a 1 μM concentration of the corresponding peptide, Gag349 [ACQGVGGPGHK], Nef73 [QVPLRPMTYK], or Nef84 [AVDLSHFLK]). Wells positive for growth after about 2 weeks were transferred to 48-well plates together with 1 ml of the cloning mixture. The clones were examined for CTL activity by the standard 51Cr release assay. All CTL clones were cultured in RPMI 1640-10% FCS supplemented with 200 U/ml recombinant human interleukin-2 and were stimulated weekly with irradiated target cells prepulsed with the appropriate HIV-1-derived peptide.
HIV-1 clones.
Infectious proviral clones of HIV-1, pNL-432, and its Nef mutant, pNL-M20A (containing a substitution of Ala for Met at residue 20 of Nef), reported previously, were used (2). For pNL-432-Nef84-2L9R, the mutation was introduced by site-directed mutagenesis (Invitrogen).
CTL assay for target cells pulsed with HIV-1 peptide.
Cytotoxicity activity was measured by the standard 51Cr release assay, as previously described (34). Target cells (2 × 105) were incubated for 60 min with 100 μCi Na251CrO4 in saline and then washed three times with RPMI 1640 medium containing 10% newborn calf serum (NCS). Labeled target cells (2 × 103/well) were added to 96-well round-bottom microtiter plates (Nunc) along with the appropriate amount of the corresponding peptide. After a 1-h incubation, effector cells were added, and the mixtures were then incubated for 4 h at 37°C. The supernatants were collected and analyzed with a gamma counter.
Intracellular cytokine (ICC) production assay.
PBMCs from HLA-A*1101-positive HIV-1-infected patients were stimulated with a given peptide (1 μM) in culture medium (RPMI 1640 medium supplemented with 10% FCS and 200 U/ml recombinant human interleukin-2). After 14 days in culture, the cells were assessed for gamma interferon (IFN-γ) production activity by using a FACSCalibur instrument. Briefly, bulk cultures were stimulated by C1R-A*1101 cells pulsed with or without the corresponding peptide (1 μM) for 2 h at 37°C. Brefeldin A (10 μg/ml) was then added, and the cultures were continued for an additional 4 h. Cells were collected and stained with 7-amino-actinomycin D (7-AAD) at room temperature for 10 min. After 2 washes with RPMI 1640 medium supplemented with 10% FCS, cells were stained with phycoerythrin (PE)-labeled anti-CD8 monoclonal antibody (MAb) (Dako Corporation, Glostrup, Denmark). After having been treated with 4% paraformaldehyde solution, the cells were permeabilized in permeabilization buffer (0.1% saponin and 20% NCS in phosphate-buffered saline) at 4°C for 10 min and stained with fluorescein isothiocyanate (FITC)-labeled anti-IFN-γ MAb (PharMingen, San Diego, CA). After a thorough washing with the permeablization buffer, the cells were analyzed by using the FACSCalibur instrument. Nonspecific binding of anti-IFN-γ MAb and nonspecific production of IFN-γ were excluded by subtracting the data of the negative control, which was the same sample stimulated with C1R-A*1101 cells without the specific peptide and stained with the same MAbs.
For ex vivo analysis, PBMCs from HLA-A*1101-positive HIV-1-infected patients were stimulated with the corresponding peptide (1 μM), and IFN-γ production was measured 6 h later, as described above.
HLA class I stabilization assay.
The binding of peptides to HLA-A*1101 molecules was tested as previously described (11). RMA-S-A*1101 cells transfected with HLA-A*1101 and human β2-microglobulin were used. These cells express a very low level of HLA class I molecules on their cell surface when they are cultured at 37°C, whereas empty HLA class I molecules are stably expressed if they are cultured at 26°C. The stabilization of HLA class I molecules is dependent on peptide binding affinity (22, 30, 40). Briefly, RMA-S-A*1101 cells were cultured at 26°C for 14 to 18 h. The cells were incubated at 26°C for 1 h with Nef84 (AVDLSHFLK), Nef84-2L (ALDLSHFLK), or Nef84-2L9R (ALDLSHFLR) peptide at various concentrations and then at 37°C for 3 h. After 2 washes with phosphate-buffered saline (PBS) supplemented with 20% FCS (PBS-20% FCS), they were subsequently incubated for 30 min on ice with an appropriate dilution of MAb TP25.99 (41). After 2 washes with PBS-20% FCS, the cells were incubated for 30 min on ice with an appropriate dilution of FITC-conjugated sheep IgG with anti-mouse Ig specificity (Silenus Laboratories, Hawthorn, Australia). Finally, they were washed three times with PBS-20% FCS, after which the fluorescence intensity was measured by using a flow cytometer (Becton Dickinson, Mountain View, CA).
Surface expression of HLA class I molecules on HIV-1-infected cells.
To assess HLA class I expression on HIV-1-infected CD4+ T cells, we stained the cells with anti-HLA-A11 MAb followed by PE-labeled anti-mouse Ig (Pharmingen International, San Diego) and thereafter fixed and permeabilized them for intracellular HIV-1 p24 staining with FITC-labeled anti-p24 MAb KC-57. The expression of HLA class I molecules on HIV-1infected CD4+ T cells was examined by using the FACSCalibur instrument with Cell Quest software (Becton Dickinson, San Jose, CA).
Suppression of HIV-1 replication by HIV-1-specific CTL clones.
The ability of HIV-1-specific CTL clones to suppress HIV-1 replication was examined as previously described (42). CD4+ T cells purified by means of anti-human CD4 MAb-coated magnetic beads (MACS beads; Miltenyi Biotec) from PBMCs of an HIV-1-seronegative individual with HLA-A*1101 were cultured and infected with HIV-1 clones. Cultured CD4+ T cells were incubated with an HIV-1 clone for 4 h at 37°C with intermittent agitation and then washed three times with RPMI 1640 medium supplemented with 10% FCS. HIV-1-infected CD4+ T cells were cocultured with an HIV-1-specific CTL clone in culture medium. From day 2 to day 7 postinfection, 10 μl of culture supernatant was collected, and the concentration of p24 antigen (Ag) in the supernatant was measured by conducting an enzyme immunoassay (HIV-1 p24 Ag enzyme-linked immunosorbent assay [ELISA] kit; ZeptoMetrix). Percent suppression was calculated as follows: (concentration of p24 Ag in the supernatant of HIV-1-infected CD4+ T cells cultured with HIV-1-specific CTLs/concentration of p24 Ag in the supernatant of HIV-1-infected CD4+ T cells cultured without the CTLs) × 100.
HLA-peptide tetrameric complexes.
The tetrameric complexes of HLA-A*1101, HLA-A*2402, and HLA-A*2601 were synthesized as previously described (3). The purified complexes were enzymatically biotinylated at a BirA recognition sequence located at the C terminus of the heavy chain and were mixed with PE- or allophycocyanin (APC)-conjugated avidin (Molecular Probes) at a molar ratio of 4:1.
Analysis of PD-1 or CD27 CD28 CD45RA expression on HIV-1-specific CD8+ T cells.
For the analysis of PD-1 expression, cryopreserved PBMCs of HIV-positive individuals were first stained with Pacific Blue-conjugated CD8 MAb (BD Bioscience) and FITC-conjugated CD3 MAb (Dako Corporation, Glostrup, Denmark) at 4°C for 30 min followed by PE-conjugated PD-1 MAb (BD Bioscience) at the room temperature for 30 min. After 2 washes with RPMI 1640 medium supplemented with 10% FCS, the cells were stained with allophycocyanin (APC)-conjugated tetramer at 37°C for 30 min. After 2 additional washes, the cells were stained with 7-AAD (BD Bioscience) at room temperature for 10 min and analyzed by using flow cytometry (FACS Canto II; BD Bioscience). For the phenotypic analysis of HIV-1-specific CD8+ T cells, the PBMCs were first stained with PE-Cy7-conjugated anti-CD3 (BioLegend), Pacific Blue-conjugated CD8 (BD Bioscience), FITC-conjugated anti-CD27 (BD Bioscience), PE-conjugated anti-CD28 (BioLegend), and phycoerythrin-Texas red (ECD)-conjugated anti-CD45RA (Beckman Coulter) MAbs at 4°C for 30 min. After 2 washes with RPMI 1640 medium supplemented with 10% FCS, the cells were stained with APC-conjugated tetramer at 37°C for 30 min. After 2 additional washes, the cells were stained with 7-AAD at room temperature for 10 min and analyzed by using flow cytometry.
Enzyme-linked immunospot (ELISPOT) assay.
Cryopreserved PBMCs of 2 HLA-A*1101+ HIV-1-infected individuals (KI-015 and KI-036) were plated out in 96-well polyvinylidene plates (Millipore, Bedford, MA) which had been precoated with 0.5 μg/ml anti-IFN-γ MAb 1-DIK (Matbech, Stockholm, Sweden). The appropriate amount of Nef73 or Nef84 peptides was added in a volume of 50 μl, and then PBMCs were added at 1 × 105 cells/well in a volume of 100 μl. The plate was incubated for 40 h at 37°C in 5% CO2 and was washed with PBS before the addition of biotinylated anti-IFN-γ MAb (Mabtech) at 0.5 μg/ml. After it was incubated at room temperataure for 100 min and then washed with PBS, streptavidin-conjugated alkaline phospatase (Mabtech) was added following a 40-min incubation at room temperature. Individual cytokine-producing cells were detected as dark spots after a 20-min reaction with 5-bromo-4-chloro-3-indolyl phosphate and Nitro Blue Tetrazolium by using an alkaline phosphatase-conjugated substrate (Bio-Rad, Richmond, CA). The spot number was counted by using an Eliphoto counter (Minerva Teck, Tokyo, Japan). The number of spots for each peptide-specific T cell response was caluculated by subtracting the negative-control spots.
RESULTS
Immunodominancy of 4 HLA-A*1101-restricted HIV-1 epitopes.
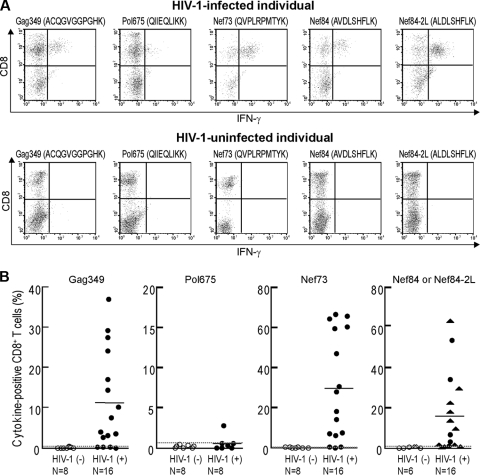
We first focused on HIV-1 CTL epitopes presented by only a given HLA allele that influences the control of HIV-1, because the effect of each epitope presented by the same HLA class I allele on the ability of specific CTLs to suppress HIV-1 replication and to select escape mutants can be compared. HLA-A*1101 is an HLA allele relatively associated with a slow progression to AIDS (32), implying that some epitope-specific CTLs may have the ability to suppress HIV-1 replication. We selected 4 out of many known HLA-A*1101-restricted HIV-1 epitopes (Gag349, ACQGVGGPGHK; Pol675, QIIEQLIKK; Nef73, QVPLRPMTYK; and Nef84, AVDLSHFLK; or Nef84-2L, ALDLSHFLK [both sequences are frequently found in clade B]), because CTLs specific for these epitopes were previously shown to be frequently detected in chronically HIV-1-infected individuals (10, 14, 19). We reevaluated whether CD8+ T cells specific for these HIV epitopes could be frequently detected in chronically HIV-1-infected Japanese individuals carrying HLA-A*1101. PBMC from these individuals and HIV-1-seronegative HLA-A*1101+ individuals were stimulated with these epitope peptides and cultured for 2 weeks. The percentage of specific CD8+ T cells in these cultures was determined by performing an intracellular cytokine (ICC) production assay using these epitope peptides (Fig. 1A). Pol675-specific CD8+ T cells were detected in only 1 of the 8 individuals, whereas Gag349-specific, Nef73-specific, and Nef84- or Nef84-2L-specific ones were detected in 12 of 16 individuals, 13 of 16 individuals, and 11 of 16 individuals, respectively (Fig. 1B). These results indicate that Gag349, Nef73, and Nef84 (or Nef84-2L) are recognized as immunodominant epitopes in HIV-1-infected Japanese individuals carrying HLA-A*1101. We therefore focused on these 3 epitopes for further studies.
FIG. 1.
Four HLA-A*1101-restricted HIV-1-specific CD8+ T cells in chronically HIV-1-infected HLA-A*1101+ individuals. (A) After PBMC from an HLA-A*1101+ HIV-1-infected and HIV-1-uninfected individuals had been stimulated singly with each of the indicated peptides for 2 weeks, HIV-1-specific CD8+ T cells were detected by measuring IFN-γ-producing CD8+ T cells in the culture after stimulation with the corresponding peptide-pulsed cells. Either Nef84 or Nef84-2L peptide was used for individuals infected with HIV carrying the corresponding sequence. A representative result is shown. (B) Summary of ICC assays for HLA-A*1101+ HIV-1-infected individuals and HIV-1-uninfected individuals. For detection of Nef84- and Nef84-2L-specific CD8+ T cells, Nef84 and Nef84-2L peptides were incubated with cells from individuals infected with the wild type or the 2F and 2L viruses, respectively. The circle symbols and the triangle symbols represent the frequency of IFN-γ-producing CD8+ T cells after stimulation with Nef84 and Nef84-2L peptides, respectively. The average + 3 SD of IFN-γ-producing CD8+ T cells in HIV-1-uninfected individuals was defined as a positive value (Gag349, >0.34%; Pol675, >0.56%; Nef73, >0.32%; Nef84 or Nef84-2L, >0.63%). Dotted lines indicate the average + 3 SD, and solid lines indicate the average in HIV-1-infected individuals.
Ability of 3 HLA-A*1101-restricted HIV-1-specific CTLs to suppress HIV-1 replication in vitro.
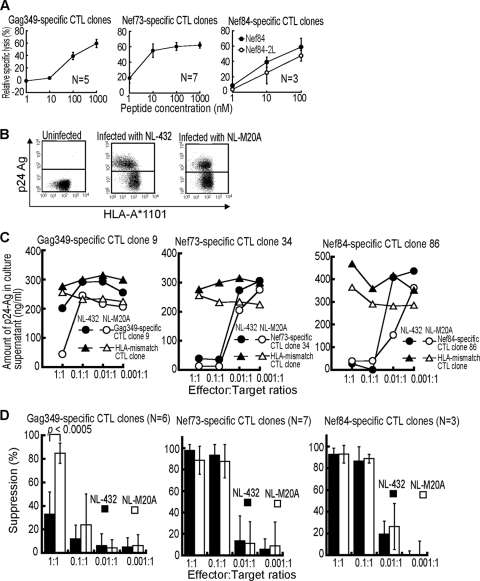
To investigate the ability of these T cells to suppress HIV-1 replication, we next established 5 Gag349-specific, 7 Nef73-specific, and 3 Nef84-specific CTL clones from PBMC of chronically HIV-1-infected individuals carrying HLA-A*1101. These CTL clones exhibited a strong cytolytic activity against C1R-A*1101 cells prepulsed with the corresponding epitope peptide (Fig. 2A) and against those infected with recombinant vaccinia virus expressing the HIV-1 SF2 Nef or Gag protein (data not shown). We investigated the ability of these CTL clones to suppress HIV-1 replication in primary CD4+ T cells infected with the NL-432 clone or its Nef mutant NL-M20A, which has the ability to downregulate the cell surface expression of CD4 but not that of HLA-class I A and B molecules, in HIV-1-infected cells (2). Indeed, NL-432-infected CD4+ T cells exhibited the downregulation of HLA-A*1101, whereas NL-M20A-infected ones did not (Fig. 2B). Both Nef73-specific and Nef84-specific CTL clones completely suppressed the replication of both NL-432 and NL-M20A at effector/target cell (E:T) ratios of 1:1 and 0.1:1 (Fig. 2C). A Gag349-specific CTL clone partially suppressed NL-432 replication and completely suppressed that of NL-M20A at an E:T ratio of 1:1 but failed to suppress the replication of either clone at an E:T ratio of 0.1:1 (Fig. 2C). Analysis using 6 Gag349-specific, 7 Nef73-specific, and 3 Nef84-specific CTL clones confirmed that the ability of the Nef73-specific and Nef84-specific CTL clones to suppress HIV-1 replication was much stronger that of the Gag349-specific ones (Fig. 2D). It also revealed that Nef-mediated HLA-class I downregulation did not affect the recognition of HIV-1-infected CD4+ T cells by Nef73-specific and Nef84-specific clones. These results together indicate that Nef73-specific and Nef84-specific CTLs have a strong ability to suppress HIV-1 replication in vitro.
FIG. 2.
Ability of HLA-A*1101-restricted CTLs to suppress HIV-1 replication in HIV-1-infected CD4+ T cells. (A) Cytolytic activities of HLA-A*1101-restricted HIV-1-specific CTLs (5 Gag349-specific, 7 Nef73-specific, and 3 Nef84 consensus B-specific CTL clones) were tested by using C1R-A*1101 cells pulsed with various concentrations of the corresponding peptide (effector-to-target-cell ratio = 2:1). (B) Surface expression of HLA class I molecules on CD4+ T cells infected with HIV-1 NL-432 or NL-M20A. CD4+ T cells infected with HIV-1 NL-432 or NL-M20A were stained with anti-HLA-A*1101 and anti-p24 MAbs and then analyzed by using flow cytometry. (C) Ability of HLA-A*1101-restricted CTLs to suppress HIV-1 replication in cultures of HIV-1-infected CD4+ T cells. CD4+ T cells from an HLA-A*1101+ healthy individal were infected with NL-432 or NL-M20A and then cocultured with HLA-A*1101-restricted CTL clones or HLA-mismatch CTL clone (HLA-B*5101) at various effector-to-target ratios. HIV-1 p24 Ags in the supernatant were measured on day 6 or 7 postinfection by conducting an enzyme immunoassay. (D) Analysis using multiple HLA-A*1101-restricted CTLs to suppress replication of NL-432 or NL-M20A.
Ex vivo analysis of Nef73-specific and Nef84-specific CTLs in chronically HIV-1-infected individuals.
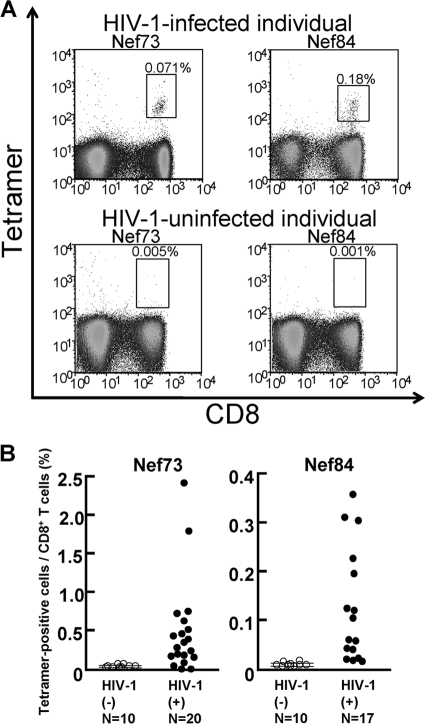
Nef73-specific and Nef84-specific CTLs could be induced from the memory T-cell pool by in vitro stimulation with the specific peptides in more than 50% of chronically HIV-1 infected individuals carrying HLA-A*1101 (Fig. 1). To clarify whether these specific T cells would be elicited in vivo, we analyzed PBMCs from chronically HIV-1-infected individuals carrying HLA-A*1101 by using the specific tetramers. Nef73-specific CD8+ T cells were detected for 16 of 20 chronically HIV-1-infected HLA-A*1101+ individuals, and Nef84-specific CD8+ T cells were detected for 13 of 17 (Fig. 3). These results together with those shown in Fig. 1 indicate that both Nef73-specific and Nef84-specific CTLs were effectively elicited in chronically HIV-1-infected HLA-A*1101+ individuals.
FIG. 3.
Frequency of HLA-A*1101-restricted Nef epitope-specific CD8+ T cells. PBMCs from HLA-A*1101+ HIV-1-infected or HIV-1-uninfected individuals were examined by using Nef73-specific or Nef84-specific tetramers and anti-CD8 MAb or by using only anti-CD8 MAb. (A) A representative result of Nef73-specific or Nef84-specific tetramer binding CD8+ T cells. (B) Summary of frequency of HLA-A*1101+-restriced Nef73-specific or Nef84-specific CD8+ T cells in HIV-1-infected individuals and HIV-1-uninfected individuals. The mean frequencies + 3 SD of Nef73-specific and Nef83-specific CD8+ T cells among total CD8+ T cells from the HIV-1-uninfected individuals were 0.032% + 0.045% and 0.009% + 0.012%, respectively. More than 0.077% and 0.021% were evaluated as showing positive binding of Nef73-specific and Nef84-specific tetramers, respectively.
Association of an HLA-A*1101 allele with mutations in the 3 CTL epitopes.
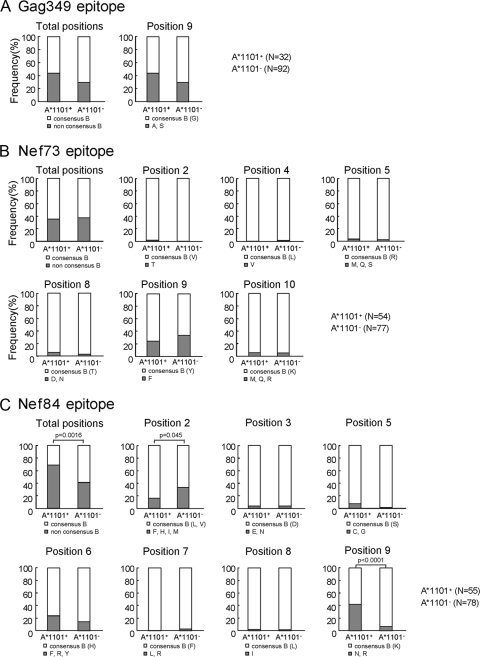
We speculated that these 2 Nef epitope-specific CTLs having a strong ability to suppress HIV-1 replication could select escape mutants but that Gag349-specific CTLs having a weak ability to suppress HIV-1 replication could not. We therefore analyzed the sequences of these epitopes and their flanking regions from HLA-A*1101+ and HLA-A*1101− individuals who had been chronically infected with HIV-1 to clarify whether they selected the escape mutations. In the Gag349 epitope, only the 9S mutation was found, but there was no significant difference in the frequency of this mutation between the HLA-A*1101-positive and -negative individuals (Fig. 4A). In the Nef73 epitope, several mutations were found at positions 2, 4, 5, 8, 9, and 10 (Fig. 4B). The 9F mutation was frequently found, but there was no significant difference in the frequency of this mutation, nor in that of the other mutations, between the HLA-A*1101-positive and -negative subjects. In the Nef84 epiope, there were several mutations, at positions 2, 3, 5, 6, 7, 8, and 9, though the mutations at positions 2, 6, and 9 were the most frequently detected ones (Fig. 4C). The frequency of the Arg mutation at position 9 was significantly higher in HLA-A*1101-positive individuals than in HLA-A*1101-negative ones (P < 0.0001) (Fig. 4C). In contrast, the mutations at position 2 were significantly more frequently detected for HLA-A*1101-negative individuals than for HLA-A*1101-positive ones (P = 0.045), suggesting that they were not selected by HLA-A*1101-restricted CTLs. There were 3 mutations (Phe, Tyr, and Arg) at position 6. The frequency of each one at position 6 was not significantly higher for HLA-A*1101-positive individuals than for HLA-A*1101-negative ones. These results together suggest that only the 9R mutation was selected by Nef84-specific CTLs.
FIG. 4.
Frequency of mutations in 3 HLA-A*1101-restricted epitopes. Three epitope sequences, Gag349 (A), Nef73 (B), and Nef84 (C), from HLA-A*1101-positive and HLA-A*1101-negative individuals chronically infected with HIV-1 were analyzed. Consensus sequences of these epitopes in clade B are as follow: Gag349, ACQGVGGPGHK; Nef73, QVPLRPMTYK; Nef84, AVDLSHFLK and ALDLSHFLK. The frequency of mutations in the total sequence of the epitopes was calculated as (number of individuals having the mutation[s]/number of individuals tested) × 100, whereas those at a given position were calculated as (number of individuals having the mutation[s] at a given position/number of individuals tested) × 100. The results were compared between HLA-A*1101-positive and HLA-A*1101-negative individuals, and the P values were determined by using Fisher's exact test.
There were several mutations in the flanking region of these epitopes, but no significant difference in them between the HLA-A*1101-positive and -negative individuals was found (data not shown).
In vitro recognition of the 9R mutation by Nef84-specific CTLs.
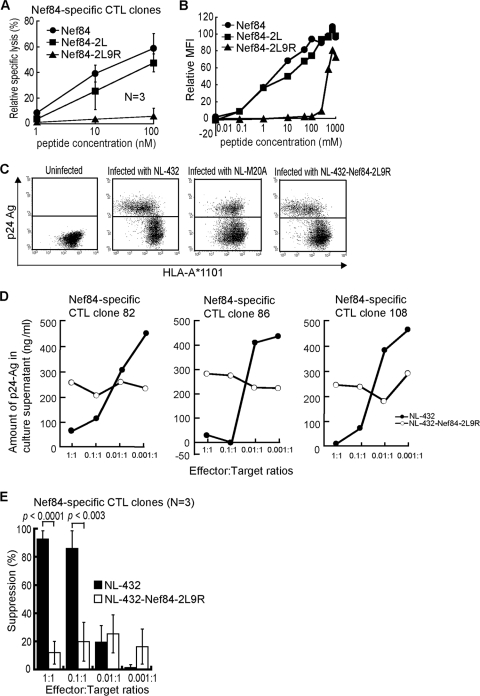
We speculated that the 9R mutant is an escape mutant from Nef84-specific CTLs because this mutation is associated with the HLA-A*1101 allele. We therefore investigated whether or not the Nef84-specific CTLs could recognize the Nef84-9R mutant epitope. We first tested the activity of Nef84-specific CTL clones in killing target cells prepulsed with the Nef84-9R mutant peptide. Three Nef84-specific CTL clones effectively killed target cells prepulsed with Nef84 or Nef84-2L wild-type peptide but failed to kill those prepulsed with Nef84-2L9R peptides (Fig. 5A). The results of an HLA class I stabilization assay showed that the affinity of the Nef84-2L9R peptide for HLA-A*1101 was much weaker than that of Nef84 or Nef84-2L for it (Fig. 5B). Taken together, these results suggest that the Nef84-2L9R peptide is very weakly presented in HIV-1 mutant virus-infected cells because of the very low affinity of Nef84-2L9R peptide for HLA-A*1101. We generated an NL-432 mutant carrying 2L and 9R mutations of Nef84 (NL-432-Nef84-2L9R) virus and infected HLA-A*1101+ CD4+ T cells with this virus. The infected cells showed downregulation of HLA-A*1101 on target cells infected with NL-432 or NL-432-Nef84-2L9R but not on those infected with NL-M20A (Fig. 5C). Thus, these result also revealed that the 2L9R mutations do not affect the downregulation of HLA class I molecules. Three Nef84-specific CTL clones failed to suppress replication of NL-432-Nef84-2L9R (Fig. 5D), whereas these T-cell clones effectively suppressed replication of NL-432 at E:T ratios of 1:1 and 0.1:1 (Fig. 5D and 5E). These results indicate that the CTL clones could not recognize cells infected with NL-432-Nef84-2L9R and confirmed 9R to be an escape mutation.
FIG. 5.
Ability of Nef84-specific CTLs to suppress replication of HIV-1-Nef84-9R mutant virus. (A) Cytolytic activities of Nef84-specific CTL clones in killing C1R-A*1101 cells pulsed with Nef84-9R peptide. C1R-A*1101 cells were prepulsed with various concentrations of Nef84, Nef84-2L, or Nef84-2L9R peptide. Cytolytic activities of Nef84-specific CTL clones were measured at an effector-to-target ratio of 2:1. (B) Ability of Nef84-2L9R peptide to bind HLA-A*1101. The affinity was measured by a stabilization assay using RMA-S-A*1101 cells. (C) Surface expression of HLA class I molecules on CD4+ T cells infected with NL-432-Nef84-2L9R. (D) Ability of each Nef84-specific CTL clone to suppress NL-432-Nef84-2L9R replication in CD4+ T cells. (E) Analysis of ability of all 3 Nef84-specific CTL clones to suppress replication of NL-432 or NL-432-Nef84-2L9R.
Different surface expression levelS of PD-1 between Nef73-specific and Nef84-specific CTLs.
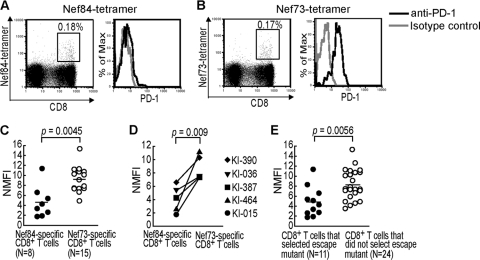
Both Nef73-specific and Nef84-specific CTL clones effectively suppressed HIV-1 replication in vitro. In contrast, the latter CTLs selected an escape mutation in vivo, whereas the former ones did not. These findings suggest the possibility that Nef73-specific CTLs cannot mediate selection of escape mutants in vivo. PD-1 expression on HIV-1-specific T cells is known to be associated with dysfunction of T cells (15, 35, 44, 47). Therefore, high expression of PD-1 on the CTL surface is a possible reason why Nef73-specific CTLs failed to select escape mutants. To clarify the PD-1 expression on Nef73-specific and Nef84-specific CTLs, we stained PBMCs from HLA-A*1101+ HIV-1-infected individuals with anti-PD-1 and anti-CD8 MAbs aND with the specific tetramer (Fig. 6A and B). Nef73-specific and Nef84-specific CD8+ T cells were, respectively, detected by the tetramers in 16 and 13 chronically HIV-1-infected individuals carrying HLA-A*1101 (Fig. 3), but only 15 and 8 individuals had a sufficient number of Nef73-specific and Nef84-specific CD8+ T cells for analysis of PD-1 expression, respectively. The Nef73-specific CD8+ T cells expressed a significantly higher level of PD-1 than the Nef84-specific ones (Fig. 6C). But only 8 individuals (2 having the 9R mutant and 6 having wild-type Nef84) had enough Nef84-specific CD8+ T cells for analysis of PD-1 expression. We did not find any difference in the expression levels of PD-1 between these 2 groups (see Fig. S1 in the supplemental material). These results suggest that the 9R mutation did not influence the level of PD-1 on Nef84-specific CD8+ T cells.
FIG. 6.
PD-1 expression on Nef84- and Nef73-specific CD8+ T cells. (A and B) PD-1 expression on Nef84- and Nef73-specific CD8+ T cells among PBMCs from an HIV-1-infected individual (KI-015). PBMCs from KI-015 were stained with anti-CD3, anti-CD8, anti-PD-1 MAb, and the tetramer. The frequency of tetramer+ CD8+ T cells in the lymphocyte population was plotted (left). The histogram shows PD-1 expression on the specific CD8+ T cells (right). (C) PD-1 expression on Nef84- and Nef73-specific CD8+ T cells in PBMCs of HIV-1-infected individuals. PD-1 expression on the cells from each individual was normalized by the mean fluorescence intensity of the isotype control (NMFI). (D) PD-1 expression on Nef84- and Nef73-specific CD8+ T cells from the same individuals (KI-015, -036, -387, -390, and -464). (E) PD-1 expression on CD8+ T cells having a strong ability to suppress HIV-1 replication in vitro and to select escape mutants. The left part of the plot shows 8 HLA-A*1101-restricted Nef84-specific and 3 HLA-A*2402-restricted Nef-138-specific CD8+ T cells that select escape mutants, and the right part shows 15 HLA-A*1101-restricted Nef73-specific and 9 HLA-A*26-restricted Gag169-specific CD8+ T cells that do not select them.
Both Nef84- and Nef73-specific CTLs, enough for the analysis of PD-1 expression, were detected in only 5 of the individuals tested. We compared the levels of PD-1 between the CTLs within the same individual. A similar difference was found between these CTLs within each individual (Fig. 6D). PD-1 is known to be upregulated on activated T cells (34). Therefore, we speculate that Nef84-specific CTLs are not activated, because the wild-type virus disappeared and the Nef84-9R escape mutant was selected in many HLA-A*1101+ individuals, resulting in downregulation of PD-1 expression on the T cells. We investigated the sequences of these Nef epitopes in HIV-1 from the 5 individuals whose Nef73-specific and Nef84-specific CD8+ T cells were analyzed for PD-1 expression. These 5 individuals were infected with HIV-1 carrying the wild-type Nef73 sequence, whereas the sequence of Nef84 was wild type (2V or 2L) in 3 of these individuals, Nef84-9R in 1, and a mixture of both in 1 individual (Table 1). The Nef84-specific CD8+ T cells from the individual infected with the Nef84-9R mutant (KI-390) expressed the highest level of PD-1 among the T cells from these 5 individuals (Fig. 6D). Together with the results showing no difference in the expression levels of PD-1 between individuals infected with the 9R mutant and those infected with the wild-type virus, these results exclude the possibility that the lower level of expression of PD-1 on Nef84-specific T cells resulted from the appearance of the Nef84-9R mutant virus in these individuals.
TABLE 1.
Sequences of Nef73 and Nef84 epitopes in HIV-1 from the 5 subjects whose Nef73- and Nef84-specific CD8 T cells were analyzed for PD-1 expression
| Patient ID or sequence description | Sequencea |
|
|---|---|---|
| Nef73 | Nef84 | |
| Wild type | QVPLRPMTYK | AV(L)DLSHFLK |
| KI-015b | - - - - - - - - - - | - - - - - - - - - |
| KI-036c | - - - - - - - - - - | - L - - - - - - - |
| KI-387b | - - - - - - - - - - | - L - - - - - - - |
| KI-390b | - - - - - - - - - - | - L - - - - - - R |
| KI-464b | - - - - - - - - - - | - L - - - - - - K/Rd |
Sequences were analyzed by the direct sequencing method. “-” indicates agreement with wild-type sequence.
The same sample was analyzed for sequencing and PD-1 expression.
This patient was analyzed for the sequence of HIV-1 on 6 October 2005 and for PD-1 expression on the T cells on 14 July 1999.
The mixture of sequences carrying K or R at position 9 was detected.
A recent study showed that PD-1 is highly expressed on effector memory T cells and that its expression is related to the differentiation of CD8+ T cells (37). Therefore, the difference in expression of PD-1 may result from the difference in differentiation status between these 2 Nef epitope-specific T cells. We analyzed the CD27 CD28 CD45RA phenotype of these T cells in the 5 individuals to clarify differentiation of the T cells. The results showed no difference in differentiation status between these 2 Nef epitope-specific T cells, although effector and late effector subsets were predominantly detected in Nef84-specific and Nef73-specific T cells from one individual (see Fig. S2 in the supplemental material). These results indicate that a difference in expression of PD-1 between these T cells was not due to the difference in differentiation status.
We speculate that there is no difference in the level of PD-1 expression between Nef73-specific and Nef84-specific CTL clones, because both CTL clones showed strong ability to suppress HIV-1 replication. To complement the ex vivo data, we analyzed the PD-1 expression on our in vitro-generated CTL clones. The results showed that both CTL clones expressed a low level of PD-1 and that there was no difference in the expression level between these CTL clones (data not shown).
We further investigated PD-1 expression on 2 CTLs having a strong ability to suppress HIV-1 replication. HLA-A*2402-restricted Nef138-specific CTLs were recently shown to have a strong ability to suppress HIV-1 replication and to select Nef138-2F escape mutants (18). HLA-A*26-restricted Gag169-specific CD8+ T cells also have a strong ability to suppress HIV-1 replication but cannot select any escape mutant (unpublished observation). PD-1 expression on Nef138-specific and Gag169-specific CD8+ T cells from chronically HIV-1-infected individuals was measured by using specific tetramers and anti-PD-1 MAbs. PD-1 expression on Nef138-specific CD8+ T cells was lower than that on the Gag169-specific ones. Taken together, these results show that PD-1 expression on CD8+ T cells that can select escape mutants is significantly lower than that on CD8+ T cells that are unable to select escape mutants (Fig. 6E).
Different functional abilities between ex vivo Nef73-specific and Nef84-specific CTLs.
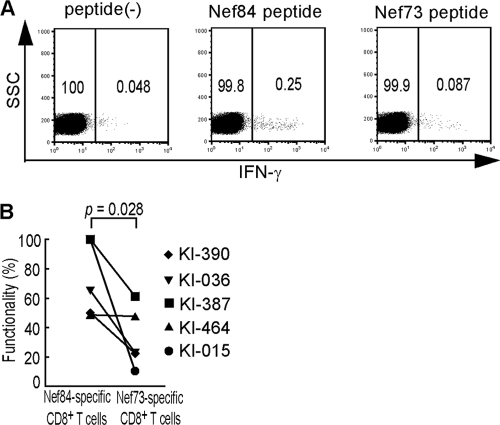
We speculated that Nef84-specific CTLs have a stronger functional ability in vivo than Nef73-specific ones. Therefore, we investigated whether Nef84-specific CTLs from ex vivo PBMC would respond to the specific epitope more effectively than Nef73-specific ones. To compare functional abilities between these 2 CTLs, we selected 5 individuals who had both Nef73-specific and Nef84-specific CTLs. IFN-γ production from these T cells among ex vivo PBMC was measured after they had been stimulated with Nef84 peptide or Nef73 peptide (Fig. 7A). The results showed that the frequency of IFN-γ-producing cells was higher for Nef84-specific CD8+ T cells than for Nef73-specific ones from each individual. That is, it is significantly higher for the former T cells than for the latter ones (Fig. 7B; see also Fig. S3 in the supplemental material). These results support the idea that Nef73-specific T cells can partially function in vivo.
FIG. 7.
Functional analysis of ex vivo Nef84- and Nef73-specific CD8+ T cells. (A) IFN-γ production of Nef84- and Nef73-specific CD8+ T cells among PBMCs from an HIV-1-infected individual, KI-036. PBMCs from KI-036 were stimulated with Nef73 peptide or Nef84 peptide and stained with anti-CD8, followed by intracellular staining for IFN-γ. The frequency of IFN-γ+ CD8+ T cells among total CD8+ T cells was plotted. (B) Frequency of Nef84- and Nef73-specific CD8+ T cells producing IFN-γ. The percent functionality was calculated as follows: (frequency of IFN-γ+ CD8+ T cells among total CD8+ T cells/that of tetramer+ CD8+ T cells among total CD8+ T cells) × 100.
DISCUSSION
Previous studies showed an inverse correlation between the plasma viral load (pVL) and the frequency of some HIV-1-specific CTLs in HIV-1-infected individuals, indicating that these CTLs control HIV-1 in vivo (5, 28, 33). However, this correlation was not found in the case of many other HIV-1-specific CTLs (16, 25, 26), suggesting the possibility that the quality of HIV-1-specific CTLs is a critical factor for the control of HIV-1 in vivo. However, it is not easy to assess the quality of HIV-1-specifc CTLs. An assay to directly measure the ability of the CTLs to suppress HIV-1 replication in vitro is a very useful method to evaluate the ability of the CTLs to control HIV-1. A previous study using this assay demonstrated that the ability of HLA-B*5101-restricted HIV-1-specific CTLs to suppress HIV-1 replication is dependent on the epitope recognized by these CTLs (43). In addition, a recent study showed that HLA-A*2402-restricted Nef138-specific CTLs have a strong ability to suppress HIV-1 replication, whereas HLA-A*2402-restricted Gag133-8-, Pol797-8-, or Gag263-10-specific CTLs showed a weak ability or no ability to suppress HIV-1 replication (18).
The Nef138-specific CTLs select the 2F escape mutation within 1 to 2 years after the start of an HIV-1 infection (18). The frequency of the Nef138-specific CTLs is inversely correlated with pVL in individuals infected with wild-type virus before the virus with the 2F mutant (the 2F virus) is selected. In contrast, it did not correlate with pVL in them after the 2F virus appeared or in individuals originally infected with the 2F virus (18). These observations strongly suggest that Nef138-specific CTLs have a strong ability to suppress the replication of wild-type HIV-1 in vivo, such that they can select the 2F escape virus. Thus, a strong ability of HIV-1-specific CTLs to suppress HIV-1 replication is necessary to select CTL escape mutants in vivo.
In the present study, we showed that 2 HLA-A*1101-restricted Nef-specific CTLs had a strong ability to suppress HIV-1 replication. Nef84-specific CTLs selected the escape mutant 9R, whereas Nef73-specific ones did not select any escape mutant. There are several hypotheses to explain the difference in the abilities of these CTLs to select escape mutants. One is that the frequency of mutations is much lower in a part of the Nef73 epitope and its flanking region than in that of the Nef84 epitope and its flanking region. This idea is not likely to be true, however, because the analysis of sequences of HIV-1 isolates reported in the Los Alamos HIV-1 Sequence Database showed that the frequency of mutations in the Nef73 epitope is almost the same as that in the Nef84 one (data not shown). Another possibility is that Nef73-specific CTLs can have a strong ability to suppress HIV-1 replication in vitro but not in vivo. We analyzed the ability of HIV-1-specific CTLs to suppress HIV-1 replication by using the specific CTL clones. Since CTL clones are established from a small part of the memory or memory effector T-cell population that can effectively proliferate, they may not reflect the CTLs in vivo.
Recent studies showed that PD-1 expression on HIV-1-specific T cells is associated with dysfunction of the T cells and disease progression (15, 35, 44, 47). PD-1 is a regulator of virus-specific T-cell survival (4, 8, 24, 31, 38). Therefore, we speculated that Nef73-specific CD8+ T cells express a higher level of PD-1 on their cell surface, such that they lose their ability to suppress HIV-1 replication in vivo. Indeed, the expression of PD-1 on Nef73-specific CD8+ T cells was significantly higher than that on Nef84-specific ones. This difference was found in the case of both Nef73-specific and Nef84-specific CD8+ T cells present in the same individuals. In addition, the ex vivo analysis of both Nef138-specific and Gag169-specific CD8+ T cells having a strong ability to suppress HIV-1 replication in vitro confirmed that PD-1 was expressed significantly at a lower level on the former T cells, which can select escape mutants, than on those unable to select escape mutants. Thus, since PD-1 expression on the latter cells was much higher than that on the former ones, it is likely that the former could not proliferate and promptly died in vivo so that they failed to select escape mutants. A recent study showed that PD-1 expression on HIV-1-specific CD8+ T cells decreased after the variation appeared in the target epitope sequences (39), suggesting that reduced signaling via T-cell receptors (TCR) decreased PD-1 expression. However, the present study showed that lower expression of PD-1 was also found in 4 individuals who had HIV-1 carrying the wild-type Nef84 epitope. Therefore, the T cells in these individuals may not indicate that reduced signaling via TCR decreased the PD-1 expression, because they have wild-type HIV-1. Recent studies suggested that PD-1 expression is a marker of homeostatic stimulation or T-cell differentiation (9, 21, 27, 29, 37). The analysis of the CD27 CD28 CD45RA phenotype of Nef73-specific and Nef84-specific T cells in the 5 individuals excluded the possibility that the difference in expression of PD-1 between these T cells was due to that in differentiation status between these T cells. On the other hand, the present study could not exclude another interpretation, i.e., that the difference between these T cells in ability to suppress HIV-1 replication in vivo is due to some mechanism other than that involving PD-1 expression. We showed that ex vivo Nef84-specific CD8+ T cells had a stronger ability to recognize the epitope than Nef73-specific ones, suggesting that Nef84-specific CD8+ T cells had a stronger ability to suppress wild-type HIV-1 in vivo. Further study of these T cells is necessary to clarify what determines a weak function of Nef73-specific T cells and a strong function of Nef84-specific T cells in vivo.
We showed in the present study that 1 of 2 HIV-1-specific CD8+ T cells having a strong ability to suppress HIV-1 replication in vitro selected escape mutants. In addition, we recently found that 1 of 2 Pol epitope-specific HLA-B*5101-restricted CD8+ T cells and 1 Nef epitope-specific HLA-A*2402-restricted CD8+ T cell having a strong ability to suppress HIV-1 replication in vitro could select escape mutants (6; our unpublished observation). Thus, half of HIV-1-specific CD8+ T cells having a strong ability to suppress HIV-1 replication in vitro, which were previously and presently analyzed, can select escape mutants in vivo, whereas the other half of these CD8+ T cells lose this ability. High expression of PD-1 on the CD8+ T cells may be one explanation for this difference. The mechanism responsible for the presence of 2 types of CD8+ T cells in HIV-1-infected individuals remains unknown.
In the present study, we showed that out of the HIV-1-specific CTLs having the ability to suppress HIV-1 replication in vitro, only those having a strong ability to recognize an HIV-1 epitope can select escape mutants. Thus, it is not true that CTL escape mutations are simply selected by CTLs having a strong ability to suppress HIV-1 replication in vitro. It is still unknown why a given HIV-1-specific CTL can have a strong ability to recognize the epitope in vivo and others cannot, even though both have a strong ability to suppress HIV-1 in vitro. Further analysis of the function of HIV-1-specific CTLs in vivo will be necessary for clarification of the immunopathogenesis of AIDS and the development of immunotherapy and an effective AIDS vaccine.
Supplementary Material
Acknowledgments
The authors have no conflicting financial interests.
This research was supported by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases and by the Global COE program “Global Education and Research Center Aiming at the control of AIDS,” launched as a project commissioned by the Ministry of Education, Science, Sports, and Culture, Japan, by a grant-in-aid for scientific research from the Ministry of Health, Japan, by a grant-in-aid (no. 18390141) for scientific research from the Ministry of Education, Science, Sports and Culture (no.20390134), Japan, and by a grant from the Japan Health Science Foundation.
We thank Yoshiko Tamura for her technical assistance and Sachiko Sakai for her secretarial assistance.
Footnotes
Published ahead of print on 24 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adnan, S., A. Balamurugan, A. Trocha, M. S. Bennett, H. L. Ng, A. Ali, C. Brander, and O. O. Yang. 2006. Nef interference with HIV-1-specific CTL antiviral activity is epitope specific. Blood 108:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., S. Arold, T. Fukumori, T. Okazaki, K. Strebel, and A. Adachi. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J. Virol. 74:2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman, J. D., P. A. Moss, P. J. Goulder, D. H. Barouch, M. G. McHeyzer-Williams, J. I. Bell, A. J. McMichael, and M. M. Davis. 1996. Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94-96. [DOI] [PubMed] [Google Scholar]
- 4.Barber, D. L., E. J. Wherry, D. Masopust, B. Zhu, J. P. Allison, A. H. Sharpe, G. J. Freeman, and R. Ahmed. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439:682-687. [DOI] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 7.Brander, C., O. O. Yang, N. G. Jones, Y. Lee, P. Goulder, R. P. Johnson, A. Trocha, D. Colbert, C. Hay, S. Buchbinder, C. C. Bergmann, H. J. Zweerink, S. Wolinsky, W. A. Blattner, S. A. Kalams, and B. D. Walker. 1999. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic-T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 73:10191-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L. 2004. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 4:336-347. [DOI] [PubMed] [Google Scholar]
- 9.Chomont, N., M. El-Far, P. Ancuta, L. Trautmann, F. A. Procopio, B. Yassine-Diab, G. Boucher, M.-R. Boulassel, G. Ghattas, J. M. Brenchley, T. W. Schacker, B. J. Hill, D. C. Douek, J.-P. Routy, E. K. Haddad, and R.-P. Sekaly. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choppin, J., W. Cohen, A. Bianco, J. Briand, F. Connan, M. Dalod, and J. Gullet. 2001. Characteristics of HIV-1 Nef regions containing multiple CD8+ T cell epitopes: wealth of HLA-binding motifs and sensitivity to proteasome degradation. J. Immunol. 166:6164-6169. [DOI] [PubMed] [Google Scholar]
- 11.Chujoh, Y., Y. Sobao, K. Miwa, Y. Kaneko, and M. Takiguchi. 1998. The role of anchor residues in the binding of peptides to HLA-A*1101 molecules. Tissue Antigens 52:501-509. [DOI] [PubMed] [Google Scholar]
- 12.Collins, K. L., and D. Baltimore. 1999. HIV's evasion of the cellular immune response. Immunol. Rev. 168:65-74. [DOI] [PubMed] [Google Scholar]
- 13.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 14.Culmann-Penciolelli, B., S. Lamhamedi-Cherradi, I. Couillin, N. Guegan, J. P. Levy, J. G. Guillet, and E. Gomard. 1994. Identification of multirestricted immunodominant regions recognized by cytolytic T lymphocytes in the human immunodeficiency virus type 1 Nef protein. J. Virol. 68:7336-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day, C. L., D. E. Kaufmann, P. Kiepiela, J. A. Brown, E. S. Moodley, S. Reddy, E. W. Mackey, J. D. Miller, A. J. Leslie, C. DePierres, Z. Mncube, J. Duraiswamy, B. Zhu, Q. Eichbaum, M. Altfeld, E. J. Wherry, H. M. Coovadia, P. J. R. Goulder, P. Klenerman, R. Ahmed, G. J. Freeman, and B. D. Walker. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350-354. [DOI] [PubMed] [Google Scholar]
- 16.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, M. E. Feeney, M. M. Addo, M. Lichterfeld, M. P. Lahaie, E. Pae, A. G. Wurcel, T. Roach, M. A. S. John, M. Altfeld, F. M. Marincola, C. Moore, S. Mallal, M. Carrington, D. Heckerman, T. M. Allen, J. I. Mullins, B. T. Korber, P. J. R. Goulder, B. D. Walker, and C. Brander. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173-178. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara, M., and M. Takiguchi. 2007. HIV-1-specific CTLs effectively suppress replication of HIV-1 in HIV-1-infected macrophages. Blood 109:4832-4838. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara, M., J. Tanuma, H. Koizumi, Y. Kawashima, K. Honda, S. Mastuoka-Aizawa, S. Dohki, S. Oka, and M. Takiguchi. 2008. Different abilities of escape mutant-specific cytotoxic T cells to suppress replication of escape mutant and wild-type human immunodeficiency virus type 1 in new hosts. J. Virol. 82:138-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukada, K., H. Tomiyama, C. Wasi, T. Matsuda, S. Kusagawa, H. Sato, S. Oka, Y. Takebe, and M. Takiguchi. 2002. Cytotoxic T-cell recognition of HIV-1 cross-clade and clade-specific epitopes in HIV-1-infected Thai and Japanese patients. AIDS 16:701-711. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 21.Hokey, D. A., et al. 2008. Activation drives PD-1 expression during vaccine-specific proliferation and following lentiviral infection in macaques. Eur. J. Immunol. 38:1435-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibe, M., Y. Ikeda-Moore, K. Miwa, Y. Kaneko, S. Yokota, and M. Takiguchi. 1996. Role of strong anchor residues in the effective binding of 10-mer and 11-mer peptides to HLA-A*2402 molecules. Immunogenetics 44:233-241. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda-Moore, Y., H. Tomiyama, M. Ibe, S. Oka, K. Miwa, Y. Kaneko, and M. Takiguchi. 1998. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope derived from HIV-1 Gag protein. AIDS 12:2073-2074. [DOI] [PubMed] [Google Scholar]
- 24.Ishida, Y., Y. Agata, K. Shibahara, and T. Honjo. 1992. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11:3887-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. James, S. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. Korber, H. M. Coovadia, B. D. Walker, and P. J. R. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-774. [DOI] [PubMed] [Google Scholar]
- 26.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 27.Kinter, A. L., et al. 2008. The common γ-chain cytokines IL-2, IL-7, IL-15 and IL-21 induce the expression of programmed death-1 and its ligands. J. Immunol. 181:6738-6746. [DOI] [PubMed] [Google Scholar]
- 28.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, S. J., C. D. Peacock, K. Bahl, and R. M. Welsh. 2007. Programmed death-1 (PD-1) defines a transient and dysfunctional oligoclonal T cell population in acute homeostatic proliferation. J. Exp. Med. 204:2321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ljunggren, H. G., N. J. Stam, C. Ohlen, J. J. Neefjes, P. Hoglund, M. T. Heemels, J. Bastin, T. N. M. Shumacher, A. Townsend, K. Karre, and H. L. Ploegh. 1990. Empty MHC class I molecules come out in the cold. Nature 346:476-480. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura, H., M. Nose, H. Hiai, N. Minato, and T. Honjo. 1999. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 11:141-151. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 33.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, M., M. Makino, I. Kitajima, I. Maruyama, and M. M. Baba. 1997. HIV-1-infected myelomonocytic cells are resistant to Fas-mediated apoptosis: effect of tumor necrosis factor-alpha on their Fas expression and apoptosis. Med. Microbiol. Immunol. 186:11-17. [DOI] [PubMed] [Google Scholar]
- 35.Petrovas, C., J. P. Casazza, J. M. Brenchley, D. A. Price, E. Gostick, W. C. Adams, M. L. Precopio, T. Schacker, M. Roederer, D. C. Douek, and R. A. Koup. 2006. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 203:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, and A. J. McMichael. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 37.Sauce, D., J. R. Almeida, M. Larsen, L. Haro, B. Autran, G. J. Freeman, and V. Appay. 2007. PD-1 expression on human CD8 T cells depends on both state of differentiation and activation status. AIDS 21:2005-2013. [DOI] [PubMed] [Google Scholar]
- 38.Sharpe, A. H., and G. J. Freeman. 2002. The B7-CD28 superfamily. Nat. Rev. Immunol. 2:116-126. [DOI] [PubMed] [Google Scholar]
- 39.Streeck, H., Z. L. Brumme, M. Anastario, K. W. Cohen, J. S. Jolin, A. Meier, C. J. Brumme, E. S. Rosenberg, G. Alter, T. M. Allen, B. D. Walker, and M. Altfeld. 2008. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 5:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuber, G., S. Modrow, P. Hoglund, L. Franksson, J. Elvin, H. Wolf, K. Karre, and G. Klein. 1992. Assessment of major histocompatibility complex class I interaction with Epstein-Barr virus and human immunodeficiency virus peptides by evaluation of membrane H-2 and HLA in peptide loading-deficient cells. Eur. J. Immunol. 22:2697-2703. [DOI] [PubMed] [Google Scholar]
- 41.Tanabe, M., M. Sekimata, S. Ferrone, and M. Takiguchi. 1992. Structural and functional analysis of monomorphic determinants recognized by monoclonal antibodies reacting with the HLA class I alpha 3 domain. J. Immunol. 148:3202-3209. [PubMed] [Google Scholar]
- 42.Tomiyama, H., H. Akari, A. Adachi, and M. Takiguchi. 2002. Different effects of Nef-mediated HLA class I down-regulation on human immunodeficiency virus type 1-specific CD8+ T-cell cytolytic activity and cytokine production. J. Virol. 76:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomiyama, H., M. Fujiwara, S. Oka, and M. Takiguchi. 2005. Epitope-dependent effect of Nef-mediated HLA class I down-regulation on ability of HIV-1-specific CTLs to suppress HIV-1 replication. J. Immunol. 174:36-40. [DOI] [PubMed] [Google Scholar]
- 44.Trautmann, L., L. Janbazian, N. Chomont, E. A. Said, S. Gimmig, B. Bessette, M. R. Boulassel, E. Delwart, H. Sepulveda, R. S. Balderas, J. Routy, E. K. Haddad, and R. Sekaly. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198-1202. [DOI] [PubMed] [Google Scholar]
- 45.Yang, O. O., P. T. Nguyen, S. A. Kalams, T. Dorfman, H. G. Gottlinger, S. Stewart, I. S. Chen, S. Threlkeld, and B. D. Walker. 2002. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. J. Virol. 76:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokomaku, Y., H. Miura, H. Tomiyama, A. Kawana-Tachikawa, M. Takiguchi, A. Kojima, Y. Nagai, A. Iwamoto, Z. Matsuda, and K. Ariyoshi. 2004. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J. Virol. 78:1324-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, J. Y., Z. Zhang, X. Wang, J. L. Fu, J. Yao, Y. Jiao, L. Chen, H. Zhang, J. Wei, L. Jin, M. Shi, G. F. Gao, H. Wu, and F. S. Wang. 2007. PD-1 up-regulation is correlated with HIV-specific memory CD8+ T-cell exhaustion in typical progressors but not in long-term nonprogressors. Blood 109:4671-4678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.