Abstract
Retroviruses are both powerful evolutionary forces and dangerous threats to genome integrity. As such, they have imposed strong selective pressure on their hosts, notably triggering the emergence of restriction factors, such as TRIM5α, that act as potent barriers to their cross-species transmission. TRIM5α orthologues from different primates have distinct retroviral restriction patterns, largely dictated by the sequence of their C-terminal PRYSPRY domain, which binds the capsid protein of incoming virions. Here, by combining genetic and functional analyses of human and squirrel monkey TRIM5α, we demonstrate that the coiled-coil domain of this protein, thus far essentially known for mediating oligomerization, also conditions the spectrum of antiretroviral activity. Furthermore, we identify three coiled-coil residues responsible for this effect, one of which has been under positive selection during primate evolution, notably in New World monkeys. These results indicate that the PRYSPRY and coiled-coil domains cooperate to determine the specificity of TRIM5α-mediated capture of retroviral capsids, shedding new light on this complex event.
The genomes of higher organisms, including primates, contain large numbers of endogenous retroviruses, remnants of past infections established in their germ lines over millions of years (13). Exogenous retroviruses are still an ongoing threat, as illustrated by the human immunodeficiency virus (HIV) pandemic. Retroviruses have thus subjected higher species to formidable selective pressures, leading to the emergence of a number of host-encoded antiviral factors (42). TRIM5α is one such factor, which can restrict a range of retroviruses in a virus- and species-specific fashion (9, 16, 31, 34, 36, 40, 49). Although the mechanism of TRIM5α action is still not fully understood, it is known that it binds to and multimerizes around incoming capsids, inducing their premature uncoating (3, 5, 30, 37). The accelerated disassembly of the capsid impairs reverse transcription (RT), and the viral genome is also prevented from migrating to the cell nucleus (1, 16, 36, 39, 43).
TRIM5α encompasses a RING finger, a B-box type II and a coiled-coil domain, which together form the so-called RBCC or tripartite motif (TRIM), upstream of a C-terminal PRYSPRY domain (24, 27). Each of these regions has been demonstrated to participate in the antiviral function of TRIM5α, and yet a full understanding of their respective contributions is still lacking. A RING domain is often found in proteins that function as E3 ubiquitin ligases and was demonstrated to promote TRIM5α auto-ubiquitination and to regulate protein turnover (4, 10, 44). Deletion of the RING domain significantly affects restriction activity, although the exact role of the ubiquitin pathway in restriction is not fully understood (1, 3, 29, 32, 43). The B-box II domain plays an essential function in TRIM5α activity partly by mediating the higher-order self-assembly of TRIM5α, which potentiates restriction (6, 11, 19, 29). The coiled-coil region promotes the formation of multimers, a process necessary for efficient capsid binding (12, 15, 17, 22, 29, 37). Coiled-coil domains are found in many cellular proteins and are characterized by heptad repeats denoted (abcdefg)n with, typically, hydrophobic residues at positions a and d and polar/charged amino acids at positions e and g. Such domains are predicted to form α-helices with amphipathic properties responsible for homomultimerization (28). Finally, the PRYSPRY region directly binds to retroviral capsids, and species-specific sequence variations or engineered changes in its sequence can modify the spectrum of retroviruses restricted by given TRIM5α orthologues (21, 25, 38, 50).
Although the prominent role of the PRYSPRY domain in dictating the range of TRIM5α-mediated restriction is well established, comparative analyses of the sequences of primate TRIM5α genes reveal that residues located not only in this but also the coiled-coil domain have been subjected to positive selection during evolution (14, 26, 33). Correspondingly, a study of interspecies TRIM5α chimeric derivatives pointed to a possible influence of the coiled-coil domain on the specificity of murine leukemia virus (MLV) restriction (50).
Here, we took human and squirrel monkey TRIM5α proteins as starting material to examine the role of the TRIM5α coiled-coil domain in dictating the specificity of retrovirus restriction. We found that this region conditions specificity toward MLV derivatives, but not toward the primate lentiviruses simian immunodeficiency virus macaque (SIVmac) and human immunodeficiency virus type 2 (HIV2). Furthermore, we identified TRIM5α coiled-coil residues influencing target specificity, one of which has been under positive selection during primate evolution.
MATERIALS AND METHODS
Cell lines and culture.
Mus dunni tail fibroblasts (MDTF) and human embryonic kidney 293T cells (HEK 293T) were purchased from the American Type Culture Collection, and the squirrel monkey cell line Pindak was kindly provided by Greg Towers. All cell lines were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics (100 U of penicillin/ml and 100 mg of streptomycin/ml).
Plasmids.
MLV-based particles were produced using packaging constructs derived from Moloney MLV (pCIGPB), N- and B-tropic MLV (pCIG3-N and pCIG3-B) and the green fluorescent protein (GFP)-encoding construct pCNCG as described previously (21). HIV1-based vectors were produced with the packaging construct pCMVR8.74 and the vector pRRLsin PGK GFP. HIV2-based vectors were produced using pHIV2RODΔenvGFP that contains the HIV2 ROD proviral sequence with the env gene replaced with GFP. SIVmac GFP vectors were generated with the packaging construct pSIV4+ and the pR4SA vector kindly provided by D. Nègre and F. L. Cosset, respectively. The env construct for all viral productions was pMD2G expressing vesicular stomatitis virus G protein. The MLV plasmids encoding human and rhesus macaque TRIM5α (pLPCX-huTRIM5α-HA and pLPCX-rhTRIM5α-HA) were kindly provided by J. Sodroski (36). The squirrel monkey TRIM5α sequence was amplified from pCXCR-sqTRIM5α (kindly provided by G. Towers), adding a 3′ influenza virus hemagglutinin (HA) coding sequence by PCR (51), and inserted in the MLV vector construct pLPCX (Clontech), allowing for puromycin selection of transduced cells. The same approach was used to generate pLPCX expressing either C-terminal HA-tagged truncated forms (SH1 and SH1 V229L ΔPRYSPRY) or full-length C-terminal FLAG-tagged versions of different TRIM5α variants. Chimeras were constructed following standard subcloning methods. All of the site-directed mutagenesis on pLPCX-SH1-HA was performed by using the XL QuikChange mutagenesis kit (Stratagene). MLV capsid mutants have already been described (21). HA-and FLAG-tagged versions of the deletion mutant sqTRIM5α Δcoiled-coil were generated using overlapping, two-stage PCR as described previously (48). All of the primers used in the present study are listed in the Table S1 in the supplemental material.
Vector production.
MLV-, HIV1 and SIVmac-based GFP vectors were produced by CaPO4-mediated transient cotransfection of the retroviral vector, gag-pol and env encoding constructs at a ratio of 2.8:1.8:1 as described previously (21). HIV2 GFP vectors were produced by cotransfecting pHIV2RODΔenvGFP with pMD2G at a ratio of 7:1. All vector-containing supernatants were centrifuged, filtrated, and in some cases concentrated by ultracentrifugation. Titrations were performed on Fv1-null MDTF cells.
Generation of stable cell lines.
Retroviral vectors encoding TRIM5α variants were produced using the pLPCX-derived plasmids as described above. A total of 5 × 104 MDTF or Pindak cells were transduced with viral supernatants containing recombinant retroviral vectors and 3 days later were selected with puromycin (Sigma) at a concentration of 5 or 10 μg/ml, respectively, and were maintained continuously in the presence of the drug. TRIM5α expression levels were determined by immunoblot as described previously (21). The HA-tagged and FLAG-tagged proteins were detected by using peroxidase-conjugated rat monoclonal (clone 3F10; Roche) and mouse monoclonal anti-FLAG M2-peroxidase (Sigma) antibodies, respectively. Actin was used as a protein loading control and was detected by using a mouse monoclonal antibody (Chemicon), followed by a secondary sheep anti-mouse antibody conjugated to horseradish peroxidase.
Infection with GFP reporter vectors.
MDTF or Pindak stable cell lines were seeded at 2.5 × 104 cells in a 24-well plate and transduced 24 h later with six or three doses (in duplicate) of 2-fold serial dilutions of GFP reporter vectors. Cells were harvested 3 days posttransduction, fixed in 1% formaldehyde-containing phosphate-buffered saline (PBS), and resuspended in PBS 1% fetal calf serum. The percentage of GFP-positive cells was determined by flow cytometry using the Becton Dickinson FACScan. The results were analyzed with FlowJo 8.1.1 software.
Detection of reverse transcripts.
MDTF cells stably expressing TRIM5α variants or stably transduced with the empty pLPCX construct as a control were seeded at 105 in a six-well plate. N, B, Mo-MLV, HIV2, and SIVmac stocks encoding GFP were treated with DNase I (20 μg/ml) in the presence of MgCl2 (10 mM) for 30 min at 37°C and then used to transduce stable cell lines in duplicate. As a PCR-negative control, MDTF stably transduced with the empty vector were treated 2 h before and during infection with azidothymidine (AZT; 62.5 μM; Calbiochem). Cells were then harvested before or 8 h posttransduction and used for DNA extraction (DNeasy tissue extraction kit from Qiagen), while some were put back in culture and used for flow cytometry 3 days later. DNA was subjected to quantitative real-time PCR (ABI Prism 7900) using TaqMan Universal master mix (Applied Biosystems) and standard procedures. Reverse transcripts were detected using primers and TaqMan probe against GFP (see Table S1 in the supplemental material), and Titin was used as a normalization gene (see Table S1 in the supplemental material). The amount of reverse transcripts per cell was calculated with a standard curve made with serial dilutions of known copy numbers of a GFP-containing plasmid and of DNA extracts from a determined number of cells.
Evolutionary analyses.
The number of nonsynonymous (KA) over synonymous (KS) substitutions was estimated for each branch of the TRIM5α gene tree, based on the accepted primate phylogeny, and using the free-ratio model of the codeml tool of the PAML program package (7, 45). For this substitutional pattern analysis, two independent runs were performed, one with the entire TRIM5α coding sequence (1,479 bp) and a second with only the TRIM5α coiled-coil coding sequence (339 bp), using 33 TRIM5α primate sequences, all available in the National Center for Biotechnology Information database, from apes, Old and New World monkeys, and prosimian sources (see Table S2 in the supplemental material for accession numbers). To examine positive selection over the entire TRIM5α sequence and specifically on coiled-coil residues in different primate families, we used a hypothesis testing analysis by comparing two null models (M1a and M7 [46, 47]), which assume two site classes (sites under purifying selection and neutrally evolving sites) with two alternative models (M2a and M8 [46, 47]), which adds a third site class that allows for sites with KA/KS > 1) using a likelihood ratio test. Evaluation with the chi-square test assumed two degrees of freedom, as suggested by (47). The null model (M1a-M7) could be rejected when the P value was described above the significance level (0.05) (see Table S3 in the supplemental material). We then used empirical Bayes methods to identify positively selected sites (Post prob., >0.95). In this second analysis, probabilities of positively selected codons were calculated by using the entire primate lineage or individual families (Hominoids-OWMs-NWMs) (see Table S3 in the supplemental material).
RESULTS
Squirrel monkey and human TRIM5α both impair the RT step.
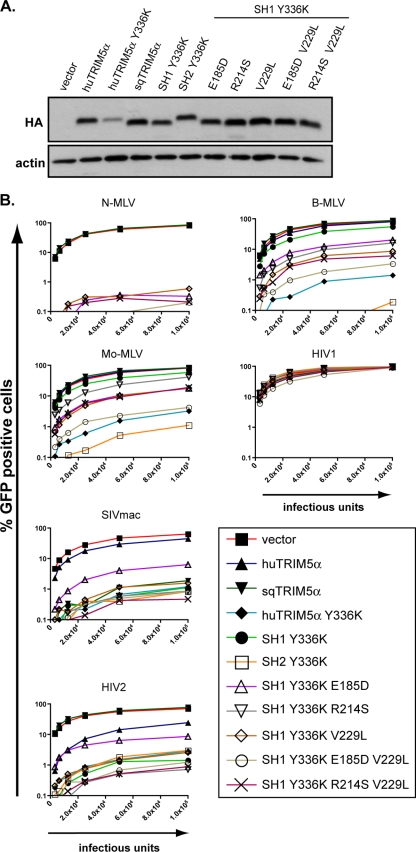
The present study was originally initiated to investigate the mechanism of TRIM5α action. It was recently reported that squirrel monkey TRIM5α (sqTRIM5α), in contrast with its human counterpart (huTRIM5α), can restrict retroviruses without significantly affecting RT (51). To examine the region of the protein involved in governing this differential phenotype, we constructed a series of chimeras between the two orthologues (Fig. 1A). Furthermore, to broaden the spectrum of retroviruses under study, we took advantage of a huTRIM5α derivative with a point mutation at tyrosine336 (Y336K) in the PRYSPRY domain that expands its range of antiretroviral activity (5, 21). Cell lines stably expressing either wild-type or Y336K huTRIM5α, sqTRIM5α or the various chimeras were generated by retroviral vector-mediated transduction of permissive Fv1-null MDTF (Fig. 1B). We then exposed these cells to GFP-expressing vectors derived from N-tropic, B-tropic, or Moloney strains of MLV or derived from SIVmac or HIV2, quantified the intermediate RT products early after infection and scored infectivity by flow cytometry 3 days later (Fig. 1C).
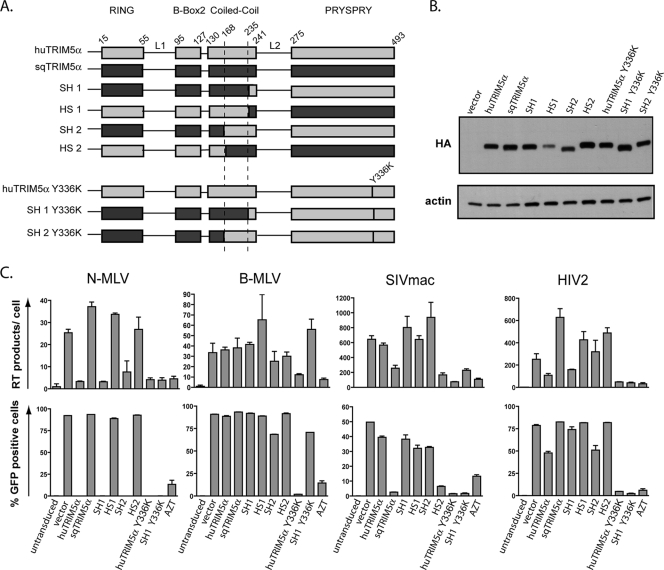
FIG. 1.
The TRIM5α RBCC influences the specificity of retroviral restriction. (A) Schematic representation of squirrel monkey (sqTRIM5α) and either human wild-type TRIM5α (huTRIM5α) or Y336K mutant (huTRIM5α Y336K), and of the derived chimeras. The four domains (RING, B-Box2, coiled-coil, and PRYSPRY) with the linker regions 1 and 2 (L1 and L2) are depicted. The numbering of positions delimiting the domains is based on the human TRIM5α sequence. (B) Western blot analysis of extracts from MDTF cells stably transduced with a retroviral control vector (vector) or with vectors expressing HA-tagged versions of TRIM5α and derivatives, using HA (top)- and actin (bottom)-specific antibodies. (C) MDTF cell lines expressing the indicated TRIM5α molecules or transduced with a control vector (vector) were challenged in duplicate with a single dose of MLV, SIVmac, HIV2-, and HIV1-based GFP vectors initially titered on permissive MDTF cells, using infection of empty vector-transduced cells in the presence of azidothymidine (AZT) as a negative control. Levels of intermediate minus-strand DNA RT products were detected by real-time PCR 8 h postinfection (top), while infectivity was scored 3 days after infection by flow cytometry (bottom). The chimera HS1 restricts SIVmac only weakly, a phenotype probably caused by its low steady-state level of expression. These results are representatives of two independent experiments.
As expected, wild-type huTRIM5α potently blocked N-MLV and modestly blocked HIV2, but not B-MLV or SIVmac, whereas sqTRIM5α inhibited SIVmac only (Fig. 1C) (9, 16, 30, 35, 49, 51). HuTRIM5α inhibited N-MLV prior to or during RT, as indicated by the decreased amount of RT products detected in its presence (Fig. 1C) (16, 39). Surprisingly, the sqTRIM5α-mediated blockade of SIVmac similarly correlated with a reduction in viral cDNA synthesis, albeit to a lesser extent than that observed for huTRIM5α Y336K despite their comparable activities against SIVmac. In addition, there was a good correlation between inhibition of infection and impairment of RT for all chimeras between sqTRIM5α and wild-type huTRIM5α (Fig. 1C). We conclude that, despite possible minor differences in the efficiencies with which human and sqTRIM5α impair RT, the two orthologues block retroviruses through similar general mechanisms.
The coil-coiled domain influences the target specificity of TRIM5α.
Restriction analysis of the chimeras between squirrel monkey and human TRIM5α led to another unexpected finding. The Y336K mutant of huTRIM5α had a spectrum of antiviral activity broadened toward B-MLV, SIVmac, and HIV2 as previously described (5, 21). However, the SH1 Y336K chimera, which harbors the modified PRYSPRY Y336K domain and the so-called linker region L2 from huTRIM5α tethered to the sqTRIM5α RBCC moiety, could no longer block B-MLV, whereas it retained full activity against SIVmac and HIV2 (Fig. 1C). This suggested that the antiviral specificity of TRIM5α is not solely dictated by the PRYSPRY domain but also depends on elements within the RBCC region.
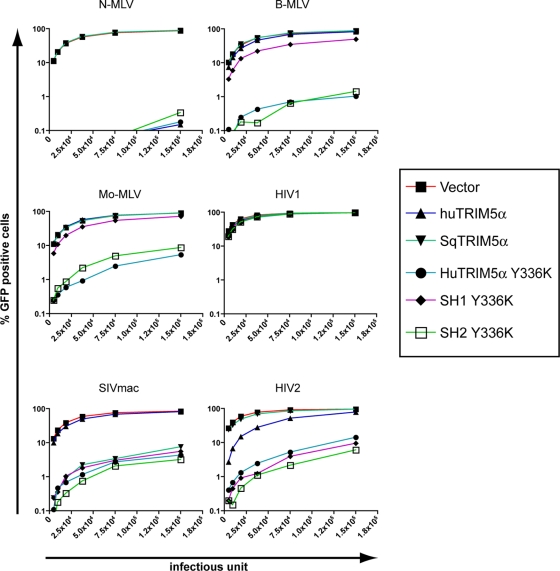
To probe this issue further, we generated a MDTF cell line expressing a chimera (termed SH2 Y336K) that only differs from SH1 Y336K in the presence of the distal two-thirds of the huTRIM5α coiled-coil domain (Fig. 1A and B). This modification was sufficient to restore full antiviral activity against B-MLV, while preserving restriction of SIVmac and HIV2 (Fig. 2). Inclusion of the huTRIM5α distal coiled-coil domain was also necessary to confer activity against Mo-MLV, previously found to be restricted by huTRIM5α Y336K (5, 21) (Fig. 2). In contrast, similar swaps between the rhesus macaque (rh) and human TRIM5α coiled-coil domains had no effect on TRIM5α Y336K target range (see Fig. S1 in the supplemental material).
FIG. 2.
The coiled-coil domain can condition TRIM5α antiretroviral specificity. MDTF cells expressing the indicated squirrel monkey and human TRIM5α Y336K chimeras (see Fig. 1A), or transduced with an empty vector (vector), were challenged with serial 2-fold dilutions of MLV-, SIVmac, HIV2-, or HIV1-based GFP vectors initially titered on parental MDTF cells. Infections were scored 72 h postinfection by flow cytometry. The data are representative of at least three independent experiments, and the restriction activities of each of the TRIM5α variants were obtained using two independent stable cell lines.
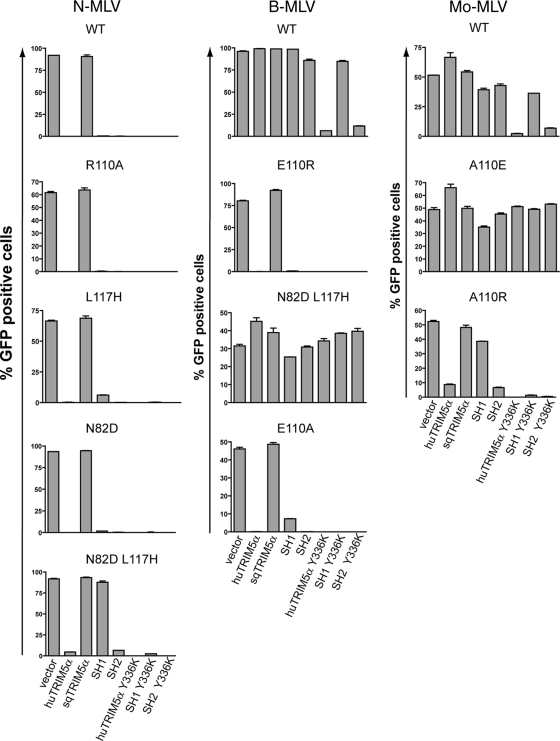
We previously demonstrated that specific amino acids within the retroviral capsid influence susceptibility to restriction (5, 21). To ask whether sequences within the coiled-coil domain influence this process, cells expressing the SH1 and SH2 chimeras (Fig. 1A) were infected with a series of N-, B-, and Mo-MLV derivatives mutated at capsid positions previously demonstrated to affect sensitivity to huTRIM5α (18, 21, 49). We found that, despite both harboring a wild-type human TRIM5α PRYSPRY domain, the SH1 chimera was less potent than SH2 in blocking B- or Mo-MLV harboring a single point mutation at position 110 (B E110A or Mo A110R) (Fig. 3). Similarly, dual mutations at positions 82 and 117 of N-MLV (N N82D L117H) conferred complete protection from SH1 but not from SH2 (Fig. 3). These results confirm the influence of the coiled-coil domain on the specificity of TRIM5α capsid recognition.
FIG. 3.
The coiled-coil domain modulates recognition of MLV derivatives by human wild-type PRYSPRY. MDTF-derived cell lines expressing indicated TRIM5α chimeras were transduced in duplicate with a single dose of N-MLV (left panels), B-MLV (middle panels), and Mo-MLV (right panels) GFP vectors harboring either a wild-type (WT) or the indicated mutant capsid. Infectivity was scored 72 h later by flow cytometry. The data are representative of two independent experiments.
Mapping coiled-coil amino acid determinants involved in restriction specificity.
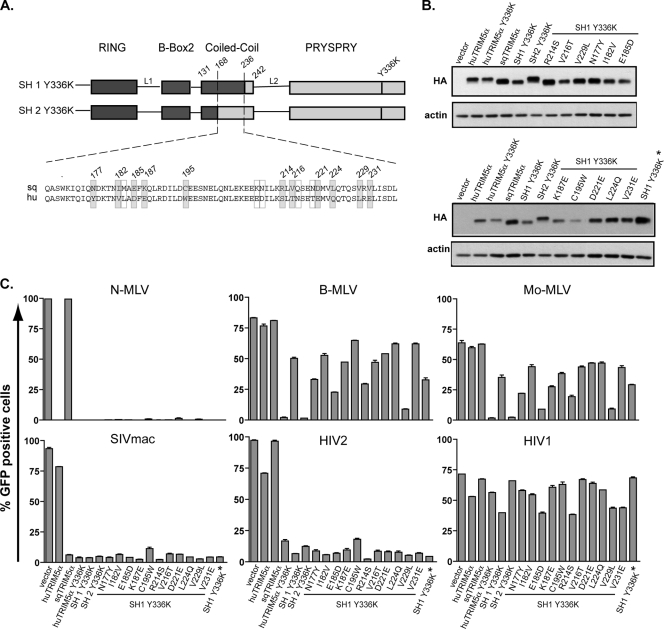
There are 17 amino acid differences between the coiled-coil domains of human and squirrel monkey TRIM5α (Fig. 4A). Interestingly, three of these (at positions 214, 216, and 229) were previously identified as having undergone positive selection during primate evolution (33). To identify the residues important for modulating PRYSPRY-conferred target specificity, we individually replaced the coiled-coil squirrel monkey TRIM5α amino acids by their human counterparts at 11 of 17 positions within the context of the SH1 Y336K chimera (Fig. 4B). All of the resulting derivatives conserved the ability to restrict N-MLV, SIVmac, and HIV2, while none could significantly block HIV1. Interestingly, single mutations at three different positions (E185D, R214S, and V229L) rendered SH1 Y336K more potent in blocking both B- and Mo-MLV, albeit to various degrees (Fig. 4C). Although introducing human-specific residues at both positions 214 and 229 did not result in any further enhancement, combining this type of modification at amino acids 185 and 229 almost completely restored the ability of the squirrel coiled-coil domain to support TRIM5α-mediated restriction of B- and Mo-MLV (Fig. 5).
FIG. 4.
Positions 186, 214, and 229 of the CC domain condition TRIM5α specificity. (A) Amino acid sequence alignment of the region encompassing positions 168 to 236 derived from squirrel monkey (sq) and human (hu) TRIM5α. Differences are framed, with residues targeted by site-directed mutagenesis in gray. Numbering is based on the sqTRIM5α sequence. (B) MDTF cell lines stably expressing indicated HA-tagged TRIM5α derivatives were engineered by retroviral transduction and analyzed by Western blotting. Here, the cell line “SH1 Y336K*” was included in this experiment to show that there is only a minor increase in restriction despite the high overexpression of this TRIM5α chimera. (C) These cells were transduced in duplicate with N-MLV-, B-MLV-, Mo-MLV-, SIVmac-, HIV2-, or HIV1-derived GFP expressing vectors, scoring infectivity 3 days later by flow cytometry. The restriction activities from the relevant point mutants were reproduced in two independent experiments.
FIG. 5.
Combined changes at critical positions in the coiled-coil domain increase the restriction activity toward specific retroviruses. (A) MDTF cell lines stably expressing the indicated HA-tagged TRIM5α derivatives were analyzed by Western blotting with HA- and actin-specific antibodies. (B) Infectivity assays using the indicated cell lines and vectors and performed as described in Fig. 2. The data are representative of two independent experiments.
The sqTRIM5α coiled-coil domain is functional for oligomerization.
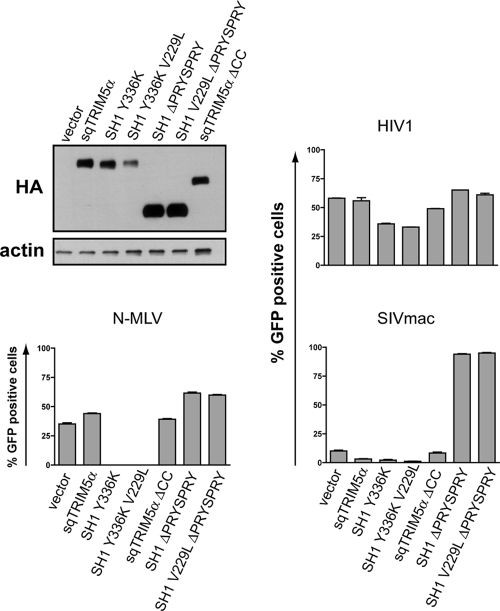
The main known function of the TRIM5α coiled-coil domain is to mediate dimerization, a prerequisite for capsid binding and, in turn, restriction (12, 15, 17, 22, 29, 37). The inability of the squirrel monkey coiled-coil to support restriction of B-and Mo-MLV is unlikely to stem from a defect in oligomerization given the potent restriction activity of wild-type sqTRIM5α against SIVmac and also the preserved activity of SH1 Y336K against SIVmac and HIV2. However, while the importance of TRIM5α oligomerization to promote capsid binding is well established, it is possible that restriction of some retroviruses such as SIVmac by some alleles of TRIM5α does not require oligomerization. To explore this possibility, we sought to determine whether the squirrel monkey coiled-coil domain was functional for oligomerization.
TRIM5α derivatives that oligomerize but lack the PRYSPRY domain can exert dominant-negative effects on retroviral restriction (29, 36). When we overexpressed such truncated mutants of both SH1 (SH1 ΔPRYSPRY) and SH1 V229L (SH1 V229L ΔPRYSPRY) in squirrel monkey cells that endogenously produce sqTRIM5α and hence inhibit SIVmac, restriction was relieved, while the infectivity of unrestricted viruses such as HIV1 and N-MLV was not affected (Fig. 6). This suggests that both wild-type and mutant forms of the sqTRIM5α coiled coil can mediate hetero-oligomerization with the endogenous sqTRIM5α proteins, although we cannot completely exclude the possibility that these truncation mutants titrate a putative cellular cofactor. We additionally confirmed that HA- and FLAG-tagged versions of wild-type sqTRIM5 could be efficiently coprecipitated, as could the SH1 Y336K and SH1 Y336K V229L chimeras (data not shown). Even though such associations may be indirect within the context, for instance, of cytoplasmic bodies, our results altogether suggest that the squirrel coiled-coil domain is functional for oligomerization.
FIG. 6.
The squirrel monkey coiled-coil domain can efficiently mediate multimerization. Squirrel monkey Pindak cells stably expressing the indicated full-length or truncated versions of TRIM5α were analyzed by Western blotting (upper left panel) and then challenged with HIV1, N-MLV, and SIVmac GFP vectors and analyzed 72 h later by flow cytometry. The figure shows infections performed in duplicate using a single dose of each GFP vector, but similar results were obtained using two other vector doses.
Position 229 of TRIM5α has been under positive selection mainly in New World monkeys.
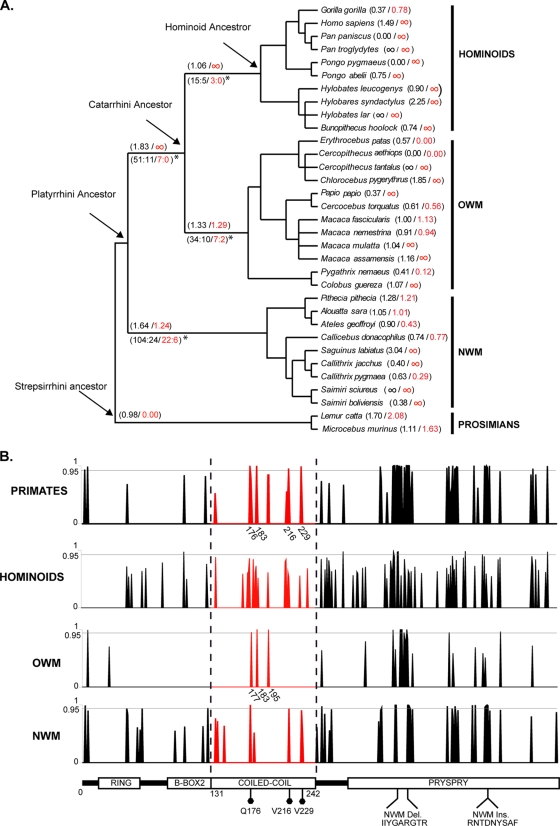
The observed coiled-coil domain-mediated modulation of TRIM5α specificity is consistent with previous evolutionary analyses (14, 26, 33). Signs of positive selection, most likely caused by antagonistic interactions between retroviruses and TRIM5α throughout primate evolution, can indeed be noted at sites distributed not only in the PRYSPRY but also in the coiled-coil domain.
To examine this positive selection within the coiled-coil domain, we performed evolutionary analyses both on full-length TRIM5α and on the coiled-coil domain alone, using genetic data from 33 different primate species (including prosimians). We used a codon-based maximum-likelihood procedure to estimate the ratio of nonsynonymous (KA) over synonymous (KS) substitutions per site. It revealed that positive selection (KA/KS > 1) has been operating on TRIM5α as previously reported (13, 25, 32) but also individually on the coiled-coil domain of TRIM5α in ancestors of the three primate families, e.g., hominoids, Old World monkeys (OWMs) and New World monkeys (NWMs) (Fig. 7A). This suggests that the coiled-coil domain has participated in a genetic conflict over at least the last 40 million years.
FIG. 7.
The coiled-coil domain has been under position selection during primate evolution. (A) Phylogenetic tree of the TRIM5α gene. The KA/KS ratios for the Hominoids, Catharrini and Platyrrhini ancestors, as well as for current species are indicated. The KA/KS ratio based on sequences of the full-length TRIM5α gene or the coiled-coil domain only are indicated in black or red font, respectively. When KA>0 and KS = 0, the lineages are labeled as “∞”. For the ancestors highlighted with an asterisk, the estimated number of nonsynonymous and synonymous substitutions are indicated. (B) Probabilities of positively selected codons performed on the entire primate lineage or by individual families (Hominoids-OWMs-NWMs). Codons in the coiled-coil domain are indicated in red. Positions in that domain that have been under positive selection in primates or in the OWM lineage are displayed in the histograms based on squirrel monkey TRIM5α numbering. At the bottom, a schematic representation of TRIM5α is shown with codons in the coiled-coil domain that have been under positive selection (filled hexagons). Specific amino acid insertions (Ins.) and deletions (Del.) that occurred in TRIM5α sequences in the New World monkey lineage are illustrated.
Using models that allow for different KA/KS rates at different sites and making use of all primate sequences, signs of positive selection were revealed at four positions in the coiled-coil domain (176, 183, 216, and 229) (Fig. 7B). In contrast, when each primate family phylogeny was analyzed separately, the NWMs presented one position 176 under strong positive selection (Post prob. > 0.97), while two additional positions (216 and 229) were identified, albeit with lower confidence (Post prob. > 0.90). Compared to the other primate families (hominoids and OWMs), it seems that selective pressures exerted on these positions occurred mainly in the NWMs, in particular for residue 229. This is consistent with our observation that this residue influences the spectrum of squirrel monkey TRIM5α-mediated retroviral restriction (Fig. 7B.). In both codon- and lineage-specific analyses, the TRIM5α coiled-coil region of NWM showed signs of positive selection.
DISCUSSION
TRIM5α-mediated restriction requires the recognition of incoming retroviral capsids, a process primarily mediated by the PRYSPRY region of the cellular protein (25, 29, 37, 38). Here, we reveal that its coiled-coil domain also partakes in TRIM5α antiviral specificity, by demonstrating that sequence divergence in this region between human and squirrel monkey TRIM5α contributes to explaining differences in the spectrum of restriction induced by these two orthologues, in a PRYSPRY-independent fashion. Furthermore, by substituting human residues for their squirrel monkey counterparts, we identified three positions in the coiled-coil (amino acids 185, 214 and 229) that influence the spectrum of restriction induced by TRIM5α. Interestingly, one of them (amino acid 229) has been subjected to intense positive selection, particularly in the New World monkey lineage.
Short of structural data on the TRIM5α-capsid complex or even on full-length TRIM5α, one can only speculate on how the coiled-coil domain affects retroviral capsid recognition. At least three models can be envisioned. First, the coiled-coil could be a site of interaction with a cellular factor required for the recognition and/or the effector function of TRIM5α toward some MLV strains and derivatives (blocked by human but not squirrel monkey coiled-coil-containing TRIM5α chimeras) but not toward SIVmac and HIV2 (blocked by both types of chimeras). Second, the coiled-coil domain could directly contact the viral capsid, either near or at a distance from the PRYSPRY binding site. Finally, the coiled-coil could indirectly influence specificity by affecting the avidity of TRIM5α for given retroviral capsids.
TRIM5α dimerization is dependent on the coiled-coil domain and is required for capsid binding (12, 37). However, the inability of the squirrel monkey coiled-coil domain to support TRIM5α-mediated restriction of B- and Mo-MLV was unlikely to be caused by a failure to self-associate, because (i) a TRIM5α chimera containing the squirrel monkey coiled-coil domain efficiently restricted SIVmac and HIV2 and (ii) overexpression of a truncated mutant bearing the squirrel monkey coiled-coil and lacking the PRYSPRY domain exerted a strong dominant-negative effect over endogenous sqTRIM5α. Although this latter experiment demonstrated that truncated proteins can hetero-oligomerize with full-length sqTRIM5α, it does not exclude the possibility that squirrel monkey coiled-coil domain displays subtle impairments in its affinity to mediate hetero-oligomerization and/or homo-oligomerization of full-length proteins. Alternatively, it may be that the formation of higher-order TRIM5α complexes, previously documented to be dependent on the B-box II domain, is additionally influenced by the coiled-coil domain (6, 19). Such higher-order TRIM5α multimers were shown to contribute to restriction activity particularly when the PRYSPRY domain has a low affinity for the capsid (20). Here, we found an influence of the coiled-coil domain on TRIM5α bearing a PRYSPRY Y336K domain capable of recognizing N-, B-, Mo-MLV, SIVmac, and HIV2. As only restriction activities against B- and Mo-MLV were affected by changes in the coiled-coil domain, and since there is no evidence suggesting that the affinity of PRYSPRY Y336K toward these two retroviruses is weaker than toward SIVmac and HIV2, it is unlikely that the coiled-coil and B-box II domains play similar roles in restriction.
Alternatively, it is tempting to postulate that changes in the coiled-coil alter the avidity of TRIM5α for the capsid by influencing the orientation and/or the spacing of the two PRYSPRY domains within a dimer. Such influence would likely be subtle, as substituting one coiled-coil region for another in our chimeras did not affect the restriction of SIVmac and HIV2 but only of B- and Mo-MLV. Similarly, fusing the squirrel monkey TRIM5α coiled-coil domain to the wild-type human PRYSPRY yielded a chimera still potently active against N-MLV displaying a wild-type capsid (CA) but not a capsid mutated at positions 82 (CA82) and 117 (CA117). This is consistent with a previous report indicating that full restriction of N-MLV harboring a mutation at CA117 required both the coiled-coil and the PRYSPRY domains of huTRIM5α, whereas HIV1 restriction by rhesus TRIM5α involved only its PRYSPRY domain (50). Here, we documented an influence of the coiled-coil domain only on MLV derivatives but not on SIVmac and HIV2, suggesting that the coiled-coil domain might be required for MLV but not for lentivirus restriction. This may reflect general differences in capsid structures with that of MLV being spherical and that of lentiviruses conical.
An influence of the coiled-coil domain on TRIM5α specificity was previously suggested by the high degree of within-species polymorphism of this factor in two OWM species, where many nonsynonymous differences clustered in both the coiled-coil and PRYSPRY domains (14, 23, 41). Some polymorphic sites were shared between the two species, suggesting their emergence before speciation and their maintenance during evolution by balancing selection. Interestingly, conservation of multiples alleles of TRIM5α in these two species is consistent with a model in which variations not only in the PRYSPRY region but also within the coiled-coil domain confer a selective advantage for the host perhaps by expanding the breath of antiretroviral protection.
Interspecies variations of TRIM5α also support a role for the coiled-coil in target specificity. Indeed, our phylogenetic analyses of 33 primate TRIM5α sequences reveal that sites in both the PRYSPRY and the coiled-coil have been under positive selection during primate evolution. Some of the positively selected coiled-coil residues identified here differ from ones outlined in a previous study (33). This most likely results from our inclusion of a higher number of sequences. Of particular importance, we found position 229 to influence restriction specificity and to have undergone positive selection almost exclusively in NWMs. This suggests a critical role for this residue in conditioning resistance to retroviruses that have been pathogenic in this lineage. Retroviruses related to the current human endogenous retroviruses (HERVs) are possible candidates as the genomes of NWMs, in contrast to those from hominids and OWMs, either lack or have few copies of HERVs, with the exception of HERV-L (2, 8). However, it is noteworthy that the high variability found at position 229 when comparing different NWM species could reflect a high degree of within-species variation as described for OWMs (23). Such polymorphism shared between closely related species would also suggest that this position of the coiled-coil domain has been involved in genetic conflicts during NWM evolution.
In sum, the present study demonstrates that the TRIM5α coiled-coil domain can influence the specificity of retroviral restriction. Further investigations into the mechanism of this phenomenon will help us understand how TRIM5α recognizes and blocks retroviruses and why polymorphisms in the coiled-coil have been selected during evolution in at least two primate species.
Supplementary Material
Acknowledgments
This study was supported by the Swiss National Science Foundation.
We thank Greg Towers and Joseph Sodroski for supplying plasmids; Amalio Telenti, Nadia Rahm, and Priscilla Turelli for advice; Helen Rowe for reading the manuscript; and Séverine Reynard and Sujana Nylakonda for technical assistance.
Footnotes
Published ahead of print on 10 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, J. L., E. M. Campbell, X. Wu, N. Vandegraaff, A. Engelman, and T. J. Hope. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80:9754-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benit, L., J. B. Lallemand, J. F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 73:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, E. M., O. Perez, J. L. Anderson, and T. J. Hope. 2008. Visualization of a proteasome-independent intermediate during restriction of HIV1 by rhesus TRIM5α. J. Cell Biol. 180:549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz-Griffero, F., X. Li, H. Javanbakht, B. Song, S. Welikala, M. Stremlau, and J. Sodroski. 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349:300-315. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Griffero, F., M. Perron, K. McGee-Estrada, R. Hanna, P. V. Maillard, D. Trono, and J. Sodroski. 2008. A human TRIM5α B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology 378:233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Griffero, F., X. R. Qin, F. Hayashi, T. Kigawa, A. Finzi, Z. Sarnak, M. Lienlaf, S. Yokoyama, and J. Sodroski. 2009. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83:10737-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman, M. 1999. The genomic record of Humankind's evolutionary roots. Am. J. Hum. Genet. 64:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenwood, A. D., A. Stengel, V. Erfle, W. Seifarth, and C. Leib-Mosch. 2005. The distribution of pol containing human endogenous retroviruses in non-human primates. Virology 334:203-213. [DOI] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. U. S. A. 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 11.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 12.Javanbakht, H., W. Yuan, D. F. Yeung, B. Song, F. Diaz-Griffero, Y. Li, X. Li, M. Stremlau, and J. Sodroski. 2006. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353:234-246. [DOI] [PubMed] [Google Scholar]
- 13.Jern, P., and J. M. Coffin. 2008. Effects of retroviruses on host genome function. Annu. Rev. Genet. 42:709-732. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, W. E., and S. L. Sawyer. 2009. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics. 61:163-176. [DOI] [PubMed] [Google Scholar]
- 15.Kar, A. K., F. Diaz-Griffero, Y. Li, X. Li, and J. Sodroski. 2008. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5α protein. J. Virol. 82:11669-11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U. S. A. 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langelier, C. R., V. Sandrin, D. M. Eckert, D. E. Christensen, V. Chandrasekaran, S. L. Alam, C. Aiken, J. C. Olsen, A. K. Kar, J. G. Sodroski, and W. I. Sundquist. 2008. Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82:11682-11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lassaux, A., M. Sitbon, and J. L. Battini. 2005. Residues in the murine leukemia virus capsid that differentially govern resistance to mouse Fv1 and human Ref1 restrictions. J. Virol. 79:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X., and J. Sodroski. 2008. The TRIM5α B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82:11495-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, X., B. Song, S. H. Xiang, and J. Sodroski. 2007. Functional interplay between the B-box 2 and the B30.2(SPRY) domains of TRIM5α. Virology 366:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillard, P. V., S. Reynard, F. Serhan, P. Turelli, and D. Trono. 2007. Interfering residues narrow the spectrum of MLV restriction by human TRIM5α. PLoS Pathog. 3:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman, R. M., L. Hall, M. Connole, G. L. Chen, S. Sato, E. Yuste, W. Diehl, E. Hunter, A. Kaur, G. M. Miller, and W. E. Johnson. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5α. Proc. Natl. Acad. Sci. U. S. A. 103:19134-19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defense. Nat. Rev. Microbiol. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 25.Ohkura, S., M. W. Yap, T. Sheldon, and J. P. Stoye. 2006. All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80:8554-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz, M., N. Guex, E. Patin, O. Martin, I. Xenarios, A. Ciuffi, L. Quintana-Murci, and A. Telenti. 2009. Evolutionary trajectories of primate genes involved in HIV pathogenesis. Mol. Biol. Evol. 26:2865-2875. [DOI] [PubMed]
- 27.Ozato, K., D. M. Shin, T. H. Chang, and H. C. Morse III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parry, D. A., R. D. Fraser, and J. M. Squire. 2008. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J. Struct. Biol. 163:258-269. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 81:2138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rold, C. J., and C. Aiken. 2008. Proteasomal degradation of TRIM5α during retrovirus restriction. PLoS Pathog. 4:e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 35.Song, B., H. Javanbakht, M. Perron, D. H. Park, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. U. S. A. 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, S. J., B. L. Webb, C. Maplanka, R. M. Newman, E. J. Verschoor, J. L. Heeney, and G. J. Towers. 2008. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 82:7243-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf, D., and S. P. Goff. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, X., J. L. Anderson, E. M. Campbell, A. M. Joseph, and T. J. Hope. 2006. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV1 reverse transcription and infection. Proc. Natl. Acad. Sci. U. S. A. 103:7465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamauchi, K., K. Wada, K. Tanji, M. Tanaka, and T. Kamitani. 2008. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 275:1540-1555. [DOI] [PubMed] [Google Scholar]
- 45.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, Z., W. S. Wong, and R. Nielsen. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107-1118. [DOI] [PubMed] [Google Scholar]
- 48.Yap, M. W., G. B. Mortuza, I. A. Taylor, and J. P. Stoye. 2007. The design of artificial retroviral restriction factors. Virology 365:302-314. [DOI] [PubMed] [Google Scholar]
- 49.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV1 and murine leukemia virus. Proc. Natl. Acad. Sci. U. S. A. 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human TRIM5α leads to HIV1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 51.Ylinen, L. M., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of human immunodeficiency virus type 2 and simian immunodeficiency virus SIVmac by TRIM5α alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.