Abstract
Complex N-glycans flank the receptor binding sites of the outer domain of HIV-1 gp120, ostensibly forming a protective “fence” against antibodies. Here, we investigated the effects of rebuilding this fence with smaller glycoforms by expressing HIV-1 pseudovirions from a primary isolate in a human cell line lacking N-acetylglucosamine transferase I (GnTI), the enzyme that initiates the conversion of oligomannose N-glycans into complex N-glycans. Thus, complex glycans, including those that surround the receptor binding sites, are replaced by fully trimmed oligomannose stumps. Conversely, the untrimmed oligomannoses of the silent domain of gp120 are likely to remain unchanged. For comparison, we produced a mutant virus lacking a complex N-glycan of the V3 loop (N301Q). Both variants exhibited increased sensitivities to V3 loop-specific monoclonal antibodies (MAbs) and soluble CD4. The N301Q virus was also sensitive to “nonneutralizing” MAbs targeting the primary and secondary receptor binding sites. Endoglycosidase H treatment resulted in the removal of outer domain glycans from the GnTI- but not the parent Env trimers, and this was associated with a rapid and complete loss in infectivity. Nevertheless, the glycan-depleted trimers could still bind to soluble receptor and coreceptor analogs, suggesting a block in post-receptor binding conformational changes necessary for fusion. Collectively, our data show that the antennae of complex N-glycans serve to protect the V3 loop and CD4 binding site, while N-glycan stems regulate native trimer conformation, such that their removal can lead to global changes in neutralization sensitivity and, in extreme cases, an inability to complete the conformational rearrangements necessary for infection.
The intriguing results of a recent clinical trial suggest that an effective HIV-1 vaccine may be possible (97). Optimal efficacy may require a component that induces broadly neutralizing antibodies (BNAbs) that can block virus infection by their exclusive ability to recognize the trimeric envelope glycoprotein (Env) spikes on particle surfaces (43, 50, 87, 90). Env is therefore at the center of vaccine design programs aiming to elicit effective humoral immune responses.
The amino acid sequence variability of Env presents a significant challenge for researchers seeking to elicit broadly effective NAbs. Early sequence comparisons revealed, however, that the surface gp120 subunit can be divided into discrete variable and conserved domains (Fig. 1A) (110), the latter providing some hope for broadly effective NAb-based vaccines. Indeed, the constraints on variability in the conserved domains of gp120 responsible for binding the host cell receptor CD4, and coreceptor, generally CCR5, provide potential sites of vulnerability. However, viral defense strategies, such as the conformational masking of conserved epitopes (57), have made the task of eliciting bNAbs extremely difficult.
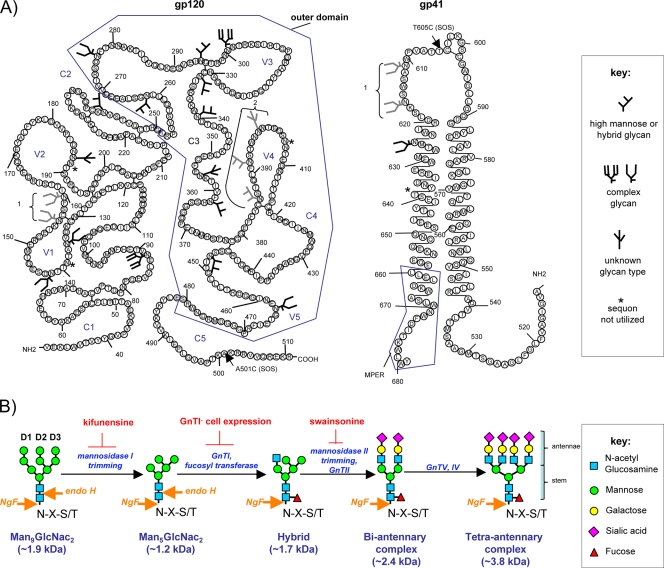
FIG. 1.
Glycan biosynthesis and distribution on gp120 and gp41. (A) Putative carbohydrate modifications are shown on gp120 and gp41 secondary structures, based on various published works (26, 42, 63, 74, 119, 128). The gp120 outer domain is indicated, as are residues that form the SOS gp120-gp41 disulfide bridge. The outer domain is divided into neutralizing and silent faces. Symbols distinguish complex, oligomannose, and unknown glycans. Generally, the complex glycans of the outer domain line the receptor binding sites of the neutralizing face, while the oligomannose glycans of the outer domain protect the silent domain (105). Asterisks denote sequons that are unlikely to be utilized, including position 139 (42), position 189 (26, 42), position 406 (42, 74), and position 637 (42). Glycans shown in gray indicate when sequon clustering may lead to some remaining unused, e.g., positions 156 and 160 (42, 119), positions 386, 392, and 397 (42), and positions 611 and 616 (42). There is also uncertainty regarding some glycan identities: glycans at positions 188, 355, 397, and 448 are not classified as predominantly complex or oligomannose (26, 42, 63, 128). The number of mannose moieties on oligomannose glycans can vary, as can the number of antennae and sialic acids on complex glycans (77). The glycan at position 301 appears to be predominantly a tetra-antennary complex glycan, as is the glycan at position 88, while most other complex glycans are biantennary (26, 128). (B) Schematic of essential steps of glycan biosynthesis from the Man9GlcNAc2 precursor to a mature multiantennary complex glycan. Mannosidase I progressively removes mannose moieties from the precursor, in a process that can be inhibited by the drug kifunensine. GnTI then transfers a GlcNAc moiety to the D1 arm of the resulting Man5GlcNAc2 intermediate, creating a hybrid glycan. Mannose trimming of the D2 and D3 arms then allows additional GlcNAc moieties to be added by a series of GnT family enzymes to form multiantennary complexes. This process can be inhibited by swainsonine. The antennae are ultimately capped and decorated by galactose and sialic acid. Hybrid and complex glycans are usually fucosylated at the basal GlcNAc, rendering them resistant to endo H digestion. However, NgF is able to remove all types of glycan.
Carbohydrates provide a layer of protection against NAb attack (Fig. 1A). As glycans are considered self, antibody responses against them are thought to be regulated by tolerance mechanisms. Thus, a glycan network forms a nonimmunogenic “cloak,” protecting the underlying protein from antibodies (3, 13, 20, 29, 39, 54, 65, 67, 74, 85, 96, 98, 117, 119, 120). The extent of this protection can be illustrated by considering the ways in which glycans differ from typical amino acid side chains. First, N-linked glycans are much larger, with an average mass more than 20 times that of a typical amino acid R-group. They are also usually more flexible and may therefore affect a greater volume of surrounding space. In the more densely populated parts of gp120, the carbohydrate field may even be stabilized by sugar-sugar hydrogen bonds, providing even greater coverage (18, 75, 125).
The process of N-linked glycosylation can result in diverse structures that may be divided into three categories: oligomannose, hybrid, and complex (56). Each category shares a common Man3GlcNAc2 pentasaccharide stem (where Man is mannose and GlcNAc is N-acetylglucosamine), to which up to six mannose residues are attached in oligomannose N-glycans, while complex N-glycans are usually larger and may bear various sizes and numbers of antennae (Fig. 1B). Glycan synthesis begins in the endoplasmic reticulum, where N-linked oligomannose precursors (Glc3Man9GlcNAc2; Glc is glucose) are transferred cotranslationally to the free amide of the asparagine in a sequon Asn-X-Thr/Ser, where X is not Pro (40). Terminal glucose and mannose moieties are then trimmed to yield Man5GlcNAc2 (Fig. 1B). Conversion to a hybrid glycan is then initiated by N-acetylglucosamine transferase I (GnTI), which transfers a GlcNAc moiety to the D1 arm of the Man5GlcNAc2 substrate (19) (Fig. 1B). This hybrid glycoform is then a substrate for modification into complex glycans, in which the D2 and D3 arm mannose residues are replaced by complex antennae (19, 40, 56). Further enzymatic action catalyzes the addition of α-1-6-linked fucose moiety to the lower GlcNAc of complex glycan stems, but usually not to oligomannose glycan stems (Fig. 1B) (21, 113).
Most glycoproteins exhibit only fully mature complex glycans. However, the steric limitations imposed by the high density of glycans on some parts of gp120 lead to incomplete trimming, leaving “immature” oligomannose glycans (22, 26, 128). Spatial competition between neighboring sequons can sometimes lead to one or the other remaining unutilized, further distancing the final Env product from what might be expected based on its primary sequence (42, 48, 74, 119). An attempt to assign JR-FL gp120 and gp41 sequon use and types, based on various studies, is shown in Fig. 1A (6, 26, 34, 35, 42, 63, 71, 74, 119, 128). At some positions, the glycan type is conserved. For example, the glycan at residue N301 has consistently been found to be complex (26, 63, 128). At other positions, considerable heterogeneity exists in the glycan populations, in some cases to the point where it is difficult to unequivocally assign them as predominantly complex or oligomannose. The reasons for these uncertainties might include incomplete trimming (42), interstrain sequence variability, the form of Env (e.g., gp120 or gp140), and the producer cell. The glycans of native Env trimers and monomeric gp120 may differ due to the constraints imposed by oligomerization (32, 41, 77). Thus, although all the potential sequons of HXB2 gp120 were found to be occupied in one study (63), some are unutilized or variably utilized on functional trimers, presumably due to steric limitations (42, 48, 75, 96, 119).
The distribution of complex and oligomannose glycans on gp120 largely conforms with an antigenic map derived from structural models (59, 60, 102, 120), in which the outer domain is divided into a neutralizing face and an immunologically silent face. Oligomannose glycans cluster tightly on the silent face of gp120 (18, 128), while complex glycans flank the gp120 receptor binding sites of the neutralizing face, ostensibly forming a protective “fence” against NAbs (105). The relatively sparse clustering of complex glycans that form this fence may reflect a trade-off between protecting the underlying functional domains from NAbs by virtue of large antennae while at the same time permitting sufficient flexibility for the refolding events associated with receptor binding and fusion (29, 39, 67, 75, 98, 117). Conversely, the dense clustering of oligomannose glycans on the silent domain may be important for ensuring immune protection and/or in creating binding sites for lectins such as DC-SIGN (9, 44).
The few available broadly neutralizing monoclonal antibodies (MAbs) define sites of vulnerability on Env trimers (reviewed in reference 52). They appear to fall into two general categories: those that access conserved sites by overcoming Env's various evasion strategies and, intriguingly, those that exploit these very defensive mechanisms. Regarding the first category, MAb b12 recognizes an epitope that overlaps the CD4 binding site of gp120 (14), and MAbs 2F5 and 4E10 (84, 129) recognize adjacent epitopes of the membrane-proximal external region (MPER) at the C-terminal ectodomain of gp41. The variable neutralizing potencies of these MAbs against primary isolates that contain their core epitopes illustrate how conformational masking can dramatically regulate their exposure (11, 118). Conformational masking also limits the activities of MAbs directed to the V3 loop and MAbs whose epitopes overlap the coreceptor binding site (11, 62, 121).
A second category of MAbs includes MAb 2G12, which recognizes a tight cluster of glycans in the silent domain of gp120 (16, 101, 103, 112). This epitope has recently sparked considerable interest in exploiting glycan clusters as possible carbohydrate-based vaccines (2, 15, 31, 70, 102, 116). Two recently described MAbs, PG9 and PG16 (L. M. Walker and D. R. Burton, unpublished data), also target epitopes regulated by the presence of glycans that involve conserved elements of the second and third variable loops and depend largely on the quaternary trimer structure and its in situ presentation on membranes. Their impressive breadth and potency may come from the fact that they target the very mechanisms (variable loops and glycans) that are generally thought to protect the virus from neutralization. Like 2G12, these epitopes are likely to be constitutively exposed and thus may not be subject to conformational masking (11, 118).
The above findings reveal the importance of N-glycans both as a means of protection against neutralization as well as in directly contributing to unique neutralizing epitopes. Clearly, further studies on the nature and function of glycans in native Env trimers are warranted. Possible approaches may be divided into four categories, namely, (i) targeted mutation, (ii) enzymatic removal, (iii) expression in the presence of glycosylation inhibitors, and (iv) expression in mutant cell lines with engineered blocks in the glycosylation pathway. Much of the available information on the functional roles of glycans in HIV-1 and simian immunodeficiency virus (SIV) infection has come from the study of mutants that eliminate glycans either singly or in combination (20, 54, 66, 71, 74, 91, 95, 96). Most mutants of this type remain at least partially functional (74, 95, 96). In some cases these mutants have little effect on neutralization sensitivity, while in others they can lead to increased sensitivity to MAbs specific for the V3 loop and CD4 binding site (CD4bs) (54, 71, 72, 74, 106). In exceptional cases, increased sensitivity to MAbs targeting the coreceptor binding site and/or the gp41 MPER has been observed (54, 66, 72, 74).
Of the remaining approaches for studying the roles of glycans, enzymatic removal is constrained by the extreme resistance of native Env trimers to many common glycosidases, contrasting with the relative sensitivity of soluble gp120 (67, 76, 101). Alternatively, drugs can be used to inhibit various stages of mammalian glycan biosynthesis. Notable examples are imino sugars, such as N-butyldeoxynojirimycin (NB-DNJ), that inhibit the early trimming of the glucose moieties from Glc3Man9GlcNAc2 precursors in the endoplasmic reticulum (28, 38, 51). Viruses produced in the presence of these drugs may fail to undergo proper gp160 processing or fusion (37, 51). Other classes of inhibitor include kifunensine and swainsonine, which, respectively, inhibit the trimming of the Man9GlcNAc2 precursor into Man5GlcNAc2 or inhibit the removal of remaining D2 and D3 arm mannoses from the hybrid glycans, thus preventing the construction of complex glycan antennae (Fig. 1B) (17, 33, 76, 104, 119). Unlike NB-DNJ, viruses produced in the presence of these drugs remain infectious (36, 76, 79, 100).
Yet another approach is to express virus in insect cells that can only modify proteins with paucimannose N-glycans (58). However, the inefficient gp120/gp41 processing by furin-like proteases in these cells prevents their utility in functional studies (123). Another option is provided by ricin-selected GnTI-deficient cell lines that cannot transfer GlcNAc onto the mannosidase-trimmed Man5GlcNAc2 substrate, preventing the formation of hybrid and complex carbohydrates (Fig. 1B) (17, 32, 36, 94). This arrests glycan processing at a well-defined point, leading to the substitution of complex glycans with Man5GlcNAc2 rather than with the larger Man9GlcNAc2 precursors typically obtained with kifunensine treatment (17, 32, 33, 104). With this in mind, here we produced HIV-1 pseudoviruses in GnTI-deficient cells to investigate the role of complex glycan antennae in viral resistance neutralization. By replacing complex glycans with smaller Man5GlcNAc2 we can determine the effect of “lowering the glycan fence” that surrounds the receptor binding sites, compared to the above-mentioned studies of individual glycan deletion mutants, whose effects are analogous to removing a fence post. Furthermore, since oligomannose glycans are sensitive to certain enzymes, such as endoglycosidase H (endo H), we investigated the effect of dismantling the glycan fence on Env function and stability. Our results suggest that the antennae of complex glycans protect against certain specificities but that glycan stems regulate trimer conformation with often more dramatic consequences for neutralization sensitivity and in extreme cases, infectious function.
MATERIALS AND METHODS
Monomeric gp120, monoclonal antibodies, soluble CD4, and the T-20 peptide.
Two types of soluble CD4 (sCD4), one consisting of all four outer domains (4D-sCD4) (49) and the other consisting of only the two outermost domains (2D-sCD4), and a protein in which four copies of two CD4 domains replace the heavy and light chains of IgG (CD4-IgG2) were provided by Progenics Pharmaceuticals (Tarrytown, NY). Anti-gp120 MAbs included the following: b12, 15e, and b6, directed to epitopes that overlap the CD4bs (14); 2G12, directed to a unique glycan cluster of the silent domain of gp120 (101, 103); E51, 17b, X5, and single-chain Fv X5 (scFv X5), directed to a CD4-inducible (CD4i) epitope (27, 62); 447-52D, 39F, and Fab 58.2, directed to the V3 loop (25, 107). Two anti-gp41 MAbs, 4E10 and 2F5, recognize the membrane-proximal ectodomain region (10).
MAbs b12, b6, and X5 were provided by D. Burton (The Scripps Research Institute, La Jolla, CA) (14). MAbs E51, 17b, 39F, and 15e were provided by J. Robinson (Tulane University, New Orleans, LA). MAbs 2G12, 4E10, and 2F5 were purchased from H. Katinger (Polymun Scientific Inc., Vienna, Austria). MAb 447-52D was purchased from S. Zolla-Pazner (NYU, New York, NY). The scFv X5 was expressed in bacteria (5), and Fab 58.2 was prepared by papain digestion (11, 80). These antibody fragments were provided by M. Zwick (The Scripps Research Institute, La Jolla, CA).
MAbs b12, 2G12, 4E10, and 2F5 are broadly and potently neutralizing (11); MAbs 17b, E51, and X5 exhibit only limited neutralizing activity against HIV-1 primary isolates as IgGs, but their activity is dramatically enhanced in the presence of sCD4 (25, 84). Fab and single-chain Fv (scFv) fragments of X5 can, however, neutralize in the absence of sCD4 (62, 83); the V3 loop-specific Abs can neutralize a subset of primary isolates (11). The remaining MAbs do not neutralize primary HIV-1 isolates effectively.
The T-20 peptide is based on residues 638 to 673 of gp41 (Fig. 1A) and was a gift from Progenics Pharmaceuticals.
Plasmas and sera.
Broadly neutralizing plasmas LTNP2 (also known as LT2 and N308) and Z23 (also known as 1688) and nonneutralizing plasma K370 have been described previously (8, 25, 30). Plasmas LTNP2 and K370 were provided by D. Richman (UCSD, La Jolla, CA). Plasma 1688 was provided by Zeptometrix (Buffalo, NY).
Viruses and plasmids.
Pseudovirus virus-like particles (VLPs) were produced by cotransfection of 293T or GnTI- 293S cells with the plasmid pNL-LucR-E- and pCAGGS-based Env-expressing plasmids by calcium phosphate precipitation, as previously described (80). The JR-FL gp160ΔCT SOS Env clone was used to generate our prototype virus (80). Other pCAGGS clones were used to express JR-FL gp160ΔCT SOS with an N301Q mutation in the V3 loop, JR-FL gp160 wild type (WT), JR-FL gp160ΔCT WT, YU2 gp160 WT, SF162 gp160ΔCT WT, THRO gp160ΔCT WT, and SIVmac316 gp160ΔCT WT (7, 23, 64).
Neutralization assays.
Pseudovirus neutralization assays were performed as described previously (7, 8, 25). Briefly, virus was incubated for 1 h with inhibitors, before adding the mixture to CF2.CD4.CCR5 target cells for 2 h (starting with a 10-min spinoculation at room temperature [RT], with the remainder of the infection in an incubator at 37°C). Cells were then washed, incubated for 2 days, lysed, and assayed for luciferase as a marker of infection.
Neuraminidase treatment of virus particles.
Transfection supernatants (30 ml) containing pseudoviruses were filtered and then pelleted by centrifugation at 50,000 × g for 1 h, followed by a second spin in microcentrifuge tubes at 25,000 × g to remove residual culture medium. Pseudoviruses were resuspended in 30 μl of phosphate-buffered saline (PBS; 1,000× the original concentration in transfection supernatants) and then split into two equal 15-μl portions. To one portion, 3 mU of α-2-3,6,8-neuraminidase (Sigma, St. Louis, MO) was added, and the other was mock treated. Samples were then incubated at 37°C for 1 h, after which particles were pelleted, washed with 1 ml PBS, and resuspended in 5 ml of culture medium. The resulting virus preparations were compared in infectivity and neutralization assays.
BN-PAGE and Western blotting.
Blue native PAGE (BN-PAGE) was performed as described previously (23-25, 80). Briefly, concentrated VLPs were incubated with or without MAb or sCD4 for 15 min at RT. VLPs were then pelleted, washed with PBS, and gently solubilized for 5 min in 2× solubilization buffer (1% Triton X-100 in 1 mM EDTA) in 1.5 M aminocaproic acid to liberate Env. Six microliters of 3× sample buffer (150 mM morpholinopropanesulfonic acid [MOPS], 150 mM Tris-HCl [pH 7.7], 60% glycerol, 0.15% Coomassie blue) was then added prior to loading onto a 4 to 12% bis-Tris NuPAGE gel (Invitrogen, Carlsbad, CA). Ferritin (Sigma, St. Louis, MO) was used as a size standard. Samples were separated at 4°C for 3 h at 100 V with 50 mM MOPS-50 mM Tris (pH 7.7) containing 0.002% Coomassie blue as cathode buffer and the same buffer without Coomassie blue as the anode buffer. The gel was then blotted onto polyvinylidene difluoride, destained, and probed with a MAb cocktail consisting of b12, 2G12, E51, 39F, 2F5, and 4E10 (24). A goat anti-human Fc-alkaline phosphatase conjugate was then used to detect the primary MAbs (Accurate Chemical, Westbury, NY). Trimer binding was assayed by the depletion of the unliganded form in Western blot assays and measured with the UN-SCAN-IT software (Silk Scientific, Orem, UT). Trimer shift 50% inhibitory concentrations (IC50s) were determined by plotting these data versus ligand concentrations. Molecular masses were also estimated using this software, using ferritin 24-mers and 12-mers with molecular masses of 440 kDa and 220 kDa, respectively, as markers.
Endo H treatment of virus particles.
For infectivity analyses, concentrated virus preparations were incubated at 37°C for various time intervals with endo H (200 U; New England BioLabs, Ipswich, MA). Particles were then centrifuged and washed to remove enzyme and resuspended in culture medium for infections. Mock virus samples were incubated and washed in a similar manner without enzyme.
For BN-PAGE analysis, particle preparations from 1,000×-concentrated transfection supernatant were incubated with an excess (200 U) of endo H at 37°C for various time periods in 50 μl PBS. Particles were then repelleted in a microcentifuge, washed, and then analyzed by BN-PAGE and Western blotting.
Modeling and visualization of glycan cargos.
To visualize the glycosylation states of the different trimers (native gp120, GnTI, and endo H treated), we used a combination of computational modeling and manual placement to generate potential glycan conformations. The initial gp120 trimer configuration was generated by using Chimera (89) to fit the crystal structure of the gp120 monomer taken from the b12-bound conformation (PDB id 2NY7) (126) into the cryo-electron tomogram density of the free gp120 trimer from Liu et al. (69). The glycan composition of the parent gp120 trimer was determined by probabilistically assigning complex or high-mannose glycan types at each glycosylation position consistent with the characterization studies of Mizuochi et al. (78) and Leonard et al. (63). Initial glycan conformations were taken from models generated by Pancera et al. (86), the glycan assignments for which were provided by Leopold Kong and Peter Kwong.
Glycan conformations in the glycosylated gp120 trimer were simulated using the Glycan Relax software. All dihedral degrees of freedom on glycans and asparagine residues linked to the glycans were allowed to move, while the rest of gp120 was held fixed. Briefly, Glycan Relax is a two-stage random optimization protocol that models glycans, considering sterics and dihedral angle preferences. The first stage is a Monte Carlo Metropolis procedure that performs random torsional moves to quickly find solutions to relieve any large or extreme clashes. The second stage is a randomized descent procedure that performs both rotameric and random torsional moves to settle the glycan conformations and is effectively a minimization. Ten independent simulations of the full two-stage protocol were performed to identify a clash-free, low-energy starting point. This starting point was subjected to a single simulation trajectory using only an altered second stage that replaced the randomized descent criteria with a Boltzmann accept-reject criterion, allowing the simulation to explore an expanded region of clash-free and near-clash-free conformational space. A single low-energy, clash-free conformation was randomly selected from this trajectory as a representative of the glycosylated parent gp120 trimer (see Fig. 9A, below).
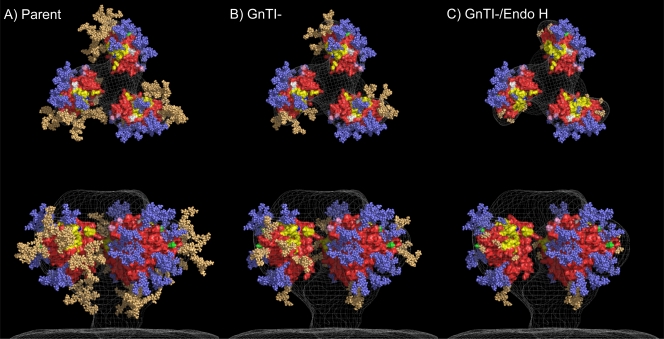
FIG. 9.
Models of Env trimers carrying different glycan cargoes. (A) Parent trimer decorated with a full complement of complex and oligomannose glycans. All complex glycans are shown in light orange, while oligomannose glycans are in slate blue. (B) GnTI- trimer in which complex glycans have been replaced by Man5GlcNac2. (C) GnTI- trimer in which oligomannose glycans of the neutralizing face have been replaced by a single GlcNac stump to simulate the effect of endo H in removing the susceptible Man5GlcNac2/oligomannose mannose glycans. In all three panels, gp120 (protein) protomers (based on the HXBc2 isolate) are shown as molecular surface representations and are primarily colored red. Colors for gp120 features are as follows: the D368 residue that is essential for binding both sCD4 and b12 is shown in dark blue, the b12 binding site is yellow, V1-V2 stems are pink, V3 stems are white, and V4 stems are green. The electron density map from reference 69 is shown as a mesh at 2 σ (2 standard deviations). The putative complex and oligomannose glycans are based on the findings reported in reference 63. The particular glycans were chosen probabilistically at each position, based on references 63 and 78. The complex carbohydrates include tetraantennary, triantennary, and biantennary structures and largely correspond to the structures in reference 78. We considered the possibility of constructing V3-containing trimer models to illustrate the large complex glycan at residue N301 and the possible effects of its replacement or removal. A major caveat, however, is that the only V3 loop-containing gp120 structure presently available is in a complex with sCD4 and X5 (47). Docking this structure into the unliganded density of the trimer produces a trimer model with a highly exposed V3 loop (see Fig. 2F in reference 69), but that cannot be the case on unliganded primary isolates, because V3 antibodies do not tend to neutralize those isolates (11).
For simplicity and visualization consistency, the representative glycosylated GnTI trimer (see Fig. 9B) was created by taking the representative glycosylated parent trimer generated above and replacing the complex glycans on its neutralizing faces with Man5GlcNac2 glycans, whose conformations were manually selected from the available Man5GlcNac2 conformations generated during the prior simulation trajectory. The representative endo H-treated trimer (see Fig. 9C) was then created by taking the representative glycosylated GnTI trimer and trimming the high-mannose glycans on its neutralizing faces to a single GlcNac, mimicking the effect of endo H treatment. Renderings of each glycosylated trimer were generated using PyMOL (W. L. DeLano; http://www.pymol.org/).
RESULTS
HIV-1 pseudoviruses produced in the absence of N-acetylglucosaminyltransferase I are infectious.
We expressed various HIV-1 and SIV Env-bearing pseudoviruses in 293T (referred hereafter as “parent” viruses) and GnTI- cells (94). Protein expression in GnTI- cells prevents the progression of Man5GlcNAc2 into hybrid and complex carbohydrates (Fig. 1B). However, any oligomannose carbohydrates that are resistant to mannosidase trimming are expected to be unaffected, since they do not require GnTI to reach their final state (Fig. 1B) (17, 94). This has been demonstrated experimentally by mass spectrometry in two prior studies (21, 32). In one of these studies, Man6-9GlcNac2 as well as Man5GlcNac2 glycans were excised from soluble HIV-1 gp140 trimers, consistent with the notion that the immature glycans of silent domain “mannose patch” remain untouched while complex glycans become Man5GlcNAc2 (32).
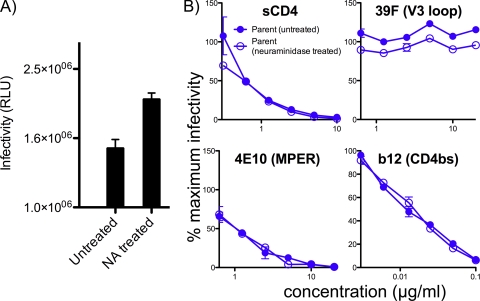
Pseudoviruses produced from both cell types were able to infect CF2.Th.CD4.CCR5 cells. However, regardless of the Env strain, the parent viruses were markedly more infectious (Fig. 2A). For example, the mean infectivities of particles bearing the JR-FL Env SOS with a gp41 cytoplasmic tail truncation (gp160ΔCT) produced in the parent and GnTI- cells differed by ∼18-fold (Fig. 2A). We chose JR-FL SOS gp160ΔCT as our prototype Env thereafter, based on its efficient expression and gp120/gp41 processing, as observed in several earlier studies (7, 23, 25, 80).
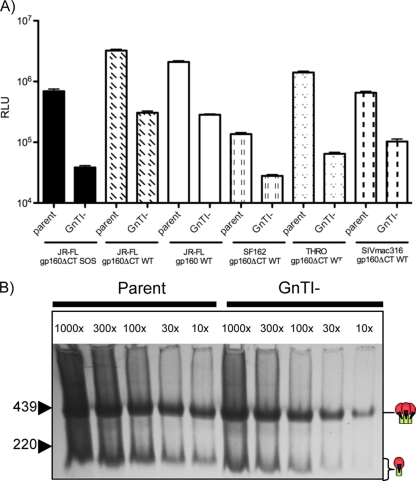
FIG. 2.
Comparison of parent and GnTI- virus infectivity and expression. (A) The infectivities of parent and GnTI- viruses were measured using CF2.CD4.CCR5 target cells. (B) The relative expression levels of parent and GnTI- VLPs were measured by titrating concentrated preparations of each by BN-PAGE and then detecting Env by Western blotting.
The lower infectivity of GnTI- cell-produced viruses could stem either from an adverse effect of replacing the complex glycans with immature glycans or simply from weaker virus expression. To investigate, we titrated the parent and GnTI- pseudovirus stocks in BN-PAGE and Western blotting assays. This revealed that parent Env trimers were expressed at approximately 10-fold-higher levels (Fig. 2B), implying that most if not all of the difference in infectivity relates to expression levels. Therefore, we can conclude that complex carbohydrates are not necessary for efficient virus infection of susceptible cells (33). Some previous studies have also reported that these GnTI- cells express lower protein levels than 293T cells (17). However, reduced expression has not been universally observed (32). The reason for this discrepancy is not clear but could relate to interlaboratory differences in cell handling and transfection conditions.
Considering the differences in the average mass of the Man5GlcNAc2 and complex glycans, the molecular masses of parent and GnTI- Env trimers might be expected to differ. The mass of complex glycans typically ranges from 2 to 4 kDa (Fig. 1B), while the Man5GlcNAc2 moieties that replace them in GnTI- cell-produced proteins have a molecular mass of only 1.2 kDa (17). Several reports suggest that the complex carbohydrates of gp120 are predominantly of the relatively simple biantennary type (Fig. 1B) (26, 32, 77, 78, 128). Thus, if we assume a mass of 2.4 kDa for complex glycans (Fig. 1A) and 1.2 kDa for Man5GlcNAc2, and that half of the estimated ∼60 N-glycosylation sites (Fig. 1A) are occupied by complex glycans (26, 63), then the molecular mass difference between parent and GnTI- trimers should be ∼40 kDa. Note, however, that mannose trimming on trimeric Env may be more restricted than on monomeric gp120 (32), resulting in fewer complex glycans on trimeric Env. An ∼40-kDa size differential may therefore be a slight overestimation.
We and others previously showed that BN-PAGE can yield reasonable estimates of the molecular masses of protein complexes (see Fig. 2 in reference 107). Using the previously estimated masses of the parent trimer and monomer of ∼420 and ∼140 kDa (23, 80) and ferritin as markers, we estimated here that GnTI- trimers have a mass of ∼394 to 390 kDa, i.e., an ∼26- to 30-kDa differential (Fig. 2B). Thus, allowing both for the limitations of molecular mass estimates by BN-PAGE and for our incomplete knowledge of sequon usage and the types of glycans that decorate these trimers, the modest differences observed matched our expectations.
Expression in GnTI- cells does not affect CD4-dependent infection or Env trimer-sCD4 binding stoichiometry.
Previous reports had shown that certain HIV-1 Env glycan deletion mutants result in CD4-independent infection (55, 61, 68, 92). It is unknown whether this altered phenotype depends on eliminating the glycan antennae or if the removal of the entire glycan is necessary. The reduced mass of the glycan shield covering GnTI- Env trimers might lower the “glycan fence” lining the receptor and coreceptor binding sites, which may affect receptor binding, possibly even allowing it to use CCR5 even in the absence of CD4. However, a comparison of the infection of parent and GnTI- viruses in CF2CD4.CCR5 and CF2.CCR5 cells revealed that the GnTI- virus still depended on CD4 for efficient infection (data not shown). Therefore, the effects of certain glycan mutants on CD4 dependency may hinge on the complete removal of a glycan(s), rather than a reduction in the mass of complex glycan antennae.
SIV isolates bearing truncated gp41 cytoplasmic tails have been found to exhibit dramatically augmented infection in the presence of soluble CD4 (1, 23, 53). This was explained by the truncated SIV Env trimers' unique propensity to bind a maximum of only one sCD4 molecule. Hypothetically, sCD4 binding induces a global conformational change in the SIV trimer, exposing three coreceptor binding sites while being encumbered by only one, not three, CD4 molecules, and as a result enjoys enhanced fusogenicity (see Fig. 7 of reference 23). This 1:1 Env trimer/sCD4 binding stoichiometry may stem in part from the glycans lining the three potential binding sites, such that, in this case, room exists for only one sCD4 molecule (23, 53). The replacement of large complex glycans that line the receptor binding sites with smaller Man5GlcNac2 glycans might conceivably relieve these constraints. However, we observed similar dramatic sCD4 enhancement for both the parent and GnTI- viruses from truncated SIVmac239 Env trimers (data not shown). Thus, the less-dense glycan shell encapsulating GnTI- trimers did not appear to allow additional CD4 molecules to bind to SIV Env trimers.
GnTI- production enhances neutralization by sCD4 and V3 MAbs.
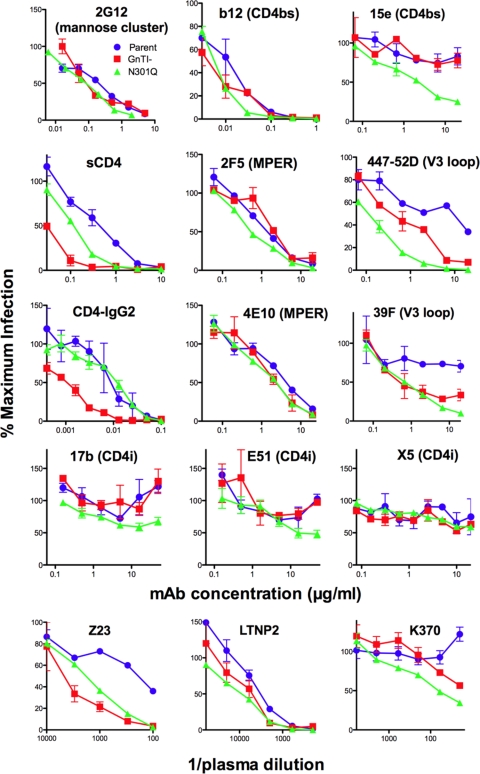
Several studies have examined the effects of eliminating Env glycans on neutralization sensitivity (3, 54, 74). Of these, N301Q is perhaps one of the most studied, eliminating a large complex glycan at the base of the V3 loop (Fig. 1A) (3, 54, 74, 106). If we envisage a fence of glycans lining the gp120 receptor binding sites, then removal of a single glycan, as in the N301Q mutant, would be equivalent to removing a fence post. Conversely, the replacement of complex glycans with Man5GlcNac2 in GnTI- virus might be compared to lowering the glycan fence. Like the N301Q mutant, this could affect antibody access to trimers. We therefore compared the susceptibilities of parent, GnTI-, and N301Q JR-FL viruses to neutralization by a panel of MAbs, sCD4, and plasmas from HIV-1-infected patients. We consider the effects on various specificities in turn below.
(i) Susceptibility to MAb 2G12.
The parent and GnTI- viruses were found to be equally sensitive to 2G12 (Fig. 3; Table 1). Thus, the replacement of complex glycans of the neutralizing face with Man5GlcNac2 does not affect the already constitutively exposed oligomannose epitope of 2G12 in the silent domain (9, 16, 25, 101, 103). Previously, it was shown that the plant alkaloid and mannose analog kifunensine, which competitively inhibits trimming of immature Man9GlcNAc2 glycans to Man5GlcNAc2 (Fig. 1B), can lead to multiple 2G12 epitopes on the surfaces of previously nonantigenic self proteins and cells (104). 2G12 epitopes can be created on the surface of yeast cells by the synthesis of compact clusters of oligomannose glycans (70). However, an increase in neutralization by 2G12 was not evident for the GnTI- cell-produced virus, probably because, unlike kifunensine, the lack of GnTI arrests glycan synthesis after the mannose trimming stage, so that complex glycans are likely to be replaced by Man5GlcNac2 rather than Man9-6GlcNAc2. This is important, because 2G12 targets the α-1-2-terminal moieties of essentially untrimmed oligomannose glycans (Fig. 1B).
FIG. 3.
Neutralization sensitivities of parent, GnTI-, and N301Q viruses. The neutralization activities of a panel of MAbs, 4D-sCD4, and HIV-1-infected donor plasmas were assayed against parent, GnTI-, and N301Q viruses all bearing the SOS mutation. Each virus is color coded and indicated by the symbol shown. Representative data from duplicate titrations are shown.
TABLE 1.
Neutralizing titers for MAbs, sCD4, and HIV-1-infected donor plasma samples against parent, GnTI-, and N301Q mutant viruses
| Ligand | Mean IC50 (μg/ml) or ID50 (1/plasma dilution)a |
||
|---|---|---|---|
| Parent | GnTI- | N301Q | |
| JR-FL gp160ΔCT SOS | |||
| 2G12 (mannose cluster) | 0.37 | 0.1 | 0.1 |
| b12 (CD4bs) | 0.01 | 0.01 | 0.009 |
| sCD4 | 0.3 | 0.03 | 0.1 |
| CD4-IgG2 | 0.015 | 0.001 | 0.016 |
| 15e (CD4bs) | >20 | >20 | 1.5 |
| 2F5 (MPER) | 1 | 2 | 0.7 |
| 4E10 (MPER) | 3 | 2 | 2 |
| X5 (CD4i) | >20 | >20 | >20 |
| E51 (CD4i) | >50 | >50 | 20 |
| 17b (CD4i) | >50 | >50 | >50 |
| 39F (V3 loop) | >30 | 0.3 | 0.8 |
| 447-52D (V3 loop) | 15 | 0.5 | 0.1 |
| T-20 peptide | 0.009 | 0.01 | 0.01 |
| THRO gp160ΔCT WT | |||
| b12 | 2 | 0.5 | |
| sCD4 | 1 | 0.4 | |
| 447-52D | >20 | >20 | |
| SF162 gp160ΔCT WT | |||
| b12 | 0.07 | 0.03 | |
| sCD4 | 0.7 | 0.3 | |
| 447-52D | 0.3 | 0.1 | |
| JR-FL gp160ΔCT WT | |||
| b12 | 0.04 | 0.04 | |
| sCD4 | 1 | 0.05 | |
| 447-52D | 25 | 5 | |
| JR-FL gp160 WT | |||
| sCD4 | 1 | 0.2 | |
| Plasma | |||
| Z23 | 600 | 3,000 | 2,300 |
| LTNP2 | 3,000 | 6,900 | 6,100 |
| K370 | <20 | 20 | 110 |
Mean IC50s are shown for MAbs and sCD4. ID50s are shown for plasma samples. Titers are the averages of at least two assays, with each run in duplicate.
The absence of the glycan at residue N301 at the N-terminal base of the V3 loop also had no effect on 2G12 neutralization, despite the fact that it resides close to a key glycan contact at N295. This again suggests that the 2G12 epitope is already optimally exposed and that modification of adjacent glycans does little to improve its binding. Furthermore, the N301 glycan does not appear to sterically restrict the maturation of the neighboring glycan at N295, which apparently remains oligommanose even in the absence of the N301 glycan.
(ii) Susceptibility to MAb b12.
Like 2G12, we observed only small differences in the neutralization sensitivities of the parent and GnTI- viruses to b12 (Fig. 3 and Table 1). Similar results were observed with JR-FL gp160ΔCT WT pseudotyped virus (Table 1). The N301Q mutation also had only a modest effect on b12 sensitivity, contrasting with the greater increase in YU2 sensitivity reported previously (54). This inconsistency is perhaps not altogether surprising, considering the already exquisite sensitivity of the JR-FL isolate to the b12 MAb, and may be related to the unusual absence of a key glycan at the base of the V2 loop at position N197 (Fig. 1A) that may cause a repositioning of the V1V2 loop (118). MAb b12 may therefore already have virtually unrestricted access to its epitope in the parent, which cannot be easily improved upon. Further studies with THRO and SF162 Env pseudotype viruses indicated a 3- to 4-fold-greater sensitivity of the GnTI- virus (Table 1).
(iii) Susceptibility to soluble CD4 and CD4-IgG2.
In contrast to the limited effects on b12 neutralization, the GnTI- virus was approximately 10-fold more sensitive than the parent virus to sCD4 (Fig. 3; Table 1). This was reflected by an analysis of sCD4 binding to parent and GnTI- Env trimers with BN-PAGE. The GnTI- Env trimer was more effectively complexed by sCD4, as indicated by the depletion of unliganded Env trimer in concert with the appearance of CD4-trimer complexes at an approximately 3-fold-lower sCD4 concentration (Fig. 4).
FIG. 4.
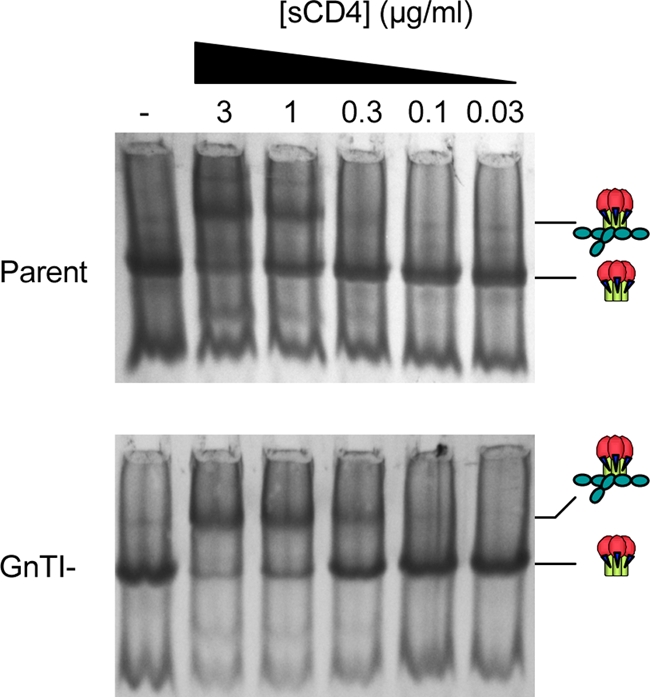
Relative affinity of soluble CD4 for parent and GnTI- trimers. Concentrated viruses were mixed with graded concentrations of sCD4 for 1 h. Samples were then resolved by BN-PAGE.
The neutralization of viruses with full-length gp160 and gp160ΔCT versions of WT JR-FL Env was similarly affected (Table 1). GnTI- versions of THRO and SF162 viruses were also more susceptible to sCD4, albeit to a lesser extent than observed with JR-FL (Table 1). The N301Q mutant was also more sensitive to sCD4 than the parent virus, but not to the same extent as the GnTI- virus.
We further investigated the exposure of the primary receptor binding site with tetrameric CD4-IgG2. Compared to monomeric sCD4, markedly stronger neutralization titers were observed against all three viruses. The enhanced sensitivity of the GnTI- virus to sCD4 was recapitulated with CD4-IgG2, but the somewhat different sensitivities of the parent and N301Q viruses to sCD4 were not (Fig. 3; Table 1). Overall, we infer that replacing complex glycans with Man5GlcNac2 glycans relieves some of the constraints for high-affinity CD4 binding to JR-FL trimers and that these constraints are also relieved, at least partially, by removing the N301 glycan.
(iv) Susceptibility to a “nonneutralizing” MAb that overlaps the CD4 binding site.
We next investigated the sensitivity of the GnTI- virus to MAb 15e, whose binding site overlaps that of CD4 but fails to effectively neutralize the JR-FL parent virus or most other primary isolates. Previously, the N301Q and other glycan deletion mutants of YU2, JR-FL, and HXBc2 all exhibited increased susceptibility to similar nonneutralizing CD4bs MAbs (54). Here, we also found that the N301Q mutant was markedly more susceptible to 15e, but the GnTI- virus remained resistant (Fig. 3; Table 1). Thus, the removal of a key glycan had a greater effect on exposing this occluded epitope than replacing complex outer domain glycans with the smaller Man5GlcNac2. Similar results were obtained using MAb b6, which is directed to a similar epitope.
(v) Susceptibility to MPER MAbs 2F5 and 4E10.
The activities of MPER MAbs 2F5 and 4E10 were equivalent against all three viruses. This suggests that the antennae of the complex glycans in the upper part of the gp41 C-helix (Fig. 1A) and those of gp120 have little or no influence on MAb access to the MPER.
(vi) Susceptibility to MAbs targeting epitopes that overlap the coreceptor binding site.
We next assessed the susceptibility of the three viruses to MAbs whose epitopes overlap the coreceptor binding site, i.e., the so-called CD4-induced (CD4i) epitope (99, 121). The parent and GnTI- viruses both resisted neutralization by E51, 17b, and X5. The same was largely true for the N301Q virus. However, some susceptibility to E51 was observed at high concentrations.
(vii) Susceptibility to V3 loop-specific MAbs.
As previously reported, the parent JR-FL virus is largely resistant to the V3-specific MAbs 39F and 447-52D (25) (Fig. 3; Table 1). However, the GnTI- virus was significantly more sensitive to MAbs 447-52D and 39F (Fig. 3; Table 1). The N301Q mutant was also more sensitive than the parent to V3 MAbs. The magnitude of the increased sensitivity was, at least in the case of 447-52D, slightly greater for the N301Q mutant virus (Fig. 3; Table 1). GnTI- versions of JR-FL 160ΔCT WT virus were also more sensitive to 447-52D (Table 1), and the susceptibility of the already-sensitive SF162 160ΔCT WT parent virus to 447-52D was also somewhat increased (Table 1). These observations suggest that antennae of complex glycans can play a significant role in protecting the V3 loop from antibody binding to Env trimers.
(viii) Susceptibility to plasma from HIV-1-infected donors.
Previously some, but not all, glycan deletions, in particular those in the vicinity of the V3 loop, were found to increase neutralization susceptibility to sera from HIV-1-infected donors (74). We therefore evaluated the activities of neutralizing plasmas against our three virus prototypes. Plasma Z23 was approximately 5- to 10-fold more effective against both the GnTI- and N301Q viruses, the former being more sensitive (Fig. 3; Table 1). In contrast, the 50% inhibitory dilution (ID50) of plasma LTNP2 was increased by only 2- to 3-fold (Fig. 3; Table 1). This might be because the increased sensitivities of the GnTI- and N301Q viruses are, in this case, diluted by the effects of a very high titer of b12-like NAbs (Fig. 3; Table 1) (8).
To allow us to better evaluate the titers of any normally nonneutralizing Abs against the two types of modified Env trimers, we next examined the activity of a plasma that does not neutralize the parent virus. Above, we observed that the GnTI- virus is sensitive to V3 MAbs that have little or no activity against the parent virus. Therefore, we anticipated that in the absence of any neutralization against the parent virus, the impact of any increased sensitivity of GnTI- virus to otherwise-nonneutralizing specificities in the plasma might be easier to detect, in contrast to LTNP2. As expected, plasma K370 did not detectably neutralize the parent virus (25) (Fig. 3; Table 1). However, the GnTI- virus was neutralized at a modest ID50 of 1:20 and the N301Q virus was even more susceptible, with an ID50 of 1:110. The difference may stem from the above observations that the N301Q virus is more sensitive to nonneutralizing CD4bs and possibly CD4i Abs, in addition to V3-specific Abs, while the GnTI- virus is more sensitive to V3 Abs only.
Summarizing our neutralization results in reference to the published literature on glycan-modified viruses, as in the previous studies, our N301Q mutant exhibited increased sensitivity to sCD4 and CD4-IgG2 (54, 74), “nonneutralizing” CD4bs MAbs (3, 54), V3 MAbs (3, 54, 74, 106), and infected subject sera (54, 74), while the sensitivity to 2F5 was unaffected (54, 74). A notable increase in b12 activity was noted in two previous studies using N301Q mutants of non-JR-FL Envs (54, 74). However, one study that also examined JR-FL reported only a modest effect, in agreement with our data (54). Our N301Q mutant was also largely resistant to CD4i MAbs, as in previous studies (54, 74). However, some weak activity of one CD4i MAb, E51, was observed, but this cannot be reconciled with the earlier studies, where it was not tested. In one study, the removal of multiple glycans was necessary to achieve susceptibility to the CD4i MAb 17b (54). Clearly, the global replacement of complex glycans with smaller Man5GlcNac2 glycans in the GnTI- virus was insufficient to achieve a similar phenotype, suggesting the importance of removing the glycan stems in reaching a CD4i MAb-sensitive phenotype. Overall, our N301Q mutant exhibited similar sensitivity patterns to those observed previously, while the GnTI- virus's acute CD4 sensitivity and increased V3 sensitivity without any concomitant effect of either of the nonneutralizing CD4bs or CD4i is unlike any previously published glycan-modified virus.
Terminal sialic acid moieties on complex glycans do not affect the neutralization sensitivity of the parent virus.
One possible explanation for the increased neutralizing sensitivity profiles of the N301Q and GnTI- viruses might relate to the fact that they lack some or all of the sialic acids present on the termini of complex glycans of the parent Env trimers. This would result in a loss of the negative charge of glycan shield, which could affect protein-protein electrostatic interactions and possibly neutralization. We investigated this by treating a concentrated preparation of the parent virus with neuraminidase, washing away the enzyme, resuspending the virus in culture medium, and assessing its infectivity and neutralizing sensitivity. We found that neuraminidase treatment led to an ∼50% increase in viral infectivity (Fig. 5A), consistent with previous reports (46, 76). In neutralization assays, the neuraminidase-treated virus exhibited sensitivities to sCD4 and 39F identical to that of the untreated parent virus (Fig. 5B), contrasting with the enhanced sensitivities of both the GnTI- and N301Q viruses to these inhibitors. The sensitivities of the treated and untreated parent virus to b12 and 4E10 were also identical, as expected. In fact, the lack of any marked increase in neutralizing sensitivity was not surprising, considering a previous report on SIV, in which either no effect or, in some cases, increased neutralization resistance was observed with neuraminidase treatments (76).
FIG. 5.
Effect of neuraminidase treatment on neutralization sensitivity. The JR-FL SOS parent virus transfection supernatant was concentrated by centrifugation and split into two batches, of which one was treated with neuraminidase and the other mock treated for 1 h at 37°C. Viruses were then diluted in culture medium and tested for their infectivity (A) and for their sensitivity to various MAbs and sCD4 (B), as indicated.
Endo H treatment of GnTI- Env trimers leads to biphasic removal of glycans.
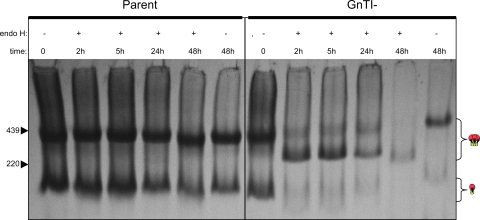
We next examined the susceptibility of Env trimers to deglycosylating enzymes. PNGase F (NgF) results in the most complete removal of glycans at their base (Fig. 1B). However, the relatively hydrophobic asparagine-linked glycan structure is converted into a hydrophilic aspartic acid that can result in protein aggregation (28). Conversely, endo H cleaves between the two N-acetylglucosamine (GlcNac) residues of the diacetylchitobiose stem of the oligosaccharide, leaving behind a single N-acetylglucosamine stump, with minimal consequences on folding (17, 101). Oligomannose and hybrid glycans are sensitive to digestion by endo H, but the addition of fucose at the base of complex glycans renders them resistant (Fig. 1B). Previously, native trimers have been found to be largely refractory to attempts to enzymatically deglycosylate them, presumably due to their compact and thus inaccessible nature, contrasting with the more enzymatically sensitive soluble gp120 and gp140 proteins (76, 101). The replacement of complex glycans with Man5GlcNAc2 in the outer domain of GnTI- virus might, however, affect the resistance of native Env trimers to glycosidases. We therefore compared the effects of endo H on parent and GnTI- viral Env over time by BN-PAGE (Fig. 6). The parent trimers were largely unaffected by endo H, consistent with a previous study describing the resistance of SIVmac316 Env trimers to various glycosidases (76). However, a tangible drop in mass occurred by 48 h, when the trimer size fell from ∼420 kDa to ∼370 kDa. In repeat experiements, however, this small change in mass was not consistently observed. In contrast, a rapid drop in size of the GnTI- trimer from ∼420 kDa to ∼240 kDa was observed (Fig. 6, right lanes), followed much later by a further drop in trimer mass from ∼240 kDa to ∼215 kDa between 24 and 48 h. This biphasic drop in mass suggests a rapid removal of easily excisable glycans followed by a much slower removal of less-accessible glycans.
FIG. 6.
Effect of endo H on Env trimers. VLPs were incubated with or without endo H for the times indicated and then pelleted, washed, and resolved by BN-PAGE and Western blotting.
An alternative explanation for the initial drop in size of GnTI- trimers from 0 to 2 h might be a shift in GnTI- trimers to dimers. However, if trimers dissociated into dimers, a band reflecting a dissociated monomer might also be expected, but we saw no evidence of this. Instead, the biphasic drop in mobility of the major Env band, coupled with loss of 2G12 staining, is more consistent with a rapid removal of the outer domain Man5GlcNAc2 glycans, in line with their relatively sparse distribution. This is followed by the much slower removal of the more tightly packed and less accessible silent domain glycans. The outer domain GnTI- glycans are removed rapidly (<2 h), while the silent domain glycans of either virus are not significantly affected until >24 h. The consistently slow kinetics of removal of silent domain glycans suggests that the presence of easily removable oligomannose glycans in the adjacent outer domain of GnTI-trimers does not markedly accelerate the removal of glycans from the neighboring silent domain. Further circumstantial evidence for a slow second phase of removal of silent domain glycans is suggested by the abrupt loss in staining intensity of the GnTI- trimer between 24 and 48 h, concomitant with the drop in the band's size (Fig. 6). This may be related to the loss in detection by MAb 2G12, one of the most effective components of the MAb cocktail used to probe Western blots. A further incubation to 72 h led to even fainter staining (data not shown).
Besides the loss in staining between 24 and 48 h, there was an initial perceptible loss in GnTI- trimer staining, even after 2 h, that did not occur with the parent trimer. Considering that the SOS mutant Env trimers are shackled with a gp120-gp41 disulfide bridge, gp120 shedding is unlikely to explain this observation. Instead, we suggest that the fainter bands reflect a partial loss of 2G12 staining. Previously, it was found that gp120 monomers expressed in the presence of kifunensine (preventing glycan maturation [Fig. 1B]) can bind more than one copy of 2G12, thanks to the formation of a novel oligomannose glycan cluster in the outer domain that serves as a second 2G12 binding site (104). Similarly, additional 2G12 binding sites may be present on GnTI- trimers, and their rapid removal by endo H could explain the initial drop in trimer staining intensity here. Another explanation may be that the removal of outer domain glycans perturbs trimer structure, such that the arrangement of the remaining silent domain glycans changes and is recognized less efficiently by 2G12.
The GnTI- gp160ΔCT monomer band was, like the corresponding trimer, rapidly affected by endo H treatment, perhaps to an even greater extent. A discernible loss in band intensity was also observed earlier with the parent monomer than with the corresponding trimer. This suggests that glycans can be removed more easily from particle-based Env monomers, as has been noted with soluble Env monomers (76).
Treatment with endo H rapidly reduces GnTI- virus infectivity.
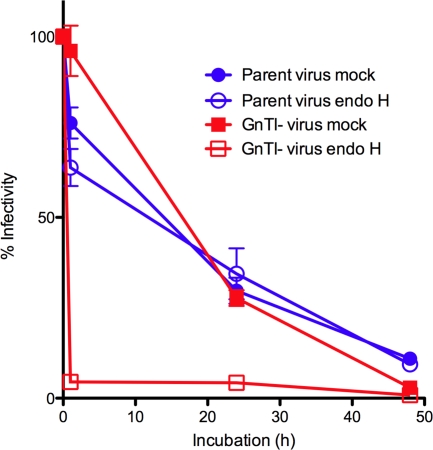
We next investigated the effect of endo H on viral infectivity. In previous reports, only enzyme treatments affecting the termini of glycan antennae affected infectivity. Removal of galactose and sialic acid moieties both increased SIV infectivity, while mannosidase reduced infectivity (76). Deglycosylation can have different consequences on sCD4 binding to gp120, depending on the experimental conditions (12, 67, 73). We found that the infectivity of the parent virus decayed at a similar rate over time, regardless of exposure to endo H (Fig. 7). This was expected, considering its relative resistance to the enzyme (Fig. 6). In contrast, GnTI- virus infectivity was highly sensitive to endo H treatment and dropped to background levels within 1 h, in contrast to a mock-treated virus that decayed at a similar rate to the parent virus (Fig. 7). Similar experiments with GnTI- THRO and SF162 pseudoviruses gave comparable outcomes, suggesting that the endo H-susceptible glycans on the outer domain of the GnTI- Env trimers play a crucial role in preserving infectious potential.
FIG. 7.
Effects of endo H on viral infectivity. Viruses were incubated for various times at 37°C in the presence or absence of 200 U of endo H, as indicated. Particles were then pelleted in microcentrifuge tubes and incubated with CF2.CD4.CCR5 cells. Infection was then later determined by a luciferase readout and was plotted as a percentage of the infectivity exhibited by the untreated parent virus at time zero.
Visualizing MAb binding to Env trimers and the effects of endo H treatment.
We next examined the binding of various MAbs and sCD4 to Env trimers by BN-PAGE, with two goals in mind: (i) to understand the basis of the differences in neutralization susceptibility between parent and GnTI- viruses, as observed in Fig. 3, and (ii) to determine what relationship, if any, exists between the loss of infectivity of GnTI- virus with endo H treatment (observed in Fig. 7) and the ability of trimers to bind receptor and coreceptor analogs.
(i) Relationship between trimer binding and MAb neutralization.
We previously examined the binding of several IgGs to parent trimers by BN-PAGE (8, 23, 25, 80). In Fig. 8, we compared MAb and sCD4 binding to parent and GnTI- trimers, using each MAb at a concentration of 30 μg/ml to ensure saturation. Clearly, to measure trimer binding, it is essential to be able to distinguish it from gp120/gp41 monomer binding. As in a previous publication (23), we observed high-molecular-mass Env-IgG complexes with MAbs b12, 15e, 4E10, and 39F at ∼500 kDa (marked with an asterisk in Fig. 8A, lanes 2, 3, and 4, and E, lane 5). If these bands constitute a trimer-IgG complex, this would suggest an IgG-induced shift of only ∼80 kDa, similar to that expected by trimer binding with three copies of the much smaller 2D-sCD4 ligand (Fig. 8E, lane 2, ∼500 kDa). However, MAbs 15e and 39F do not neutralize the parent virus (Fig. 3) and therefore would not be expected to bind to trimers. In fact, neither of these MAbs perturbs the native trimer, which remains undepleted and comparable to the control lane with no MAb added (compare the trimer band in Fig. 8A, lane 1, to that in A, lane 3, and E, lane 5). In contrast, the trimer band is depleted by the neutralizing ligands b12, 4E10, 2F5, and sCD4 (Fig. 8A, lanes 2, 4, and 5, and E, lane 2). Therefore, the band marked with an asterisk in Fig. 8 does not represent MAb complexing with native Env trimers, which is instead better assessed by depletion of native trimer (8, 23, 25, 80). Well-defined IgG-Env trimer complexes are in fact rarely observed, probably because IgGs tend to be flexible molecules that can adopt various conformations, which generally leads them to separate as smears on BN-PAGE gels. Complexes of IgGs and trimers adopt the properties of the IgGs and are also poorly resolved. In contrast, monovalent Fab fragments, sCD4, and 2G12 produce well-defined Env trimer complexes due to their inherently limited flexibility, as we discussed previously (see Fig. 6 of reference 23). Overall, as we previously reported, the ability of a MAb to complex with and deplete Env trimers on BN-PAGE depends on its ability to neutralize (8, 23, 25, 80).
FIG. 8.
Effect of endo H on Env trimer-ligand binding. We evaluated the binding of various ligands to parent and GnTI- trimers with or without endo H treatment, as indicated. Ligands (each at 30 μg/ml) included MAbs b12, 15e, 4E10, 2F5, E51, and 39F, and 2D-sCD4, scFv X5, and Fab 58.2 were also evaluated. After incubation with ligand, VLPs were then pelleted, washed, and resolved by BN-PAGE and Western blot assays.
GnTI- Env trimers were recognized by the panel of ligands in a largely comparable pattern to that of the parent trimers (Fig. 8, compare A and B), except that the band marked with an asterisk was absent. MAbs 4E10 and 2F5 completely depleted the unliganded GnTI- trimer. Importantly, the lack of MAb 15e neutralization (Fig. 3) was reflected by its lack of trimer binding (Fig. 8B, lane 3). Although the GnTI- virus was sensitive to MAb 39F (Fig. 3), surprisingly, there was no trimer depletion at 30 μg/ml (Fig. 8F, lane 5). In fact, we observed previously that the MAb IC50s required to neutralize can be somewhat lower than those required to mediate trimer binding in BN-PAGE (23). An alternative possibility is that the GnTI- virus might fuse more slowly than the parent, exposing the V3 loop for a longer time. However, this is unlikely, considering that the two viruses are both resistant to CD4i MAbs and equally sensitive to the T-20 peptide (Table 1), together implying unchanged fusion kinetics.
One hour of endo H treatment was accompanied by a significant drop in GnTI- trimer mass (Fig. 8C), as described above (Fig. 6). These smaller trimers became partially sensitive to MAbs 15e and 39F, which had no effect against the untreated trimers (Fig. 8C, lane 3, and G, lane 5). In addition, MAbs 2F5, 4E10, and b12 completely depleted the unliganded trimer (Fig. 8C, lanes 2, 4, and 5). Prolonged treatment with endo H led to an even more dramatic effect. Here, MAb 15e depleted the trimer far more convincingly, consistent with the removal of constraints to its recognition (Fig. 8D, lane 3). At the same time, b12 binding remained strong.
(ii) Effects of endo H on Env trimer binding receptor and coreceptor analogs.
We next studied Env binding to various receptor and coreceptor analogs to try to better understand the effects of endo H on GnTI- virus infectivity. Soluble CD4 bound more effectively to GnTI- than parent trimers, as evidenced by the more pronounced depletion of unliganded trimer (Fig. 8E and F, lane 2), consistent with our observations in Fig. 4. Importantly, sCD4 was also able to bind to endo H-treated trimers, even after 48 h (Fig. 8G and K, lane 2), suggesting that the removal of glycans that line the receptor binding site of the neutralizing face does not affect CD4 binding, eliminating this as a possible explanation for the rapid loss in GnTI- virus infectivity in the presence of endo H that we observed in Fig. 7.
The epitopes of CD4i and V3 loop-specific MAbs are largely obscured on primary isolate Env trimers but become exposed by the conformational changes induced by CD4 binding. Here, we used these MAbs as markers for coreceptor binding, bearing in mind the obvious limitations. As expected, neither of MAbs 39F or E51 bound to native Env trimers (Fig. 8E and F, compare lanes 1, 3, and 5). In the presence of sCD4, it was difficult to unequivocally determine IgG binding, since sCD4 alone already depletes trimers (Fig. 8E and F, lane 2). An additional problem is that, as mentioned above, well-resolved trimer complexes usually do not occur with IgGs. Nevertheless, a comparison of the patterns of lanes containing sCD4 alone and MAb alone with those containing both MAb and sCD4 suggests that CD4-induced MAb binding probably occurs (Fig. 8E, lanes 2 to 6). These patterns held true, even when the GnTI- virus was treated for 1 h with endo H. In fact, a trimer complex formed with sCD4 and 39F was particularly prominent (Fig. 8G, lane 6). To better resolve the binding of V3 and CD4i MAbs, we performed similar experiments using scFv X5 (a CD4i MAb) in place of E51 and Fab 58.2 (a V3 MAb) in place of 39F. Here, clear trimer-sCD4-MAb complexes were seen with parent and GnTI- trimers (Fig. 8H and I, compare lanes 2 with lanes 5 and 6). These shifts occurred even in the face of a 1- or 48-h incubation with endo H (Fig. 8J and K). Together, this suggests that CD4-induced coreceptor binding probably remains intact, even under conditions where infection is ablated by endo H treatment. In fact, scFv X5 was able to partially bind even in the absence of sCD4 after a 48-h endo H digestion (Fig. 8K, lane 3), suggesting that as for MAb 15e binding (Fig. 8D, lane 3), some constraints on binding had been lifted by the complete removal of glycans that line the receptor binding sites. In summary, glycan removal by endo H does not appear to affect either CD4 binding or CD4-induced structural rearrangements.
DISCUSSION
Here we set out to investigate the effects of replacing the complex glycans on the neutralizing face of HIV-1 Env trimers with oligomannose glycans. We looked at infectivity of the modified trimers, their susceptibility to a deglycosylating enzyme, its effects on infectivity, and the basis for that effect. We also compared the neutralization sensitivity of viruses bearing modified glycans with a mutant virus that eliminates a key glycan of the V3 loop.
We initially observed that GnTI- cells can express functional virus. This finding is mirrored by the observation that soluble gp120 and gp140 trimers produced from GnTI- cells retain an ability to be recognized by sCD4 and CD4i MAbs (32). It is also consistent with earlier observations that functional virus can be produced in the presence of glycan analogs such as kifunensine and swainsonine (36, 79, 100) that inhibit stages of glycan processing immediately preceding and following that of GnTI enzyme (Fig. 1). Thus, only inhibitors that affect very early stages of glycan maturation, such as NB-DNJ, appear to dramatically impact infectious function (37, 114). The infectious potential per Env expression in GnTI- cells appeared to be proportional to that of the corresponding virus prepared in parent 293T cells, suggesting that expression in GnTI- cells has no ill effects on Env function (32). Thus, the GnTI- cell effect of replacing complex glycans with Man5GlcNac2 did not adversely affect the folding and oligomerization of the gp160 precursor into trimers or its processing into gp120/gp41. The lack of effect on oligomerization was expected, considering that this step in Env synthesis occurs in the endoplasmic reticulum and precedes that of ablating GnTI in the cis-Golgi complex. However, gp160 processing is a very late step in Env synthesis that occurs in the trans-Golgi network (81, 82). The proper gp160 processing in GnTI- cells therefore suggests that the complex glycan antennae play little or no role in modifying the oligomer's sensitivity to furin or related proteases that mediate this late maturation step.
Another way to investigate the importance of glycans in HIV-1 infection is to treat virus with glycosidases. However, despite the sensitivity of soluble Env proteins to these enzymes, studies until now have found native Env trimers to be resistant to all but those that target the outer glycan moieties (32, 76, 101). In contrast to the modest effects of mannosidase (reduced infection) and sialidase (enhanced infection) (76), here we found that endo H rapidly ablated the infectivity of GnTI- but not the parent virus. In BN-PAGE, GnTI- trimers rapidly dropped in size with endo H treatment, while the parent trimers remained largely endo H resistant. Three possibilities might explain this observation: (i) that N-glycans are directly involved in conformational changes necessary for fusion; (ii) that the removal of glycans exposes Env trimers to proteases and therefore leads to degradation; and (iii) that the removal of glycans exposes hydrophobic protein domains, leading to aggregation. We do not favor the latter two possibilities. Although a significant loss of the GnTI- trimer band staining in BN-PAGE (Fig. 6) could suggest degradation, it required prolonged endo H treatment and was much slower than would be expected if this were to account for the rapid loss of infectivity (Fig. 7). The retention of binding by various MAbs in Fig. 8 suggests that the loss in trimer staining relates to a slow loss in 2G12 staining, rather than to degradation. We did not observe significant aggregation in our BN-PAGE analysis, although we note that the gentle detergents we used to release Env from the viral membrane may have masked any aggregating effects.
To further understand the basis of the effect of endo H on infectivity, it is useful to consider whether all or only a fraction of glycans are removed. In BN-PAGE, there appeared to be a much slower second phase of digestion after the rapid initial phase of glycan removal. Similarly, endo H treatment of JR-FL gp120 produced from Drosophila melanogaster cells was previously shown to remove only 90% of the oligomannose carbohydrate (58). This probably reflects the different sensitivities of the glycans of the neutralizing and silent faces. The former is decorated more sparsely with glycans that are therefore more accessible, while the latter domain exhibits closely packed and therefore enzyme-resistant glycans. In a previous study, the glycans that decorate soluble gp140 trimers produced in GnTI- cells were found to comprise a mixture of Man5-9GlcNac2 sugars (32), in contrast to the uniform Man5GlcNac2 sugars that decorate most proteins produced in these cells. Therefore, the steric constraints that limit the trimming of oligomannose precursors of the silent domain appear to be unaffected by expression in GnTI- cells. This is perhaps not surprising, considering that mannose trimming is necessary to create the Man5GlcNac2 substrate of the GnTI enzyme (Fig. 1B). It also explains the difficulty of removing these glycans from native Env trimers, even when they are expressed in GnTI- cells (17, 32, 94). Overall, our results suggest the loss in infectivity due to endo H treatment is due to removal of glycans from the neutralizing face.
Further BN-PAGE experiments (Fig. 8) revealed that endo H did not affect trimer-sCD4 binding or its ability to induce the CD4i and V3 epitopes, suggesting that receptor and coreceptor binding is competent. This is consistent with previous studies in which these ligands were able to recognize deglycosylated forms of gp120 (12, 67). Thus, the lack of function of endo H-treated trimer appears to stem from a post-CD4/CCR5 binding entry block.
The above observations have been modeled in Fig. 9. This figure represents our best imagining of the scenario, while acknowledging the many gaps and caveats in the available information from which these images are constructed. HXBc2 gp120 structures (in red) are derived from a b12 complex (126) and are fit into the cryoelectron tomogram of native unliganded trimers of the BaL isolate (69). The complex glycans of the neutralizing face of parent trimers (Fig. 9A, orange glycans) are replaced by Man5GlcNac2 in GnTI- trimers (Fig. 9B). Endo H treatment of these trimers results in the rapid removal of Man5GlcNac2 and other high-mannose glycans from the neutralizing face, leaving behind single GlcNac stumps (Fig. 9C, four orange glycans and one slate blue glycan), while the tightly packed oligomannose moieties on the silent face are relatively endo H resistant and therefore remain intact. Figure 9C shows a heavily depleted trimer that retains its gross architecture, as supported by BN-PAGE (Fig. 6 and 8). In one scenario, the lack of neutralizing face glycans lining both receptor binding sites may limit the ability of trimers to transmit the effects of receptor/coreceptor binding by undergoing further conformational changes, perhaps involving gp41. Possibly, the removal of these glycans could affect gp120 dissociation from gp41 and subsequent exposure of the fusion peptide (124). Previously, NB-DNJ-treated virus Env was also found to be capable of binding to CD4, despite its lack of infectivity (37). Furthermore, some single glycan mutants can ablate infectivity, for example, the N301 mutant in SF162 was not functional in one study (74). Thus, receptor-induced conformational changes may depend in part on proper glycan processing, as well as their frank presence, so that they can properly orchestrate post-receptor binding refolding events in fusion.
Our neutralization results suggested that the effect of “lowering the glycan fence” (as modeled by the GnTI- virus) compared to “knocking out a fence post” (as modeled by the N301Q mutant) was relatively subtle: while the N301Q virus was more sensitive to V3, nonneutralizing CD4bs mAbs sCD4, and at least one of the CD4i MAbs, the GnTI- virus was more sensitive only to V3 MAbs and sCD4. Previous studies suggest a largely predictable order in which glycan deletion mutants become neutralization sensitive, with increases in V3 and CD4bs exposure being frequently observed, often together, while increases in CD4i and MPER epitope exposure occur only for exceptionally sensitive mutants (54, 74). Exposure of CD4i epitopes may require larger holes in the protective shell, provided only by the removal of three glycans in one study (54) or by the complete removal of glycans from the neutralizing face (Fig. 8K, lane 3). The N301Q mutant falls into the latter “exceptionally sensitive” category, in keeping with the loss of a particularly large glycan (3, 54, 106). However, the GnTI- virus does not fit into either category, as, despite its V3 sensitivity, it remained resistant to nonneutralizing CD4bs MAbs and other specificities.
Two possibilities might explain the slightly increased sensitivity of the N301Q mutant to CD4i MAbs: (i) an altered steady-state trimer conformation or (ii) slower fusion kinetics that might grant prolonged access to the transition state targeted by these MAbs. The former possibility is supported by the observation of increased sensitivity to MAb 15e. This MAb is expected to block primary receptor binding, and its exposure should therefore stem from an altered ground state conformation. This explanation is also consistent with the lack of change in sensitivity to MPER MAbs or the T-20 peptide, whose epitopes become better exposed during fusion (7, 25) and therefore might be expected to exhibit a greater potency against Envs that fuse more slowly. It has been suggested that “nonpotent” CD4bs and CD4i MAbs generally fail to neutralize, due to the quaternary constraints of trimers that restrict the conformational reorganization required for binding (57). This model was generated from observing the unusually high entropic penalties associated with binding of “nonpotent” CD4bs and CD4i MAbs to monomeric gp120, compared to the lower penalty of b12 binding. Applying this model to the present scenario, the replacement of the N301 glycan might alter the steady-state conformation of the trimer or else may relieve the restrictions on conformational change that normally prevent the binding of these MAbs.
The GnTI- virus remained resistant to nonneutralizing CD4bs and CD4i MAbs, suggesting that the glycan stems or “fence posts” affected by the N301Q mutation, but not by GnTI- expression, are important for maintaining trimers in a native conformation in which these specificities remain occluded. Furthermore, in contrast to the decreased CD4 dependency of some glycan point mutants, CD4i epitope exposure (a surrogate for coreceptor binding) on the GnTI- virus was still dependent on sCD4 binding, in keeping with the retention of native conformation.
Although the GnTI- virus does not embody any conformational changes that grant access to otherwise-nonneutralizing ligands, it does allow greater access to those that do recognize the parent trimers. The dramatically increased sensitivity of GnTI- viruses to sCD4 (and to a lesser extent b12) suggests a relief of the steric constraints to accessing the CD4 binding loop in the GnTI- trimers. We have modeled this in Fig. 9. Here, the yellow area on each red gp120 protomer represents the b12 binding site, which closely approximates the CD4 binding site. For example, comparing the lower panels of Fig. 9A and B, it appears that the complex carbohydrate at position N276 of the parent gp120, as part of the “fence” that surrounds the receptor binding sites, may impinge on the b12 binding site, and its replacement with the smaller Man5GlcNac2 in Fig. 9B opens up the b12 binding site, ostensibly allowing increased b12/sCD4 access. Positions N397 and N463 may offer a lesser though similar scenario, depending on the angle of entry of b12/sCD4.
Studies with monomeric gp120 suggest that certain mutations can increase the binding of sCD4 but not CD4bs MAbs, while other mutations have the reverse effect (122). Therefore, the differential sensitivity profiles of N301Q and GnTI- viruses may be a simple reflection of alternative trimer steady-state conformations that generally favor the binding of one or the other of these ligands.
In marked contrast to the GnTI- virus's resistance to nonneutralizing CD4bs and CD4i MAbs, its increased V3 loop MAb sensitivity was almost as dramatic as that of the N301Q mutant. In the absence of a conformational change (as described above), one interpretation is that the large complex antennae of the N301Q mutant protects the V3 loop from MAb binding. An alternative explanation is that V3 loop exposure may be a result of disrupting the global network glycan-glycan hydrogen bonds, whose role may in part be to protect the V3 loop. The fact that we observed a V3-sensitive phenotype with the GnTI- virus without exposure of other conformationally occluded epitopes suggests, however, that increased V3 sensitivity does not require gross changes in trimer conformation.
Perhaps the closest precedent of the present analysis of GnTI- virus neutralization sensitivity is a study of an SIV isolate expressed in the presence of swainsonine. However, the sensitivity of this virus to SIV-infected macaque sera was not significantly affected (76). The difference compared to the generally increased sensitivity we observed with GnTI- and HIV-1-infected plasmas may relate to the fact that swainsonine inhibits a later step in glycan synthesis: the conversion of hybrid into complex glycans, leaving behind a “higher fence” than expression in GnTI- cells (Fig. 1B). Alternatively, the difference may be related to the difficulties in detecting increased sensitivity to certain epitopes amid a background of neutralizing activity targeting another epitope(s), as we observed with plasma LTNP2 (Fig. 3).
Overall, our neutralization analysis suggests that lowering the glycan fence and knocking out a fence post have markedly different consequences, with the major difference being that only the latter can alter trimer conformation. This is logical, considering that glycans participate in earliest stages of Env folding, so that deletion of one may have a greater consequence on conformation than the ablation of GnTI, which affects a later stage in glycoprotein maturation (67, 119). Indeed, eliminating glycans can affect gp160 processing and the exposure of distal epitopes, supporting a conformational effect (74). That N301Q mutation has no effect on CD4bs MAb recognition of monomeric gp120 (54) further supports the idea that N301 glycan removal does not directly expose the CD4bs. It is possible that the removal of the N301 glycan affects its interaction with the V1V2 loop, whose repositioning may lead to increased exposure of the CD4bs (54, 127). In comparison, the replacement of complex glycans with Man5GlcNAc2 has a more subtle effect on the glycan shell.
From a broader perspective, our results may have some relevance for ongoing attempts to crystallize native Env trimers. The replacement of complex glycans with simpler glycans that can then be removed by glycosidases has been a key tool in facilitating the formation and analysis of diffractable crystals. The ability of endo H-treated trimers to still bind to receptor and coreceptor analogs suggests that the fundamental properties of the trimer remain intact, despite these treatments, and therefore that a structure of such a molecule would largely reflect that of the untreated trimer.
Our findings also have some bearing on attempts to design an Env-based vaccine to elicit broad NAb responses. Further investigation of the effects of glycan modification on the immunogenicity of candidate vaccines is clearly warranted (4, 20, 66, 93, 96, 108, 109). However, a balance may need to be struck between increasing the exposure of neutralizing epitopes and avoiding the exposure of nonneutralizing epitopes that are not available on the native Env spike. Specifically, GnTI- Env trimers retain the same conformation as their native counterparts and relieve the constraints on sCD4 and possibly b12 binding without exposing nonneutralizing epitopes, with the important exception of the V3 loop. Thus, if GnTI- trimers are considered as immunogens, modifications may need to be engineered to dampen the sensitivity to V3 loop MAbs (88, 108, 111), so that antibody responses may instead focus on the more-exposed CD4 binding site. Finally, GnTI- viruses may also be useful tools to help us to better understand the specificities of neutralizing MAbs that depend partly or wholly on glycans (45, 115, 119).
Acknowledgments
This study was supported by NIH RO1 AI58763, R21 AI084714 (J.M.B.), Bill and Melinda Gates Collaboration for AIDS Vaccine Discovery/Vaccine Immune Monitoring Consortium grant 38619, and the AIDS and the Infectious Disease Science Center at the Torrey Pines Institute for Molecular Studies (J.M.B.). R.W.S. is a recipient of a VENI fellowship from the Netherlands Organization for Scientific Research and a Mathilde Krim research fellowship from the American Foundation for AIDS Research.
We thank D. Burton and M. Zwick for providing MAbs b6, b12, Fab 58.2, and scFv and whole IgG forms of X5, J. Robinson for providing MAbs 39F, 15e, 17b, and E51, D. Richman for providing human plasmas LTNP2 and K370, Bill Olson and Ken Kang for sCD4 and CD4-IgG2, and P. Reeves for the GnTI- cell line. We also thank Laura Walker, Ann Hessell, Max Crispin, and Chris Scanlan for useful discussions and Irina Zharkikh for technical assistance.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Allan, J. S., J. Strauss, and D. W. Buck. 1990. Enhancement of SIV infection with soluble receptor molecules. Science 247:1084-1088. [DOI] [PubMed] [Google Scholar]
- 2.Astronomo, R. D., H. K. Lee, C. N. Scanlan, R. Pantophlet, C. Y. Huang, I. A. Wilson, O. Blixt, R. A. Dwek, C. H. Wong, and D. R. Burton. 2008. A glycoconjugate antigen based on the recognition motif of a broadly neutralizing human immunodeficiency virus antibody, 2G12, is immunogenic but elicits antibodies unable to bind to the self glycans of gp120. J. Virol. 82:6359-6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back, N. K., L. Smit, J. J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, K., S. Andjelic, P. J. Klasse, Y. Kang, R. W. Sanders, E. Michael, R. J. Durso, T. J. Ketas, W. C. Olson, and J. P. Moore. 2009. Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 389:108-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, et al. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 6.Biller, M., A. Bolmstedt, A. Hemming, and S. Olofsson. 1998. Simplified procedure for fractionation and structural characterisation of complex mixtures of N-linked glycans, released from HIV-1 gp120 and other highly glycosylated viral proteins. J. Virol. Methods 76:87-100. [DOI] [PubMed] [Google Scholar]
- 7.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. S. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of HIV-1 plasmas from subtype B and C chronic infections. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binley, J. M., S. Ngo-Abdalla, P. Moore, M. Bobardt, U. Chatterji, P. Gallay, D. R. Burton, I. A. Wilson, J. H. Elder, and A. de Parseval. 2006. Inhibition of HIV Env binding to cellular receptors by monoclonal antibody 2G12 as probed by Fc-tagged gp120. Retrovirology 3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion- associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binley, J. M., R. Wyatt, E. Desjardins, P. D. Kwong, W. Hendrickson, J. P. Moore, and J. Sodroski. 1998. Analysis of the interaction of antibodies with a conserved enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 13.Bolmstedt, A., S. Sjolander, J. E. Hansen, L. Akerblom, A. Hemming, S. L. Hu, B. Morein, and S. Olofsson. 1996. Influence of N-linked glycans in V4-V5 region of human immunodeficiency virus type 1 glycoprotein gp160 on induction of a virus-neutralizing humoral response. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 12:213-220. [DOI] [PubMed] [Google Scholar]
- 14.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 15.Calarese, D. A., H. K. Lee, C. Y. Huang, M. D. Best, R. D. Astronomo, R. L. Stanfield, H. Katinger, D. R. Burton, C. H. Wong, and I. A. Wilson. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc. Natl. Acad. Sci. U. S. A. 102:13372-13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 17.Chang, V. T., M. Crispin, A. R. Aricescu, D. J. Harvey, J. E. Nettleship, J. A. Fennelly, C. Yu, K. S. Boles, E. J. Evans, D. I. Stuart, R. A. Dwek, E. Y. Jones, R. J. Owens, and S. J. Davis. 2007. Glycoprotein structural genomics: solving the glycosylation problem. Structure 15:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 19.Choi, B. K., P. Bobrowicz, R. C. Davidson, S. R. Hamilton, D. H. Kung, H. Li, R. G. Miele, J. H. Nett, S. Wildt, and T. U. Gerngross. 2003. Use of combinatorial genetic libraries to humanize N-linked glycosylation in the yeast Pichia pastoris. Proc. Natl. Acad. Sci. U. S. A. 100:5022-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole, K. S., J. D. Steckbeck, J. L. Rowles, R. C. Desrosiers, and R. C. Montelaro. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crispin, M., D. J. Harvey, V. T. Chang, C. Yu, A. R. Aricescu, E. Y. Jones, S. J. Davis, R. A. Dwek, and P. M. Rudd. 2006. Inhibition of hybrid- and complex-type glycosylation reveals the presence of the GlcNAc transferase I-independent fucosylation pathway. Glycobiology 16:748-756. [DOI] [PubMed] [Google Scholar]
- 22.Crispin, M. D., G. E. Ritchie, A. J. Critchley, B. P. Morgan, I. A. Wilson, R. A. Dwek, R. B. Sim, and P. M. Rudd. 2004. Monoglucosylated glycans in the secreted human complement component C3: implications for protein biosynthesis and structure. FEBS Lett. 566:270-274. [DOI] [PubMed] [Google Scholar]
- 23.Crooks, E. T., P. Jiang, M. Franti, S. Wong, M. B. Zwick, J. A. Hoxie, J. E. Robinson, P. L. Moore, and J. M. Binley. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crooks, E. T., P. L. Moore, M. Franti, C. S. Cayanan, P. Zhu, P. Jiang, R. P. de Vries, C. Wiley, I. Zharkikh, N. Schulke, K. H. Roux, D. C. Montefiori, D. R. Burton, and J. M. Binley. 2007. A comparative immunogenicity study of HIV-1 virus-like particles bearing various forms of envelope proteins, particles bearing no envelope and soluble monomeric gp120. Virology 366:245-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crooks, E. T., P. L. Moore, D. Richman, J. Robinson, J. A. Crooks, M. Franti, N. Schulke, and J. M. Binley. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101-113. [PMC free article] [PubMed] [Google Scholar]
- 26.Cutalo, J. M., L. J. Deterding, and K. B. Tomer. 2004. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J. Am. Soc. Mass Spectrom. 15:1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darbha, R., S. Phogat, A. F. Labrijn, Y. Shu, Y. Gu, M. Andrykovitch, M. Y. Zhang, R. Pantophlet, L. Martin, C. Vita, D. R. Burton, D. S. Dimitrov, and X. Ji. 2004. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry 43:1410-1417. [DOI] [PubMed] [Google Scholar]
- 28.Davis, S. J., E. A. Davies, A. N. Barclay, S. Daenke, D. L. Bodian, E. Y. Jones, D. I. Stuart, T. D. Butters, R. A. Dwek, and P. A. van der Merwe. 1995. Ligand binding by the immunoglobulin superfamily recognition molecule CD2 is glycosylation-independent. J. Biol. Chem. 270:369-375. [DOI] [PubMed] [Google Scholar]
- 29.Derdeyn, C. A., and E. Hunter. 2008. Viral characteristics of transmitted HIV. Curr. Opin. HIV AIDS 3:16-21. [DOI] [PubMed] [Google Scholar]
- 30.Dhillon, A. K., H. Donners, R. Pantophlet, W. E. Johnson, J. M. Decker, G. M. Shaw, F. H. Lee, D. D. Richman, R. W. Doms, G. Vanham, and D. R. Burton. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 81:6548-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunlop, D. C., A. Ulrich, B. J. Appelmelk, D. R. Burton, R. A. Dwek, N. Zitzmann, and C. N. Scanlan. 2008. Antigenic mimicry of the HIV envelope by AIDS-associated pathogens. AIDS 22:2214-2217. [DOI] [PubMed] [Google Scholar]
- 32.Eggink, D., M. Melchers, M. Wuhrer, T. van Montfort, A. K. Dey, B. Naaijkens, K. B. David, V. LeDouce, A. M. Deelder, K. Kang, W. C. Olson, B. Berkhout, C. H. Hokke, J. P. Moore, and R. W. Sanders. 19 March 2010, posting date. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 33.Elbein, A. D., J. E. Tropea, M. Mitchell, and G. P. Kaushal. 1990. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J. Biol. Chem. 265:15599-15605. [PubMed] [Google Scholar]
- 34.Fenouillet, E., I. Jones, B. Powell, D. Schmitt, M. P. Kieny, and J. C. Gluckman. 1993. Functional role of the glycan cluster of the human immunodeficiency virus type 1 transmembrane glycoprotein (gp41) ectodomain. J. Virol. 67:150-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenouillet, E., and I. M. Jones. 1995. The glycosylation of human immunodeficiency virus type 1 transmembrane glycoprotein (gp41) is important for the efficient intracellular transport of the envelope precursor gp160. J. Gen. Virol. 76:1509-1514. [DOI] [PubMed] [Google Scholar]
- 36.Fenouillet, E., R. Miquelis, and R. Drillien. 1996. Biological properties of recombinant HIV envelope synthesized in CHO glycosylation-mutant cell lines. Virology 218:224-231. [DOI] [PubMed] [Google Scholar]
- 37.Fischer, P. B., M. Collin, G. B. Karlsson, W. James, T. D. Butters, S. J. Davis, S. Gordon, R. A. Dwek, and F. M. Platt. 1995. The alpha-glucosidase inhibitor N-butyldeoxynojirimycin inhibits human immunodeficiency virus entry at the level of post-CD4 binding. J. Virol. 69:5791-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer, P. B., G. B. Karlsson, R. A. Dwek, and F. M. Platt. 1996. N-Butyldeoxynojirimycin-mediated inhibition of human immunodeficiency virus entry correlates with impaired gp120 shedding and gp41 exposure. J. Virol. 70:7153-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagneux, P., and A. Varki. 1999. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9:747-755. [DOI] [PubMed] [Google Scholar]
- 41.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760-11767. [PubMed] [Google Scholar]
- 42.Go, E. P., J. Irungu, Y. Zhang, D. S. Dalpathado, H. X. Liao, L. L. Sutherland, S. M. Alam, B. F. Haynes, and H. Desaire. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 7:1660-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes, B. F., and D. C. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong, P. W., S. Nguyen, S. Young, S. V. Su, and B. Lee. 2007. Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J. Virol. 81:8325-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honnen, W. J., C. Krachmarov, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81:1424-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu, H., T. Shioda, C. Moriya, X. Xin, M. K. Hasan, K. Miyake, T. Shimada, and Y. Nagai. 1996. Infectivities of human and other primate lentiviruses are activated by desialylation of the virion surface. J. Virol. 70:7462-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irungu, J., E. P. Go, Y. Zhang, D. S. Dalpathado, H. X. Liao, B. F. Haynes, and H. Desaire. 2008. Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J. Am. Soc. Mass Spectrom. 19:1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobson, J. M., R. J. Israel, I. Lowy, N. A. Ostrow, L. S. Vassilatos, M. Barish, D. N. Tran, B. M. Sullivan, T. J. Ketas, T. J. O'Neill, K. A. Nagashima, W. Huang, C. J. Petropoulos, J. P. Moore, P. J. Maddon, and W. C. Olson. 2004. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 48:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston, M. I., and A. S. Fauci. 2007. An HIV vaccine: evolving concepts. N. Engl. J. Med. 356:2073-2081. [DOI] [PubMed] [Google Scholar]
- 51.Karlsson, G. B., T. D. Butters, R. A. Dwek, and F. M. Platt. 1993. Effects of the imino sugar N-butyldeoxynojirimycin on the N-glycosylation of recombinant gp120. J. Biol. Chem. 268:570-576. [PubMed] [Google Scholar]
- 52.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143-155. [DOI] [PubMed] [Google Scholar]
- 53.Kim, M., B. Chen, R. E. Hussey, Y. Chishti, D. Montefiori, J. A. Hoxie, O. Byron, G. Campbell, S. C. Harrison, and E. L. Reinherz. 2001. The stoichiometry of trimeric SIV glycoprotein interaction with CD4 differs from that of anti-envelope antibody Fab fragments. J. Biol. Chem. 276:42667-42676. [DOI] [PubMed] [Google Scholar]
- 54.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 55.Kolchinsky, P., E. Kiprilov, and J. Sodroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 57.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 58.Kwong, P. D., R. Wyatt, E. Desjardins, J. Robinson, J. S. Culp, B. D. Hellmig, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1999. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274:4115-4123. [DOI] [PubMed] [Google Scholar]
- 59.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310- 10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 64.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li, Y., L. Luo, N. Rasool, and C. Y. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li, Y., M. A. Rey-Cuille, and S. L. Hu. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res. Hum. Retrovir. 17:1473-1479. [DOI] [PubMed] [Google Scholar]
- 69.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luallen, R. J., J. Lin, H. Fu, K. K. Cai, C. Agrawal, I. Mboudjeka, F. H. Lee, D. Montefiori, D. F. Smith, R. W. Doms, and Y. Geng. 2008. An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies. J. Virol. 82:6447-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matthews, T. J., K. J. Weinhold, H. K. Lyerly, A. J. Langlois, H. Wigzell, and D. P. Bolognesi. 1987. Interaction between the human T-cell lymphotropic virus type IIIB envelope glycoprotein gp120 and the surface antigen CD4: role of carbohydrate in binding and cell fusion. Proc. Natl. Acad. Sci. U. S. A. 84:5424-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 76.Means, R. E., and R. C. Desrosiers. 2000. Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J. Virol. 74:11181-11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J. Biol. Chem. 265:8519-8524. [PubMed] [Google Scholar]
- 78.Mizuochi, T., M. W. Spellman, M. Larkin, J. Solomon, L. J. Basa, and T. Feizi. 1988. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem. J. 254:599-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montefiori, D. C., W. E. Robinson, Jr., and W. M. Mitchell. 1988. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 85:9248-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moore, P. L., E. T. Crooks, L. Porter, P. Zhu, C. S. Cayanan, H. Grise, P. Corcoran, M. B. Zwick, M. Franti, L. Morris, K. H. Roux, D. R. Burton, and J. M. Binley. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morikawa, Y., E. Barsov, and I. Jones. 1993. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J. Virol. 67:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 83.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. U. S. A. 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olofsson, S., and J. E. Hansen. 1998. Host cell glycosylation of viral glycoproteins: a battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30:435-440. [DOI] [PubMed] [Google Scholar]
- 86.Pancera, M., S. Majeed, Y. E. Ban, L. Chen, C. C. Huang, L. Kong, Y. D. Kwon, J. Stuckey, T. Zhou, J. E. Robinson, W. R. Schief, J. Sodroski, R. Wyatt, and P. D. Kwong. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739-769. [DOI] [PubMed] [Google Scholar]
- 88.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 90.Phogat, S., R. T. Wyatt, and G. B. Karlsson Hedestam. 2007. Inhibition of HIV-1 entry by antibodies: potential viral and cellular targets. J. Intern. Med. 262:26-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pikora, C., C. Wittish, and R. C. Desrosiers. 2005. Identification of two N-linked glycosylation sites within the core of the simian immunodeficiency virus glycoprotein whose removal enhances sensitivity to soluble CD4. J. Virol. 79:12575-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puffer, B. A., S. Pohlmann, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinones-Kochs, M. I., L. Buonocore, and J. K. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reeves, P. J., N. Callewaert, R. Contreras, and H. G. Khorana. 2002. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U. S. A. 99:13419-13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reitter, J. N., and R. C. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 97.Rerks-Ngarm, S., P. Pitisuttithum, S. Nitayaphan, J. Kaewkungwal, J. Chiu, R. Paris, N. Premsri, C. Namwat, M. de Souza, E. Adams, M. Benenson, S. Gurunathan, J. Tartaglia, J. G. McNeil, D. P. Francis, D. Stablein, D. L. Birx, S. Chunsuttiwat, C. Khamboonruang, P. Thongcharoen, M. L. Robb, N. L. Michael, P. Kunasol, and J. H. Kim. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209-2220. [DOI] [PubMed] [Google Scholar]
- 98.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson, J. E., D. H. Elliott, E. A. Martin, K. Micken, and E. S. Rosenberg. 2005. High frequencies of antibody responses to CD4 induced epitopes in HIV infected patients started on HAART during acute infection. Hum. Antibodies 14:115-121. [PubMed] [Google Scholar]
- 100.Robinson, W. E., Jr., D. C. Montefiori, and W. M. Mitchell. 1987. Evidence that mannosyl residues are involved in human immunodeficiency virus type 1 (HIV-1) pathogenesis. AIDS Res. Hum. Retrovir. 3:265-282. [DOI] [PubMed] [Google Scholar]
- 101.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scanlan, C. N., J. Offer, N. Zitzmann, and R. A. Dwek. 2007. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature 446:1038-1045. [DOI] [PubMed] [Google Scholar]
- 103.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scanlan, C. N., G. E. Ritchie, K. Baruah, M. Crispin, D. J. Harvey, B. B. Singer, L. Lucka, M. R. Wormald, P. Wentworth, Jr., N. Zitzmann, P. M. Rudd, D. R. Burton, and R. A. Dwek. 2007. Inhibition of mammalian glycan biosynthesis produces non-self antigens for a broadly neutralising, HIV-1 specific antibody. J. Mol. Biol. 372:16-22. [DOI] [PubMed] [Google Scholar]
- 105.Schief, W. R., Y. E. Ban, and L. Stamatatos. 2009. Challenges for structure-based HIV vaccine design. Curr. Opin. HIV AIDS 4:431-440. [DOI] [PubMed] [Google Scholar]
- 106.Schonning, K., B. Jansson, S. Olofsson, J. O. Nielsen, and J. S. Hansen. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 107.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Selvarajah, S., B. A. Puffer, F. H. Lee, P. Zhu, Y. Li, R. Wyatt, K. H. Roux, R. W. Doms, and D. R. Burton. 2008. Focused dampening of antibody response to the immunodominant variable loops by engineered soluble gp140. AIDS Res. Hum. Retrovir. 24:301-314. [DOI] [PubMed] [Google Scholar]
- 109.Shan, M., P. J. Klasse, K. Banerjee, A. K. Dey, S. P. Iyer, R. Dionisio, D. Charles, L. Campbell-Gardener, W. C. Olson, R. W. Sanders, and J. P. Moore. 2007. HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PloS Pathog. 3:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Starcich, B. R., B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. Wolf, E. S. Parks, W. P. Parks, S. F. Josephs, R. C. Gallo, et al. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 111.Tobin, G. J., J. D. Trujillo, R. V. Bushnell, G. Lin, A. R. Chaudhuri, J. Long, J. Barrera, L. Pena, M. J. Grubman, and P. L. Nara. 2008. Deceptive imprinting and immune refocusing in vaccine design. Vaccine 26:6189-6199. [DOI] [PubMed] [Google Scholar]
- 112.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Voynow, J. A., R. S. Kaiser, T. F. Scanlin, and M. C. Glick. 1991. Purification and characterization of GDP-L-fucose-N-acetyl beta-D-glucosaminide alpha 1-6 fucosyltransferase from cultured human skin fibroblasts. Requirement of a specific biantennary oligosaccharide as substrate. J. Biol. Chem. 266:21572-21577. [PubMed] [Google Scholar]
- 114.Walker, B. D., M. Kowalski, W. C. Goh, K. Kozarsky, M. Krieger, C. Rosen, L. Rohrschneider, W. A. Haseltine, and J. Sodroski. 1987. Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc. Natl. Acad. Sci. U. S. A. 84:8120-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang, S. K., P. H. Liang, R. D. Astronomo, T. L. Hsu, S. L. Hsieh, D. R. Burton, and C. H. Wong. 2008. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc. Natl. Acad. Sci. U. S. A. 105:3690-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 118.Wu, X., T. Zhou, S. O'Dell, R. T. Wyatt, P. D. Kwong, and J. Mascola. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody b12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu, Z., S. C. Kayman, W. Honnen, K. Revesz, H. Chen, S. Vijh-Warrier, S. A. Tilley, J. McKeating, C. Shotton, and A. Pinter. 1995. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J. Virol. 69:2271-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 121.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 122.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yamshchikov, G. V., G. D. Ritter, M. Vey, and R. W. Compans. 1995. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology 214:50-58. [DOI] [PubMed] [Google Scholar]
- 124.Yang, X., E. Mahony, G. H. Holm, A. Kassa, and J. Sodroski. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117-125. [DOI] [PubMed] [Google Scholar]
- 125.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]
- 126.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhu, C. B., L. Zhu, S. Holz-Smith, T. J. Matthews, and C. H. Chen. 2001. The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc. Natl. Acad. Sci. U. S. A. 98:15227-15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]
- 129.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]