Abstract
Nonenveloped viral capsids are metastable structures that undergo conformational changes during virus entry that lead to interactions of the capsid or capsid fragments with the cell membrane. For members of the Caliciviridae, neither the nature of these structural changes in the capsid nor the factor(s) responsible for inducing these changes is known. Feline junctional adhesion molecule A (fJAM-A) mediates the attachment and infectious viral entry of feline calicivirus (FCV). Here, we show that the infectivity of some FCV isolates is neutralized following incubation with the soluble receptor at 37°C. We used this property to select mutants resistant to preincubation with the soluble receptor. We isolated and sequenced 24 soluble receptor-resistant (srr) mutants and characterized the growth properties and receptor-binding activities of eight mutants. The location of the mutations within the capsid structure of FCV was mapped using a new 3.6-Å structure of native FCV. The srr mutations mapped to the surface of the P2 domain were buried at the protruding domain dimer interface or were present in inaccessible regions of the capsid protein. Coupled with data showing that both the parental FCV and the srr mutants underwent increases in hydrophobicity upon incubation with the soluble receptor at 37°C, these findings indicate that FCV likely undergoes conformational change upon interaction with its receptor. Changes in FCV capsid conformation following its interaction with fJAM-A may be important for subsequent interactions of the capsid with cellular membranes, membrane penetration, and genome delivery.
The interactions between viruses and receptors on the surface of host cells strongly influence viral pathogenesis and regulate morbidity and mortality in the host. Virus-receptor interactions determine the types of cells that can be infected, the pathway of entry into the cell, and the efficiency of productive infection. Interactions between nonenveloped virus capsids and their receptor(s) trigger one or more steps required for infectious entry. These steps can include interaction with other receptors, exposure to low pH or endosomal proteases, or other factors. Ultimately, one or more of these interactions induce changes in capsid conformation that result in the exposure of hydrophobic regions or release of a lipid-seeking factor that can interact with and disrupt the limiting cellular membrane to allow the capsid and/or the genome to be delivered to the interior of the cell (reviewed in reference 60).
The Caliciviridae are small nonenveloped viruses containing a positive-sense RNA genome (∼7 to 8 kb). Several important disease-causing members of the Caliciviridae, including human noroviruses and rabbit hemorrhagic disease virus, cannot be propagated in tissue culture systems (19, 56). This has slowed progress on studies of the mechanisms of cellular entry of these viruses. In contrast, feline caliciviruses (FCVs) propagate readily in tissue culture, and two cell surface receptor molecules, feline junctional adhesion molecule A (fJAM-A) and α2,6 sialic acid, are known (29, 55).
The FCV receptor, fJAM-A, is a type I transmembrane glycoprotein (molecular size of 36 to 41 kDa) member of the immunoglobulin superfamily (IgSF); it consists of an amino-terminal signal peptide, an extracellular domain (composed of two Ig-like domains—a membrane-distal D1 and a membrane-proximal D2), a transmembrane domain, and a short cytoplasmic domain that contains a type II PDZ domain-binding motif (11, 30). We have previously shown that the D1 domain of the fJAM-A ectodomain is necessary and sufficient for FCV binding and that preincubation of FCV with soluble fJAM-A (sfJAM-A) results in virus neutralization (35). Additional roles that fJAM-A might play in FCV entry, however, have not been investigated.
Caliciviruses are composed of 180 copies of a single capsid protein. Atomic resolution structures of recombinant virus-like particles of Norwalk virus (genus Norovirus) and native San Miguel sea lion virus (SMSV) virions (genus Vesivirus) indicate that the virion consists of 90 dimers of the capsid protein arranged in T=3 icosahedral symmetry (5, 41). Each capsid monomer contains three structural domains—an N-terminal arm (NTA), the shell (S), and a protruding domain (P) that is further subdivided into P1 and P2 subdomains. The distal subdomain, P2, is structurally conserved between Norwalk virus and SMSV, but there is little sequence conservation. In the primary sequence of the FCV capsid, there are two hypervariable regions that contain neutralizing epitopes (18, 34, 58). The corresponding hypervariable regions (HVRs) of the SMSV capsid structure map to surface-exposed loops. Surface residues at the dimeric interface between two capsid monomers are conserved within individual calicivirus genera, and it has been suggested that this interface is involved in receptor binding (5). A cryo-electron microscopy (cryo-EM) reconstruction of the FCV vaccine strain F9 complexed with the ectodomain of fJAM-A (modeled on the crystal structures of SMSV and human JAM-A, respectively) shows that fJAM-A engages the top of the P2 domain and that binding causes a rotation in the P dimer (1). However, the relatively low resolution and the lack of atomic resolution structures of FCV and fJAM-A prevented precise identification of residues on the viral capsid that contact fJAM-A.
A classical approach for identifying virus residues that directly bind or modulate the binding of a receptor is to select for mutant viruses resistant to neutralization with soluble receptors (6, 23, 46). Soluble receptor-resistant (srr) mutants of poliovirus revealed that both surface-exposed and internal residues regulate receptor attachment and conformational changes in the capsid (6, 42). Here, we report 24 srr mutants and the location of their capsid mutations on a 3.6-Å structure of FCV. In addition, we describe the growth kinetics and receptor-binding properties of a subpanel of eight srr mutants and examine changes in capsid hydrophobicity concurrent with the interaction of FCV capsids with sfJAM-A.
MATERIALS AND METHODS
Cells and viruses.
Crandell-Reese feline kidney (CRFK; ATCC CCL-94) cells were grown in Eagle's minimal essential medium (EMEM; CellGro) supplemented with 5% fetal bovine serum (FBS; HyClone), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B, 1 mM sodium pyruvate, and nonessential amino acids (CellGro). Suspension Chinese hamster ovary (CHO-S) cells (Invitrogen) were grown in CHO serum-free medium (Gibco).
The F9 vaccine strain (VR-782) of FCV was obtained from the ATCC. The viral isolates FCV-5, Deuce, Kaos, FCV-127, FCV-131, and FCV-796 were previously characterized (36). FCV-Urbana viral stock was generated from a full-length infectious clone as previously described (50, 51). Briefly, CRFK cells were infected with vaccinia virus expressing T7 RNA polymerase (MVA/T7) and then transfected using FuGene 6 (Roche) 1 h later with the FCV-Urbana infectious clone (pQ14). Samples were lysed at 18 h postinfection (h pi) by freeze-thawing, and viral plaques were selected. The viral capsid gene was sequenced to verify that FCV-Urbana was present. Third-passage viral stocks were prepared from twice-plaque-purified viruses amplified in CRFK cells. Plaque assays were performed as previously described (36).
Antibodies and reagents.
A rabbit antiserum against the purified fJAM-A ectodomain was previously described (35). Virus was detected with either an anti-FCV mouse monoclonal antibody (MAb) (S1-9; Custom Monoclonal Antibodies International) or rabbit anti-FCV-5 antiserum. The fJAM-A extracellular domain (amino acid residues 26 to 232) was purified as a glutathione S-transferase (GST)-tagged fusion as previously described (35). Recombinant protein was purified by glutathione affinity chromatography. When GST-free fJAM-A was desired, N-terminal GST and His tags were cleaved from the recombinant protein by using a human rhinovirus (HRV) 3C protease (Novagen) according to the manufacturer's directions.
Neutralization of FCV by fJAM-A and selection of soluble receptor-resistant (srr) mutants of FCV-5.
To investigate the neutralization of FCV by fJAM-A, ∼1 × 105 PFU of virus in Dulbecco's modified Eagle's medium (DMEM; CellGro) plus 0.1% bovine serum albumin (BSA; Calbiochem) were incubated in either the presence or absence of sfJAM-A (concentrations of receptor, temperatures of incubation, and lengths of incubation varied by experiment; please refer to Results for experimental details). The infectious titer of the samples was determined by plaque assay. srr mutants of FCV-5 were selected by plaque titrating virus neutralized as described above on CRFK cells in six-well plates and overlaid with EMEM containing 5% FBS and 1% Bacto agar. Following incubation at 37°C in humidified 5% CO2 for 48 h, individual plaques were selected and amplified without selection on CRFK cells in 12-well plates (Corning). Once wells showed 100% cytopathic effect (CPE), a small aliquot from each well was again incubated with GST-fJAM-A and used to infect CRFK cells in 12-well plates. In wells that developed CPE, a sample of the lysate was subjected to an additional round of GST-fJAM-A neutralization, and samples were amplified on CRFK cells in a 96-well plate (Corning). Twenty-four wells showing CPE were chosen at random and twice plaque-purified. To verify that the selected srr mutants were resistant to neutralization by soluble receptor, a subpanel of eight srr mutants was incubated with a soluble receptor, and the infectious titer of the samples was determined by plaque assay.
Sequencing the major (VP1) and minor (VP2) capsid proteins of the srr mutants.
Recovery of viral RNA, reverse transcription-PCR (RT-PCR), and sequencing of the major capsid protein of the srr mutants were performed as described previously (36). Briefly, total RNA was extracted from infected cell lysates (RNeasy minikit; Qiagen), and first-strand cDNA synthesis of the capsid region of the genome was performed using a degenerate primer and AccuScript high-fidelity reverse transcriptase (Stratagene). The major capsid open reading frame (ORF) was then amplified from the first-strand cDNA template (primers available upon request). The resulting ∼2.1-kb PCR products, encompassing the entirety of the capsid ORF, were purified and sequenced directly. First-strand cDNA synthesis of the region of the genome containing VP2 was performed using RNA extracted from infected cell lysates as described above and using the 3′ RACE system for amplification of cDNA ends (Invitrogen). The minor capsid ORF was amplified from the first-strand cDNA template, and the resulting PCR products were purified and sequenced directly. The VP2 sequence of FCV-5 was submitted to GenBank (accession no. HM001263).
Purification of FCV.
Roller bottles (Corning) containing confluent CRFK monolayers were infected with FCV (multiplicity of infection [MOI] = 5) and incubated for 8 h at 37°C. At 8 h pi, cells were removed from the plastic surface by physical agitation, and the cells were collected by centrifugation of the medium at 500 × g. The cell pellet was resuspended in 250 mM NaCl and 85 mM Tris base (pH 7.5) and frozen at −80°C. The pellet was thawed and briefly sonicated to lyse the cells, and trichlorotrifluoroethane was used to extract the virus from the cell lysate. To clarify the phases, the trichlorotrifluoroethane-lysate mixture was centrifuged for 10 min at 5,850 × g. The aqueous phase was removed and the trichlorotrifluoroethane extraction step repeated. Following the second centrifugation, the aqueous phase was placed on a CsCl gradient (1.30 to 1.45 g/ml) in an ultracentrifuge tube (Beckman) and centrifuged at 97,000 × g for 16 h. Typically, two bands were visible—a lower band containing infectious particles and an upper band containing noninfectious particles. Occasionally, a third minor band would be visible between the upper and lower bands; infectivity of this band when present was variable. Each band was collected separately and dialyzed exhaustively against virion buffer (150 mM NaCl, 10 mM Tris base, 15 mM MgCl2 [pH 7.2]) for 48 to 72 h at 4°C. Purified virus was stored at 4°C.
Crystallization.
“Full” infectious particles of FCV-5 were used for crystallization. Crystallization trials were carried out using the hanging drop vapor diffusion method at room temperature. Crystal screens 1 and 2 from Hampton Research were used to explore various conditions. Typically, for each condition, 2 μl of the virus solution, at a concentration of 3 mg/ml, was mixed with 2 μl of the well solution and equilibrated with 0.5 ml of the well solution. Initial screening produced crystals under a few conditions, including the crystal screen 1 condition 3 (0.4 M ammonium phosphate) and condition 13 (30% polyethylene glycol [PEG] 400, 0.1 M Tris HCl [pH 8.5], 0.2 M sodium citrate). After optimization of crystallization conditions and synchrotron on-site screening, crystals of the best diffraction quality were obtained using a well solution containing 0.45 M ammonium phosphate with additive screen reagent C10 (1.0 M glycine). Cryo-protection of crystals was carried out by soaking the crystals in the mother liquor with increasing concentrations of PEG 400 through six steps, as follows: 5%, 10%, 15%, 20%, 22.5%, and 25%. The equilibration time at each solution was at least 10 min. The crystals were then flash frozen in liquid nitrogen and shipped to synchrotron sites for data collection.
Data collection and analysis.
The diffraction data from single crystals were collected under cryo-EM conditions at an APS 19ID station with monochromatic X-rays (wavelength = 0.9795 Å) and a detector to crystal distance of 480 mm on an ADSC 3-by-3 charge-coupled-device (CCD) detector, using an oscillation angle of 0.3° and an exposure time of 3 s. Indexing, integration, scaling, postrefinement, and reduction of the data were carried out using the HKL-2000 (37) and d*TREK (40) packages. Analysis of the diffraction data clearly indicated that the FCV-5 crystal belonged to the orthorhombic space group I222 (Table 1). The higher-resolution reflections were generally weak and appeared to be sensitive to radiation damage. A similar trend was observed with several other crystals that were used for data collection. The best crystal, which diffracted to ∼3.6 Å, was used for data collection. The data between 30 and 3.6 Å from 300 frames were scaled with an Rmerge factor of about 22%. The overall completeness of the data to 3.6-Å resolution is about 90.1%. To determine the precise orientation of the virus particle in the unit cell, self-rotation functions (45) were calculated using the program GLRF (59). These calculations showed all the peaks corresponding to 5-fold (κ = 72°), 3-fold (κ = 120°), and 2-fold (κ = 180°) axes, which are expected from a particle with icosahedral symmetry. In the I222 space group, consistent with the unit cell dimensions and the particle radius, which is estimated to be 200 Å, the particle position is uniquely defined by the intersection of the crystallographic 2-fold axes. In such a setting, an icosahedral particle can assume one of the two possible orientations. This ambiguity in orientation was clearly resolved by the self-rotation function calculations by use of the X-ray diffraction data. Each crystallographic asymmetric unit is composed of one-fourth of the virion with a 15-fold noncrystallographic redundancy due to one set of icosahedral 5- and 3-fold symmetry.
TABLE 1.
Data collection and refinement statistics
| Parameter | Data |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.97925 |
| Space group | I222 |
| Cell dimensions | |
| a, b, c (Å) | 427.08, 450.73, 467.59 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 30-3.6 (3.73-3.60)a |
| Rsym or Rmerge (%) | 22 (55) |
| Completeness (%) | 90.1 (87.0) |
| Total no. of reflections | 1,233,681 |
| No. of unique reflections | 462,277 |
| Refinement | |
| Resolution (Å) | 30-3.6 |
| No. of reflections | 462,277 |
| Rwork/Rfree | 0.37/0.36 |
Values in parentheses are for the highest-resolution shell.
Crystal structure determination and refinement.
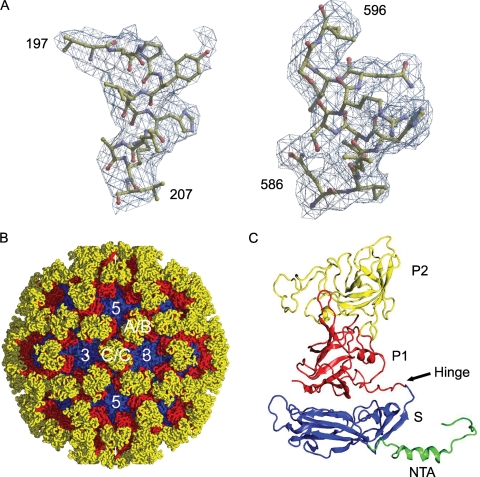
A properly positioned and oriented SMSV-4 capsid was placed in an FCV-5 unit cell. One initial model at a resolution of 10 Å was calculated from the 15 copies of SMSV-4 (Protein Data Bank [PDB] ID 2GH8) capsid protein related first by icosahedral 5-fold symmetry and then by icosahedral 3-fold symmetry. Phase refinement and extension were carried out by iterative cycles of real-space electron density averaging, solvent flattening, and back transformation with the RAVE and CCP4 (3, 25) program packages. The phases were extended to the final 3.6-Å resolution with each step being less than one reciprocal space point. An initial mask was constructed from the cryo-EM reconstruction with the program MAMA, and the mask was edited at each step to avoid truncation of the density. The averaged 3.6-Å density map is of good quality and readily interpretable (see Fig. 2A). After initial model building using COOT (12) and the final refinement using CNS (3), the structure has an Rfree (2) of 0.39, calculated using 10% of the data that were not included in the refinement, and a final R factor of 0.37. The atomic coordinates and associated structure factors have been deposited into the PDB (www.pdb.org) (PDB ID 3M8L).
FIG. 2.
X-ray structure of FCV (FCV-5) capsid. (A) Two sample regions in the calculated electron density map with modeled amino acid residues 197 to 207 (S domain) and 586 to 596 (P domain), respectively. (B) X-ray structure of FCV-5 viewed along the icosahedral 2-fold axis. Location of a set of A/B and C/C dimers and icosahedral 5-fold and 3-fold axes are shown. (C) Ribbon representation of the B subunit structure. The NTA (green), S domain (blue), and P1 (red) and P2 (yellow) subdomains are indicated.
Virus-binding assay by flow cytometry.
FCV binding to CHO-S cells transiently expressing fJAM-A was detected as previously described (35). Briefly, CHO-S cells (106 cells) were transiently transfected with a plasmid encoding fJAM-A. At 24 h posttransfection, the cells were washed and incubated with FCV (MOI = 5) in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 [pH 7.5]) plus 1% BSA (PBSA) on ice for 30 min. After being washed to remove unbound virions, the cells were incubated with fJAM-A rabbit antiserum and anti-FCV mouse MAb in PBSA on ice for 30 min. The cells were then washed in PBSA, fixed in 2% paraformaldehyde in PBS, and incubated with Alexa 647-conjugated goat anti-rabbit and Alexa 488-conjugated goat anti-mouse IgG for 30 min at room temperature. The cells were analyzed using a FACSCalibur (Becton Dickinson). Cells expressing fJAM-A on their surface were gated and analyzed for virus binding (10,000 cells were gated for each sample).
Plaque reduction of srr mutants by fJAM-A antiserum.
Confluent CRFK monolayers in 60-mm dishes (Corning) were incubated in PBSA with or without fJAM-A rabbit antiserum for 30 min at room temperature. Virus (∼100 PFU per dish) was adsorbed for 1 h at room temperature. The monolayers were then overlaid with EMEM containing 5% FBS and 1% Bacto agar. After incubation at 37°C for 48 h in humidified 5% CO2, the overlay was removed, and the cells were fixed with 10% buffered formalin and stained with 1% (wt/vol) crystal violet in 20% ethanol. The plaques were counted, and percent reduction from controls was calculated.
Single- and multiple-cycle growth kinetics of srr mutants.
CRFK cell monolayers were inoculated with srr mutants at MOIs of 5 and 0.01 for single- and multiple-cycle growth curves, respectively. Virus was adsorbed to cells for 1 h at room temperature in DMEM plus 0.1% BSA; EMEM plus 5% FBS supplemented as described above was then added, and the cells were incubated at 37°C in 5% CO2. At various times postinfection, samples were collected and stored at −80°C for later titration. Prior to plaque assay, all samples were frozen and thawed three times. Virus yield at each time point was calculated by subtracting the log10(PFU ml−1) at T = 0 from the log10(PFU ml−1) measured at the time point.
ELISA binding of FCV to fJAM-A.
Purified fJAM-A ectodomain (1 μg ml−1) in carbonate/bicarbonate buffer (Sigma) was bound to 96-well enzyme-linked immunosorbent assay (ELISA) plates (100 ng per well) at 4°C overnight. The wells were blocked with PBS containing 0.05% Tween 20 (PBS-T) plus 0.5% BSA for 1 h at room temperature and washed three times with PBS-T. Twofold dilutions of purified virus (starting concentration, 200 μg ml−1) were prepared in PBS-T plus BSA; 100 μl of each dilution of the virus was bound to plates for 3 h on ice. After the plates were washed, bound virus was detected with rabbit anti-FCV-5, followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG. Antibodies diluted in PBS-T plus BSA were incubated with the plates for 90 min on ice. Following washing, substrate 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Sigma) in a citric acid-sodium phosphate buffer was added to each well and incubated for 60 min at room temperature. Absorbance was measured at 595 nm on a Microplate biokinetics reader (BioTek Instruments). The mean ± standard error (SE) of three plates (two replicates per plate) is shown.
ELISA competitive binding of FCV in the presence of sfJAM-A.
Purified fJAM-A ectodomain (1 μg ml−1) in carbonate/bicarbonate buffer was bound to 96-well ELISA plates (100 ng per well) at 4°C overnight. The wells were blocked with PBS-T plus BSA for 1 h at room temperature and washed three times with PBS-T. Twofold dilutions of fJAM-A ectodomain were prepared in PBS-T plus BSA with 750 ng purified virus (starting fJAM-A concentration, 5.3 μM); 100 μl of each dilution of the fJAM-A-virus mixture was bound to plates for 3 h on ice. Plates were washed, and virus binding was detected as described above for the assay to determine virus binding to fJAM in the absence of soluble receptor. Absorbance readings for each virus were normalized, and the mean ± SE of three plates (two replicates per plate) is shown.
Detection of bis-ANS binding to FCV by fluorescence spectroscopy.
Purified virus (60 μg ml−1, a 0.5 μM concentration of the FCV VP1 dimer) or 1.5 μM purified receptor (fJAM-A or hJAM-A ectodomain) diluted in virion buffer was incubated in a 400-μl fluorometer cell (Varian) at 4° or 37°C for 10 min to allow temperature equilibration. To detect bis-(8-anilinonaphthalene-1-sulfonate) (bis-ANS) binding to native virus and receptor, bis-ANS was added to the sample to a final concentration of 3 μM. Bis-ANS was excited at 395 nm (slit width, 5 nm) and emission collected at 495 nm (slit width, 10 nm) at 2-s intervals for 10 min on a Cary Eclipse spectrofluorometer (Varian). To investigate the binding of bis-ANS to preincubated virus and receptor, virus and receptor were mixed and incubated for 10 min at 4°or 37°C. Bis-ANS was added (3 μM) and sample readings were performed as detailed above. All spectrofluorometer readings were taken with the photomultiplier tube set at 700 V.
Statistical analyses.
The Analyze-it (Analyze-it Software) statistical analysis add-in for Microsoft Excel was used to perform analysis of variance (ANOVA) where necessary. Graphs were prepared using Kaleidagraph (Synergy Software) and Adobe Illustrator CS3 (Adobe). Protein images were prepared using PyMOL (DeLano Scientific) (8).
RESULTS
FCV-5 is neutralized by preincubation with the sfJAM-A ectodomain at 37°C.
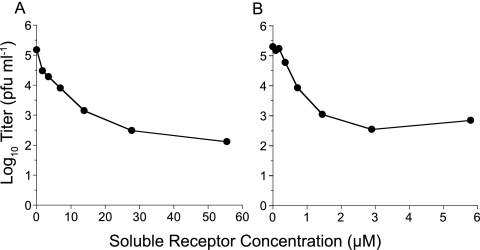
Preincubation of nonenveloped viruses, such as rhinoviruses and polioviruses, with their functional receptors neutralizes virus infectivity (17, 23). In a previous study, we showed that preincubation on ice of FCV-5 with a purified soluble GST-fused form of the ectodomain of fJAM-A modestly reduced infectivity (35); FCV-5 is a field isolate responsible for highly virulent systemic disease in cats. During these experiments, we noted that when the virus-receptor preincubation was carried out at 37°C rather than 4°C, substantially more infectivity was lost. To quantify the extent to which FCV-5 was neutralized by preincubation with soluble receptor at 37°C and to assess the influence of receptor concentration and the form of sfJAM-A ectodomain on neutralization, we preincubated ∼105 PFU of FCV-5 with increasing concentrations of purified GST-fJAM-A or with a soluble form of fJAM-A in which the GST had been removed (sfJAM-A) for 30 min at 37°C. We found a clear relationship between the concentration of soluble receptor and the extent of virus neutralization with concentrations of GST-fJAM-A of >27.5 μM (Fig. 1A) and of sfJAM-A of >2.8 μM (Fig. 1B), causing an ∼250-fold loss of viral titer. Cleavage of GST from the N terminus of the fJAM-A ectodomain enhanced the capacity of the protein to neutralize FCV-5 10-fold. As GST was fused to the D1 portion of the ectodomain, which we have shown is necessary for binding to FCV, it seems likely that GST sterically hindered the FCV-ectodomain interaction.
FIG. 1.
Neutralization of FCV-5 by sfJAM-A. FCV-5 (∼1 × 105 PFU) was incubated with various concentrations of soluble GST-fJAM-A (A) or cleaved fJAM-A at 37°C for 30 min (B). The virus samples were then plaque titrated on CRFK cell monolayers. Each data point represents the average of two replicates from a single representative experiment.
As we had previously found only a modest reduction in FCV-5 infectivity when virus and GST-fJAM-A were preincubated at 4°C, we assessed the loss of infectivity following preincubation of virus with sfJAM-A (4.3 μM) at 4°C for 3 h, followed by an additional incubation at 4, 16, 20, or 37°C for 30 min. We found a temperature dependence with the loss of infectivity, with a 4-fold loss of infectivity at 4°C compared to ∼250-fold loss of infectivity at 37°C (Table 2). We conclude that soluble forms of the fJAM-A ectodomain can substantially neutralize FCV infectivity when preincubated at 37°C and that neutralization is temperature and concentration dependent and saturable.
TABLE 2.
Temperature dependence of FCV-5 neutralization
| Incubation temp (°C)a | Avg log titer (PFU ml−1) after incubation withb: |
Change in log titer ± SD | |
|---|---|---|---|
| Buffer | 4.4 μM sfJAM-A | ||
| 4 | 4.8 | 4.2 | −0.6 ± 0.2 |
| 16 | 5.2 | 3.8 | −1.4 ± 0.3 |
| 24 | 4.7 | 2.6 | −2.1 ± 0.2 |
| 37 | 4.4 | 2.0 | −2.4 ± 0.1 |
All samples were incubated at 4°C for 3 h and then for 30 min at the temperature indicated.
Average log titer (as determined by plaque assay on CRFK cells) of at least three replicates.
Selection of soluble receptor-resistant (srr) mutants of FCV.
Because some residual infectivity always remained after preincubation of FCV-5 with sfJAM-A at 37°C, we hypothesized that resistant mutant viruses were being selected. As we expected that analysis of these mutants would shed light on possible capsid residues important in binding to fJAM-A or receptor-induced conformational changes in the capsid associated with neutralization, we selected 24 mutants that were resistant to preincubation with GST-fJAM-A (55 μM) at 37°C for 30 min. All 24 mutants contained one or two point mutations within the major capsid protein (Table 3), giving a total of 20 unique mutations. Three of these mutations were present in the leader sequence of the capsid, which is cleaved from the capsid precursor protein during capsid assembly by a virus-encoded proteinase (4, 33, 54); however, the mutants that contained mutations in the leader sequence also had an additional mutation in another region of the capsid protein. Several residue changes were found in more than one mutant. These changes were at positions G329 (three mutants), T438 (two mutants), K480 (three mutants), V516 (five mutants), and K572 (two mutants). Only two mutants had mutations within the minor capsid protein, VP2 (Table 3), and both of these mutants also contained mutations in VP1.
TABLE 3.
Capsid mutations in srr mutants
| VP1 mutation(s)a | VP2 mutationa | No. of mutants |
|---|---|---|
| A168S + F486L | 1 | |
| T202A + T447I | 1 | |
| G329D | 1 | |
| G329S | 1 | |
| D15N + G329S | 1 | |
| E401K | I15V | 1 |
| D434N + I661T | 1 | |
| T438I | N13D | 1 |
| T438A | 1 | |
| N443S | 2 | |
| I94T + S465N | 1 | |
| K480R | 1 | |
| K480Q | 1 | |
| K480E + N483S | 1 | |
| V516I | 2 | |
| V516A | 3 | |
| P116T + I535V | 1 | |
| K572E | 1 | |
| K572Q | 1 | |
| I94T + Y575H | 1 |
Mutations are identified by the wild-type amino acid position in ORF 2 (VP1; including leader sequence) or ORF 3 (VP2) of FCV-5.
Overview of the crystal structure.
To identify the location of srr mutations on the capsid, we determined the crystal structure of FCV-5 at 3.6-Å resolution by molecular replacement and real-space averaging (Fig. 2). The structure was refined to a crystallographic R factor of 0.37 (Rfree = 0.39) at 3.6-Å resolution. As in other T=3 viruses, the icosahedral asymmetrical unit consisted of three independent but chemically identical subunits (Fig. 2C). These subunits are traditionally designated A, B, and C. The present model of the FCV-5 icosahedral asymmetrical unit consists of residues 129 to 662 of the A and C subunits and residues 133 to 662 of the B subunit.
Polypeptide fold.
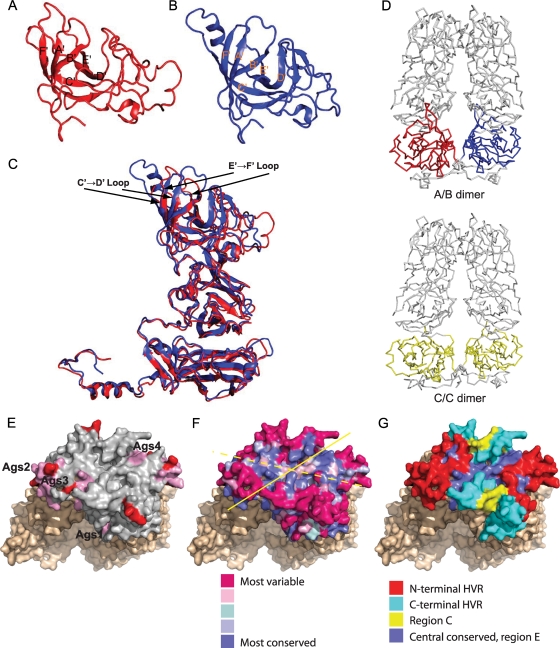
The structure of each subunit comprises an NTA, an S domain, and a P domain (Fig. 2C), similar to that seen in the recombinant Norwalk virus capsid (rNV) and SMSV-4 (Fig. 3C) (5, 41). Residues 170 to 329 fold into a canonical eight-stranded β-barrel structure similar to that found in rNV, SMSV-4, and many other viral capsid proteins. These eight strands, conventionally denoted as B to I, form two four-stranded sheets, BIDG and CHEF. Two α helices are situated between strands C and D and strands E and F, respectively. Structural comparison of the S domains of FCV-5 and SMSV-4 revealed that the most significant variations were located in the loop region connecting strands E and F (Fig. 3C).
FIG. 3.
Ribbon representation of the P2 subdomain in FCV-5 (A) and SMSV-4 (B). The β-strands are labeled from A′ to F′ in each case. (C) The superposition of the P domain of FCV-5 (red) and SMSV-4 (blue) shows that the six β-strands in the P2 subdomains are similarly spatially arranged into a compact barrel, despite the low sequence similarity. Arrows indicate the loops connecting β-strands C and D, and E and F, of the FCV-5 P2 subdomain. (D) The FCV-5 A/B dimer (top) as viewed along the line joining the icosahedral 3- and 5-fold axes and the C/C dimer (bottom) as viewed along the line joining the adjacent icosahedral 3-fold axes demonstrate the bent and flat conformations, respectively, assumed by the subunits. In both cases, the dimeric 2-fold axis is vertical. (E to G) Positions of antigenic sites and conserved residues on the P2 subdomain of the C/C dimer of FCV-5. (E) Neutralizing epitopes mapped on P2 (gray). Antigenic sites (Ags1 to -4; colored pink) represent B-cell linear epitopes identified by Radford et al. (43). MAb escape mutations identified by Tohya et al. are colored red (58). (F) Degree of conservation of surface-exposed residues on the P2 subdomain as predicted by Consurf 3.0. The solid yellow line indicates dimer interface; the dashed yellow line indicates the axis of conservation. (G) Locations of N-terminal and C-terminal HVRs, region C and central conserved portion of region E mapped on surface of P2 subdomain.
The P domain of FCV-5 is comprised of residues 330 to 662 and is linked to the S domain by a flexible hinge. The P domain is subdivided into P1 and P2 subdomains. The P1 subdomain, consisting of two segments of polypeptide (residues 330 to 381 and 551 to 662), folds into a structure similar to that seen in SMSV-4.
The P2 subdomain, the outermost portion of the capsid protein, is a large insertion into the P1 subdomain. The P2 subdomains of SMSV-4 and FCV-5 form a compact barrel structure consisting of six strands, sequentially labeled as A′ to F′ (Fig. 3A and B). However, the six strands in FCV-5 vary in length and relative orientations, compared to their counterparts in SMSV-4. These six strands are connected by loops of various lengths. Structure superimposition showed that loop C′D′ in FCV-5 is significantly shorter than that of SMSV-4 (Fig. 3C). The P2 subdomain consists of residues 382 to 550 and contains seven fewer residues than that of SMSV-4. Structural comparison showed that those “inserted” residues are mainly distributed in the loop connecting strands C′ and D′.
Structural comparison between different subunits.
Pairwise superposition calculations indicated that the structure of the P domain of FCV-5 adopts similar structures in all three subunits. The S domain, however, shows larger variations between the different subunits to facilitate the assembly of a T=3 capsid. In addition, the A and C subunits have four more disordered N-terminal residues than the B subunit.
Superposition of the whole subunit structure revealed that the A subunit forms a more compacted structure, as evidenced by the closer association of the S and P domains. The interactions between the S and P domains are less prominent in the B subunit, resulting in a more relaxed structure. The C subunit has the most open conformation, with the fewest interactions between the S and P domains. These differences contribute to the bent and flat conformations of the A/B and C/C dimers, respectively (Fig. 3D).
Mapping of neutralizing epitopes and conserved surface regions on the P2 subdomain of FCV-5.
Hypervariable regions (C and E regions) of the FCV linear capsid sequence were previously identified, and neutralizing epitopes were mapped predominantly to residues within the E region (Fig. 3G) (43, 48, 57, 58). Using the FCV-5 structure, we mapped the locations of four linear B-cell epitopes identified in FCV-F9 (43). All of these sites lay on the margins of the P2 subdomain, with one site, Ags4, at the dimer interface (Fig. 3E). Similarly, antibody escape mutations found in mutants of FCV-F4 are also located on the margins of the P2 subdomain within hypervariable regions (Fig. 3E) (58). To more accurately assess the conservation of individual surface-exposed residues on the P2 subdomain, we used Consurf 3.0, which maps the rate of evolution as determined by an empirical Bayesian estimation among homologous proteins onto the three-dimensional (3-D) structure (26). As input, we used an alignment of 47 different FCV capsid sequences to derive an evolutionary conservation score that was used to color code individual residues on the surface of the FCV-5 capsid structure (Fig. 3F). This analysis revealed a patchwork of conserved surface residues along the dimeric interface of the P domain that also extended laterally at a 45° angle to the dimer interface on the surface of each monomer. Highly variable residues on the margins of the P2 subdomain surrounded the more central conserved residues.
Mapping srr mutations on the FCV-5 structure.
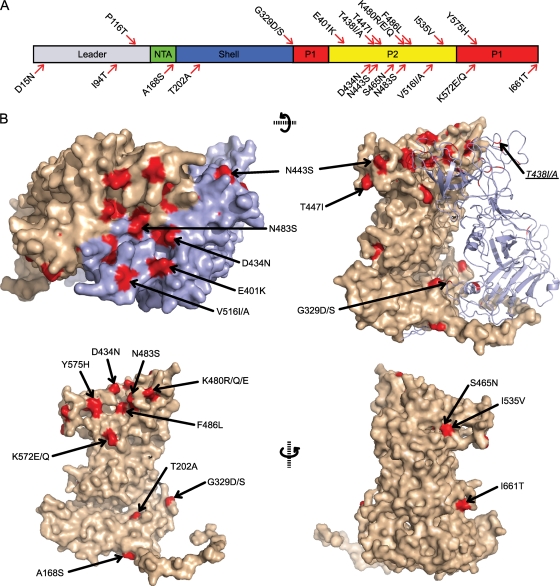
Sequencing of the capsid protein of the 24 srr mutants revealed 17 unique amino acid changes from the parental FCV-5 mature capsid protein sequence. We mapped the locations of these changed residues on the structure of FCV-5 (Fig. 4). Three mutations were found in the NTA and S domains, and 14 mutations were present in the P domain (Fig. 4A). Of the 24 mutants, 21 (87.5%) contained mutations that mapped to the P domain of the capsid, and 18 of those mutations were to residues within the P2 subdomain.
FIG. 4.
Positions of mutations found in srr mutants in the structure of FCV-5. (A) The residues mutated in the srr mutants are illustrated on a schematic of the FCV capsid protein. The locations of the different structural domains in the primary capsid sequence are indicated. (B) Locations of the mutations in the FCV-5 atomic resolution structure.
Of the mutations found in the NTA and S domains, one change was to a residue (A168S) within the NTA, but this mutation was accompanied by a second amino acid change in the P2 domain (F486L). Similarly, a mutant with a change in the shell domain (T202A) also had a change in the P2 domain (T447I). Three srr mutants contained mutations at residue G329 in the shell domain on a loop that connects the shell with the protrusion domain. Residue G329 is located at the point of the flexible hinge, and mutations at G329 were present alone or in addition to a change in the leader sequence of the capsid (D15N) (Fig. 4B and Table 3). Mutations to residue G329 were the only changes identified in the S domain that occurred independently of an additional mutation within the P domain.
Three of the 21 mutated residues identified in the P domain were present within the P1 subdomain (K572E/Q, Y575H, and I661T). Residues K572 and Y575 were buried at the dimer interface of the capsid structure (Fig. 4B). Residue I661 was located at the base of the P1 subdomain, in close proximity to the S domain (Fig. 4B); the mutant containing the I661T change also had a second change within the P2 domain (D434N). The other 11 P domain mutations were in the P2 subdomain. Eight of these mutated residues were within surface-exposed loops of P2: E401K, D434N, T438A, N443S, T447I, S465N, K480R/Q/E, V516I/A, and I535V (Fig. 4B). Three of the eight surface-exposed mutations were present in more than one srr mutant: N443 (two mutants), K480 (three mutants), and V516 (five mutants). Two mutants had mutations to residues at the P2 dimer interface; one mutant possessed a change in the surface-exposed residue N483, while another mutant contained a change to the buried residue F486. Lastly, two mutants had mutations to residue T438 that is buried in the present structure and maps to a loop near the surface of the P2 subdomain. In summary, the location of mutations present in srr isolates mapped to three general regions of the structure of FCV-5: (i) surface-exposed residues on the P2 domain; (ii) residues that are buried or not easily accessible from the surface of the capsid; and (iii) the hinge region of the capsid.
srr mutants are resistant to neutralization by sfJAM-A but retain the capacity to bind fJAM-A and require cellular fJAM-A for infection.
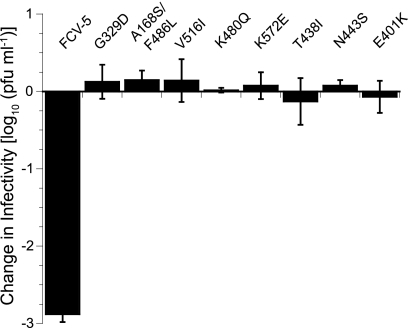
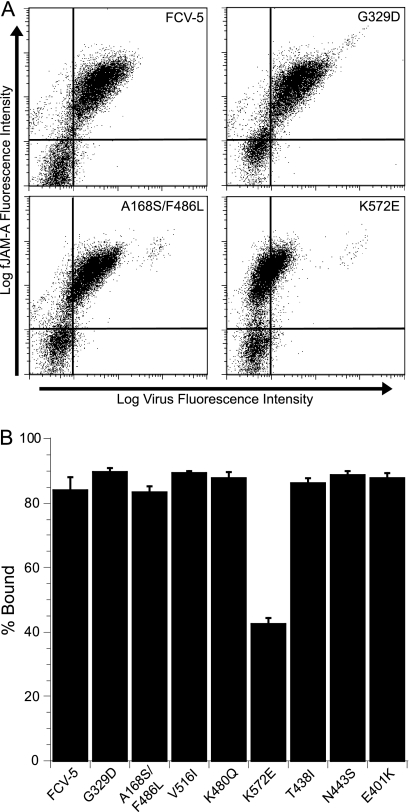
From the original 24 srr mutants, we selected eight mutant viruses to further characterize. We selected these mutants based on the location of their mutated residues in an effort to have representative mutations from all regions of VP1. Additionally, we selected mutants based on the frequency that particular residues were mutated, with overrepresented mutations to residues included. All of the eight mutant viruses in the subpanel were resistant to neutralization with sfJAM-A under the conditions in which they were selected (Fig. 5). To verify that the eight selected srr mutants still bound to fJAM-A expressed on the cell surface, we used flow cytometry to measure the capacity of each of the mutants to bind to CHO-S cells expressing fJAM-A on their surface (Fig. 6A and B). We used CHO-S cells transiently expressing fJAM-A rather than a stable fJAM-A-expressing CHO-K1 cell line that we previously described (35), because of technical difficulties using adherent CHO-K1 cells in flow cytometry assays. We found that all eight mutants bound to CHO-S cells expressing fJAM-A. One mutant, the K572E mutant, however, bound to 50% fewer cells than did the parental FCV-5. In addition, all eight mutants still required fJAM-A as a functional receptor, as pretreatment of CRFK cells with a rabbit antiserum against fJAM-A completely blocked infection (data not shown). We conclude from these findings that the subpanel of eight srr mutants still binds and requires cellular fJAM-A for cell infection.
FIG. 5.
Neutralization of srr mutants by sfJAM-A. A subpanel of eight srr mutants and FCV-5 (1 × 105 PFU) was incubated in the presence or absence of soluble GST-fJAM-A (55 μM) at 37°C for 30 min. The infectivity of each sample was assayed by plaque titration. The change in log titer was calculated by subtracting the titer of samples incubated with receptor from that of samples incubated without receptor. The mean change in log10 titer ± standard deviation (SD) of three replicates of a representative experiment is shown.
FIG. 6.
Binding of srr mutants to CHO cells expressing fJAM-A. (A) Nonpermissive CHO-S cells were transfected with a DNA construct encoding fJAM-A. At 24 h posttransfection, FCV (MOI = 5) was adsorbed to the cells on ice for 30 min. After being washed with cold PBS to remove unbound virus, the cells were fixed and bound virus and cell surface fJAM-A were detected with mouse anti-FCV MAb and rabbit anti-fJAM-A antibodies, followed by Alexa 488-conjugated goat anti-mouse IgG and Alexa 647-conjugated goat anti-rabbit IgG. Virus binding and receptor expression were analyzed by flow cytometry. Flow cytometry dot plots for four representative samples are shown. (B) Virus binding was assayed by determining the percentage of fJAM-A-positive cells that bound virus. The means of the results for three independent experiments (two samples of 1 × 104 cells per experiment) ± SE are shown.
At low MOI, the growth of some srr mutants differs from that of parental FCV-5.
To evaluate the growth properties of the srr mutants, we generated single- and multiple-cycle growth curves (Fig. 7A and B, respectively) for each of the eight selected srr mutants. The growth kinetics and final yields following a single replicative cycle (MOI = 5) for the mutant viruses were similar to those of the parental FCV-5 (Fig. 7A and data not shown). However, we found that during multiple replicative cycles (MOI = 0.01) six of the mutants (G329D, V516I, K572E, T438I, N443S, and E401K mutants) had significantly decreased infectivity at 4 h pi compared to parental FCV-5 as determined by ANOVA (P < 0.005) (Fig. 7B and data not shown). Two of the eight mutants (K480Q and A168S/F486L mutants) were similar to FCV-5, producing new infectious virions by 4 h pi. By 8 h pi, all of the viruses had attained similar increases in titer, and the final yields were similar. We conclude that six of the panel of eight srr mutants have slower growth kinetics at early times postinfection than the parental FCV-5 in cells infected at a low multiplicity.
FIG. 7.
Growth of srr mutants. CRFK monolayers were infected with FCV-5 and the indicated srr mutants under single-cycle (MOI = 5) (A) or multiple-cycle (MOI = 0.01) (B) conditions. For clarity, the data from only four mutants have been included in each graph. The change in virus titer was determined by plaque assay. The mean log10 titer (log10 titer at each time point − log10 titer at T = 0) for each time point is shown. The error bars shown at 4 and 8 h pi for the multiple-cycle growth represent the standard deviation of four replicates; for other data points, error bars have been omitted from the figure for clarity. The asterisks indicate significant differences between viral isolates as determined by ANOVA at these time points.
Binding of srr mutants to fJAM-A.
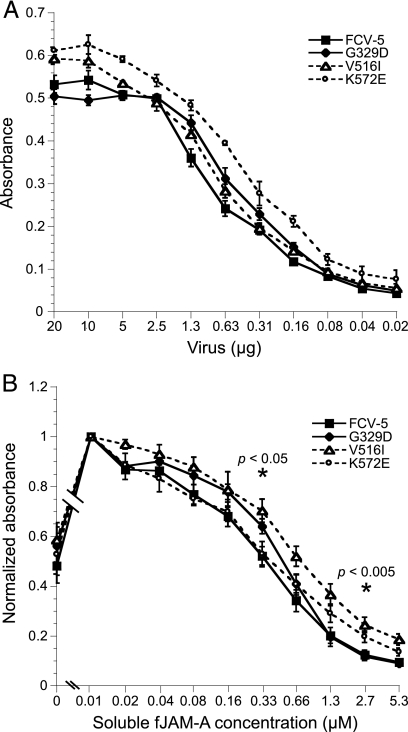
In order to assess the kinetics of binding of the srr mutants to fJAM-A, we purified three mutants (G329D, V516I, and K572E mutants) and compared their in vitro binding to plate-bound sfJAM-A by ELISA to that of parental FCV-5 (Fig. 8A). All the viruses showed saturable concentration-dependent binding to fJAM-A. The binding kinetics of FCV-5 and the G329D mutant were similar, and saturation of binding to fJAM-A occurred at similar concentrations of virus. The binding of V516I and K572E mutants to plate-bound fJAM-A saturated at slightly higher virus concentrations than did those of FCV-5 and the G329D mutant. One possible explanation for this difference is differential detection by the polyclonal rabbit FCV-5 antisera. We conclude that G329D, V516I, and K572E srr mutants and FCV-5 all bind immobilized recombinant fJAM-A with relatively similar kinetics under in vitro conditions.
FIG. 8.
Binding of srr mutants to immobilized fJAM-A in the absence (A) or presence (B) of increasing concentrations of sfJAM-A. ELISA plates were coated with 100 ng of sfJAM-A ectodomain. Various concentrations of purified FCV-5 and G329D, V516I, and K572E srr mutants were incubated with the immobilized protein for 3 h on ice. To investigate the effect of sfJAM-A on the binding of virus to immobilized receptor, 750 ng of each virus and various concentrations of soluble receptor were incubated together with plate-bound fJAM-A for 3 h on ice. Bound FCV was detected with rabbit anti-FCV serum, followed by HRP-conjugated goat anti-rabbit IgG. Colorimetric HRP substrate was added, and the amount of bound FCV-5 was quantified by absorbance at 595 nm. The means of three and four plates, respectively (two replicates per plate), ± SE are shown. ANOVA was performed on three concentrations of soluble receptor (0.04, 0.33, and 2.7 μM) to determine statistical differences; significant differences are indicated by asterisks.
Relative affinities of srr mutants for fJAM-A.
To determine if the V516I, K572E, and G329D srr mutants differed from FCV-5 in their affinity for fJAM-A, we performed a competition ELISA (Fig. 8B). Different concentrations of sfJAM-A were used to compete for binding of virus to plate-bound fJAM-A at 4°C. We found that binding of FCV to the plate-bound receptor was strongly inhibited at the highest concentrations of sfJAM-A. The binding of FCV to plate-bound fJAM-A correlated inversely to the concentration of soluble receptor. A 50% decrease in binding of all the tested viruses to plate-bound fJAM-A occurred at concentrations of sfJAM-A between 0.33 and 0.66 μM. All the viruses exhibited similar competition curves; however, binding of the V516I mutant to plate-bound fJAM-A was 50% inhibited at a slightly higher concentration of sfJAM-A, as can be seen by the rightward shift in the competition binding curve (Fig. 8B). Statistical analysis (ANOVA) at three concentrations of soluble receptor (0.04, 0.33, and 2.7 μM) showed significantly more viral binding of the V516I mutant than that of FCV-5 for all but the lowest concentration of receptor (P < 0.05). We found that preincubation of virus with low concentrations of sfJAM-A significantly enhanced binding to plate-bound fJAM-A compared to virus alone. As fJAM-A can form dimers, and FCV binding does not seem to require or occlude the fJAM-A dimerization interface (1, 35), this enhanced binding may be mediated by fJAM-fJAM interactions between immobilized and virus-bound soluble receptor. An alternative explanation is that the binding of sfJAM-A to FCV capsids enhances the binding to the immobilized receptor allosterically. We conclude that the G329D, V516I, and K572E srr mutants demonstrate similar affinities for fJAM-A.
Bis-ANS binding indicates a change in FCV surface hydrophobicity following preincubation with sfJAM-A.
We hypothesized that the interaction of soluble receptor with FCV caused a change in conformation of the viral capsid. We, therefore, used binding of bis-ANS as a probe to investigate changes in hydrophobicity that occurred upon interaction of FCV with fJAM-A in solution. Bis-ANS is a fluorophore that binds to exposed hydrophobic regions of protein molecules (44). The sequestration of bis-ANS in hydrophobic pockets of proteins is accompanied by a large increase in the fluorescence quantum yield of the probe. Thus, bis-ANS can be used to probe for structural changes in viral proteins that are accompanied by changes in surface hydrophobicity (10, 14, 15). We first preincubated virus or receptor alone in a cuvette in the spectrofluorometer at 37°C for 10 min and then added bis-ANS and collected fluorescence intensity readings over a period of 10 min at 37°C (Fig. 9A to C). To quantify differences in bis-ANS fluorescence, we waited until the fluorescence emission readings had stabilized and then averaged the fluorescence intensity measurements over 1 min. Fluorescence intensity increased during the first 5 to 10 min after addition of bis-ANS to any of the protein samples, but it stabilized after 10 min. This increase in fluorescence is likely caused by bis-ANS binding to surface-exposed hydrophobic pockets on the proteins (Fig. 9A to C).
FIG. 9.
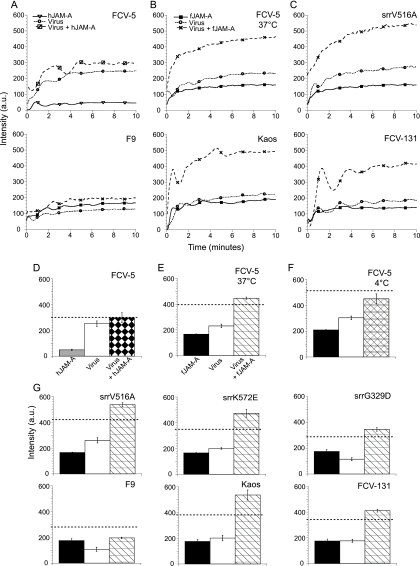
Changes in bis-ANS binding upon mixing of FCV with sfJAM-A. At time zero, bis-ANS was added to the indicated purified virions, soluble JAM-A (human [A] or feline [B and C]), or virions preincubated with JAM-A. Fluorescence was recorded every 2 s continuously for 10 min at excitation and emission wavelengths of 395 and 495 nm, respectively. (D to G) Stabilized fluorescence intensities measured during the last minute for each sample were averaged. Broken lines indicate the additive average fluorescence intensity measured for both native virus and receptor. The means ± SE (n = 3) are shown.
To determine if the interaction of FCV with fJAM-A triggered a conformational change that would change the binding of bis-ANS to the proteins, we preincubated FCV with either hJAM-A (hJAM-A does not bind FCV [35; Fig. 9A and D]) or fJAM-A (Fig. 9B and E) at 37°C for 10 min and then added bis-ANS and monitored changes in fluorescence intensity over 10 min. We found that the average stabilized bis-ANS fluorescence intensity of samples containing FCV-5 plus hJAM-A was equal to the sum of the intensities associated with incubation of bis-ANS with FCV-5 and hJAM-A individually (Fig. 9D). These findings were expected and indicate that the overall binding of bis-ANS to either protein was unchanged when they were mixed. In contrast, the average stabilized fluorescence intensity of samples containing FCV-5 plus fJAM-A was greater than the sum of the bis-ANS fluorescence intensities of the two proteins when incubated individually (Fig. 9E). This finding suggests that an interaction between virus and fJAM-A changed the conformation of one or both proteins, resulting in the exposure of more hydrophobic patches on the protein and thus an increase in the amount of bis-ANS binding and resulting fluorescence.
As we found that FCV-5 neutralization by sfJAM-A was temperature dependent (Table 2), we investigated if changes in bis-ANS binding to the fJAM-A-FCV-5 mixture were affected by temperature (compare Fig. 9E and F). We found that, unlike our findings at 37°C (Fig. 9E), the fluorescence intensity of samples of FCV-5 and fJAM-A preincubated together at 4°C was less than the sum of the bis-ANS intensities of virus and receptor incubated at 4°C with the fluorescent probe alone (Fig. 9F). Our interpretation of these findings is that binding of fJAM-A to FCV-5 at 4°C blocks hydrophobic patches previously available for bis-ANS binding on the virus and/or fJAM-A. In contrast, the increase in binding of bis-ANS to the fJAM-A-FCV mixture at 37°C indicates a change in conformation of the viral capsid and/or the receptor.
The binding of bis-ANS at 37°C to V516A, K572E, and G329D srr mutants and to field isolates Kaos and FCV-131 was also investigated. Following incubation of virus and receptor at 37°C, these samples showed an increase in fluorescence intensities similar to that seen for FCV-5 (Fig. 9C and G). In contrast, incubation of the vaccine strain F9 with fJAM-A led to a fluorescence intensity maximum that was less than what would be expected from the sum of the intensities associated with the receptor and virus alone; this pattern was similar to that of FCV-5 incubated with receptor at 4°C. From these findings, we conclude that interaction of fJAM-A with FCV at 37°C leads to changes in the binding of bis-ANS that are indicative of changes in hydrophobicity of virus and/or receptor. For FCV-5, these changes in bis-ANS binding are dependent upon temperature, as virus and receptor incubated at 4°C did not display the same increase in fluorescence intensity.
Not all FCV isolates are neutralized by sfJAM-A.
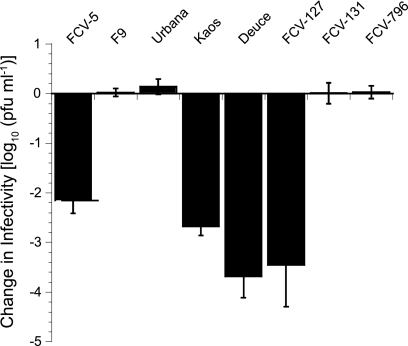
The FCV-5 isolate that we used to select for srr mutants was recovered from a cat suffering from severe systemic disease. As this form of FCV disease is uncommon, we hypothesized that not all isolates of FCV would be neutralized by incubation with soluble receptor at 37°C. To test this hypothesis, we incubated ∼105 PFU of six additional FCV isolates with 45 μM GST-fJAM-A at 37°C; these virus isolates included the vaccine strain F9, two additional isolates recovered from cats with virulent systemic disease, the FCV-Urbana isolate, and three other field isolates recovered from cats with mild- to moderate-severity FCV disease. Several additional FCV isolates were neutralized by preincubation with GST-fJAM-A at 37°C; however, the vaccine strain F9, FCV-Urbana, and the two isolates recovered from cats with milder disease were not neutralized under these conditions (Fig. 10). We conclude that there are natural differences among FCV isolates in their responses to incubation with sfJAM-A at 37°C.
FIG. 10.
Ability of sfJAM-A to neutralize select FCV isolates. A panel of six FCV field isolates (FCV-5, Kaos, Deuce, FCV-127, FCV-131, and FCV-796) and two tissue culture-adapted strains (F9 and Urbana) (1 × 105 PFU) was incubated in the presence or absence of soluble GST-fJAM-A (45 μM) at 37°C for 30 min. The remaining infectivity in each sample was assayed by plaque titration. The change in log titer was calculated by subtracting the titer of samples incubated with receptor from the titer of samples incubated without receptor. The mean change in log10 titer ± SD (n = 4) is shown.
DISCUSSION
Several nonenveloped viruses are neutralized by preincubation with their functional receptors (23, 31, 49). However, the extent to which infectivity is lost at physiologic temperatures depends upon the mechanism of receptor-mediated neutralization. For polioviruses and some HRVs, preincubation with their receptors induces irreversible inactivating conformational changes in the virus (6, 17). We found that some but not all isolates of FCV were neutralized by preincubation with a soluble form of fJAM-A. Although it is possible that neutralization was due to steric hindrance or occupation of receptor-binding sites, the prominent temperature dependence of neutralization strongly suggests that receptor-induced conformational changes are involved. Receptor-induced conformational changes in both HRVs and poliovirus are temperature dependent (16, 20). Polioviruses undergo an irreversible neutralizing transition from a 160S particle to a 135S particle when they are incubated with soluble receptor at 37°C, but this transition does not occur at temperatures below 33°C (16, 61). In contrast, although FCV-5 receptor-mediated neutralization was clearly temperature dependent, no obvious temperature threshold was observed. Rather, a gradual diminishment of the neutralizing effect as the temperature decreased from 37°C to 4°C was found (Table 2). We propose that some FCV isolates undergo a temperature-dependent, fJAM-A-mediated neutralization following incubation at 37°C; however, as not all FCV isolates undergo neutralization under these conditions, an additional cellular factor or environmental condition is likely required by some FCV isolates for neutralization.
Analysis of soluble receptor-resistant (srr) mutants in poliovirus and murine coronavirus identified virus residues that either directly or indirectly facilitated receptor binding (6, 23, 46). Of the 24 srr mutants of FCV-5, mutations to 20 unique VP1 residues were identified; six mutations mapped to surface-exposed residues on the P2 domain of the capsid that potentially could directly interact with the receptor. Several mutated residues were buried or inaccessible from the surface of the structure and could possibly indirectly affect binding. Similarly, poliovirus srr mutants contained mutations to both internal and surface-exposed residues. The mutated residues for poliovirus mapped to a hydrophobic pocket in the structure. It is believed that changes to residues in this hydrophobic pocket regulate receptor attachment and conformational changes within the virus particle (6, 42).
One to two copies of the VP2 protein are found in FCV virions, and VP2 is essential for the synthesis and maturation of infectious FCV virions (52, 53). It was, therefore, important to ensure that resistance to neutralization of the srr mutants was not due to changes to VP2. Sequencing of the ORF encoding the minor capsid protein of FCV revealed that only two mutants of the 24-mutant panel contained coding mutations to VP2. As both mutants possessing VP2 mutations also had VP1 mutations, and the remainder of the srr mutants contained only VP1 mutations, changes to VP2 cannot explain the resistance of the mutants to receptor-mediated neutralization.
Our analysis of the FCV srr mutants indicates that residues within critical regions of the capsid are necessary for structural flexibility or receptor interactions. In particular, changes to residue G329 (mutated in three srr mutants) suggest that movement between the S and P domains of the capsid protein occurs upon receptor interaction and is involved in receptor-mediated neutralization. The glycine residue at position 329 is 100% conserved in the FCV capsid sequence and is located in a short loop between the S and P domains of the capsid protein. A similar glycine residue is found in this hinge region of the capsid in all caliciviruses. Cryo-EM reconstructions of murine norovirus in complex with neutralizing Fab fragments show that this hinge region is extended, moving the P domain radially away from the surface of the shell domain (24). In addition, a cryo-EM structure of FCV-F9 complexed with the fJAM-A ectodomain showed that the P domain of the capsid moved with respect to the S domain when bound to fJAM-A, indicating that movement in this region occurs (1). Our findings further support the hypothesis that flexibility in this hinge region is important to permit conformational changes of the structural domains of the calicivirus capsid in relation to each other and indicate that this region actively moves during receptor interactions. Moreover, our findings indicate that changes at residue G329 substantially alter the susceptibility of FCV to neutralization by sfJAM-A.
Several srr mutants had mutated residues at the P domain dimer interface. These mutations could alter receptor binding indirectly, or these residues could become accessible for receptor binding following a conformational change of the capsid. The altered residue in the K572E srr mutant is in P1 and is unlikely to be accessible from the surface of the capsid. This mutant had decreased binding to fJAM-A on cells. However, the K572E mutant had binding kinetics similar to that of parental FCV-5 in in vitro ELISA binding assays. A possible explanation for this difference is that unlike fJAM-A purified from bacteria, JAM-A on cells is N-glycosylated (32).
Poliovirus srr mutants had reduced binding affinity for their receptor (6). It was, therefore, unexpected that only one mutant (the K572E mutant) had decreased cell surface fJAM-A binding and that none of the FCV srr mutants we examined in detail had a substantive change in binding to fJAM-A in vitro. In fact, the V516I mutant showed higher affinity for immobilized fJAM-A (Fig. 8B). These findings indicate that resistance of the srr mutants to receptor-induced neutralization cannot be explained by changes in their binding affinity for fJAM-A.
Single-step growth kinetics of poliovirus srr mutants were similar, but all mutants had lower titers than did wild-type P1/Mahoney poliovirus at early time points postinfection. This delay in replication was attributed to an assembly defect, as all the srr mutants bound as efficiently as wild-type poliovirus to cells at room temperature (6). In contrast, we found that the FCV srr mutants and parental FCV-5 had similar single-step growth kinetics and titers throughout the replicative cycle. We found a difference during multiple-cycle growth experiments, where six of the eight srr mutants investigated had significantly decreased titers in comparison to parental FCV-5 at 4 h pi. One explanation for these findings is decreased efficiency of viral entry (receptor binding, cellular trafficking, and/or uncoating) of the mutants with a consequent lag in the onset of viral replication. As small differences in the efficiency of entry may be masked when a high MOI is utilized, experiments using low input titers of virus can be more sensitive. The K572E mutant demonstrated decreased binding to cell surface-expressed fJAM-A and also had delayed multiple-cycle growth kinetics. Therefore, the decreased early growth kinetics of this mutant could be due to decreased receptor binding. However, G329D and V516I srr mutants bound cell-expressed and plate-bound fJAM-A to the same extent as did parental FCV-5 or had increased affinity for plate-bound fJAM-A and also had delayed early growth kinetics. We previously showed that lower-virulence-field or tissue culture-adapted FCV isolates had delayed multiple cycle growth kinetics compared to more-virulent isolates (including FCV-5) (36). Although the virulence of these srr mutants is currently unknown, it is possible that by selecting for srr mutants, we may have also selected for viruses with altered virulence.
Following attachment, nonenveloped viruses must undergo conformational alterations that mediate membrane penetration and deliver their genetic material (60). The temperature dependence of the sfJAM-A neutralization of FCV and the identification of srr mutant residues buried in the capsid structure suggested that fJAM-A induces a conformational change in the FCV capsid. Our finding that mixtures of FCV-5 and sfJAM-A incubated at 37°C had bis-ANS fluorescence intensities greater than the sum of the fluorescence intensities associated with virus and sfJAM-A individually provided further support for this concept. Bis-ANS binding has previously been used to detect conformational changes associated with changes in hydrophobicity in the envelope glycoproteins of Newcastle disease virus and HIV-1 upon incubation with gangliosides and CD4, respectively (14, 22). Additionally, our finding that preincubation of FCV-5 and fJAM-A at 4°C resulted in lower bis-ANS fluorescence than the sum of the intensities of the two components is consistent with the idea that binding of fJAM-A to FCV-5 at 4°C occludes potential bis-ANS binding sites and that exposure of new hydrophobic patches as a consequence of conformational change does not occur or is decreased at 4°C (Fig. 9E and F). An alternative explanation is that the increases in hydrophobicity we observed were due to changes in the receptor, not the virus. However, as there is substantial evidence for viral protein conformational changes following interaction with cellular receptors, and such conformational changes are likely a requirement of all nonenveloped viruses, this explanation is less likely. For those viruses that when mixed with sfJAM-A showed an increase in total bis-ANS fluorescence intensity, the intensity was 11 to 30% greater for the mixtures of FCV and sfJAM-A than the sum of the bis-ANS fluorescence intensities of each component incubated separately. Although this is an in vitro assay, we believe that taken together with our other data, these changes are biologically significant. Additional investigation will be required to determine the nature of the conformational changes and their functional significance.
Upon incubation with soluble receptor, the srr mutants and parental FCV-5 demonstrated similar increases in hydrophobicity. These findings raise the possibility that FCV neutralization by soluble receptor does not involve a conformational change in the capsid. However, a more likely possibility is that more than one intermediate capsid conformer forms upon interaction of FCV with fJAM-A. Following interaction of picornaviruses with their functional receptors, three intermediate conformers of the capsid are formed—the 135S altered particle and two forms of the 80S empty particle (7, 9, 13, 27, 28). Therefore, our working model is that mutants resistant to neutralization undergo an initial conformational change upon interacting with fJAM-A that either is reversible or results in a stable, infectious intermediate.
A cryo-EM reconstruction of the FCV-F9 strain complexed with fJAM-A at 18 Å revealed that fJAM-A interacted with the top of the P2 subdomain of the F9 capsid, causing an ∼13° counterclockwise rotation of the P dimers about their 2-fold axes of symmetry (1). Interestingly, we found that in contrast to the other FCV isolates we investigated, the F9-sfJAM-A complex bound less bis-ANS than did the F9 virus alone (Fig. 9G). Given the structural evidence for a conformational change in the F9 capsid, one possible explanation is that fJAM-A binding induces a conformational change that decreases capsid surface hydrophobicity. In the face of our observations of increased bis-ANS binding to other FCV isolates when preincubated with fJAM-A, it is possible that the capsid conformational changes reported by Bhella et al. (1) for the vaccine isolate F9 differ from changes that occur in the capsids of other FCV field isolates.
We found that some FCV isolates, including low-passage field isolates and tissue culture-adapted lab strains, were not susceptible to fJAM-A neutralization (Fig. 10). A natural isolate that was resistant to receptor neutralization, FCV-131, also exhibited increased hydrophobicity following incubation with soluble receptor. These findings suggest that similar to the srr mutants, field isolates undergo a conformational alteration following interaction with fJAM-A but do not lose infectivity. Other isolates, however, may undergo more drastic, global conformational alterations that result in either a complete loss of or significant decrease in infectivity. Interestingly, we found that sensitivity of different FCV isolates to neutralization correlated with their virulence in cats. FCV is a common pathogen of cats that typically causes mild or unapparent disease, with fatal disease being unusual. However, highly virulent isolates of FCV cause systemic signs of disease and result in mortality rates as high as 50% (21, 38, 39, 47). In our panel, the most virulent isolates were susceptible to fJAM-A neutralization, while low-virulence isolates were resistant. It is possible that the in vitro assay of receptor neutralization is highlighting an important determinant of in vivo virulence.
Acknowledgments
We thank Christian Nelson, Meg Crapster-Pregont, Sarah Caddy, and Rachel Mays for excellent technical assistance. We thank Terry Dermody and Kristen Guglielmi for the generous gift of reagents and Stanislav Sosnovtsev for technical advice. We acknowledge the SBC-CAT 19ID beamline at the Advanced Photon Source (supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no.W-31-109-Eng-38) and its staff for their help during data collection.
This work was supported by grants from The Cornell Feline Health Center and the Winn Foundation to J.S.L.P., National Institutes of Health (PO1 AI057788), and R. Welch Foundation (Q-1279) to B.V.V.P. R.J.O. is the recipient of a scholarship from Cornell University.
Footnotes
Published ahead of print on 31 March 2010.
REFERENCES
- 1.Bhella, D., D. Gatherer, Y. Chaudhry, R. Pink, and I. G. Goodfellow. 2008. Structural insights into calicivirus attachment and uncoating. J. Virol. 82:8051-8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 3.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Carter, M. J., I. D. Milton, P. C. Turner, J. Meanger, M. Bennett, and R. M. Gaskell. 1992. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 122:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, R., J. D. Neill, M. K. Estes, and B. V. Prasad. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colston, E., and V. R. Racaniello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowell, R. L., and L. Philipson. 1971. Specific alterations of coxsackievirus B3 eluted from HeLa cells. J. Virol. 8:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA.
- 9.De Sena, J., and B. Mandel. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554-566. [DOI] [PubMed] [Google Scholar]
- 10.de Sousa, P. C., Jr., R. Tuma, P. E. Prevelige, Jr., J. L. Silva, and D. Foguel. 1999. Cavity defects in the procapsid of bacteriophage P22 and the mechanism of capsid maturation. J. Mol. Biol. 287:527-538. [DOI] [PubMed] [Google Scholar]
- 11.Ebnet, K., C. U. Schulz, M. K. Meyer Zu Brickwedde, G. G. Pendl, and D. Vestweber. 2000. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 275:27979-27988. [DOI] [PubMed] [Google Scholar]
- 12.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick, M. L., and P. D. Cooper. 1962. Early interactions between poliovirus and ERK cells: some observations on the nature and significance of the rejected particles. Virology 18:212-223. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira, L., E. Villar, and I. Munoz-Barroso. 2004. Conformational changes of Newcastle disease virus envelope glycoproteins triggered by gangliosides. Eur. J. Biochem. 271:581-588. [DOI] [PubMed] [Google Scholar]
- 15.Gaspar, L. P., J. E. Johnson, J. L. Silva, and A. T. Da Poian. 1997. Partially folded states of the capsid protein of cowpea severe mosaic virus in the disassembly pathway. J. Mol. Biol. 273:456-466. [DOI] [PubMed] [Google Scholar]
- 16.Gómez Yafal, A., G. Kaplan, V. R. Racaniello, and J. M. Hogle. 1993. Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology 197:501-505. [DOI] [PubMed] [Google Scholar]
- 17.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hoover-Litty, D. Wunderlich, and A. McClelland. 1991. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiver, M., E. Littler, E. O. Caul, and A. J. Fox. 1992. The cloning, sequencing and expression of a major antigenic region from the feline calicivirus capsid protein. J. Gen. Virol. 73(Pt. 9):2429-2433. [DOI] [PubMed] [Google Scholar]
- 19.Guix, S., M. Asanaka, K. Katayama, S. E. Crawford, F. H. Neill, R. L. Atmar, and M. K. Estes. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81:12238-12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoover-Litty, H., and J. M. Greve. 1993. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol. 67:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley, K. E., P. A. Pesavento, N. C. Pedersen, A. M. Poland, E. Wilson, and J. E. Foley. 2004. An outbreak of virulent systemic feline calicivirus disease. J. Am. Vet. Med. Assoc. 224:241-249. [DOI] [PubMed] [Google Scholar]
- 22.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, G., D. Peters, and V. R. Racaniello. 1990. Poliovirus mutants resistant to neutralization with soluble cell receptors. Science 250:1596-1599. [DOI] [PubMed] [Google Scholar]
- 24.Katpally, U., C. E. Wobus, K. Dryden, H. W. Virgin IV, and T. J. Smith. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J. Virol. 82:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleywegt, G. J., and T. A. Jones. 1999. Software for handling macromolecular envelopes. Acta Crystallogr. D Biol. Crystallogr. 55:941-944. [DOI] [PubMed] [Google Scholar]
- 26.Landau, M., I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, and N. Ben-Tal. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33:W299-W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, H. C., M. Bostina, D. J. Filman, and J. M. Hogle. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonberg-Holm, K., and B. D. Korant. 1972. Early interaction of rhinoviruses with host cells. J. Virol. 9:29-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino, A., M. Shimojima, T. Miyazawa, K. Kato, Y. Tohya, and H. Akashi. 2006. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 80:4482-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandell, K. J., and C. A. Parkos. 2005. The JAM family of proteins. Adv. Drug Deliv. Rev. 57:857-867. [DOI] [PubMed] [Google Scholar]
- 31.Marlin, S. D., D. E. Staunton, T. A. Springer, C. Stratowa, W. Sommergruber, and V. J. Merluzzi. 1990. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature 344:70-72. [DOI] [PubMed] [Google Scholar]
- 32.Naik, U. P., Y. H. Ehrlich, and E. Kornecki. 1995. Mechanisms of platelet activation by a stimulatory antibody: cross-linking of a novel platelet receptor for monoclonal antibody F11 with the Fc gamma RII receptor. Biochem. J. 310(Pt. 1):155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neill, J. D., I. M. Reardon, and R. L. Heinrikson. 1991. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 65:5440-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neill, J. D., S. V. Sosnovtsev, and K. Y. Green. 2000. Recovery and altered neutralization specificities of chimeric viruses containing capsid protein domain exchanges from antigenically distinct strains of feline calicivirus. J. Virol. 74:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ossiboff, R. J., and J. S. Parker. 2007. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 81:13608-13621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ossiboff, R. J., A. Sheh, J. Shotton, P. A. Pesavento, and J. S. Parker. 2007. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 88:506-517. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen, N. C., J. B. Elliott, A. Glasgow, A. Poland, and K. Keel. 2000. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 73:281-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesavento, P. A., N. J. MacLachlan, L. Dillard-Telm, C. K. Grant, and K. F. Hurley. 2004. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 41:257-263. [DOI] [PubMed] [Google Scholar]
- 40.Pflugrath, J. W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55:1718-1725. [DOI] [PubMed] [Google Scholar]
- 41.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 42.Racaniello, V. R. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. U. S. A. 93:11378-11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radford, A. D., K. Willoughby, S. Dawson, C. McCracken, and R. M. Gaskell. 1999. The capsid gene of feline calicivirus contains linear B-cell epitopes in both variable and conserved regions. J. Virol. 73:8496-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosen, C. G., and G. Weber. 1969. Dimer formation from 1-amino-8-naphthalenesulfonate catalyzed by bovine serum albumin. A new fluorescent molecule with exceptional binding properties. Biochemistry 8:3915-3920. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G., and D. M. Blow. 1962. The detection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 15:24-31. [Google Scholar]
- 46.Saeki, K., N. Ohtsuka, and F. Taguchi. 1997. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 71:9024-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schorr-Evans, E. M., A. Poland, W. E. Johnson, and N. C. Pedersen. 2003. An epizootic of highly virulent feline calicivirus disease in a hospital setting in New England. J. Feline Med. Surg. 5:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seal, B. S., and J. D. Neill. 1995. Capsid protein gene sequence of feline calicivirus isolates 255 and LLK: further evidence for capsid protein configuration among feline caliciviruses. Virus Genes 9:183-187. [DOI] [PubMed] [Google Scholar]
- 49.Silberstein, E., G. Dveksler, and G. G. Kaplan. 2001. Neutralization of hepatitis A virus (HAV) by an immunoadhesin containing the cysteine-rich region of HAV cellular receptor-1. J. Virol. 75:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 51.Sosnovtsev, S., S. Sosnovtseva, and K. Y. Green. 1997. Proceedings of the First International Symposium on Caliciviruses, Reading, United Kingdom.
- 52.Sosnovtsev, S. V., G. Belliot, K. O. Chang, O. Onwudiwe, and K. Y. Green. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 79:4012-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 54.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart, A. D., and T. D. Brown. 2007. Alpha2,6-linked sialic acid acts as a receptor for feline calicivirus. J. Gen. Virol. 88:177-186. [DOI] [PubMed] [Google Scholar]
- 56.Thiel, H. J., and M. Konig. 1999. Caliciviruses: an overview. Vet. Microbiol. 69:55-62. [DOI] [PubMed] [Google Scholar]
- 57.Tohya, Y., K. Masuoka, E. Takahashi, and T. Mikami. 1991. Neutralizing epitopes of feline calicivirus. Arch. Virol. 117:173-181. [DOI] [PubMed] [Google Scholar]
- 58.Tohya, Y., N. Yokoyama, K. Maeda, Y. Kawaguchi, and T. Mikami. 1997. Mapping of antigenic sites involved in neutralization on the capsid protein of feline calicivirus. J. Gen. Virol. 78(Pt. 2):303-305. [DOI] [PubMed] [Google Scholar]
- 59.Tong, L., and M. G. Rossmann. 1997. Rotation function calculations with GLRF. Methods Enzymol. 276:594-611. [PubMed] [Google Scholar]
- 60.Tsai, B. 2007. Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23:23-43. [DOI] [PubMed] [Google Scholar]
- 61.Tsang, S. K., B. M. McDermott, V. R. Racaniello, and J. M. Hogle. 2001. Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol. 75:4984-4989. [DOI] [PMC free article] [PubMed] [Google Scholar]