Abstract
Most class I fusion proteins exist as trimers of dimers composed of a receptor binding and a fusion subunit. In their postfusion forms, the three fusion subunits form trimers of hairpins consisting of a central coiled coil (formed by the N-terminal helices), an intervening sequence, and a region containing the C helix (and flanking strands) that runs antiparallel to and packs in the grooves of the N-terminal coiled coil. For filoviruses and most retroviruses, the intervening sequence includes a “chain reversal region” consisting of a short stretch of hydrophobic residues, a Gly-Gly pair, a CX6CC motif, and a bulky hydrophobic residue. Maerz and coworkers (A. L. Maerz, R. J. Center, B. E. Kemp, B. Kobe, and P. Poumbourios, J. Virol. 74:6614-6621, 2000) proposed a model for this region of human T-cell leukemia virus type 1 (HTLV-1) Env in which expulsion of the final bulky hydrophobic residue is important for early conformational changes and specific residues in the chain reversal region are important for forming the final, stable trimer of hairpins. Here, we used mutagenesis and pseudovirus entry assays to test this model for the avian retrovirus avian sarcoma/leukosis virus (ASLV) and the filovirus ebolavirus Zaire. Our results are generally consistent with the model proposed for HTLV-1 Env. In addition, we show with ASLV EnvA that the bulky hydrophobic residue following the CX6CC motif is required for the step of prehairpin target membrane insertion, whereas other residues are required for the foldback step of fusion. We further found that a His residue that is unique to the chain reversal region of ASLV EnvA controls the pH at which ASLV entry occurs.
Class I fusion proteins are trimeric glycoproteins that project from the viral membrane surface. Most harbor the host cell receptor binding and membrane fusion functions within distinct subunits (16, 41). Each class I fusion subunit contains a hydrophobic fusion peptide (or fusion loop), two heptad repeats separated by an intervening sequence, a membrane-spanning sequence, and a cytoplasmic tail. Class I fusion proteins are primed for fusion by one or more proteolytic events that, in most cases, separate the receptor binding and fusion subunits, leaving the fusion peptide/loop at or near the N terminus of the fusion subunit, and most sit on the viral membrane surface in a metastable state in which the receptor binding subunit “clamps” the fusion subunit (21, 42). Receptor binding, decreasing pH (during endocytosis), disulfide exchange, or a combination of these factors triggers release of this clamp (41). Once triggered, the fusion subunit undergoes conformational changes that first extend the fusion peptide/loop for interaction with the target membrane and then bend the fusion subunit roughly in half, forming a trimer of hairpins that brings the fusion peptides and the membrane-spanning domains, and hence the two membranes they are tethered to, together to create a fusion pore. For class I hairpins, the N-heptad repeats form a trimeric coiled coil, and C-terminal regions containing the helical C-heptad repeats pack (antiparallel) in the grooves of the N-terminal coiled coil. Additional zippering of the N- and C-terminal regions likely facilitates fusion pore formation (26).
The intervening sequence between the N- and C-heptad repeats is of variable length. For the fusion proteins of coronaviruses and paramyxoviruses, it is quite long (∼150 to 250 residues) (1, 43), whereas for the influenza virus HA2 subunit, it contains only 7 residues. The conversion of these 7 HA2 residues from a helix to a loop during fusion activation reverses the direction of the C-terminal end of HA2 (1, 43). For the human T-cell leukemia virus type 1 (HTLV-1) Env glycoprotein, the intervening sequence is 30 residues long and is comprised of a “chain reversal region” at the apex of the bend and a strand that extends to the C helix (20). The chain reversal region contains a CX6CC motif (20, 23) in which the first two Cys residues form an intramolecular disulfide bond, whereas the last Cys is bonded to the last Cys of a CXXC thiol-disulfide exchange motif in the receptor binding subunit. Reduction of the intersubunit disulfide bond by the free Cys of the CXXC motif is an integral part of the fusion-triggering process (22, 30). Many retroviral and all filoviral fusion subunits contain a CX6CC motif (Fig. 1A) (32). However, the filovirus glycoproteins (GPs) and alpharetrovirus Envs lack a thiol exchange motif, and at least for the avian sarcoma/leukosis virus subtype A (ASLV-A) Env (EnvA), the receptor binding and fusion subunits remain covalently attached throughout the fusion process (35).
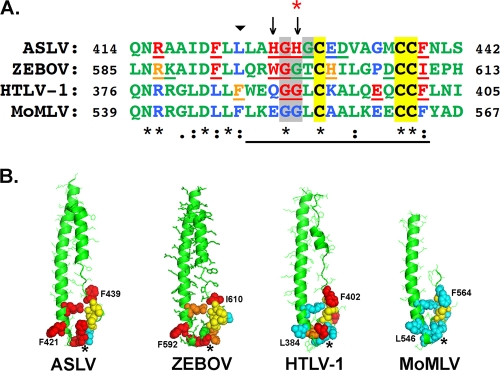
FIG. 1.
Alignment of residues in the chain reversal regions of the ASLV-A, ZEBOV, HTLV-1, and MoMLV fusion subunits. The sequence of ASLV EnvA is available at http://home.ncifcrf.gov/hivdrp/RCAS/RCASBPa_provisional_sequence.txt. ZEBOV, accession number U31033; HTLV-1, accession number NP_955621; MLV, accession number NP_955589. The numbers of the first and last residues in the sequence are given. For ASLV EnvA, the numbering begins after signal peptide cleavage; for all others, the numbering is from the start Met. Residues were aligned using the Clustal W PBIL multiple-alignment algorithm; output symbol designations are as follows: *, identical; :, strongly similar; ., weakly similar. The last residue of the N helix is denoted by an inverted triangle above the sequences. The chain reversal region as defined by Maerz, et al. (23) is underlined. The two unique His residues in the ASLV chain reversal region are denoted by arrows. Backbone sequences are in green. The three Cys residues of the CX6CC motif are colored black and highlighted in yellow. The Gly-Gly pair is highlighted in gray. Mutated residues are underlined. Residues for which infectivity compared to the wild type is decreased by 1 to 2 log units (this study) or cell-cell fusion is 20% of wild type (23) are colored orange; those for which infectivity is decreased by ≥2 log units (this study) or cell-cell fusion is less than 10% of wild type (23) are colored red. Corresponding untested residues in the other sequences are colored blue. Residues that were tested but were nearly wild type in infectivity are left green. The His residue that affects the pH of ASLV entry is denoted with a red asterisk. (B) Postfusion structures of CX6CC-bearing fusion subunits. The cores of the postfusion structures of the ZEBOV GP (1EBO), HTLV-1 Env (1MG1), and MoMLV Env (1MOF) are presented, together with a model of the postfusion ASLV TM core obtained by threading the corresponding EnvA sequence onto the ZEBOV GP postfusion structure using SwissPDB Viewer. Residues are colored as in panel A, except that the Cys residues of the CX6CC motif are colored yellow and the blue residues in panel A are now cyan. Residues with relevance to this study are shown as spheres. The first Gly of the Gly-Gly pair is denoted by a star. The structures were rendered in Pymol.
Maerz and coworkers (23) assessed the role of the chain reversal region for HTLV-1 Env (which contains a thiol exchange motif in its receptor binding subunit) by analyzing the ability of mutant HTLV-1 Env proteins to induce cell-cell fusion. Based on their findings, they presented a model in which a hydrophobic residue immediately after the CX6CC motif is extracted from a hydrophobic pocket during fusion triggering and then a Gly-Gly pair preceding the CX6CC motif acts as an “activation hinge” redirecting the C-terminal portion of the molecule so that it runs antiparallel to the N helix. It was further postulated that a salt bridge (between a charged residue within the CX6CC motif and an Arg residue near the C terminus of the N helix), as well as hydrophobic interactions among aromatic residues at the C terminus of the N helix, stabilizes the hairpin (see Fig. 6 in reference 23).
FIG. 6.
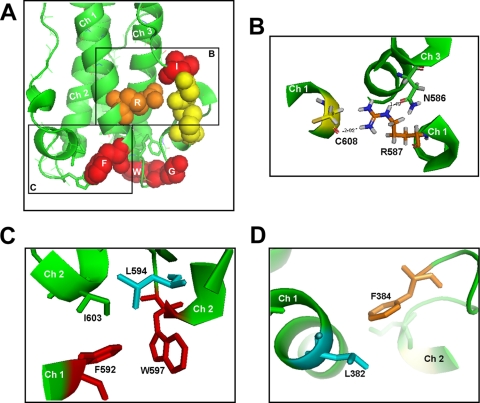
Location of critical residues at the base of the postfusion structures of ZEBOV GP2 and HTLV-1 TM. (A) An enlarged view of the base of the ZEBOV GP postfusion trimer of hairpins. The three chains are colored green. Residues on chain 1 are colored as in Fig. 1 and, where applicable, shown as spheres. (B) Enlarged and reoriented view of the neighborhood of R587 (box B in panel A) showing its interaction with the backbone carbonyl of the second Cys of the CX6CC motif on its own chain and N586 of a neighboring chain. (C) Enlarged and reoriented view of the interchain hydrophobic pocket (box C in panel A) showing the interactions of F592 with L594, W597, and I603 of a neighboring chain. Note that R587 and F592 interact with different neighboring chains. (D) Interaction of the HTLV-1 TM L382, which aligns with ZEBOV GP2 F592 (Fig. 1A), with F384 (which aligns with L592) on a neighboring chain. The figures were generated in Pymol using PDB 1EBO (A to C) and PDB 1MG1 (D).
In this study, we asked what elements of the model proposed by Maerz and coworkers (23) extend to CX6CC motif-bearing fusion proteins that do not contain a thiol exchange motif in their receptor binding subunits. We did this by analyzing analogous and additional mutations in the Zaire ebolavirus (ZEBOV) GP and in the ASLV EnvA. Each system provided an advantage. The structure of ZEBOV GP is known in both its native and postfusion states (21, 40), but its fusion trigger has not yet been elucidated. Conversely, no structures are available for ASLV EnvA, but its “2-step” fusion-triggering mechanism has been characterized in detail (2, 6, 25, 28, 36). Our findings are generally consistent with the proposed model (23) and reveal requirements for analogous and additional residues in the chain reversal regions of ASLV EnvA and ZEBOV GP. In particular, we show, for EnvA, that the bulky hydrophobic residue following the CX6CC motif is required for receptor-triggered target membrane association of the prehairpin, whereas specific residues in the chain reversal region are critical for forming the hairpin. In addition we identify a His residue, unique to the chain reversal region of EnvA, as being a critical determinant of the low-pH dependence of ASLV entry.
MATERIALS AND METHODS
Cells and reagents.
293T, PG950, DF-1, BHK, and Vero e6 cells and their growth conditions have been described previously (8, 33). Dulbecco's modified Eagle's medium (DMEM), Opti-MEM, fetal bovine serum (FBS), Penn/Strep, l-glutamine, and sodium pyruvate were from Gibco. The anti-GP1 polyclonal antibody (PAb), was a gift from Paul Bates. Anti-A-tail and anti-murine leukemia pseudovirus (MLV) gag p30 have been described previously (8, 17).
Preparation and characterization of pseudotyped viruses.
The preparation of murine leukemia pseudovirus bearing EnvA (MLV-EnvA) and vesicular stomatitis virus pseudovirus bearing GPΔ (VSV-GPΔ) has been described previously (17, 33). For MLV-EnvA pseudovirions, culture supernatants were used directly for infectivity assays or pelleted through 20% (wt/vol) sucrose in HMS buffer (20 mM HEPES, 20 mM MES [morpholineethanesulfonic acid], 150 mM NaCl, pH 7.8) and resuspended in 200 μl HM buffer (20 mM HEPES, 20 mM MES, pH 7.8). The virus was kept on ice until it was used. Infectivity assays were performed as described previously (8, 18) using PG950 cells, a mouse cell line that stably expresses the ASLV-A receptor, Tva (15). The assays for the incorporation of EnvA into MLV pseudovirions, receptor binding, and the flotation of soluble Tva (sTva)-triggered MLV-EnvA pseudoparticles were performed as previously described (6, 7, 9). Peptide-N-glycosidase F (PNGase) treatment was performed according to the manufacturer's instructions (New England Biolabs).
VSV-GPΔ was produced by polyethyleneimine (PEI) transfection of BHK cells with 12 μg of plasmids encoding wild-type (WT) or mutant ebolavirus GP per 10-cm dish. Twenty-four hours after transfection, 100 μl of helper virus was added, and 24 h later (48 h post transfection), the supernatants were collected, frozen at −80°C for 20 to 30 min, thawed at 37°C for 10 min, and then kept at 4°C throughout the purification. Cellular debris was removed by two centrifugations for 10 min each at 1,080 × g. The cleared supernatants were transferred to a 300,000 molecular weight cutoff (MWCO) Vivaspin concentrator (Sartorius) and concentrated by centrifugation for 45 min at 2,760 × g and then multiple times to a final volume of 0.75 to 1.0 ml. The virus was then banded between 25 and 60% (wt/vol) sucrose in HN buffer (25 mM HEPES, 130 mM NaCl, pH 7.4) by centrifugation for 16 h at 77,175 × g. The banded virus was collected and pelleted through 20% sucrose in HN buffer for 2 h at 100,000 × g. The pellet was resuspended in 125 μl HN buffer containing 10% (wt/vol) sucrose, and the virus was either used directly for an infection assay or aliquoted and stored at −80°C until it was used. Infectivity assays and generation of 19 kDa GP1 were performed as previously described (10, 33).
HIV pseudovirions were prepared by transfection of plasmids bearing EnvA (pCBC/EnvA), BlaM-VPr (pMM10), and HIV provirus lacking the Env gene (p8.2Δ) into 293T cells by the calcium phosphate method using 6 μg of pCB6/EnvA and 10 μg each of pMM10 and p8.2Δ; 24 h later, the cells were induced with 6 mM sodium butyrate and harvested 48 h posttransfection. The culture supernatants were clarified, pelleted through 20% (wt/vol) sucrose in HMN buffer for 2 h at 100,000 × g, and resuspended in 150 μl HM buffer with an additional 50 μl of 20% (wt/vol) sucrose in HMN buffer. Aliquots were stored at −80°C until they were used. The virus concentration was assessed by p24 enzyme-linked immunosorbent assay (ELISA).
Entry assay.
The β-lactamase (BlaM) entry assay (3) was adapted for assessing the pH dependence of EnvA entry as follows. DF-1 cells (1 × 106 to 1.25 × 106/well of a 24-well dish) were plated and incubated for approximately 20 h. The cells (except controls) were pretreated with 100 nM bafilomycin for 30 min prior to the addition of virus and maintained in 100 nM bafilomycin throughout the experiment. The cells were cooled to 4°C, and aliquots of virus in fusion buffer (70 μl virus/ml FB [100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 20 mM HEPES, 20 mM succinate, 0.2% glucose, pH 7.4]) were added and incubated at 4°C for 1 h and then at 37°C for 20 min. The pH was then adjusted to the indicated value by adding a predetermined amount of 0.3 M acetic acid. After 6 min, the cells were reneutralized with a predetermined amount of 2 M Tris, pH 8.0, washed with dye-free Opti-MEM (Gibco), and returned to the incubator for 2 to 2.5 h. The cells were then stained with CCF2/AM dye (Gene Blazer loading kit; Invitrogen) for 1 h in the dark at room temperature (RT), washed with fresh medium, incubated overnight in the dark at RT, and fixed for fluorescence-activated cell sorter (FACS) analysis. FACS analysis was performed the day of fixing using a CyAn LX 9 Color flow cytometer (DakoCytomation) and analyzed with FloJo. The percentage of infected cells was calculated as the ratio of the number of blue cells to the total number of cells gated. The value at pH 5.0 was set to 1, and the fraction of this value at each other pH was calculated. Three independent experiments were each performed in duplicate, and all values at a given pH were averaged.
RESULTS
Comparison of the chain reversal regions of ASLV-A, HTLV-1, and MoMLV Envs and ZEBOV GP.
Structures have been solved for the cores of the postfusion trimers for three fusion subunits that contain a CX6CC motif in their chain reversal regions: those for ZEBOV GP and the HTLV-1 and Moloney murine leukemia virus (MoMLV) Envs (12, 20, 40). The fusion subunit of ASLV-A Env (EnvA TM) was predicted to adopt a structure similar to that of ZEBOV GP2 (14), and this prediction was supported by a threading analysis (Fig. 1B). These structures have a common fold, with the N helix extending to the base of the hairpin, followed by a tight turn and the CX6CC motif, which packs against the C-terminal end of the N helix (Fig. 1B) (12, 20, 40). Alignment of these regions (Fig. 1A) revealed absolutely conserved residues, as well as residues with conservative substitutions. In addition to the conserved Cys residues of the CX6CC motif, these included an Asn-Arg pair, an Asp followed by three or four hydrophobic residues, a Gly-Gly pair (interrupted in EnvA TM), the CX6CC motif with a bulky hydrophobic residue at position X3, and a bulky hydrophobic residue immediately following the CX6CC motif. The ASLV TM differed in that it contained two His residues, one immediately preceding and one interrupting the Gly-Gly pair.
Mutations at similar positions at the base of the hairpin of ASLV EnvA and ZEBOV GP inhibit pseudovirion infectivity.
We mutated selected residues (underlined in Fig. 1A) at the base of the N helix and within the chain reversal regions of the fusion subunits of ASLV-A EnvA and ZEBOV GP. We first tested the ability of MLV-pseudotyped virions bearing EnvAs and VSV-pseudotyped virions bearing ZEBOV GPs to infect PG950 or Vero cells, respectively. The results are presented in Table 1. For EnvA, the mutants displayed three phenotypes: those whose infectivity was decreased by ≤1 log unit (G429A, E431A, and D432A), 2 to 3 log units (H426A, G427A, and H428A), or >3 log units (R416A, R416K, F421A, and F439A). The fact that the EnvA mutant with Lys rather than Ala substituted for R416 was also severely debilitated (infectivity decreased by >5 log units) suggests a specific need for Arg at this position, not simply a need for a positive charge. The results for GP were qualitatively similar (Table 1). Ala substitutions at GP R587, F592, W597, G588, and I610 and the G588A G589A double mutation (GG) inhibited infectivity by more than 1 log unit. While the extents of inhibition were similar for most of these GP mutants, mutations at R587 and G599 were less severe while those for W597 and G598 were more severe than their EnvA analogues. Nevertheless, similar residues were important: residues at the base of the N helix, the first Gly of the Gly-Gly pair, the residue preceding the Gly-Gly pair, and the hydrophobic residue following the CX6CC motif (orange or red in Fig. 1A). Also similar to EnvA, both the K588A and the D607A mutant GPs exhibited wild-type infectivity, suggesting that a (putative) salt bridge involving these residues is not important for either GP- or EnvA-mediated fusion.
TABLE 1.
Infectivity of ASLV (EnvA) and ZEBOV (GP) chain reversal region mutantsa
| EnvA mutant | Infectivity (IU/ml) | Log10 decrease in infectivity | GP mutant | Infectivity (IU/ml) | Log10 decrease in infectivity |
|---|---|---|---|---|---|
| WTA | 1.35 ± 1.92 × 107 | NA | GPΔ | 8.05 ± 2.62 × 107 | NA |
| R416A | 2.84 ± 3.16 × 102 | −4.7 | R587A | 4.71 ± 2.47 × 106 | −1.2 |
| R416K | 6.83 ± 6.91 × 101 | −5.3 | |||
| K588A | 9.38 ± 0.98 × 107 | +0.1 | |||
| F421A | 3.09 ± 4.20 × 103 | −3.6 | F592A | 2.09 ± 2.40 × 105 | −2.6 |
| H426A | 4.83 ± 1.58 × 104 | −2.4 | W597A | 1.14 × 104 | −3.8 |
| G427A | 3.22 ± 1.88 × 104 | −2.6 | G598A | 2.86 ± 0.81 × 104 | −3.4 |
| H428A | 6.86 ± 3.78 × 104 | −2.3 | G599A | 4.10 ± 1.77 × 107 | −0.3 |
| G429A | 2.00 ± 1.41 × 106 | −0.8 | T600A | 5.01 ± 2.15 × 107 | −0.2 |
| E431A | 2.28 ± 2.29 × 106 | −0.8 | H602A | 9.63 ± 5.06 × 106 | −0.9 |
| D432A | 3.60 ± 4.32 × 106 | −0.6 | D607A | 6.30 ± 0.37 × 107 | −0.1 |
| F439A | 3.93 ± 1.54 × 102 | −4.5 | I610A | 2.44 ± 2.74 × 105 | −2.5 |
| GG | 1.24 ± 0.39 × 102 | −5.0 | GG | 5.15 ± 0.81 × 104 | −3.2 |
Spaces are left in each column so that mutations at equivalent positions along the chain reversal region are presented in the same row. GG denotes the double-Gly-to-Ala mutants for EnvA (G427AG429A) and GP (G598AG599A). NA, not applicable.
Characterization of EnvA and GP pseudovirions.
As seen in Fig. 2A, each EnvA mutant was incorporated into pseudovirions to levels similar to those of wild-type EnvA (71 to 118%). Since the banding patterns of D432A and F439A looked somewhat different than the others, we subjected them to PNGase treatment. As seen in Fig. 2B, all EnvAs tested comigrated at ∼25 kDa following PNGase treatment, suggesting that D432A and F439A have subtly different levels of glycosylation. Since EnvA D432A exhibited nearly wild-type infectivity (Table 1), this small difference in glycosylation is not likely to account for the 4.5-log-unit decrease in infectivity observed for F439A.
FIG. 2.
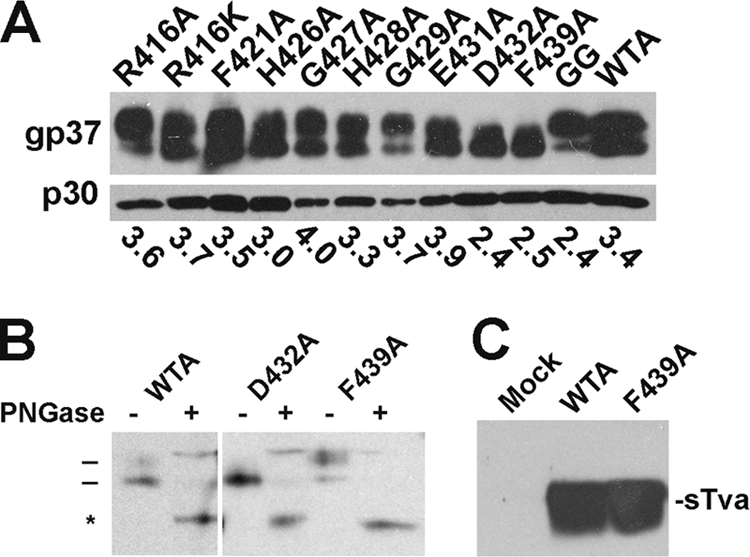
Characterization of ASLV EnvA mutants. (A) Incorporation into MLV pseudovirions. Equal volumes of MLV pseudovirions bearing the indicated EnvA mutants were run on duplicate gels and transferred to nitrocellulose. One immunoblot was probed with anti-A-tail IgG (gp37) and visualized with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (p30). The other gel was probed with anti-MLV capsid and visualized with HRP-conjugated anti-rat IgG. The bands were quantitated using a Rosenman Pixel Quantification plug-in for Adobe Photoshop. The numbers reported are for EnvA/gag ratios. (B) PNGase treatment of WTA, D432A, and F439A. Virions were treated with PNGase (+) or mock treated (−), resolved by SDS-PAGE, and transferred to nitrocellulose, and the gp37 band was probed as described for panel A. Bars, glycosylated gp37; *, deglycosylated gp37. (C) Immunoprecipitation of sTva with WTA and F439A pseudovirions. Biotinylated sTva was incubated with virions in 1% NP-40 lysis buffer, immunoprecipitated with anti-A tail IgG bound to protein A-conjugated beads, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with HRP-conjugated streptavidin. Mock, no virus added.
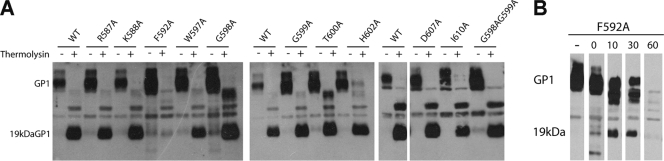
All ZEBOV GP mutants were also well incorporated into pseudovirions (Fig. 3A, − thermolysin). Moreover, all mutant GPs could be primed to the 19-kDa form of GP1 by thermolysin (10, 33), although GP F592A was not converted to the 19-kDa form either as robustly or as stably as WT GP or the other mutants (Fig. 4A and B).
FIG. 3.
Cleavage of ZEBOV GPΔ mutants with thermolysin. VSV-GPΔ (5 μg) was treated with 5 μg of thermolysin at 37°C for 1 h (A) or the indicated times (in min) (B), and the reaction was terminated by adding EDTA to a 10 mM final concentration on ice. Half of each sample was then run on an SDS-PAGE gel, transferred to nitrocellulose, and probed with an anti-GP1 PAb, followed by HRP-conjugated anti-rabbit IgG.
FIG. 4.
Receptor-induced membrane association of EnvA. MLV-EnvA-pseudotyped virus particles were incubated for 30 min at 4°C in the presence or absence of sTva, and liposomes were added and incubated for 30 min at 37°C. Samples were run on sucrose step flotation gradients, four fractions were collected, and the proteins were resolved by SDS-PAGE and immunoblotted for gp37 as described in the legend to Fig. 2A. T and B represent the top and bottom of the gradient, respectively. Data for log reduction of infectivity (from Table 1) are given for each mutant. The lower-molecular-weight band seen in lanes 4 (+ sTva and − sTva) in some of the samples is a background band that does not bind to liposomes; it has also been seen periodically in samples of WTA (B. La and S. E. Delos, unpublished results).
EnvAs bearing mutations at the base of the N helix and in their chain reversal regions undergo sTva-triggered membrane association.
Maerz and colleagues (reference 23, Fig. 6) suggested that residues at the base of the hairpin are important for forming and/or stabilizing the hairpin, but their results did not rule out a role at an earlier stage, such as membrane insertion of the prehairpin intermediate. Since we can assess this stage for EnvA, we tested pseudovirions bearing each EnvA mutant for the ability to undergo sTva-induced membrane association (9, 11, 17, 29). As seen in Fig. 4, wild-type EnvA (WTA) floated to the top of the gradient when sTva was present (left, lane 1), but not in its absence (right, lane 4). For most of the mutants, the majority of the virions floated. The exceptions were F439A, which did not float, and G429A and GG, which floated only poorly. The defect for G429A was surprising, since it is impaired for infectivity by only ∼0.8 log unit. By extrapolation, the partial defect in flotation for the GG mutant is insufficient to account for its 5-log-unit decrease in infectivity. The flotation results show that, with the exception of F439A, all EnvA mutants tested were able to undergo receptor-triggered target membrane association, the first step of fusion. To ensure that the inability of F439A to undergo receptor-mediated membrane association is not due to a lack of receptor binding, we assessed its ability to bind sTva in an immunoprecipitation experiment. As seen in Fig. 2C, F439A is fully competent to bind sTva. Thus, the defect in infectivity of EnvA F439A is most likely due to a defect in receptor-triggered target membrane binding, whereas those of the other mutants are likely to be at the foldback step of fusion.
H428A in the chain reversal region affects the pH of EnvA-mediated entry.
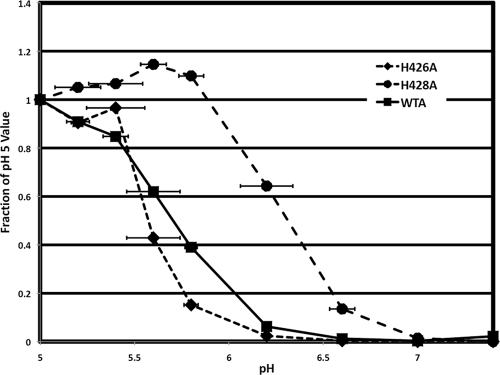
As seen in Fig. 1A, EnvA has two unique His residues (H426 and H428) in its chain reversal region, one preceding and one separating the otherwise conserved Gly-Gly pair. Since the foldback step of fusion is low pH dependent for EnvA, but not for the MoMLV or HTLV-1 Env (and is not yet known for ZEBOV GP), we asked if either of these His residues affected the pH dependence of EnvA-mediated entry. To determine this, we “forced fusion” by pulsing cells bearing prebound HIV-EnvA-pseudotyped virions containing β-lactamase-Vpr with medium of a specified pH and then measured the amount of β-lactamase-Vpr delivered to the cytosol of the target cells (3). As seen in Fig. 5, WTA mediated entry with a midpoint pH of 5.7, consistent with values reported for WTA-mediated content mixing (6, 24, 27, 28). The pH profile for EnvA H426A-mediated entry was not significantly different from that of WTA (midpoint pH, 5.6). In striking contrast, the pH profile for EnvA H428A was significantly different, having a pH of onset at pH 6.6 and of half maximal fusion at pH 6.3, the latter being a full 0.6 pH units higher than that for WTA. These results show that H428 plays a critical role in determining the low pH at which EnvA mediates cellular entry.
FIG. 5.
pH dependence of entry for wild-type, H426A, and H428A EnvA. DF-1 cells were pretreated with 100 nM bafilomycin, chilled to 4°C, and incubated with HIV pseudoparticles containing β-lactamase-Vpr and bearing the indicated EnvA for 1 h at 4°C in the continued presence of 100 nM bafilomycin. The temperature was then raised to 37°C for 20 min, whereupon a predetermined amount of acetic acid was added to adjust the pH to the indicated value. After 6 min at 37°C, the fusion buffer was reneutralized, medium containing bafilomycin was added, and the cells were returned to the incubator for 2 h. The β-lactamase substrate, CCF2A, was then added, and its conversion from a green substrate to a blue product was allowed to proceed at RT overnight. The percentage of cells that were converted from green to blue was determined by flow cytometry. Three independent experiments were performed in duplicate for each virus. The values presented here represent the average of all 6 values at each pH. The error bars represent the standard deviation for each averaged point.
DISCUSSION
Maerz and coworkers proposed a model for the role of residues in the chain reversal region in fusion activation of the HTLV-1 Env protein (23). In this study, we tested their model for two additional class I fusion proteins that, like HTLV-1 Env, contain a CX6CC motif but, unlike HTLV Env, lack a thiol exchange motif in their receptor binding subunit: GP of the filovirus ZEBOV and EnvA of the retrovirus ASLV-A. Our findings support the model and thereby suggest a common role for the chain reversal regions of CX6CC motif-containing class I fusion proteins. Our findings with EnvA extend the model by showing that one residue functions for, whereas the others function after, the step at which the prehairpin intermediate binds to target membranes. In addition, we identified a His residue, unique to this region of EnvA, that influences the pH at which ASLV entry occurs.
The hydrophobic residue following the CX6CC motif is important for receptor-triggered membrane association.
ASLV EnvA F439A is strongly impaired in mediating infectivity (Table 1) yet is processed into receptor binding and fusion subunits, incorporated into pseudovirions, and capable of binding receptor, similar to WTA (Fig. 2). Further analysis revealed that EnvA F439A was unable to bind to target membranes under fusion-triggering conditions (Fig. 4). Collectively, these findings indicate that EnvA F439A is blocked at a very early stage of fusion, most likely at the stage of receptor-triggered membrane binding. As the analogous ZEBOV GP mutant, I610A, could be primed to its fusion-competent, 19-kDa form by treatment with thermolysin (Fig. 3A) yet was significantly impaired in its ability to support infectivity (Table 1), we hypothesize that it, too, is most likely defective at an early step of fusion. Confirmation of this hypothesis awaits identification of the ZEBOV GP fusion trigger. By extension, the bulky hydrophobic residue following the CX6CC motif in other class I fusion proteins may provide a similar function.
The first Gly of the Gly-Gly pair and the residue preceding it are important for infectivity.
Maerz and coworkers (23) mutated each Gly of the Gly-Gly pair to Pro and found that the resulting HTLV-1 Envs were defective in cell-cell fusion and that maltose-binding protein (MBP)-gp21 chimeras bearing these mutations were impaired in forming trimers of hairpins. They concluded that the conserved Gly-Gly pair imparts flexibility required to reverse the C-terminal chain for foldback into the trimer of hairpins. Since the Gly-Gly pair is preceded and interrupted by His residues in EnvA, we mutated each residue of the HGHG in ASLV EnvA and the corresponding WGGT in ZEBOV GP to Ala. For ZEBOV GP, we found that replacements of the first Gly of the Gly-Gly pair and the Trp immediately preceding it impaired infectivity, whereas those at the second Gly of the pair and the residue following the Gly-Gly pair did not. A similar pattern was seen for EnvA: mutating the analogous first His and first Gly residues impaired infectivity, but mutating the second Gly of the Gly-Gly pair did not. The second His residue (interrupting the Gly-Gly pair in EnvA) has an additional function (see below). The WG sequence of GP and the first HG sequence of EnvA are found at the point of maximum curvature of the chain reversal turn (Fig. 1B, black stars). These findings support a common role for the first Gly of the Gly-Gly pair in fusion. Since the double-Gly (GG) mutants in both EnvA and GP were more debilitated than either of the single-Gly mutants, the second Gly of the Gly-Gly pair appears to play a supporting role.
Charged residues in the CRR of GP and EnvA are not important for fusion.
Maerz and coworkers found that mutation of a charged residue (E398) within the CX6CC motif to Asn resulted in 90% loss in cell-cell fusion (23). They predicted that this residue forms a salt bridge with a conserved Arg (R380). They further predicted that residues D607 and K588 would play a similar role for ZEBOV GP. We mutated each of these residues in GP (to Ala) and found that the resulting GPs retained wild-type levels of infection. ASLV EnvA has an Ala at the position analogous to GP K588 and HTLV-1 Env R380 (Fig. 1A), suggesting that an analogous salt bridge could not exist for EnvA. Nevertheless, we tested the importance of the charged residues in the CX6CC motif of EnvA and found only a minimal effect on infectivity (Table 1). Thus, salt bridges at the base of the hairpin do not appear to be critical for either ZEBOV GP- or ASLV EnvA-mediated fusion.
The conserved Arg at the base of the N helix may stabilize the postfusion hairpin.
An Arg residue near the end of the N helix is conserved among CX6CC motif-bearing fusion proteins (first R in Fig. 1A). Mutating this conserved Arg (to Ala, and/or Lys for EnvA) inhibited infectivity for both EnvA and GP, albeit more severely for EnvA (Table 1); these mutants appeared wild type in all other aspects tested, including receptor-induced target membrane association for EnvA (Fig. 4). These Arg residues are found in the “eye of the hairpin” in the postfusion structures (Fig. 1B). The structure of the base of the ZEBOV GP trimer of hairpins is shown in Fig. 6A, and the section of box B is expanded and reoriented in Fig. 6B to show that the conserved Arg of GP (R587) forms hydrogen bonds with the backbone carbonyl of the second Cys of the CX6CC motif on its own strand and an Asn residue in a neighboring strand. The Asn-Arg hydrogen bond is at the central nitrogen of the guanyl moiety of the Arg, a stabilizing interaction that could not be provided by a Lys residue. Thus, through these interactions, the Arg residue appears to play a critical role in stabilizing the trimer of hairpins (Fig. 1B and 6A and B).
The hydrophobic stretch at the base of the N helix may stabilize the trimer of hairpins.
The base of the N helix is terminated by a stretch of three or four hydrophobic residues (Fig. 1). Mutation of the Phe in this stretch of the HTLV-1 Env reduced cell-cell fusion activity by 80%. Maerz et al. proposed that interaction of the Phe with a Leu residue in an adjacent monomer stabilizes the postfusion structure (23). We found that the Phe that is conserved in this hydrophobic stretch between ZEBOV GP and ASLV EnvA is also critical. In the postfusion GP2 structure, this Phe, F592, sits in a hydrophobic pocket that is formed by F592 of one strand and L594, W597, and I603 of an adjacent strand (Fig. 6C), thus stabilizing the trimer base. In HTLV-1 Env, the Phe and Leu are located in reciprocal positions in the primary sequence (Fig. 1A), but their interaction in the postfusion trimer is maintained (Fig. 6D). Interestingly, although the HTLV-1 Phe and Leu are capable of hydrophobic interactions, the complex interaction with additional hydrophobic residues seen for GP is not observed. Perhaps the salt-bridge suggested for the postfusion structure of the HTLV-1 Env (23) provides a sufficient compensating interaction. Our findings that mutation of this Phe in EnvA to Ala (EnvA F421A) does not impair receptor-triggered binding to target membranes (Fig. 4) yet severely debilitates infectivity further support the prediction that Phe residues in the hydrophobic stretch at the base of the N helix are important to stabilize the trimer of hairpins formed during the foldback step of fusion.
The Phe residues in the hydrophobic stretch at the base of the N helix also appear to be important for the native structures of GP and EnvA. GP bearing the F592A mutation was not robustly primed to a stable 19-kDa form by in vitro treatment with thermolysin (Fig. 3). In the native structure, GP2 F592 interacts with GP1 N57 (21), an interaction that may position GP1 for more optimal priming. The analogous EnvA mutant (EnvA F421A) was also more sensitive to protease digestion (data not shown), suggesting a possible additional common role for these Phe residues in maintaining the optimal native fusion protein structure.
The role of His 428 in EnvA fusion and ASLV entry.
His residues, which are sensors for many pH-controlled conformational switches, are utilized by several viral fusion proteins as part of their fusion triggers (reviewed in references 19 and 38). Mutation of some of these His residues inhibits fusion altogether (4, 5, 13, 38), while mutation of others alters the pH dependence of fusion (31, 34, 37, 39). EnvA H428A is shifted +0.6 pH units for fusion, which is a large pH shift compared to those recorded for His mutations in other viral fusion proteins. Moreover, mutation of many His residues in viral fusion proteins shifts the pH needed for activation in the acidic direction (i.e., more protons are required to affect the conformational change otherwise caused by protonation of the His residue). In contrast, the fact that the pH of fusion for EnvA H428A is higher than that for WTA suggests that H428 is part of the fusion clamp that prevents fusion until the optimal compartment for entry is reached. H428 is absolutely conserved in all ASLV sequences available in the database, highlighting the likely importance of the residue for fusion of multiple ASLV subtypes.
Closing thoughts.
In summary, our results extend the model put forth by Maerz and colleagues for the role of the chain reversal region in fusion for HTLV-1 Env (23) to two fusion proteins, those of ebolavirus and ASLV, that contain CX6CC motifs but do not have CXXC thiol exchange motifs in their receptor binding subunits. We highlighted common roles of specific residues, including an Arg and a Phe at the base of the N helix, the first Gly of the Gly-Gly pair, and a hydrophobic residue following the CX6CC motif. In particular, we showed for EnvA that the hydrophobic residue following the CX6CC motif is important for receptor triggering of target membrane binding and that other residues function later in fusion. In addition, we determined a unique role (as a pH sensor) for a His residue that is found only in the EnvA chain reversal region. Collectively, our results highlight the pivotal position of the chain reversal region in the fusion pathway, with different residues being important early and late in the process. Our results support the concept that interactions at the membrane-distal end of the hairpin, not just helix bundles and membrane-proximal and membrane-embedded sequences, are important for late steps in fusion.
Acknowledgments
We thank Edward Park for expert technical assistance and Pengxiang Huang and Binyong Liang for helpful discussions regarding structural interpretations.
This work was supported by NIH R01 AI22470 to J.M.W. and S.E.D., U54 AI57168 to J.M.W., and a UVA Harrison Award to A.G.
Footnotes
Published ahead of print on 24 March 2010.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Barnard, R. J., S. Narayan, G. Dornadula, M. D. Miller, and J. A. Young. 2004. Low pH is required for avian sarcoma and leukosis virus Env-dependent viral penetration into the cytosol and not for viral uncoating. J. Virol. 78:10433-10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 4.Chanel-Vos, C., and M. Kielian. 2004. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J. Virol. 78:13543-13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanel-Vos, C., and M. Kielian. 2006. Second-site revertants of a Semliki Forest virus fusion-block mutation reveal the dynamics of a class II membrane fusion protein. J. Virol. 80:6115-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delos, S. E., M. B. Brecher, Z. Chen, D. C. Melder, M. J. Federspiel, and J. M. White. 2008. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J. Virol. 82:3131-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delos, S. E., M. J. Burdick, and J. M. White. 2002. A single glycosylation site within the receptor-binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354-363. [DOI] [PubMed] [Google Scholar]
- 8.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 74:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delos, S. E., and J. M. White. 2000. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J. Virol. 74:9738-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dube, D., M. B. Brecher, S. E. Delos, S. C. Rose, E. W. Park, K. L. Schornberg, J. H. Kuhn, and J. M. White. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earp, L. J., S. E. Delos, R. C. Netter, P. Bates, and J. M. White. 2003. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 77:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 13.Fritz, R., K. Stiasny, and F. X. Heinz. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 183:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallaher, W. R. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85:477-478. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, J. M., P. Bates, H. E. Varmus, and J. M. White. 1994. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J. Virol. 68:5623-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, S. C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, L. D., and J. M. White. 1998. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J. Virol. 72:3259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kampmann, T., D. S. Mueller, A. E. Mark, P. R. Young, and B. Kobe. 2006. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14:1481-1487. [DOI] [PubMed] [Google Scholar]
- 20.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. U. S. A. 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. E., M. L. Fusco, A. J. Hessell, W. B. Oswald, D. R. Burton, and E. O. Saphire. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K., S. Zhang, M. Kronqvist, M. Wallin, M. Ekstrom, D. Derse, and H. Garoff. 2008. Intersubunit disulfide isomerization controls membrane fusion of human T-cell leukemia virus Env. J. Virol. 82:7135-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maerz, A. L., R. J. Center, B. E. Kemp, B. Kobe, and P. Poumbourios. 2000. Functional implications of the human T-lymphotropic virus type 1 transmembrane glycoprotein helical hairpin structure. J. Virol. 74:6614-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markosyan, R. M., P. Bates, F. S. Cohen, and G. B. Melikyan. 2004. A study of low pH-induced refolding of Env of avian sarcoma and leukosis virus into a six-helix bundle. Biophys. J. 87:3291-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuyama, S., S. E. Delos, and J. M. White. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201-8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melikyan, G. B. 2008. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology 5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., R. J. Barnard, R. M. Markosyan, J. A. Young, and F. S. Cohen. 2004. Low pH is required for avian sarcoma and leukosis virus Env-induced hemifusion and fusion pore formation but not for pore growth. J. Virol. 78:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 29.Netter, R. C., S. M. Amberg, J. W. Balliet, M. J. Biscone, A. Vermeulen, L. J. Earp, J. M. White, and P. Bates. 2004. Heptad repeat 2-based peptides inhibit avian sarcoma and leukosis virus subgroup a infection and identify a fusion intermediate. J. Virol. 78:13430-13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, Z. L., Y. Zheng, and M. Kielian. 2009. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J. Virol. 83:4670-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders, D. A. 2000. Sulfhydryl involvement in fusion mechanisms. Subcell. Biochem. 34:483-514. [DOI] [PubMed] [Google Scholar]
- 33.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schowalter, R. M., A. Chang, J. G. Robach, U. J. Buchholz, and R. E. Dutch. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 83:1511-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. G., and J. M. Cunningham. 2007. Receptor-induced thiolate couples Env activation to retrovirus fusion and infection. PLoS Pathog. 3:e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, J. G., W. Mothes, S. C. Blacklow, and J. M. Cunningham. 2004. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 78:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinhauer, D. A., J. Martin, Y. P. Lin, S. A. Wharton, M. B. Oldstone, J. J. Skehel, and D. C. Wiley. 1996. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc. Natl. Acad. Sci. U. S. A. 93:12873-12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stiasny, K., R. Fritz, K. Pangerl, and F. X. Heinz. 2009. Molecular mechanisms of flavivirus membrane fusion. Amino Acids doi: 10.1007/s00726-009-0370-4. [DOI] [PubMed]
- 39.Thoennes, S., Z. N. Li, B. J. Lee, W. A. Langley, J. J. Skehel, R. J. Russell, and D. A. Steinhauer. 2008. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology 370:403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 41.White, J. M., S. E. Delos, M. Brecher, and K. Schornberg. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 43.Yan, Z., K. V. Holmes, and R. S. Hodges. 2009. Expression and characterization of recombinant S2 subunit of SARS-coronavirus S fusion protein. Adv. Exp. Med. Biol. 611:153-154. [DOI] [PMC free article] [PubMed] [Google Scholar]