Abstract
In this study, we used an RNA polymerase I (Pol I) transcription system for development of a reverse genetics protocol to produce hepatitis C virus (HCV), which is an uncapped positive-strand RNA virus. Transfection with a plasmid harboring HCV JFH-1 full-length cDNA flanked by a Pol I promoter and Pol I terminator yielded an unspliced RNA with no additional sequences at either end, resulting in efficient RNA replication within the cytoplasm and subsequent production of infectious virions. Using this technology, we developed a simple replicon trans-packaging system, in which transient transfection of two plasmids enables examination of viral genome replication and virion assembly as two separate steps. In addition, we established a stable cell line that constitutively produces HCV with a low mutation frequency of the viral genome. The effects of inhibitors of N-linked glycosylation on HCV production were evaluated using this cell line, and the results suggest that certain step(s), such as virion assembly, intracellular trafficking, and secretion, are potentially up- and downregulated according to modifications of HCV envelope protein glycans. This Pol I-based HCV expression system will be beneficial for a high-throughput antiviral screening and vaccine discovery programs.
Over 170 million people worldwide have been infected with hepatitis C virus (HCV) (22, 33, 37), and persistence of HCV infection is one of the leading causes of liver diseases, such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma (16, 25, 38). The HCV genome is an uncapped 9.6-kb positive-strand RNA sequence consisting of a 5′ untranslated region (UTR), an open reading frame encoding at least 10 viral proteins (Core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B), and a 3′UTR (46). The structural proteins (Core, E1, and E2) reside in the N-terminal region.
The best available treatment for HCV infection, which is pegylated alpha interferon (IFN-α) combined with ribavirin, is effective in only about half of patients and is often difficult to tolerate (25). To date, a prophylactic or therapeutic vaccine is not available. There is an urgent need to develop more effective and better tolerated therapies for HCV infection. Recently, a robust system for HCV production and infection in cultured cells has been developed. The discovery that some HCV isolates can replicate in cell cultures and release infectious particles has allowed the complete viral life cycle to be studied (23, 49, 53). The most robust system for HCV production involves transfection of Huh-7 cells with genomic HCV RNA of the JFH-1 strain by electroporation. However, using this RNA transfection system, the amount of secreted infectious viruses often fluctuate and mutations emerge in HCV genome with multiple passages for an extended period of time (54), which limits its usefulness for antiviral screening and vaccine development.
DNA-based expression systems for HCV replication and virion production have also been examined (5, 15, 21). With DNA-based expression systems, transcriptional expression of functional full-length HCV RNA is controlled by an RNA polymerase II (Pol II) promoter and a self-cleaving ribozyme(s). DNA expression systems using RNA polymerase I (Pol I) have been utilized in reverse genetics approaches to replicate negative-strand RNA viruses, including influenza virus (12, 29), Uukuniemi virus (11), Crimean-Congo hemorrhagic fever virus (10), and Ebola virus (13). Pol I is a cellular enzyme that is abundantly expressed in growing cells and transcribes rRNA lacking both a 5′ cap and a 3′ poly(A) tail. Thus, viral RNA synthesized in cells transfected with Pol I-driven plasmids containing viral genomic cDNA has no additional sequences at the 5′- or 3′ end even in the absence of a ribozyme sequence (28). The advantages of DNA-based expression systems are that DNA expression plasmids are easier to manipulate and generate stable cell lines that constitutively express the viral genome.
We developed here a new HCV expression system based on transfection of an expression plasmid containing a JFH-1 cDNA clone flanked by Pol I promoter and terminator sequences to generate infectious HCV particles from transfected cells. The technology presented here has strong potential to be the basis for trans-encapsidation system by transient transfection of two plasmids and for the establishment of an efficient and reliable screening system for potential antivirals.
MATERIALS AND METHODS
DNA construction.
To generate HCV-expressing plasmids containing full-length JFH1 cDNA embedded between Pol I promoter and terminator sequences, part of the 5′UTR region and part of the NS5B to the 3′UTR region of full-length JFH-1 cDNA were amplified by PCR using primers containing BsmBI sites. Each amplification product was then cloned into a pGEM-T Easy vector (Promega, Madison, WI) and verified by DNA sequencing. Both fragments were excised by digestion with NotI and BsmBI, after which they were cloned into the BsmBI site of the pHH21 vector (a gift from Yoshihiro Kawaoka, School of Veterinary Medicine, University of Wisconsin-Madison [29]), which contains a human Pol I promoter and a mouse Pol I terminator. The resultant plasmid was digested by AgeI and EcoRV and ligated to JFH-1 cDNA digested by AgeI and EcoRV to produce pHHJFH1. pHHJFH1/GND having a point mutation at the GDD motif in NS5B to abolish RNA-dependent RNA polymerase activity and pHHJFH1/R783A/R785A carrying double Arg-to-Ala substitutions in the cytoplasmic loop of p7 were constructed by oligonucleotide-directed mutagenesis. To generate pHHJFH1/ΔE carrying in-frame deletions of parts of the E1 and E2 regions (amino acids [aa] 256 to 567), pHHJFH1 was digested with NcoI and AscI, followed by Klenow enzyme treatment and self-ligation. To generate pHH/SGR-Luc carrying the bicistronic subgenomic HCV reporter replicon and its replication-defective mutant, pHH/SGR-Luc/GND, AgeI-SpeI fragments of pHHJFH1 and pHHJFH1/GND were replaced with an AgeI-SpeI fragment of pSGR-JFH1/Luc (20). In order to construct pCAG/C-NS2 and pCAG/C-p7, PCR-amplified cDNA for C-NS2 and C-p7 regions of the JFH-1 strain were inserted into the EcoRI sites of pCAGGS (30). In order to construct stable cell lines, a DNA fragment containing a Zeocin resistance gene excised from pSV2/Zeo2 (Invitrogen, Carlsbad, CA) was inserted into pHH21 (pHHZeo). Full-length JFH-1 cDNA was then inserted into the BsmBI sites of pHHZeo. The resultant construct was designated pHHJFH1/Zeo.
Cells and compounds.
The human hepatoma cell line, Huh-7, and its derivative cell line, Huh7.5.1 (a gift from Francis V. Chisari, The Scripps Research Institute), were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with nonessential amino acids, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 incubator. N-Nonyl-deoxynojirimycin (NN-DNJ) and kifunensine (KIF) were purchased from Toronto Research Chemicals (Ontario, Canada), castanospermine (CST) and 1,4-dideoxy-1,4-imino-d-mannitol hydrochloride (DIM) were from Sigma-Aldrich (St. Louis, MO), 1-deoxymannojirimycin (DMJ) and swainsonine (SWN) were from Alexis Corp. (Lausen, Switzerland), and N-butyl-deoxynojirimycin (NB-DNJ) was purchased from Wako Chemicals (Osaka, Japan). BILN 2061 was a gift from Boehringer Ingelheim (Canada), Ltd. These compounds were dissolved in dimethyl sulfoxide and used for the experiments. IFN-α was purchased from Dainippon-Sumitomo (Osaka, Japan).
DNA transfection and selection of stable cell lines.
DNA transfection was performed by using FuGENE 6 transfection reagent (Roche, Mannheim, German) in accordance with the manufacturer's instructions. To establish stable cell lines constitutively producing HCV particles, pHHJFH1/Zeo was transfected into Huh7.5.1 cells within 35-mm dishes. At 24 h posttransfection (p.t.), the cells were then divided into 100-mm dishes at various cell densities and incubated with DMEM containing 0.4 mg of zeocin/ml for approximately 3 weeks. Selected cell colonies were picked up and amplified. The expression of HCV proteins was confirmed by measuring secreted core proteins. The stable cell line established was designated H751JFH1/Zeo.
In vitro synthesis of HCV RNA and RNA transfection.
RNA synthesis and transfection were performed as previously described (26, 49).
RNA preparation, Northern blotting, and RNase protection assay (RPA).
Total cellular RNA was extracted with a TRIzol reagent (Invitrogen), and HCV RNA was isolated from filtered culture supernatant by using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). Extracted cellular RNA was treated with DNase (TURBO DNase; Ambion, Austin, TX) and cleaned up by using an RNeasy minikit, which includes another step of RNase-free DNase digestion (Qiagen). The cellular RNA (4 μg) was separated on 1% agarose gels containing formaldehyde and transferred to a positively charged nylon membrane (GE Healthcare, Piscataway, NJ). After drying and cross-linking by UV irradiation, hybridization was performed with [α-32P]dCTP-labeled DNA using Rapid-Hyb buffer (GE Healthcare). The DNA probe was synthesized from full-length JFH-1 cDNA using the Megaprime DNA labeling system (GE Healthcare). Quantification of positive- and negative-strand HCV RNA was performed using the RPA with biotin-16-uridine-5′-triphosphate (UTP)-labeled HCV-specific RNA probes, which contain 265 nucleotides (nt) complementary to the positive-strand (+) 5′UTR and 248 nt complementary to the negative-strand (−) 3′UTR. Human β-actin RNA probes labeled with biotin-16-UTP were used as a control to normalize the amount of total RNA in each sample. The RPA was carried out using an RPA III kit (Ambion) according to the manufacturer's procedures. Briefly, 15 μg of total cellular RNA was used for hybridization with 0.3 ng of the β-actin probe and 0.6 ng of either the HCV (+) 5′UTR or (−) 3′UTR RNA probe. After digestion with RNase A/T1, the RNA products were analyzed by electrophoresis in a 6% polyacrylamide-8 M urea gel and visualized by using a chemiluminescent nucleic acid detection module (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions.
Reverse transcriptase PCR (RT-PCR), sequencing, and rapid amplification of cDNA ends (RACE).
Aliquots (5 μl) of RNA solution extracted from filtered culture supernatant were subjected to reverse transcription with random hexamer and Superscript II reverse transcriptase (Invitrogen). Four fragments of HCV cDNA (nt 129 to 2367, nt 2285 to 4665, nt 4574 to 7002, and nt 6949 to 9634), which covers most of the HCV genome, were amplified by nested PCR. Portions (1 or 2 μl) of each cDNA sample were subjected to PCR with TaKaRa LA Taq polymerase (Takara, Shiga, Japan). The PCR conditions consisted of an initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 3 min. The amplified products were separated by agarose gel electrophoresis and used for direct DNA sequencing. To establish the 5′ ends of the HCV transcripts from pHHJFH1, a synthetic 45-nt RNA adapter (Table 1) was ligated to RNA extracted from the transfected cells 1 day p.t. using T4 RNA ligase (Takara). The viral RNA sequences were then reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) with a primer, RT (Table 1). The resultant cDNA sequences were subsequently amplified by PCR with 5′RACEouter-S and 5′RACEouter-R primers, followed by a second cycle of PCR using 5′RACEinner-S and 5′RACEinner-R primers (Table 1). To establish the terminal 3′-end sequences, extracted RNA sequences were polyadenylated using a poly(A) polymerase (Takara), reverse transcribed with CAC-T35 primer (Table 1), and amplified with the primers 3X-10S (Table 1) and CAC-T35. The amplified 5′ and 3′ cDNA sequences were then separated by agarose gel electrophoresis, cloned into the pGEM-T Easy vector (Promega), and sequenced.
TABLE 1.
Oligonucleotides used for RT-PCR and RACE of the JFH-1 genome
| Method or segment | Oligonucleotide | Sequences (5′-3′) |
|---|---|---|
| 5′RACE | RT | GTACCCCATGAGGTCGGCAAAG |
| 45-nt RNA adapter | GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA | |
| 5′RACEouter-S | GCTGATGGCGATGAATGAACACTG | |
| 5′RACEouter-R | GACCGCTCCGAAGTTTTCCTTG | |
| 5′RACEinner-S | GAACACTGCGTTTGCTGGCTTTGATG | |
| 5′RACEinner-R | CGCCCTATCAGGCAGTACCACAAG | |
| 3′RACE | CAC-T35 | CACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| 3X-10S | ATCTTAGCCCTAGTCACGGC | |
| nt 129-2367 | 44S (1st PCR) | CTGTGAGGAACTACTGTCTT |
| 2445R | TCCACGATGTTCTGGTGAAG | |
| 17S (2nd PCR) | CGGGAGAGCCATAGTGG | |
| 2367R | CATTCCGTGGTAGAGTGCA | |
| nt 2285-4665 | 2099S (1st PCR) | ACGGACTGTTTTAGGAAGCA |
| 4706R | TTGCAGTCGATCACGGAGTC | |
| 2285S (2nd PCR) | AACTTCACTCGTGGGGATCG | |
| 4665R | TCGGTGGCGACGACCAC | |
| nt 4574-7002 | 4547S (1st PCR) | AAGTGTGACGAGCTCGCGG |
| 7027R | CATGAACAGGTTGGCATCCACCAT | |
| 4594S (2nd PCR) | CGGGGTATGGGCTTGAACGC | |
| 7003R | GTGGTGCAGGTGGCTCGCA | |
| nt 6949-9634 | 6881S (1st PCR) | ATTGATGTCCATGCTAACAG |
| 3X-75R | TACGGCACTCTCTGCAGTCA | |
| 6950S (2nd PCR) | GAGCTCCTCAGTGAGCCAG | |
| 3X-54R | GCGGCTCACGGACCTTTCAC |
Western blotting.
The proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA) after separation by SDS-PAGE. After blocking, the membranes were probed with a mouse monoclonal anti-HCV core antibody (2H9) (49), a rabbit polyclonal anti-NS5B antibody, or a mouse monoclonal GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Chemicon, Temecula, CA), followed by incubation with a peroxidase-conjugated secondary antibody and visualization with an ECL Plus Western blotting detection system (Amersham, Buckinghamshire, United Kingdom).
Quantification of HCV core protein.
HCV core protein was quantified by using a highly sensitive enzyme immunoassay (Ortho HCV antigen ELISA kit; Ortho Clinical Diagnostics, Tokyo, Japan) in accordance with the manufacturer's instructions.
Sucrose density gradient analysis.
Samples of cell culture supernatant were processed by low-speed centrifugation and passage through a 0.45-μm-pore-size filter. The filtrated supernatant was then concentrated ∼30-fold by ultrafiltration by using an Amicon Ultra-15 filter device with a cutoff molecular mass of 100,000 kDa (Millipore), after which it was layered on top of a continuous 10 to 60% (wt/vol) sucrose gradient, followed by centrifugation at 35,000 rpm at 4°C for 14 h with an SW41 rotor (Beckman Coulter, Fullerton, CA). Fractions of 1 ml were collected from the bottom of the gradient. The core level and infectivity of HCV in each fraction were determined.
Quantification of HCV infectivity.
Infectious virus titration was performed by a 50% tissue culture infectious dose (TCID50) assay, as previously described (23, 26). Briefly, naive Huh7.5.1 cells were seeded at a density of 104 cells/well in a 96-well flat-bottom plate 24 h prior to infection. Five serial dilutions were performed, and the samples were used to infect the seeded cells (six wells per dilution). At 72 h after infection, the inoculated cells were fixed and immunostained with a rabbit polyclonal anti-NS5A antibody (14), followed by an Alexa Fluor 488-conjugated anti-rabbit secondary antibody (Invitrogen).
Labeling of de novo-synthesized viral RNA and immunofluorescence staining.
Labeling of de novo-synthesized viral RNA was performed as previously described with some modifications (40). Briefly, cells were plated onto an eight-well chamber slide at a density of 5 × 104 cells/well. One day later, the cells were incubated with actinomycin D at a final concentration of 10 μg/ml for 1 h and washed twice with HEPES-saline buffer. Bromouridine triphosphate (BrUTP) at 2 mM was subsequently transfected into the cells using FuGENE 6 transfection reagent, after which the cells were incubated for 15 min on ice. After the cells were washed twice with phosphate-buffered saline (PBS), they were incubated in fresh DMEM supplemented with 10% FBS at 37°C for 4 h. The cells were then fixed with 4% paraformaldehyde for 20 min and permeabilized with PBS containing 0.1% Triton X-100 for 15 min at room temperature. Immunofluorescence staining of NS5A and de novo-synthesized HCV RNA was performed as previously described (26, 40). The nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) solution (Sigma-Aldrich). Confocal microscopy was performed using a Zeiss confocal laser scanning microscope LSM 510 (Carl Zeiss, Oberkochen, Germany).
Luciferase assay.
Huh7.5.1 cells were seeded onto a 24-well cell culture plate at a density of 3 × 104 cells/well 24 h prior to inoculation with 100 μl of supernatant from the transfected cells. The cells were incubated for 72 h, followed by lysis with 100 μl of lysis buffer. The luciferase activity of the cells was determined by using a luciferase assay system (Promega). All luciferase assays were done at least in triplicate. For the neutralization experiments, a mouse monoclonal anti-CD81 antibody (JS-81; BD Pharmingen, Franklin Lakes, NJ) and a mouse monoclonal anti-FLAG antibody (Sigma-Aldrich) were used.
Flow cytometric analysis.
Cells detached by treatment with trypsin were incubated in PBS containing 1% (vol/vol) formaldehyde for 15 min. A total of 5 × 105 cells were resuspended in PBS and treated with or without 0.75 μg of anti-CD81 antibody for 30 min at 4°C. After being washed with PBS, the cells were incubated with an Alexa Fluor 488-conjugated anti-mouse secondary antibody (Invitrogen) at 1:200 for 30 min at 4°C, washed repeatedly, and resuspended in PBS. Analyses were performed by using FACSCalibur system (Becton Dickinson, Franklin Lakes, NJ).
RESULTS
Analysis of the 5′ and 3′ ends of HCV RNA sequences generated from Pol I-driven plasmids.
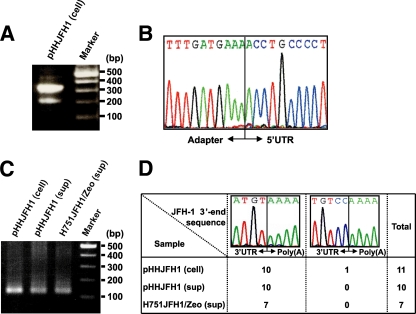
To examine whether the HCV transcripts generated from Pol I-driven plasmids had correct nucleotides at the 5′ and 3′ ends, we extracted RNA from Huh-7 cells transfected with pHHJFH1, which carries a genome-length HCV cDNA with a Pol I promoter/terminator, as well as from the culture supernatants. After this, the nucleotide sequences at both ends were determined using RACE and sequence analysis. A 328-nt fragment corresponding to cDNA from the 5′ end of HCV RNA was detected in the cell samples (Fig. 1A). Cloning of amplified fragments confirmed that the HCV transcripts were initiated from the first position of the viral genome in all of the clones sequenced (Fig. 1B). Similarly, a 127-nt amplification fragment was detected in each sample by 3′RACE (Fig. 1C), and the same 3′-end nucleotide sequence was observed in all clones derived from the culture supernatant (Fig. 1D, left). An additional two nucleotides (CC) were found at the 3′ end of the HCV transcript in a limited number of sequences (1 of 11 clones) derived from the cell sample (Fig. 1D, right), which were possibly derived from the Pol I terminator sequence by incorrect termination. These results indicate that most HCV transcripts generated from the Pol I-based HCV cDNA expression system are faithfully processed, although it is not determined whether the 5′ terminus of the viral RNA generated from Pol I system is triphosphate or monophosphate. It can be speculated that viral RNA lacking modifications at the 5′ and 3′ ends is preferentially packaged and secreted into the culture supernatant.
FIG. 1.
Determination of the nucleotide sequences at the 5′-and 3′ ends of HCV RNA produced by the Pol I system. (A and B) 5′RACE and sequence analysis. A synthesized RNA adapter was ligated to RNA extracted from cells transfected with pHHJFH1. The positive-strand HCV RNA was reverse transcribed, and the resulting cDNA was amplified by nested PCR. The amplified 5′-end cDNA was separated by agarose gel electrophoresis (A), cloned, and sequenced (B). (C and D) 3′RACE and sequence analysis. RNA extracted from pHHJFH1-transfected cells, the culture supernatant of transfected cells, and the culture supernatant of H751JFH1/Zeo cells were polyadenylated, reverse transcribed, and amplified by PCR. The amplified 3′-end cDNA was separated by agarose gel electrophoresis (C), cloned, and sequenced (D).
Production of HCV RNA, proteins, and virions from cells transiently transfected with Pol I-driven plasmids.
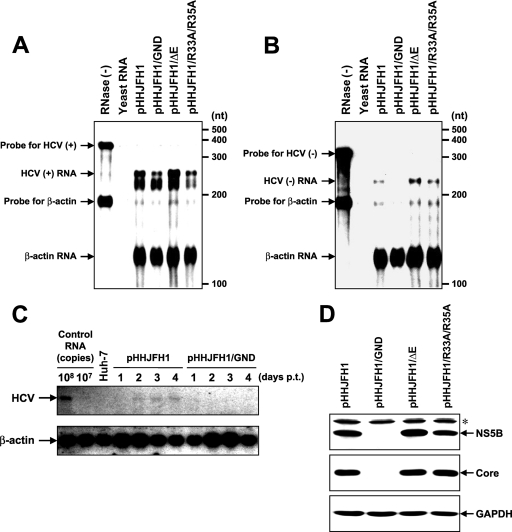
To examine HCV RNA replication and protein expression in cells transfected with pHHJFH1, pHHJFH1/GND, or virion production-defective mutants, pHHJFH1/ΔE and pHHJFH1/R783A/R785A, which possess an in-frame deletion of E1/E2 region and substitutions in the p7 region, respectively (19, 42, 49), RPA and Western blotting were performed 5 days p.t. (Fig. 2A, B, and D). Positive-strand HCV RNA sequences were more abundant than negative-strand RNA sequences in these cells. Positive-strand RNA, but not negative-strand RNA, was detected in cells transfected with the replication-defective mutant pHHJFH1/GND (Fig. 2A and B). Northern blotting showed that genome-length RNA was generated in pHHJFH1-transfected cells but not in pHHJFH1/GND-transfected cells (Fig. 2C). As shown in Fig. 2D, the intracellular expression of core and NS5B proteins was comparable among cells transfected with pHHJFH1, pHHJFH1/ΔE, and pHHJFH1/R783A/R785A. Neither viral protein was detected in pHHJFH1/GND-transfected cells, suggesting that the level of viral RNA generated transiently from the DNA plasmid does not produce enough HCV proteins for detection and that ongoing amplification of the HCV RNA by the HCV NS5B polymerase allows a high enough level of viral RNA to produce detectable levels of HCV proteins.
FIG. 2.
HCV RNA replication and protein expression in cells transfected with Pol I-driven plasmids. (A and B) Assessment of HCV RNA replication by RPA. Pol I-driven HCV-expression plasmids were transfected into Huh-7 cells. Total RNA was extracted from the cells on day 5 p.t. and positive (A)- and negative (B)-strand HCV RNA levels were determined by RPA as described in Materials and Methods. In the RNase (−) lanes, yeast RNA mixed with RNA probes for HCV and human β-actin were loaded without RNase A/T1 treatment. In the yeast RNA lanes, yeast RNA mixed with RNA probes for HCV and human β-actin were loaded in the presence of RNase A/T1. (C) Northern blotting of total RNAs prepared from the transfected cells. Huh-7 cells transfected with pHHJFH1 or pHHJFH1/GND were harvested for RNA extraction through days 1 to 4 p.t. Control RNA, given numbers of synthetic HCV RNA; Huh-7, RNA extracted from naive cells. Arrows indicate full-length HCV RNA and β-actin RNA. (D) HCV protein expression in the transfected cells. Pol I-driven HCV-expression plasmids were transfected into Huh-7 cells, harvested, and lysed on day 6 p.t. The expression of NS5B, core, and GAPDH was analyzed by Western blotting as described in Materials and Methods. The asterisk indicates nonspecific bands.
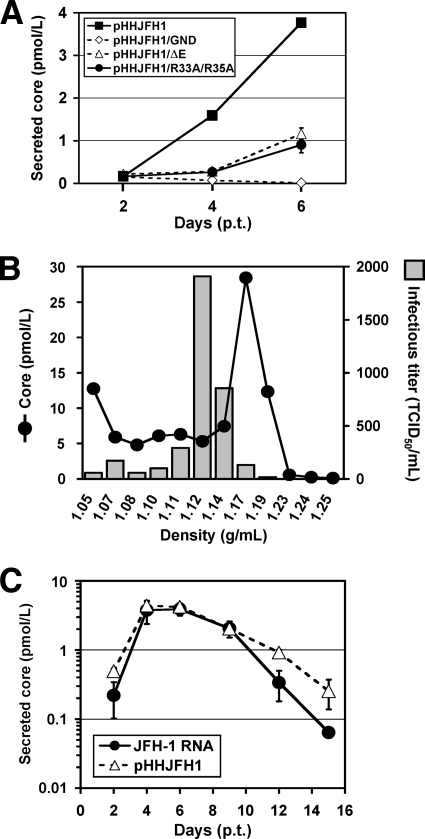
To assess the release of HCV particles from cells transfected with Pol I-driven plasmids, core protein was quantified in culture supernatant by enzyme-linked immunosorbent assay (ELISA) or sucrose density gradient centrifugation. Core protein secreted from pHHJFH1-transfected cells was first detectable 2 days p.t., with levels increasing up to ∼4 pmol/liter on day 6 (Fig. 3A). This core protein level was 4- to 6-fold higher than that in the culture supernatant of pHHJFH1/ΔE- or pHHJFH1/R783A/R785A-transfected cells, despite comparable intracellular core protein levels (Fig. 2D). Core protein was not secreted from cells transfected with pHHJFH1/GND (Fig. 3A). In another experiment, a plasmid expressing the secreted form of human placental alkaline phosphatase (SEAP) was cotransfected with each Pol I-driven plasmid. SEAP activity in culture supernatant was similar among all transfection groups, indicating comparable efficiencies of transfection (data not shown). Sucrose density gradient analysis of the concentrated supernatant of pHHJFH1-transfected cells indicated that the distribution of core protein levels peaked in the fraction of 1.17 g/ml density, while the peak of infectious titer was observed in the fraction of 1.12 g/ml density (Fig. 3B), which is consistent with the results of previous studies based on JFH-1-RNA transfection (23).
FIG. 3.
HCV released from cells transfected with Pol I-driven plasmids. (A) HCV particle secretion from the transfected cells. The culture supernatant of Huh-7 cells transfected with Pol I-driven plasmids containing wild-type or mutated HCV genome were harvested on days 2, 4, and 6 and assayed for HCV core protein levels. The data for each experiment are averages of triplicate values with error bars showing standard deviations. (B) Sucrose density gradient analysis of the culture supernatant of pHHJFH1-transfected cells. Culture supernatant collected on day 5 p.t. was cleared by low-speed centrifugation, passed through a 0.45-μm-pore-size filter, and concentrated ∼30-fold by ultrafiltration. After fractionating by sucrose density gradient centrifugation, the core protein level and viral infectious titer of each fraction were measured. (C) Kinetics of core protein secretion from cells transfected with pHHJFH1 or with JFH-1 genomic RNA. A total of 106 Huh-7 cells were transfected with 3 μg of pHHJFH1 or the same amount of in vitro-transcribed JFH-1 RNA by electroporation. The cells were passaged every 2 to 3 days before reaching confluence. Culture supernatant collected on the indicated days was used for core protein measurement. The level of secreted core protein (pmol/liter) is expressed on a logarithmic scale. The data for each experiment are averages of triplicate values with error bars showing standard deviations.
We next compared the kinetics of HCV particle secretion in the Pol I-driven system and RNA transfection system. Huh-7 cells, which have limited permissiveness for HCV infection (2), were transfected with either pHHJFH1 or JFH-1 RNA, and then cultured by passaging every 2 or 3 days. As shown in Fig. 3C, both methods of transfection demonstrated similar kinetics of core protein levels until 9 days p.t., after which levels gradually fell. However, significantly greater levels of core protein were detected in the culture of pHHJFH1-transfected cells compared to the RNA-transfected cells on day 12 and 15 p.t. This is likely due to an ongoing production of positive-strand viral RNA from transfected plasmids since RNA degradation generally occurs more quickly than that of circular DNA.
Establishment of stable cell lines constitutively producing HCV virion.
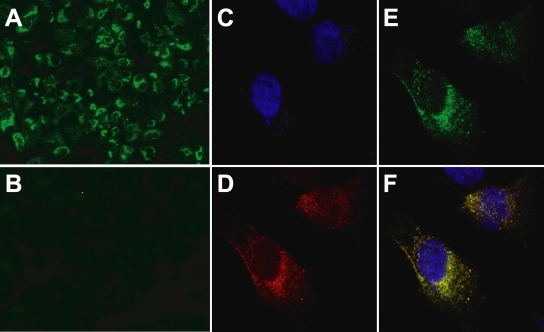
To establish cell lines with constitutive HCV production, pHHJFH1/Zeo carrying HCV genomic cDNA and the Zeocin resistance gene were transfected into Huh7.5.1 cells. After approximately 3 weeks of culture with zeocin at a concentration of 0.4 mg/ml, cell colonies producing HCV core protein were screened by ELISA, and three clones were identified that constitutively produced the viral protein (H751JFH1/Zeo cells). Core protein levels within the culture supernatant of selected clones (H751-1, H751-6, and H751-50) were 2.0 × 104, 2.7 × 103, and 1.4 × 103 fmol/liter, respectively. Clone H751-1 was further analyzed. Indirect immunofluorescence with an anti-NS5A antibody showed fluorescent staining of NS5A in the cytoplasm of almost all H751JFH1/Zeo cells (Fig. 4A), whereas no signal was detected in parental Huh7.5.1 cells (Fig. 4B). To determine where HCV RNA replicates in H751JFH1/Zeo cells, labeling of de novo-synthesized HCV RNA was performed. After interfering with mRNA production by exposure to actinomycin D, BrUTP-incorporated de novo-synthesized HCV RNA was detected in the cytoplasm of H751JFH1/Zeo cells (Fig. 4D) colocalized with NS5A in the perinuclear area (Fig. 4E and F).
FIG. 4.
Indirect immunofluorescence analysis of H751JFH1/Zeo cells. (A and B) H751JFH1/Zeo cells (A) and parental Huh7.5.1 cells (B) were immunostained with an anti-NS5A antibody. (C to F) The subcellular colocalization of de novo-synthesized HCV RNA and NS5A in H751JFH1/Zeo cells was analyzed. The cells were stained with DAPI (C), an anti-bromodeoxyuridine antibody (D), and an anti-NS5A antibody (E). The merge panel is shown in panel F.
Low mutation frequency of the viral genome in a long-term culture of H751JFH1/Zeo cells.
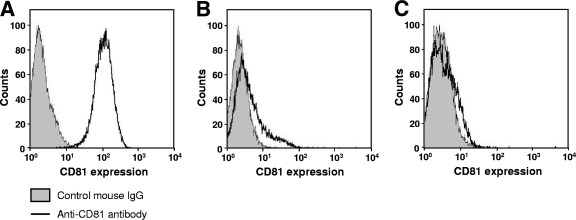
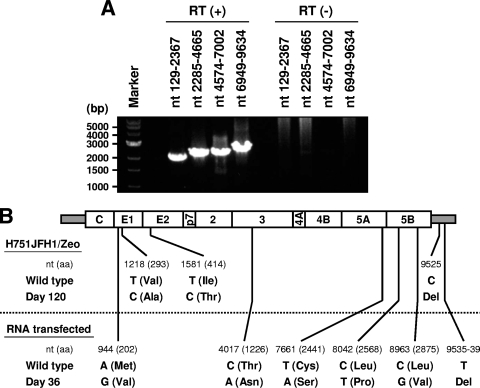
The production level of infectious HCV from H751JFH1/Zeo cells at a concentration of ∼103 TCID50/ml was maintained over 1 year of culture (data not shown). It has been shown that both virus and host cells may adapt during persistent HCV infection in cell cultures, such that cells become resistant to infection due to reduced expression of the viral coreceptor CD81 (54). As shown in Fig. 5, we analyzed the cell surface expression of CD81 on the established cell lines by flow cytometry and observed markedly reduced expression on H751JFH1/Zeo cells compared to parental Huh7.5.1 cells. It is therefore possible that only a small proportion of HCV particles generated from H751JFH1/Zeo cells enter and propagate within the cells. The H751JFH1/Zeo system is thought to result in virtually a single cycle of HCV production from the chromosomally integrated gene and thus may yield a virus population with low mutation frequencies. To further examine this, we compared HCV genome mutation rates following production from H751JFH1/Zeo cells compared to cells constitutively infected with HCV after serial passages. RNAs were extracted from the supernatant of H751JFH1/Zeo cells cultured for 120 days, and cDNA sequences were amplified by nested PCR with four sets of primers encompassing almost the entire HCV genome (Table 1). PCR products with expected sizes of 2 to 2.5 kb were obtained [Fig. 6A, RT(+)] and subjected to direct sequencing. No amplified product was detected in samples without reverse transcription [Fig. 6A, RT(−)], suggesting no DNA contamination in culture supernatants or extracted RNA solutions. As shown in Fig. 5B (upper panel), three nucleotide mutations, including two substitutions in the E1 (nt 1218) and E2 (nt 1581) regions, and one deletion in the 3′ UTR (nt 9525) were found within the HCV genome with the mutation rate calculated at 9.6 × 10−4 base substitutions/site/year. These mutations were not detected in the chromosomally integrated HCV cDNA (data not shown). The present results also indicate that no splicing of the viral RNA occurred in the Pol I-based HCV JFH-1 expression system. The HCV genome sequence produced by JFH-1 virus-infected Huh7.5.1 cells was analyzed in the same way using culture supernatant 36 days after RNA transfection. As shown in Fig. 6B (lower panel), 10 mutations, including five substitutions throughout the open reading frame and five deletions in the 3′UTR, were detected, and the mutation rate was calculated at 1.1 × 10−2 base substitutions/site/year.
FIG. 5.
Loss of CD81 expression in H751JFH1/Zeo cells. The cell surface expression of CD81 on Huh7.5.1 cells (A), H751JFH1/Zeo clone H751-1 (B), and clone H751-50 (C) was analyzed by flow cytometry after being stained with anti-CD81 antibody.
FIG. 6.
Genome mutations of HCV secreted from H751JFH1/Zeo cells. (A) RT-PCR of HCV genome extracted from the culture supernatant of H751JFH1/Zeo cells. Viral RNA sequences were reverse transcribed [RT (+)] or not [RT (−)], followed by amplification with primer pairs encompassing the specified HCV genome regions. (B) Comparison of the genome mutations of HCV secreted from H751JFH1/Zeo cells cultured for 120 days (upper panel) and JFH-1 RNA-transfected cells cultured for 36 days (lower panel). The positions of original (wild-type) and mutated (day 120, day 36) nucleotides are indicated under the schematic diagram of the HCV genome. Amino acid residues and their positions are marked in parentheses. Del, deletion.
Effects of glycosylation inhibitors on HCV production.
It is known that N-linked glycosylation and oligosaccharide trimming of a variety of viral envelope proteins including HCV E1 and E2 play key roles in the viral maturation and virion production. To evaluate the usefulness of the established cell line for antiviral testing, we determined the effects of glycosylation inhibitors, which have little to no cytotoxicity at the concentrations used, on HCV production in a three day assay using H751JFH1/Zeo cells. The compounds tested are known to inhibit the endoplasmic reticulum (ER), Golgi-resident glucosidases, or mannosidases that trim glucose or mannose residues from N-linked glycans. Some are reported to be involved in proteasome-dependent or -independent degradation of misfolded or unassembled glycoproteins to maintain protein integrity (4, 8, 27, 35).
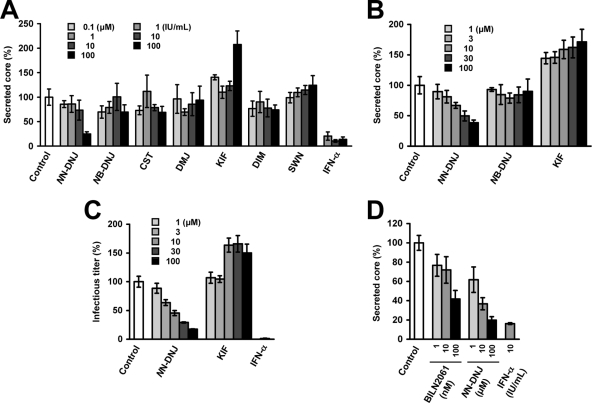
As shown in Fig. 7A and B, treatment of H751JFH1/Zeo cells with increasing concentrations of NN-DNJ, which is an inhibitor of ER α-glucosidases, resulted in a dose-dependent reduction in secreted core protein. NN-DNJ was observed to have an IC50 (i.e., the concentration inhibiting 50% of core protein secretion) of ∼20 μM. In contrast, KIF, which is an ER α-mannosidase inhibitor, resulted in a 1.5- to 2-fold increase in secreted core protein compared to control levels. The other five compounds did not significantly change core protein levels. We further determined the effects of NN-DNJ and KIF on the production of infectious HCV (Fig. 7C). As expected, NN-DNJ reduced the production of infectious virus in a dose-dependent manner, while production increased in the presence of KIF at 10 to 100 μM. Since NN-DNJ and KIF did not significantly influence viral RNA replication, as determined using the subgenomic replicon (data not shown), the present results suggest that some step(s), such as virion assembly, intracellular trafficking, and secretion, may be up- or downregulated depending on glycan modifications of HCV envelope proteins within the ER. Inhibitory effect of NN-DNJ was reproducibly observed using the cell line after 1 year of culturing (Fig. 7D). Under the same condition, the core protein secretion was inhibited by 28 and 58% with 10 and 100 nM BILN 2061, an NS3 protease inhibitor, respectively (Fig. 7D).
FIG. 7.
Effects of glycosylation inhibitors on HCV production from H751JFH1/Zeo cells. (A and B) Effects of glycosylation inhibitors on the secretion of HCV core protein. H751JFH1/Zeo cells were seeded at a density of 1 × 104 cells/well in a 96-well culture plate (A) or 3 × 104 cells/well in a 12-well cell culture plate (B). One day later, each compound was added to the cell culture supernatant at the indicated concentrations. The culture supernatant was collected after a further 3-day culture and processed by core protein-specific ELISA. The control represents an untreated cell culture. The level of secreted core protein was normalized by setting the control value at 100%. The data for each experiment are averages of triplicate values with error bars showing standard deviations. (C) Effects of NN-DNJ and KIF on infectious HCV production. The culture supernatant obtained in panel B was used to infect naive Huh7.5.1 cells. At 72 h after infection, the inoculated cells were fixed and immunostained as described in Materials and Methods for titration of virus infectivity. The infectious titer was normalized by setting the control value at 100%. Cells were treated with INF-α at 100 IU/ml as a positive control. The data for each experiment are averages of triplicate values with error bars showing standard deviations. The control represents an untreated cell culture. (D) After 1 year of culturing H751JFH1/Zeo cells, antiviral effects of NN-DNJ and BILN 2061 were evaluated. H751JFH1/Zeo cells were seeded at a density of 3 × 104 cells/well in a 12-well cell culture plate. One day later, each compound was added to the cell culture supernatant at the indicated concentrations. The culture supernatant was collected after a further 3-day culture and processed by core protein-specific ELISA. The control represents an untreated cell culture. The level of secreted core protein was normalized by setting the control value at 100%. The data for each experiment are averages of triplicate values with error bars showing standard deviations.
Replicon trans-packaging system.
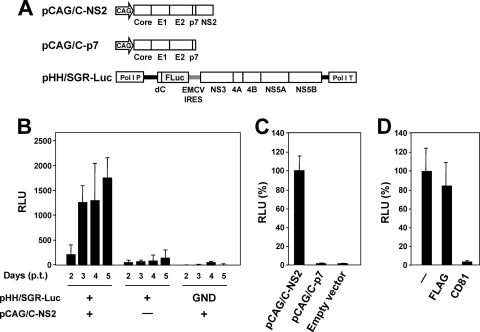
Recently, ourselves and others have developed a packaging system for HCV subgenomic replicon RNA sequences by providing trans viral core-NS2 proteins (1, 17, 41). Since viral structural proteins are not encoded by the subgenomic replicon, progeny virus cannot be produced after transfection. Thus, the single-round infectious HCV-like particle (HCV-LP) generated by this system potentially improves the safety of viral transduction. Here, in order to make the trans-packaging system easier to manipulate, we used a Pol I-driven plasmid to develop a transient two-plasmid expression system for the production of HCV-LP. pHH/SGR-Luc, which carries a bicistronic subgenomic reporter replicon with a Pol I promoter/terminator, or its replication-defective mutant, were cotransfected with or without a core-NS2 expression plasmid (Fig. 8A). The culture supernatant was then collected between days 2 and 5 p.t. and used to inoculate naive Huh7.5.1 cells. Reporter luciferase activity, as a quantitative measure of infectious virus production, was assessed in the cells 3 days postinoculation. As shown in Fig. 8B, reporter replication activity was easily detectable in cells inoculated with culture supernatant from cells cotransfected with pHH/SGR-Luc and pCAG/C-NS2, with an ∼10-fold increase in activity observed at 2 to 5 days p.t. In contrast, luciferase signal in the Huh7.5.1 cells inoculated from supernatant of cells transfected with pHH/SGR-Luc with polymerase-deficient mutation (GND) showed background levels. There was a faint luciferase signal in the cells inoculated from supernatant of cells transfected with pHH/SGR-Luc in the absence of pCAG/C-NS2, suggesting carryover of a low level of cells with the supernatants. Transfer of supernatant from infected cells to naive Huh7.5.1 cells did not result in infection, as judged by undetectable luciferase activity (data not shown). To examine whether NS2 is important for HCV production as previously demonstrated (17-19, 52), we compared the expression of core-NS2 versus core-p7 in the packaged cells (Fig. 8C). The reporter activity in cells inoculated with virus trans-packaged by core-p7 was ∼100-fold lower than the virus trans-packaged by core-NS2, indicating that NS2 needs to be expressed with the structural proteins for efficient assembly and/or infectivity. CD81-dependent infection of HCV-LP was further confirmed by demonstrating reduced reporter activity in the presence of anti-CD81 antibody (Fig. 8D). Thus, we developed a simple trans-encapsidation system based on transient two-plasmid transfection, which permits experimental separation of HCV genome replication and virion assembly.
FIG. 8.
Establishment of a trans-packaging system involving two-plasmid transfection. (A) Schematic representation of the plasmids used for the production of HCV-LP. HCV polyproteins are indicated by the open boxes. Bold lines indicate the HCV UTR. EMCV IRES is denoted by gray bars. The firefly luciferase gene (F Luc) is depicted as a gray box. CAG, CAG promoter; Pol I P, Pol I promoter; dC, 5′ region of Core gene; Pol I T, Pol I terminator. (B) Luciferase activity in Huh7.5.1 cells inoculated with culture supernatant from cells transfected with the indicated plasmids. Luciferase activity is expressed in terms of relative luciferase units (RLU). The data for each experiment are averages of triplicate values with error bars showing standard deviations. (C) Culture supernatant from cells cotransfected with pHH/SGR-Luc and the indicated plasmids were collected 4 days p.t. The luciferase activity in Huh7.5.1 cells inoculated with culture supernatant was determined 3 days postinoculation and expressed as relative luciferase units (RLU). The RLU was normalized according to the luciferase activity observed in the pCAG/C-NS2-transfected sample (C-NS2), which was set at 100%. The data for each experiment are averages of triplicate values with error bars showing standard deviations. (D) Huh7.5.1 cells were inoculated with HCVLP in the absence (−) or presence of 5 μg of anti-CD81 or anti-FLAG antibody/ml. The luciferase activity was determined 72 h postinoculation and is expressed as relative luciferase units (RLU). The RLU was normalized to the level of luciferase activity observed in the antibody-untreated sample (−), which was set at 100%. The data for each experiment are averages of triplicate values with error bars showing standard deviations.
DISCUSSION
Here, we exploited Pol I-derived vectors for expression of the HCV genome, a strategy that generates viral RNAs from the Pol I promoter and terminator. We demonstrated that the HCV JFH-1 RNA produced using this system is unspliced with precise sequences at both ends and that it is replicated in the cytoplasm of transfected cells to produce infectious particles. This approach was used to establish a replicon trans-packaging system based on transient two-plasmid transfection and enables the production of a stable cell line capable of constitutive HCV production. The cell line produced using this method can be used to screen a large number of potential antiviral agents by assessing their ability to interfere with HCV replication and/or virion formation. The Pol I-mediated transcription system was originally developed to perform reverse genetics on influenza A viruses (12, 29) which replicate in the nucleus. This system has also been shown useful in the development of reverse genetics for negative-strand RNA viruses having a cytoplasmic replication cycle (3, 10, 11, 31). The results of the present study suggest that the Pol I system can also be used to perform reverse genetics on a cytoplasmically replicating positive-strand RNA virus.
Although viral RNA transfection by electroporation is the most commonly used method to perform reverse genetics on HCV (23, 49, 53), it is comparatively difficult to manipulate. RNA electroporation requires high-quality in vitro-synthesized RNA and a large quantity of exponential-growth-phase cells, which may be hard to provide when a number of different RNA constructs are being examined in the same experiment. In addition to the Pol I system, other DNA expression systems have been examined with regard to HCV particle production (5, 15, 21). These systems require ribozyme sequences to be inserted at either end of the HCV genomic cDNA sequence in order to generate appropriately processed viral RNA. However, Heller et al. have reported that the HCV RNA generated by in vitro transcription of a HCV-ribozyme plasmid contains uncleaved or prematurely terminated forms of HCV RNA. These authors have also demonstrated that HCV RNA from the culture supernatant of HCV-ribozyme plasmid-transfected cells possesses nucleotide changes at the 5′ and 3′ ends (15), suggesting that the ribozyme is less reliable at generating correct transcripts compared to our Pol I system. In fact, there is evidence to suggest that a mouse Pol I terminator is significantly more effective than an HDV ribozyme in generating precise 3′ ends of RNA, as demonstrated in a plasmid-based influenza virus rescue system (9). Recently, it has been demonstrated that Pol I-catalyzed rRNA transcription is activated in Huh-7 cells following infection with JFH-1 or transfection with a subgenomic HCV replicon (34). HCV NS5A has been shown to upregulate the transcription of Pol I, but not Pol II, through phosphorylation of an upstream binding factor, a Pol I DNA binding transcription factor. These observations indicate that a Pol I-mediated expression system is suitable for efficient production of infectious HCV by DNA transfection.
We established a stable cell line, H751JFH1/Zeo, that constitutively and efficiently produced infectious HCV particles by introducing a Pol I-driven plasmid containing a selection marker into Huh7.5.1 cells. Interestingly, the established cell clones exhibited little to no surface expression of CD81, one of the key features of HCV glycoprotein-mediated infection (Fig. 5). Defective expression of receptor molecules might be advantageous in generating stable cell lines for robust production of HCV. HCV-induced cytotoxicity has been reported (7, 45, 54). Persistent HCV infection was established after electroporation of JFH-1 genomic RNA, and a variable cytopathic effect was observed at the peak of acute HCV infection, as well as during the persistent phase of infection (54). A recent study has demonstrated that the cytopathic effect triggered by HCV RNA transfection and viral infection is characterized by massive apoptotic cell death with expression of several ER stress markers, such as GRP78 and phosphorylated eIF2-α (39). Therefore, in the present study, it is likely that selective forces to evade cell death during high levels of HCV replication produced cell populations resistant to virus infection. As a consequence, H751JFH1/Zeo cells maintained robust production of infectious HCV particles over a long period of time without gross cytopathic effects or changes in cell morphology.
Substantial evidence demonstrates that the mutation rate of the HCV genome produced in H751JFH1/Zeo was low (Fig. 6) presumably because of consistent expression of wild-type HCV RNA from the chromosomally integrated gene. Nevertheless, a considerable proportion of the genome was mutated, with two nonsynonymous mutations in the E1 (V293A) and E2 (I414T) regions identified in the culture supernatant of H751JFH1/Zeo cells after 4 months of passages (Fig. 6). A I414T mutation has also been reported after long-term propagation of HCV in culture after JFH-1-RNA transfection (54). This mutation is located between the hypervariable regions 1 and 2 within the N terminus of E2 (51). Adaptive mutations in this region have been shown to enhance virus expansion, presumably by enabling more efficient virus entry (6, 36, 54). A possible CD81-independent mechanism for cell-to-cell transmission of HCV has been proposed (48, 50). However, the mechanisms governing cell-to-cell spread of HCV are not well understood. Further investigation into the importance of envelope protein mutations in HCV transmission independent of CD81 provide a better understanding of the complex interactions required for HCV infection.
In the present study we assessed the effects of N-linked glycosylation inhibitors on HCV production using H751JFH1/Zeo (Fig. 7) and found that an α-glucosidase inhibitor NN-DNJ inhibits the production of infectious HCV, which has also been observed in previous studies (43, 47). In contrast, HCV production is increased in the presence of an ER α-mannosidase inhibitor KIF, but not in the presence of the Golgi α-mannosidase inhibitors DMJ, DIM, and SWN. KIF inhibits α-mannosidase I, which primarily functions to remove the middle mannose branch from Man9GlcNAc2 to form Man8GlcNAc2 after the removal of glucose residues by glucosidases I and II (8, 24). Experiments to elucidate the role of mannose trimming of N-glycans in the HCV life cycle are currently under way.
It has recently been demonstrated that subgenomic replicons or defective genomes of HCV that have the potential of translation and self-replication can be encapsidated into infectious viruslike particles by trans-complementation of the viral structural proteins (1, 17, 32, 41, 44). In these studies, the viral RNAs were generally generated by in vitro transcription from linearized corresponding plasmids, followed by electroporation into the cells. Structural proteins or Core to NS2 proteins were then provided by DNA or RNA transfection, viral-vector-based transduction, or stable packaging cell lines established. Here, we achieved the replicon trans-encapsidation via transient cotransfection with two DNA plasmids. This system, which is apparently easier to manipulate and allows production of trans-encapsidated materials more rapidly compared to the systems published, can be applied to the study for understanding phenomenon and biological significance of a variety of naturally occurring HCV subgenomic deletion variants that possibly circulate in hepatitis C patients.
In summary, we have established a Pol I-based reverse-genetics system for the efficient production of infectious HCV. This methodology can be applied to develop (i) a stable HCV-producing cell line with a low mutation frequency of the viral genome and (ii) a simple trans-encapsidation system with the flexibility of genome packaging and improved biosafety. This may be useful for antiviral screening and may assist in the development of a live-attenuated HCV vaccine.
Acknowledgments
We are grateful to Francis V. Chisari (The Scripps Research Institute) for providing Huh7.5.1 cells and to Y. Kawaoka (School of Veterinary Medicine, University of Wisconsin-Madison) for providing the pHH21 vector. We thank A. Murayama and T. Date for their help in sequence and Northern blot analyses and our coworkers for their helpful discussions. We also thank S. Yoshizaki, T. Shimoji, M. Kaga, and M. Sasaki for their technical assistance and T. Mizoguchi for secretarial work.
This study was supported by grants-in-aid from the Ministry of Health, Labor, and Welfare; by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion, and Product Review of Japan (01-3); and by Research on Health Sciences focusing on Drug Innovation from the Japan Health Sciences Foundation, Japan.
Footnotes
Published ahead of print on 17 March 2010.
REFERENCES
- 1.Adair, R., A. H. Patel, L. Corless, S. Griffin, D. J. Rowlands, and C. J. McCormick. 2009. Expression of hepatitis C virus (HCV) structural proteins in trans facilitates encapsidation and transmission of HCV subgenomic RNA. J. Gen. Virol. 90:833-842. [DOI] [PubMed] [Google Scholar]
- 2.Akazawa, D., T. Date, K. Morikawa, A. Murayama, M. Miyamoto, M. Kaga, H. Barth, T. F. Baumert, J. Dubuisson, and T. Wakita. 2007. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 81:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billecocq, A., N. Gauliard, N. Le May, R. M. Elliott, R. Flick, and M. Bouloy. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabral, C. M., P. Choudhury, Y. Liu, and R. N. Sifers. 2000. Processing by endoplasmic reticulum mannosidases partitions a secretion-impaired glycoprotein into distinct disposal pathways. J. Biol. Chem. 275:25015-25022. [DOI] [PubMed] [Google Scholar]
- 5.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgrange, D., A. Pillez, S. Castelain, L. Cocquerel, Y. Rouille, J. Dubuisson, T. Wakita, G. Duverlie, and C. Wychowski. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495-2503. [DOI] [PubMed] [Google Scholar]
- 7.Deng, L., T. Adachi, K. Kitayama, Y. Bungyoku, S. Kitazawa, S. Ishido, I. Shoji, and H. Hotta. 2008. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J. Virol. 82:10375-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science 286:1882-1888. [DOI] [PubMed] [Google Scholar]
- 9.Feng, L., F. Li, X. Zheng, W. Pan, K. Zhou, Y. Liu, H. He, and L. Chen. 2009. The mouse Pol I terminator is more efficient than the hepatitis delta virus ribozyme in generating influenza-virus-like RNAs with precise 3′ ends in a plasmid-only-based virus rescue system. Arch. Virol. 154:1151-1156. [DOI] [PubMed] [Google Scholar]
- 10.Flick, R., K. Flick, H. Feldmann, and F. Elgh. 2003. Reverse genetics for Crimean-Congo hemorrhagic fever virus. J. Virol. 77:5997-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flick, R., and R. F. Pettersson. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J. Virol. 75:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groseth, A., H. Feldmann, S. Theriault, G. Mehmetoglu, and R. Flick. 2005. RNA polymerase I-driven minigenome system for Ebola viruses. J. Virol. 79:4425-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79:13473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. U. S. A. 102:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, K., K. Murakami, S. S. Hmwe, B. Zhang, J. Li, M. Shirakura, K. Morikawa, R. Suzuki, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Trans-encapsidation of hepatitis C virus subgenomic replicon RNA with viral structure proteins. Biochem. Biophys. Res. Commun. 371:446-450. [DOI] [PubMed] [Google Scholar]
- 18.Jirasko, V., R. Montserret, N. Appel, A. Janvier, L. Eustachi, C. Brohm, E. Steinmann, T. Pietschmann, F. Penin, and R. Bartenschlager. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546-28562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 43:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, T., T. Matsumura, T. Heller, S. Saito, R. K. Sapp, K. Murthy, T. Wakita, and T. J. Liang. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 81:4405-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., P. Choudhury, C. M. Cabral, and R. N. Sifers. 1999. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J. Biol. Chem. 274:5861-5867. [DOI] [PubMed] [Google Scholar]
- 25.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meusser, B., C. Hirsch, E. Jarosch, and T. Sommer. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7:766-772. [DOI] [PubMed] [Google Scholar]
- 28.Neumann, G., and Y. Kawaoka. 2001. Reverse genetics of influenza virus. Virology 287:243-250. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, Y., K. Sugiura, K. Kato, Y. Tohya, and H. Akashi. 2007. Rescue of Akabane virus (family Bunyaviridae) entirely from cloned cDNAs by using RNA polymerase I. J. Gen. Virol. 88:3385-3390. [DOI] [PubMed] [Google Scholar]
- 32.Pacini, L., R. Graziani, L. Bartholomew, R. De Francesco, and G. Paonessa. 2009. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J. Virol. 83:9079-9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 362:2095-2100. [DOI] [PubMed] [Google Scholar]
- 34.Raychaudhuri, S., V. Fontanes, B. Barat, and A. Dasgupta. 2009. Activation of rRNA transcription by hepatitis C virus involves upstream binding factor phosphorylation via induction of cyclin D1. Cancer Res. 69:2057-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruddock, L. W., and M. Molinari. 2006. N-glycan processing in ER quality control. J. Cell Sci. 119:4373-4380. [DOI] [PubMed] [Google Scholar]
- 36.Russell, R. S., J. C. Meunier, S. Takikawa, K. Faulk, R. E. Engle, J. Bukh, R. H. Purcell, and S. U. Emerson. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:4370-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeff, L. B., and J. H. Hoofnagle. 2003. Appendix: National Institutes of Health Consensus Development Conference Management of Hepatitis C 2002. Clin. Liver Dis. 7:261-287. [DOI] [PubMed] [Google Scholar]
- 38.Seeff, L. B., and J. H. Hoofnagle. 2002. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology 36:S1-S2. [DOI] [PubMed] [Google Scholar]
- 39.Sekine-Osajima, Y., N. Sakamoto, K. Mishima, M. Nakagawa, Y. Itsui, M. Tasaka, Y. Nishimura-Sakurai, C. H. Chen, T. Kanai, K. Tsuchiya, T. Wakita, N. Enomoto, and M. Watanabe. 2008. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology 371:71-85. [DOI] [PubMed] [Google Scholar]
- 40.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinmann, E., C. Brohm, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 82:7034-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinmann, E., F. Penin, S. Kallis, A. H. Patel, R. Bartenschlager, and T. Pietschmann. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinmann, E., T. Whitfield, S. Kallis, R. A. Dwek, N. Zitzmann, T. Pietschmann, and R. Bartenschlager. 2007. Antiviral effects of amantadine and iminosugar derivatives against hepatitis C virus. Hepatology 46:330-338. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama, K., K. Suzuki, T. Nakazawa, K. Funami, T. Hishiki, K. Ogawa, S. Saito, K. W. Shimotohno, T. Suzuki, Y. Shimizu, R. Tobita, M. Hijikata, H. Takaku, and K. Shimotohno. 2009. Genetic analysis of hepatitis C virus with defective genome and its infectivity in vitro. J. Virol. 83:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, T., K. Ishii, H. Aizaki, and T. Wakita. 2007. Hepatitis C viral life cycle. Adv. Drug Deliv. Rev. 59:1200-1212. [DOI] [PubMed] [Google Scholar]
- 47.Tani, H., Y. Komoda, E. Matsuo, K. Suzuki, I. Hamamoto, T. Yamashita, K. Moriishi, K. Fujiyama, T. Kanto, N. Hayashi, A. Owsianka, A. H. Patel, M. A. Whitt, and Y. Matsuura. 2007. Replication-competent recombinant vesicular stomatitis virus encoding hepatitis C virus envelope proteins. J. Virol. 81:8601-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timpe, J. M., Z. Stamataki, A. Jennings, K. Hu, M. J. Farquhar, H. J. Harris, A. Schwarz, I. Desombere, G. L. Roels, P. Balfe, and J. A. McKeating. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17-24. [DOI] [PubMed] [Google Scholar]
- 49.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witteveldt, J., M. J. Evans, J. Bitzegeio, G. Koutsoudakis, A. M. Owsianka, A. G. Angus, Z. Y. Keck, S. K. Foung, T. Pietschmann, C. M. Rice, and A. H. Patel. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 52.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]