Abstract
The hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) has been proposed to change conformations in association with RNA synthesis and to interact with cellular proteins. In vitro, the RdRp can initiate de novo from the ends of single-stranded RNA or extend a primed RNA template. The interactions between the Δ1 loop and thumb domain in NS5B are required for de novo initiation, although it is unclear whether these interactions are within an NS5B monomer or are part of a higher-order NS5B oligomeric complex. This work seeks to address how polymerase conformation and/or oligomerization affects de novo initiation. We have shown that an increasing enzyme concentration increases de novo initiation by the genotype 1b and 2a RdRps while primer extension reactions are not affected or inhibited under similar conditions. Initiation-defective mutants of the HCV polymerase can increase de novo initiation by the wild-type (WT) polymerase. GTP was also found to stimulate de novo initiation. Our results support a model in which the de novo initiation-competent conformation of the RdRp is stimulated by oligomeric contacts between individual subunits. Using electron microscopy and single-molecule reconstruction, we attempted to visualize the low-resolution conformations of a dimer of a de novo initiation-competent HCV RdRp.
Polymerases undergo a series of conformational changes at different stages of nucleic acid synthesis (14). Of the template-dependent polymerases, the RNA-dependent RNA polymerases (RdRps) are the least understood in terms of their mechanism of action. RdRps are of increasing interest since cellular RdRps play important roles in the defense against nonself RNAs (44). In addition, virus-encoded RdRps are important targets for the development of antivirals. A better understanding of RNA-dependent RNA polymerases is thus important for both basic and applied science.
Several model systems for biochemical study of viral RNA-dependent RNA synthesis exist (4, 19, 20, 25, 37, 42). Well-characterized RdRps include those from the hepatitis C virus (HCV) and poliovirus (5, 17). In the host, the RdRps are complexed with other viral and/or cellular proteins that are usually associated with membranous intracellular structures. The replicases are usually difficult to study biochemically, but the catalytic RdRp subunits of several viruses can be purified for functional and structural analyses (53). These recombinant proteins can reproduce some of the activities of the replicases, including the ability to initiate RNA synthesis by a de novo mechanism (22, 47-49). Furthermore, recombinant RdRps can affect the activities of other replicase subunits in vitro, suggesting that the recombinant RdRp is useful for an in-depth understanding of RNA synthesis by HCV (45, 60).
RdRps form a right-hand-like structure with thumb, finger, and palm subdomains. The metal-coordinating residues important for nucleotide binding are positioned within the palm subdomain (26). An interesting feature of viral RdRps is that they tend to exist in a closed conformation, even in the absence of template, in contrast to DNA-dependent RNA polymerases, which transition from open to closed complexes upon template recognition (13). The closed form of the phage φ6 RdRp has been proposed to allow specific recognition of the single-stranded viral RNA (7). The template channel formed by the closed structure, however, is too narrow to accommodate the partially duplexed RNA that forms during RNA synthesis, and hence, the closed conformation needs to undergo significant rearrangements in the ternary complex. Biswal et al. (3) have captured an X-ray crystallographic structure of a partially open conformation of the HCV RdRp. Bovine viral diarrhea virus (BVDV) RdRp was also shown to exist in a partially open conformation (11). Ranjith-Kumar and Kao (49) demonstrated that the HCV RdRp could initiate RNA synthesis from a circular RNA template, and thus, the threading of a single-stranded RNA into the template channel is not required for de novo-initiated RNA synthesis. Altogether, these results raise the possibility that the HCV RdRp can undergo rearrangements from the closed conformation seen in the crystal structure prior to de novo initiation.
A secondary structure that extends from the finger to the thumb subdomains, named the Δ1 loop, has been proposed to serve as a gate to cover the template channel and regulate the switch from de novo initiation to elongation (5, 10). Mutations that affect the interaction between the Δ1 loop and the mostly hydrophobic residues that it contacts have resulted in polymerases that are defective for de novo initiation but can bind to partially duplexed RNA and can extend from the 3′ terminus of an RNA primer (10).
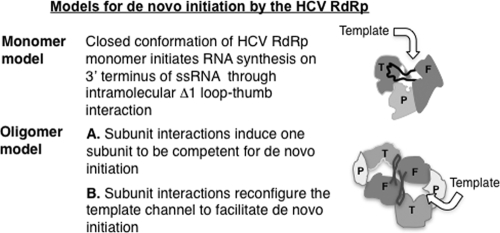
Two general models for RNA synthesis by the HCV RdRp can be proposed (Fig. 1). The first posits that the HCV RdRp functions as a monomer at least during de novo initiation because the closed template channel is needed for specific recognition of the template (5, 7, 10). It was presumed that the Δ1 loop and thumb domain interaction in the HCV RdRp is stable and mutations that disrupted this interaction would render the enzyme catalytically inactive (5, 24). However, a deletion of five residues in the tip of the Δ1 loop did not prevent RNA synthesis from a primed template by the polymerase (10). Furthermore, a genotype 2a RdRp was crystallized in a form with altered interaction between the Δ1 loop and thumb domain in comparison to the 1b RdRp (3). Interestingly, a low-affinity GTP binding site exists on the thumb domain close to the base of the Δ1 loop binding pocket. GTP binding at this site has been proposed to stabilize the Δ1 loop and thumb domain interactions, favoring the closed monomer model (6). A second model is based on the reports that HCV RdRp can oligomerize and that oligomerization increases its activity (12, 16, 46, 54). The dimer could be active due to either the second subunit increasing the stability of the Δ1 loop and thumb interactions in the first subunit to increase de novo initiation or the two subunits forming a common template-binding domain (Fig. 1). Here we have attempted to determine whether monomers or oligomers of the HCV RdRp can better perform de novo initiation using biochemical and biophysical analyses.
FIG. 1.
Models for RNA synthesis by the HCV RdRp. The monomer model is based on the central tenet that intramolecular interactions within an RdRp molecule regulate the modes of RNA synthesis. The curved arrow represents the possible orientation of the template RNA. The oligomer model is an adaptation from the dimer model of the norovirus RdRp (18). T, P, and F represent the thumb, palm, and finger domains, respectively, in different shades of gray, and the thick black line connecting the thumb and finger domains represents the Δ1 loop.
MATERIALS AND METHODS
Production of the HCV RdRp.
The prototype HCV RdRp used in the study is derived from HCV 1b isolate strain BK. The hydrophobic C-terminal tail was truncated 21 and 51 residues from the C terminus to increase solubility of the proteins. All proteins were expressed in Escherichia coli with the 6×-His tag at the C terminus of the protein. The culture was shaken at 37°C until the optical density at 600 nm (OD600) reached 0.8 to 1.0, and then the temperature was changed to 16°C and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.5 mM. After a 12- to 16-h period, the cells were pelleted at 8,000 × g for 15 min, and the pellets were frozen at −70°C until needed.
RdRp purification used cells suspended in lysis buffer (20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 10 mM imidazole, a protease inhibitor cocktail at the concentration suggested by the manufacturer [Sigma Inc.], 5 mM beta-mercaptoethanol [BME], and 0.1 mg/ml lysozyme). The suspension was sonicated for 2 min and clarified by centrifugation at 16,000 × g for 30 min. The supernatant was loaded onto nickel-nitrilotriacetic acid (Ni-NTA) columns equilibrated with 20 mM Tris, pH 7.5, 300 mM NaCl, 10% glycerol, 10 mM imidazole, and 5 mM BME. The columns were washed with the same buffer containing 20 mM imidazole, followed by elution of the bound proteins with lysis buffer containing 500 mM imidazole. Δ51 and some preparations of Δ21 along with the I432V mutant (used in differential scanning fluorimetry [DSF] assays) were further purified on a poly(U) agarose column as described previously for Δ21 (10).
Δ21 purified from the Ni-NTA column was dialyzed into buffer consisting of 20 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, and 10 mM BME using a desalting column and then loaded onto a MonoS column (HR 100/10; GE Healthcare) which was preequilibrated with desalting buffer. Δ21 was eluted with a gradient of 100 to 600 mM NaCl in desalting buffer. The proteins generated from these two columns were highly pure and did not contain other bands in Coomassie blue-stained gels.
A version of Δ21 that could have the C-terminal histidine tag removed was engineered with a TEV protease cleavage site (Glu-Asn-Leu-Tyr-Phe-Gln-Gly) immediately N-terminal to the 6-His tag sequence. After initial purification through a Ni-NTA column and later on a MonoS column, the protein was exchanged into a buffer containing 100 mM NaCl and 20 mM imidazole and then incubated with the TEV protease for 48 h. The cleaved Δ21 was passed through a second Ni-NTA column, and the flowthrough was collected, concentrated, and buffer exchanged before use for biochemical analysis.
Blue native PAGE (BN-PAGE).
Approximately 250 ng of proteins were mixed with the loading dye supplied by the manufacturer (Invitrogen Inc.) and electrophoresed in 3 to 12% native PAGE gels (Invitrogen Inc.) with a Coomassie blue-containing cathode buffer at 4°C for 2 h according to the manufacturer's instructions. The gel was stained with silver using a kit and protocols from the manufacturer (Invitrogen Inc.).
HCV RdRp assays.
LE19 is a template typically used to measure several RNA synthesis activities in one reaction (47): de novo initiation, primer extension, template switch, and nontemplated nucleotide addition. Standard RdRp reaction mixtures contained 50 nM RNA, 0.1 mM ATP and UTP, 33 nM (α-32P)CTP and 0.01 mM GTP (unless stated otherwise), and the amount of NS5B as specified. The assays also contained 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 20 mM NaCl, and 1 mM MnCl2. The reactions were incubated for 1 h at 25°C before extracting the products with a 1:1 mixture of phenol-chloroform and precipitated with ethanol in the presence of 0.3 M NaOAc. The RNA was electrophoresed on 20% urea denaturing polyacrylamide gels, and the gels were wrapped in plastic and exposed to phosphor screens prior to scanning by a PhosphorImager (Molecular Dynamics).
DSF.
Differential scanning fluorimetry (DSF) was carried out in a Stratagene MX3005P PCR machine with filter sets for excitation at 492 nm and emission at 610 nm. Each sample of 25 μl contained a 2 μM final concentration of protein in 300 mM Tris (pH 7.5), 80 mM NaCl, and SYPRO orange (Molecular Probes) at a 2.5× final concentration (the actual concentration cannot be revealed by Molecular Probes). The samples were heated at a rate of 0.5°C/min, from 25 to 75°C, and the MxPro software program (Stratagene) was used to calculate the Tmapp by plotting the negative derivative of the observed fluorescence change as a function of temperature.
DLS analysis.
Dynamic light scatter (DLS) measurements were made with Zetasizer-NanoS instrument (Malvern Instruments, United Kingdom) in a buffer containing 300 mM Tris (pH 7.5) and a 1 μM final protein concentration. The buffer components were filtered through a 0.2-μm-pore filter (Nalgene) to avoid suspended particles. The measurements were made at 10-s intervals, with the data points collected after a 1-s scan. At least three measurements were made per sample, and the average intensity for the corresponding peaks was plotted.
Gel filtration analysis.
Analytical gel filtration used a Superdex-200 HR column (GE Healthcare) equilibrated with the running buffer (50 mM Tris [pH 7.5], 100 mM KCl, 5% glycerol, and 1 mM BME) at a flow rate of 0.25 ml/min. About 100 μg of the respective proteins were either mixed with 20 mM GTP or not and were injected into the 24-ml Superdex 200 column before the run. This high concentration of GTP was used to maintain the NS5B:GTP ratio used in the RdRp assays. One-ml fractions were collected, and the fractions obtained were analyzed by SDS-PAGE. The column was calibrated with molecular mass standards (Bio-Rad Inc.): bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.35 kDa).
Electron microscopy and image reconstruction.
The protein in a buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 5% glycerol, and 1 mM BMEΕ was adjusted to ∼5 ng/μl, adsorbed to freshly glow-discharged carbon-coated copper grids (EMS), and stained with uranyl acetate (1% [wt/vol] aqueous solution). Electron microscopy was performed using a transmission electron microscope operated at 80 kV (Jeol 1010). Dimeric particles of the HCV RdRp were selected using the BOXER routine in the EMAN software program (29), filtered, and centered. Class averages were generated with these filtered and centered particles without imposed symmetry. Centered particles (about 4,000) were subjected to analysis using the Multirefine software program to sort the particles into an initial model. A molecular mass of 130 kDa was used for the surface-rendering threshold of the three-dimensional (3D) structures of the dimers. 3D reconstructions were visualized using the UCSF Chimera software package (43).
RESULTS
Analysis of Δ21 conformations and oligomerization.
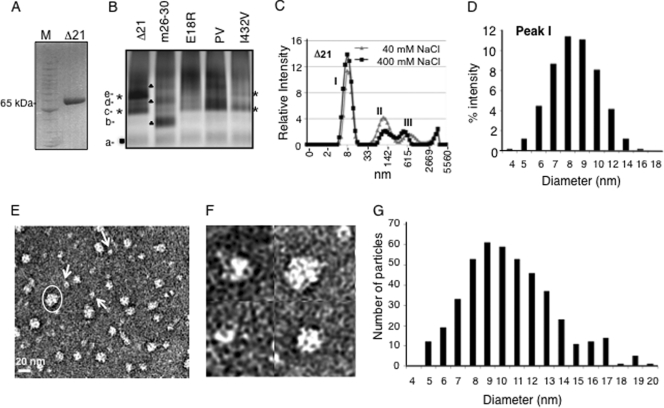
Blue-native gel electrophoresis (BN-PAGE) was used to examine the oligomeric and/or conformational state of the HCV Δ21 protein. The proteins were coated with Coomassie blue and separated in an acrylamide gel based on a combination of sizes, shapes, and/or oligomeric states (33, 57). We examined Δ21 (Fig. 2A) along with several mutant derivatives that are affected for oligomerization (E18R mutant [46]), de novo initiation (m26-30 and I432V mutants [10, 27]), or in the GTP-binding allosteric site (P495/V499A mutant, named PV [6]) (Fig. 2B). While Δ21 migrated at the expected molecular mass of 65 kDa in SDS-PAGE, it existed as multiple bands of different intensities in a BN gel (Fig. 2B). m26-30, with a five-amino-acid deletion in the Δ1 loop which renders it defective for de novo initiation but not primer extension (10), had a different banding pattern. The E18R and PV mutants deviated significantly from the banding pattern of Δ21 and exhibited some poorly resolved high-molecular-weight complexes (Fig. 2B). The I432V mutant had bands in a position similar to that of Δ21. The variety of bands exhibited by Δ21 and mutant RdRps in the BN-PAGE suggests that each of the RdRps exists in complex arrays of oligomeric states and/or conformations.
FIG. 2.
Analysis of the conformations and oligomeric states of the HCV RdRp. (A) SDS-PAGE demonstrating the mobility of the Δ21 protein at ∼65 kDa under denaturing conditions. The molecular mass markers are from Invitrogen (Benchmark protein ladder). (B) Blue-native gel of Δ21 and its variants that are affected for RNA synthesis. The different bands that were distinct in mobility and intensity are labeled from a to e. Bands that were common to all proteins are indicated by a filled box; bands common to the Δ21 and I432V proteins are shown with asterisks; bands in m26-30 are shown with filled triangles. (C) DLS analysis of Δ21 at 40 mM and 400 mM NaCl. (D) Polydisperse nature of the peak I from panel C. (E) A representative EM image of 5 ng/μl Δ21 in a Tris buffer containing 100 mM NaCl. The particles were stained with uranyl acetate after adsorption onto carbon grids. The scale bar is shown at bottom left. The arrows point to the likely monomers based on the estimated sizes of the particles. Note that some monomeric particles may represent different orientations attached to the grid, hence appearing in different shapes. One of the oligomers is indicated with an oval. (F) Magnified view of micrographs showing putative Δ21 monomers and oligomers. (G) A histogram of the dimensions of 440 particles of Δ21 picked from different micrographs. The measurements were made manually using the image processing software ImageJ (NCBI) at a 2.35-Å/pixel ratio at the specimen level. Two measurements were taken each at the longest and widest dimensions of each particle at roughly perpendicular angles, and the averages of the two measurements are plotted as histograms.
Dynamic light scatter spectroscopy (DLS) was used to measure the hydrodynamic diameter of Δ21 to estimate the dimensions of the protein. In a Tris buffer containing 40 mM NaCl, Δ21 existed in three peaks (Fig. 2C). The first has a mean diameter of more than 8 nm and represents the majority of the protein based on volume distribution of the peak. This hydrodynamic diameter was higher than expected based on the X-ray structure of the monomeric HCV RdRp, indicating polydispersity (25%). The other two peaks are likely to be high-molecular-weight oligomers. Higher salt concentrations slightly increased the abundance of peak I and altered the abundance and positions of the other two peaks in DLS (Fig. 2C). This is consistent with the idea that the three peaks are in equilibrium in a manner that can be affected by the monovalent salt concentration, as reported previously (12). Finally, Δ21 eluted as three distinct peaks in a Superdex-200 gel filtration column when in a buffer containing 100 mM monovalent salt. However, it showed a significantly delayed elution pattern, with the most abundant peak eluting at a mass smaller than 65 kDa, compared with molecular mass markers (see below). Similar observations were made by Gu et al. (16).
Next, we visualized Δ21 by electron microscopy after staining with uranyl acetate. Δ21 existed in particles of different sizes that are likely monomers (∼6-nm average diameter), dimers, and higher-order complexes, with the longest diameters being up to 20 nm (Fig. 2E and F). In several independent experiments, we did not observe particles whose average diameters were larger than 20 nm. The diameters of ∼450 Δ21 particles that could be selected from randomly picked sections of five micrographs are plotted in Fig. 2G. The distribution of Δ21 particles on electron microscopy (EM) grids matched with the polydisperse nature of peak 1 observed in DLS (compare Fig. 2D and G). We have previously reported that Δ21 is primarily a monomer in solution based on results from analytical centrifugation (10), while others have shown using the same technique that the Δ21 apoenzyme is a monomer that can form dimers and tetramers in the presence of an RNA that can direct de novo initiation (24). The reason for the discrepancy is unclear.
RNA synthesis as a function of enzyme concentration.
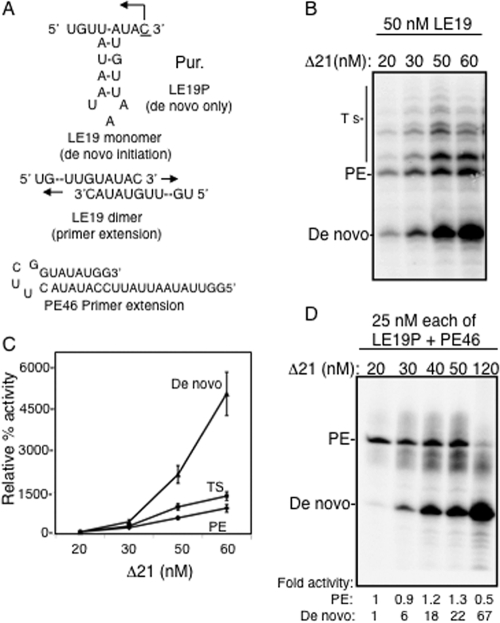
Since Δ21 can be present as an oligomer, we wanted to examine whether oligomerization can affect a particular mode of RNA synthesis. For this analysis, we used a template, LE19, that can report on both de novo initiation (producing a 19-mer RNA) or primer extension products (producing a 32-mer RNA, Fig. 3A). The reaction mixtures had a 10 μM concentration of the initiating nucleotide GTP, 50 nM template RNA, and from 20 to 60 nM Δ21. Over this concentration range, we observed a geometric increase in de novo initiation but a linear increase in primer extension and template switch products (Fig. 3B and C). This suggested that conditions that favor higher-order RdRp complexes favored de novo initiation.
FIG. 3.
Higher enzyme concentrations selectively stimulated de novo initiation by Δ21. (A) The templates used in the analysis. Template LE19 exists as a monomer and directs the synthesis of a 19-nucleotide (nt) de novo initiation product from Δ21, while it can also form a dimer with another LE19 molecule and directs the synthesis of a primer extension product of 32 nt. The capital P represents a puromycin covalently attached to the 3′ terminus of the RNA LE19P. PE46 is a stem-loop structure that can direct a primer extension product of 46 nt. (B) A representative gel image of the RdRp assay described using different concentrations of Δ21 with a constant concentration of 50 nM LE19 and 0.1 mM ATP, UTP, 0.01 mM GTP, and 33 nM (α-32P)CTP. PE and Ts denote primer extension and template switch, respectively. (C) Quantification of products of RdRp reaction. The 19-nt product is quantified as a de novo initiation product, the 32-nt product as a primer extension (PE) product, and all other higher-molecular-weight products as template switch (indicated as Ts in the left of the image). The error bars represent standard deviations from three reactions. The respective activities at 20 nM were normalized to 100%, and the relative increase at different concentrations is plotted. (D) A gel image of the products of the RdRp assay carried out as in panel B but with the templates LE19P and PE46. PE, primer extension. Each of the RNAs was at a concentration of 25 nM. The de novo and primer extension activitiess at different enzyme concentrations were normalized and compared to that of a 20 nM enzyme concentration and are indicated below respective lanes.
To confirm the results without the complex template switch products, we used LE19P, which can direct de novo initiation but not primer extension due to a puromycin being attached to the 3′-most hydroxyl of the template. A second RNA, named PE46, which can only direct primer extension, was also present to allow comparative analysis of the two modes of RNA synthesis (Fig. 3A). Both RNAs were at a 25 nM final concentration. Again, Δ21 concentrations from 20 to 120 nM yielded a geometric increase in de novo-initiated RNAs, while the primer extension product was inhibited at the highest concentration tested (Fig. 3C). These results show that the mode of RNA synthesis by Δ21 is dependent on the Δ21 concentration. The divalent cation manganese stimulated de novo initiation in all of these reactions, while primer extension was inhibited at higher enzyme concentrations in a divalent metal-independent manner (not shown). This is consistent with our previous findings that manganese has a selective effect on de novo initiation (47).
De novo initiation-defective mutant proteins can stimulate de novo initiation by Δ21.
To examine the relationship between oligomerization and de novo-initiated RNA synthesis, we needed a mutant RdRp that was defective for de novo initiation but could retain interaction with Δ21. The protein m26-30, which had been previously characterized (10), met these criteria and was selected for this activity.
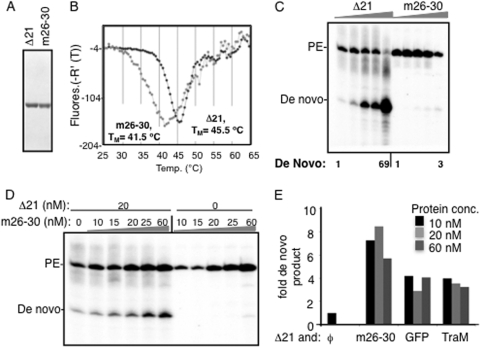
First, we examined whether highly purified m26-30 (Fig. 4A) was altered in conformation/oligomerization compared to Δ21 using the differential scanning fluorimetry (DSF) assay. The assay uses the dye SYPRO orange, which fluoresces when bound to hydrophobic regions of the protein, to report on its denaturation profile (38). A clear difference was observed between the melting profiles of Δ21 and m26-30 (Fig. 4C and D). Δ21 had a relatively symmetric melting curve with a maximum Tmapp of 45.5°C; m26-30 had a more complex melting profile and a maximum Tmapp of 41.5°C.
FIG. 4.
Intermolecular interactions in the RdRp can stimulate de novo initiation. (A) Image of SDS-PAGE of the Δ21 and m26-30 proteins used in these studies. m26-30 has a five-amino-acid deletion at the tip of the Δ1 loop (10). (B) DSF analysis of 2 μM Δ21 or m26-30. The fluorescence increase due to binding of SYPRO orange dye to hydrophobic patches in the protein is plotted as a negative derivative on the y axis. The plots were generated from the software package MxPro supplied with the real-time PCR machine (Stratagene). The minimal point on the y axis for each plot is considered the Tmapp. (C) Products of RNA synthesis assays as shown by Δ21 and m26-30 using an equal mix of the LE19P and PE46 templates. The primer extension (PE) and de novo-initiated products are marked to the left of the gel image. (D) Effects of m26-30 on de novo initiation by 20 nM Δ21. This concentration of Δ21 was selected since de novo initiation products are at a low level. The addition of m26-30, which is debilitated for de novo initiation but could retain protein-protein interaction, was able to stimulate de novo initiation by Δ21. (E) Quantification of the increase in de novo initiation by 20 nM Δ21 in the presence of m26-30 or two unrelated proteins. Δ21 and the second protein were mixed and incubated on ice for 30 min before the RNA synthesis reaction was initiated as described in Materials and Methods. The unrelated proteins were added to test for the effects of molecular crowding.
We next compared RNA synthesis by Δ21 and m26-30 over a range of protein concentrations. While de novo initiation increased by 69-fold over the range of concentrations tested, m26-30 produced only a small increase in de novo-initiated products over the same concentration range tested (Fig. 4C). Next, Δ21 at a constant 20 nM (a concentration that produces only a faint amount of de novo-initiated products) was preincubated with increasing amounts of m26-30 before the enzyme mix was used in RdRp assays. m26-30 was found to cause a concentration-dependent increase in the de novo products when it was present with Δ21 (Fig. 4D). These results suggest that the intermolecular interaction between m26-30 and Δ21 led to increased de novo initiation by Δ21 or by m26-30. Due to the deletion in m26-30, we consider rescue of de novo initiation in m26-30 to be less likely.
To test whether the interaction is specific or could be due to a molecular crowding effect, recombinant green fluorescent protein (GFP) and E. coli TraM proteins were added to Δ21 at the same concentrations as m26-30. Both had a more modest effect on de novo initiation by Δ21 than m26-30 assayed in the same reaction (Fig. 4E). These results suggest that while molecular crowding did contribute to higher de novo initiation by Δ21, m26-30 had a larger effect.
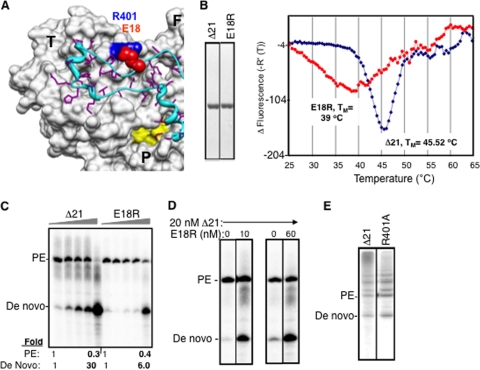
We wanted an additional de novo initiation-defective mutant to confirm that intermolecular interactions could stimulate de novo initiation by Δ21. The E18 residue was a good candidate, since it was previously claimed to be involved in a homomeric pair with H502 (46). E18 may also be needed for Δ1 loop and thumb interactions, since it formed a salt bridge with R401 in the thumb domain (Fig. 5A). More than 200 sequences from the 1b HCV strains revealed that E18 and R401 (or E18-K401) were highly conserved (data not shown). When the purified E18R mutant protein was subjected to the DSF assay, it was found to exhibit a broader denaturation peak and also a lower Tmapp than Δ21 (Fig. 5B). When tested for RNA synthesis with LE19P and PE46 templates, the E18R mutant was also defective for de novo initiation but not primer extension, except at very high concentrations of the protein (Fig. 5C). When it was coincubated with a 20 nM Δ21 protein, increasing de novo initiation was observed (Fig. 5D), thus representing a second set of interactions where the presence of an initiation-compromised derivative and Δ21 could increase de novo initiation, presumably by wild-type Δ21. An R401A mutant was not adversely affected for de novo initiation or primer extension activity from the LE19 template, confirming our previous results involving mutations in the template channel (10) (Fig. 5E). These results suggest that the ionic interaction between R401 and E18 is not required during RNA synthesis by the polymerase.
FIG. 5.
The E18R mutant can stimulate de novo initiation by Δ21. (A) A surface and ribbon representation of the X-ray crystal structure of HCV RdRp (PDB identifier 1QUV) to highlight the location of E18 (red spheres) and its interaction with R401 (blue spheres). The complete thumb domain (T), a part of the palm (P) domain, and a part of the finger (F) domain are marked. The active site metal coordinating residues in the active site are in yellow. The Δ1 loop that connects the fingers and thumb domain is in ribbon representation and is colored cyan, while the side chains of its residues are in purple. (B) SDS-PAGE of proteins used in the reactions and their behavior in a DSF reaction. (C) Representative image of the products of the RdRp assay with templates LE19P and PE46 with increasing concentrations of the Δ21 or E18R protein. The ratio of increase in the de novo-initiated product has been quantified below the gel image. (D) Effects of amending an RNA synthesis where Δ21 was kept constant at 20 nM while the E18R protein was at 10 or 60 nM. The two enzymes were preincubated on ice for 30 min before the RNAs were added. (E) R401A mutation does not affect wild-type activity of NS5B. The template LE19 was used in this assay, and the gel image of a representative reaction is shown, indicating the 19-nt de novo product and the 32-nt primer extension product.
Effect of GTP on conformation and de novo initiation by Δ21.
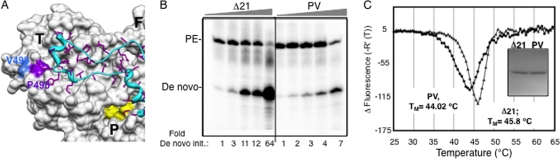
GTP can stimulate de novo initiation while inhibiting primer extension (28, 48). However, it is not clear if GTP acts at the initiation site (NTPi site [21]), the allosteric site on the thumb domain, or both (6) (Fig. 6A). The low-affinity GTP-binding pocket is at the base where the Δ1 loop interacts with the thumb domain, suggesting a relationship with the conformations needed for de novo initiation (Fig. 6A). To examine whether the allosteric site has an effect on de novo initiation, we tested a double mutant, the P495/V499A mutant (referred to as PV), for RNA synthesis using LE9P and PE46 as templates. Both Δ21 and PV can extend from PE46 to similar levels, but the PV mutant synthesized only a fraction of the de novo-initiated product made by Δ21 while producing more of the primer-extended product (Fig. 6B).
FIG. 6.
The allosteric GTP binding site and de novo initiation of RNA synthesis. (A) The location of the low-affinity GTP binding site on the surface of the thumb domain of HCV RdRp (PDB identifier 1QUV) as shown by Bressanelli et al. (6). T, F, and P, thumb, finger, and palm domains, respectively. The Δ1 loop is in cyan, and the residues P495 and V499, which were shown to bind GTP on the thumb domain, are shown in different colors. The side chains of S29 and R32, which are located in the Δ1 loop, are not highlighted, while the surfaces of P495 and V499 are in purple and blue, respectively, as indicated. Residue R503, which is part of the putative allosteric pocket, is also not highlighted. (B) Gel image of the RdRp reaction products using LE19P and PE46 with increasing protein concentrations. The two proteins were at 20, 30, 40, 50, and 120 nM in the assays. The amounts of de novo-initiated product at different enzyme concentrations were normalized and compared to that of a 20 nM concentration of either the PV mutant or Δ21, as indicated below respective lanes. PE, primer extension. (C) DSF analysis of Δ21 and PV mutant as for Fig. 4D to examine conformational differences between the two proteins. The inset shows SDS-PAGE of the two proteins used.
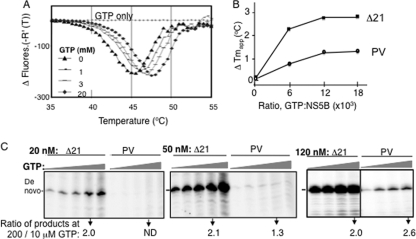
The DSF assay (38) was used to examine whether PV exists in an altered conformation relative to Δ21 in the absence of GTP. PV has a broader melting curve than Δ21, and the Tmapp of PV was also 2°C lower than that of Δ21 (Fig. 7A). Next, the effect of increasing GTP concentrations on the Tmapp of Δ21 and the PV mutant was determined. GTP increased the thermal stability of Δ21 by ∼2.5°C in a concentration-dependent manner (Fig. 7A and B). A similar increase in stability was observed with UTP (not shown). These results likely represent the combination of the effects at both the allosteric site and the NTPi site. In contrast, the PV protein had a reduced response to either GTP or UTP at all concentrations tested (Fig. 7B and data not shown). Given that the PV mutant has a defect in the low-affinity site, the response to GTP likely represents an effect associated with GTP binding at the NTPi site.
FIG. 7.
GTP can stabilize Δ21 conformation and increase de novo initiation. (A) Tmapp of a 2 μM concentration of Δ21 in the absence or presence of increasing concentrations of GTP. The Tmapp was 45.5, 46.05, 47.02, and 47.52°C at 0, 1, 3, and 20 mM GTP, respectively. (B) Increase in Tmapp of PV mutant in comparison to Δ21 at increasing GTP concentrations as indicated in the x axis. The derivative of the Tmapp at different GTP concentrations is plotted. (C) Gel image of the products of RNA synthesis by Δ21 and the PV mutant at three enzyme concentrations along with increasing GTP concentrations. LE19P was the template, and GTP was included at 0.01, 0.05, 0.1, 0.2, and 0.5 mM, while ATP and UTP were at 0.1 mM along with 33 nM (α-32P)CTP. A gel image of the products of the RdRp assay of Δ21 and the PV mutant at a 120 nM concentration with increasing GTP concentrations (0.01, 0.05, 0.1, and 0.2 mM GTP, as represented by the gray triangles), is shown. The fold increases in de novo initiation activity for different concentrations of Δ21 and the PV mutant when GTP was increased from 0.01 mM to 0.2 mM are shown at the bottom of the gel image.
Last, we examined whether increasing GTP concentrations can rescue de novo initiation by the PV mutant (Fig. 7C). Given that the PV mutant was significantly reduced in de novo initiation relative to Δ21, we measured the amount of stimulation at a range of GTP concentrations and looked at the ratio of products at 200 versus 10 μM GTP. At 20 nM Δ21 or PV, the ratio of de novo initiation products synthesized at the two GTP concentrations was 2 for Δ21, but no products were made by the PV mutant at either GTP concentration. At 120 nM, both the Δ21 and PV proteins had an increase of de novo-initiated product as a function of the GTP concentration (Fig. 7C). Altogether, these results suggest that the allosteric GTP binding site is required for optimal de novo initiation and that some rescue of de novo-initiated RNA synthesis by the PV mutant can take place under conditions that favor intermolecular interactions.
The C terminus of NS5B does not affect intermolecular interactions.
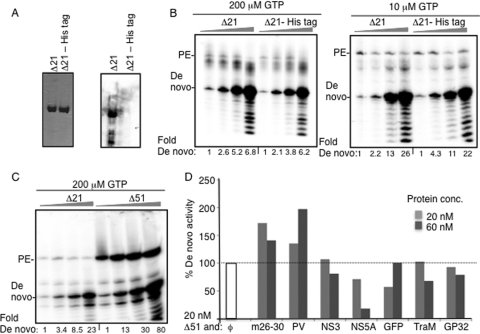
The Δ21 protein used in the present study contained a C-terminal histidine tag to aid in purification. To rule out the effect of the tag on the ability of NS5B to oligomerize, we expressed and purified a Δ21 protein with a TEV protease cleavage site immediately N-terminal to the histidine tag and subjected it to TEV protease treatment. The TEV protease-treated protein was passed through a Ni-NTA column, and the protein flowthrough was used for analysis. Western blot analysis using a monoclonal antibody specific to the histidine tag confirmed its removal (Fig. 8A). The resulting protein was functional for the production of both the de novo-initiated and primer extension products (Fig. 8B). Furthermore, a nonlinear increase in the amount of de novo-initiated product was evident as a function of the protein concentration. The protein lacking the histidine tag also had a Tmapp nearly identical to that of the protein with the tag when examined by DSF (data not shown). These results demonstrate that the C-terminal His tag did not affect de novo initiation by Δ21.
FIG. 8.
The C-terminal His tag in Δ21 does not affect de novo initiation. (A) SDS-PAGE showing the Δ21 protein with and without the His tag used in RNA synthesis assays. The gel image on the left was stained with Coomassie blue, and the one to the right was a comparable set of proteins transferred onto a nitrocellulose membrane probed in a Western blot with monoclonal antibody (MAb) specific for the histidine tag (Santa Cruz Biotechnology). (B) Representative gel images of an RdRp reaction with 20, 40, 60, and 120 nM (shown by the flattened triangle) of Δ21 or the Δ21-His tag protein (Δ21 minus the 6-His tag) at two GTP concentrations. The products (19 nt) were quantified, the intensity at 20 nM was set as 1, and the relative increase in intensities in shown below each lane. (C) Representative gel images of an RdRp reaction with 20, 40, 60, and 120 nM (shown by the flattened triangle) of the Δ21 and Δ51 enzymes. (D) Quantification of the effect of a second protein on the amount of de novo initiation products made by 20 nM Δ51. Δ51 was incubated with the respective amounts of each protein on ice for 30 min before the mix was added in RdRp assays as described in Materials and Methods. The amount of de novo-initiated 19-nt product made in the absence of a second protein (white bar) was normalized to 100%. The amounts of the 19-nt product produced in the presence of a 20 or 60 nM concentration of the other proteins are shown as gray bars.
To examine further whether the C-terminal tail contributed to de novo initiation resulting from intermolecular interactions, we used a version of NS5B that lacked the C-terminal 51 residues. The Δ51 enzyme was more active in both primer extension and de novo initiation than the Δ21 enzyme; however, a nonlinear increase in de novo initiation activity was observed as a function of higher enzyme concentrations (Fig. 8C) even with this enzyme. These results confirm that the C-terminal portion of the HCV NS5B protein does not affect the stimulation of de novo initiation associated with higher enzyme concentrations. Interestingly, the inhibition of primer extension was not seen at higher Δ51 concentrations (Fig. 8C), perhaps due to a larger template channel that can accommodate double-stranded RNA better.
The specificity of intermolecular interactions between Δ51 subunits was examined by keeping Δ51 at a constant 20 nM concentration while adding a 1 or 3 molar excess of several other proteins. The de novo initiation-defective m26-30 and PV stimulated de novo activity by Δ51 when complexed with Δ51. The other proteins (HCV NS3, NS5A [a gift from Craig Cameron], and proteins unrelated to HCV) either had no effect or had an inhibitory effect on de novo initiation by Δ51. The inhibitory effects could be due to binding to RNA or effects on nucleoside triphosphates (NTPs). These results are consistent with those seen with Δ21 (Fig. 4E) and further imply that specific interactions between NS5B subunits contribute to a stimulation of de novo initiation activity.
Comparison of activities of Δ21 from genotypes 1b and 2a.
The HCV genotype 2a JFH1 strain can establish cell culture infections, and its subgenomic replicon shows faster replication kinetics in cell culture than the replicon from genotype 1b (2). NS5B from strain JFH1 was recently demonstrated to be more competent for de novo initiation than primer extension due to polymorphisms in the thumb domain that make it a more closed structure than NS5B from the 1b HCV genotype (data not shown) (51). We examined whether de novo initiation by JFH1 Δ21 was affected by conditions that favored oligomerization. Consistent with the reports of Simister et al. (51), we observed that the 2a Δ21 protein produced more de novo initiation products than the primer extension reaction (data not shown). However, consistent with the effects of 1b Δ21, increasing the 2a Δ21 concentration resulted in a stimulation of de novo-initiated RNA synthesis without a comparable effect on primer extension.
Reconstruction of RdRp dimers.
All of the characterizations so far show that there are complex interactions and conformational changes associated with de novo initiation by the HCV RdRps. The dynamic nature of the RdRp will make it a challenge to define all of the changes necessary for RdRp function, especially by X-ray crystallography. However, 3D reconstruction from negatively stained particles obtained by electron microscopy (EM) can inform us about major changes in protein conformation. This approach has already been used to define conformations of larger molecules, such as ribosomes, GroEL, and innate immune receptors (30, 36, 52). With the immune receptors, we have determined that ligands can induce dramatically different conformations, including oigomerization (36, 52).
To capture the de novo initiation-competent structure, we collected more than 3,000 individually selected dimers/oligomers of Δ21 from negatively stained electron micrographs. However, the final models generated did not yield a good match for the class averages of the data set, and the Fourier transform over the eight rounds of refinement showed that convergence was suboptimal (data not shown). Therefore, we analyzed several other mutant RdRps and found that the I432V mutant was a good choice for reconstruction.
The I432V substitution was originally identified in a mutant HCV replicon that was resistant to cyclosporin A and subsequently was shown to be better at de novo initiation than the wild-type (WT) Δ21 protein, especially at higher enzyme concentrations (27) (Fig. 9A and B). Gel filtration chromatography profiles of the two proteins showed that the elution profile of the I432V mutant lacked a second peak seen with the WT Δ21 protein in the absence of GTP (Fig. 9C, see asterisks) while the differences were negligible when the elution was performed with GTP (Fig. 9C). In the DSF assay, noticeable differences in the thermostabilities of the Δ21 and I432V proteins were also observed as a function of increasing GTP concentrations (Fig. 9D). Finally, the dimensions of the I432V particles were more homogenous than that for Δ21, especially measured in the absence of GTP (compare Fig. 9E and F). Together these results supported our contention that the Δ21 enzyme exists in a heterogeneous state and further suggest that the I432V protein has a lesser degree of heterogeneity. Given that the I432V mutant was competent for de novo initiation and less heterogenous than Δ21, it was selected for single-particle reconstruction.
FIG. 9.
Selection of I432V for reconstruction of dimeric RdRp molecules. (A) The location of I432 in the thumb domain of NS5B. I432 is colored black, and the Δ1 loop is shown as a ribbon structure. The thumb and palm domains are labeled T and P, respectively. (B) Effects of increasing concentrations of the I432V, Δ21, and E18R proteins on the production of de novo initiation products. The assays were performed with LE19P and PE46 as described in the legend to Fig. 3D. The amount of the 19-nt de novo-initiated product was normalized to that at 20 nM for each of the three enzymes. The I432V protein produced more de novo initiation products than the other two enzymes at 120 nM compared to results at 20 nM enzyme. (C) The elution profiles of the Δ21 and I432V proteins from Superdex-200 columns. About 100 μg of each protein in the presence or absence of 20 mM GTP was injected into the column before gel filtration was performed. The calibration showed that a 44-kDa chicken ovalbumin protein elutes at the 15.1-ml position. The fractions collected were subjected to SDS-PAGE and then stained with Coomassie blue for visualization. The asterisk denotes peak fractions that contain complexes which are more heterogeneous in the Δ21 sample than in the I432V sample. (D) The effects of GTP on the Tmapp of the Δ21 and I432V proteins. The ratios of the GTP to protein are plotted against the change in Tmapp. (E) The distribution of dimensions for Δ21 with and without GTP. Δ21 (20 nM) without or with 2 mM GTP was spread on carbon-coated grids and then stained with uranyl acetate. The samples were imaged and sorted into class averages as described in Materials and Methods. The lengths of the longest (dimension 1) and shortest (dimension 2) dimensions for each of the subclasses are plotted, as well as the number of particles in each subclass. (F) The distribution of dimensions for the I432V mutant with and without GTP.
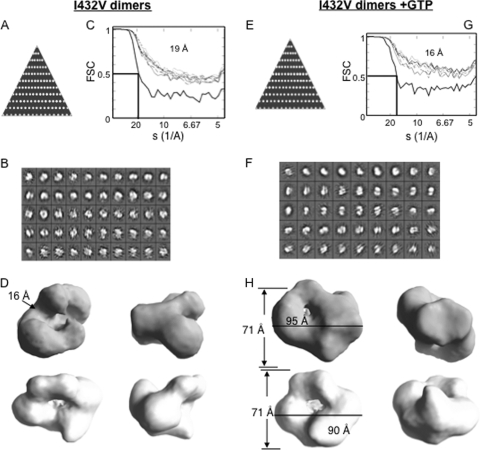
More than 4,000 dimer-like particles of the I432V mutant without GTP or with GTP were picked and sorted into class averages. The particles selected had a good coverage of the potential orientations, as determined by their distributions in the asymmetry triangle (Fig. 10A and E). The class averages generated are shown in Fig. 10B and F. Several initial models were selected and used for reconstructions without symmetry and with C2 symmetry, and the resulting models were congruous, but the model generated with C2 symmetry had higher resolution. They are shown in Fig. 10D. The final model for the complex was at 19-Å resolution without GTP and at 16 Å with GTP. The I432V molecule has the density set to the mass of 130 kDa (the mass of a Δ21 dimer) and reveals a central channel with a width of 16 Å that could accommodate RNA (Fig. 10D). The results are consistent with the model that two interacting RdRp subunits could form one template channel. However, the individual monomers were difficult to delineate in the models due to a possible combination of extensive interactions between the monomers and/or a low resolution. Interestingly, in the presence of GTP, the densities over the central channel closed and the central channel became more constricted (Fig. 10H), showing that a conformational change can reconfigure the central channel in the dimeric RdRp.
FIG. 10.
Single-particle analysis and reconstruction of I432V dimers in the presence or absence of GTP. (A) Asymmetry triangle showing distribution of orientation of the particles that constitute the model for I432V dimers. (B) Class averages as generated for the I432V dimer complex with no presumed symmetry using the EMAN program's refine2d command. (C) Fourier shell correlation (FSC) showing the convergence of the final model for the I432V protein to 19 Å as per the 0.5 σ criterion. (D) Different orientations of the final model for I432V dimers. The model measures 71 Å by 95 Å with a groove of about 16 Å. (E) Asymmetry triangle that shows the particle orientation for the final model of I432V dimers in the presence of GTP. (F) Class averages for I432V dimers complexed with GTP with no presumed symmetry (see Materials and Methods). (G) Fourier shell correlation for I432V dimers with GTP showing that the model has converged to a 16-Å resolution. (H) Different views of the final model for I432V dimers plus GTP complex. The model has dimensions of 71 Å by 90 Å.
DISCUSSION
Many viral RdRps need to initiate genomic RNA synthesis de novo from the termini of their RNA genomes in order to avoid the loss of genomic information. This mechanism is fundamentally different from that of DNA-dependent polymerases and may ensure the specificity of synthesis from viral templates. In this report, we show that factors such as GTP and interactions between intermolecular subunits of a higher-order complex can modulate the conformations of the HCV RdRp and influence the mode of RNA synthesis.
The conformational changes in viral RdRps are just starting to be defined. However, it is important to put this in the context of RNA-dependent RNA synthesis. RNA synthesis by an RdRp in vitro can be divided into discrete steps: (i) formation of productive initiation complexes (de novo initiation or primer extension complexes), (ii) formation of the first or first few phosphodiester bonds, (iii) elongation of the stable primer along the template, and (iv) termination. The first two stages are rate limiting and are collectively called initiation events. The Kms for NTPs for initiation nucleotides are significantly lower for elongation than for initiation (15, 47), consistent with the idea of a conformational change in the RdRp between initiation and elongation in RNA synthesis.
The template channel in Δ21 seems to be a region whose dimensions can change depending on the enzyme concentration and the presence of NTPs. This will have a direct effect on the initiation events in RNA synthesis. A recent report has shown that the degree of “closedness” of the enzyme depends on the amino acid composition in the thumb domain (51). This implies that a weak Δ1 loop and thumb domain interaction will result in inefficient de novo initiation (51) but efficient primer extension, at least from short RNA templates (10). In this work, we propose that the interactions between RdRp subunits can promote the de novo initiation in a manner that involves the Δ1 loop and the thumb domain.
Given that de novo initiation by Δ21 and Δ51 can be induced by the presence of a de novo initiation-defective HCV RdRp (Fig. 4D and 8D), the results suggest that the role of a second HCV RdRp subunit is to influence the conformation of the de novo-initiating subunit in the complex. Preliminary reconstruction of the de novo initiation-competent dimer of the I432V mutant also suggests that the two subunits may form a shared template channel that can be induced to close into separate entities in the monomers upon the addition of GTP (Fig. 10D and G). The changes in the template channel in the presence of GTP suggest that GTP binding at either the active site or the allosteric pocket can induce another significant conformational change in the complex that may activate the complex for template binding and RNA synthesis. A number of intermediate structures of the HCV RdRp thus remain to be better characterized in addition to the initially closed structure of the HCV RdRp monomer.
Our results supporting a dimer/oligomer of HCV RdRp being the active form for RNA synthesis (Fig. 1) are consistent with the results and interpretations of Qin et al. (46) and Wang et al. (54). Wang et al. (54) showed the presence of two broad interacting interfaces in the thumb region of one molecule and the finger region of another molecule. However, Cramer et al. (12) reported that a monomeric RdRp is active. This difference may be a matter of the sensitivity in detecting the de novo-initiated products, since we show that the HCV RdRp could exist in an array of conformations and/or oligomeric states, even at the nanomolar enzyme concentrations used in this study. Our results confirm and extend the results from the two reports of Qin et al. and Wang et al. (46, 54). Contact between the oligomerized polymerase subunits has been reported to increase the cooperativity in RNA synthesis in poliovirus 3Dpol (31, 40) and more recently for the Norovirus RdRp (18). Another proposal is that the interface contacts between poliovirus 3D polymerase monomers (thumb interface) and the poliovirus 3C protease could affect RNA synthesis through regulating protein priming (41). While the mechanism needs to be examined further for each species of RdRp, a higher-order structure of viral RdRps as a functional unit in RNA synthesis may be a shared property of the positive-strand RNA viruses (1, 45, 60). It should also be emphasized that the entire complex of replication machinery for viral RNA synthesis will likely take the form of highly ordered complexes that will involve specific targeting of viral and cellular proteins to specific sites on intracellular membranes (31, 34, 58). In this context, the association of the polymerase would also be facilitated by the cellular membrane environment.
The Δ1 loop and its interaction with the thumb domain could be interacting as part of an interface between two RdRp subunits. This would help to explain how mutations in the Δ1 loop, such as m26-30, and even the PV mutants were either incapable of or showed only a linear increase in de novo initiation activity with increasing enzyme concentrations. The residues in HCV NS5B identified to be critical for de novo initiation activity in this study, such as E18, L26-L30, and P495/V499, have all been documented to be needed for HCV replication in cells, underlying their biological significance (8, 10, 32). The interdomain interactions may also be needed for productive RNA binding when longer RNA templates are used. This is consistent with previous reports showing a cooperative effect of NS5B oligomerization on RNA synthesis using longer primer extension templates (54). Whether the elongation of RNA synthesis will require oligomeric contacts between subunits remains to be explored. However, our results indicate that in vitro, lower concentrations of RdRp (where the enzyme is not in excess of template concentrations) are capable of elongative RNA synthesis (Fig. 3).
The host-encoded prolyl isomerases, cyclophilins, could influence the rate of de novo initiation and, by extension, the level of HCV RNA replication through changes in RdRp conformation (50, 55). I432V, which confers cyclosporin A resistance to HCV (27, 50), is also more competent for de novo initiation at higher enzyme concentrations than at lower enzyme concentrations. This suggests that cyclophilins are likely to modulate conformational changes and possibly the oligomerized states of the RdRp. Chatterji et al. (9) have recently documented that the active site of cyclophylin A is required for HCV replication. NS5B phosphorylation in cells was reported to be needed for HCV replication (23), and phosphorylation may serve to “remodel” the oligomeric complexes of NS5B to assist in template loading.
Genotypic differences in HCV isolates could also impact the equilibrium between the different conformation/oligomeric states of the HCV RdRp and affect the rate of de novo initiation (56). The results reported by Simister et al. (51) demonstrating that the 2a RdRp is more active for de novo initiation are consistent with this model, although it should be considered that the increase in de novo initiation by the 2a RdRp could be by favoring a higher-order conformation rather than a more closed conformation.
Finally, it is interesting to note a difference between viruses that use a polyprotein processing mechanism (including HCV) and viruses that use different RNAs to express the replication proteins. The former class of viruses should, theoretically, have more comparable stoichiometries of all the viral proteins. In contrast, viruses that use distinct RNAs to express the replication proteins could have dramatically reduced expression of the RdRp without affecting the overall level of RNA synthesis (39, 59). These two strategies will certainly allow selection for additional activities of the RdRp (such as binding of the retinoblastoma protein [35]), as well as distinct ways to deal with the host's innate immunity mechanisms.
Acknowledgments
We thank C. T. Ranjith-Kumar and the Kao lab members for helpful discussions, Barry Stein for assistance in EM, and especially Laura Kao for editing the manuscript.
This work was funded by National Institute of Allergy and Infectious Diseases grant 1RO1AI073335 to C.C.K.
Footnotes
Published ahead of print on 7 April 2010.
REFERENCES
- 1.Ahlquist, P., S. X. Wu, P. Kaesberg, C. C. Kao, R. Quadt, W. DeJong, and R. Hershberger. 1994. Protein-protein interactions and glycerophospholipids in bromovirus and nodavirus RNA replication. Arch. Virol. Suppl. 9:135-145. [DOI] [PubMed] [Google Scholar]
- 2.Binder, M., D. Quinkert, O. Bochkarova, R. Klein, N. Kezmic, R. Barthenschlager, and V. Lohmann. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 81:5270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. James. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 4.Blumenthal, T., and G. G. Carmichael. 1979. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48:525-548. [DOI] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 8.Cai, Z., M. Yi, C. Zhang, and G. Luo. 2005. Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 79:11607-11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterji, U., M. Bobardt, S. Selvarajah, F. Yang, H. Tang, N. Sakamoto, G. Vuagniaux, T. Parkinson, and P. Gallay. 2009. The isomerase active site is critical for hepatitis C virus replication. J. Biol. Chem. 284(25):16998-17005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnaswamy, S., I. Yarbrough, S. Palaninathan, C. T. Kumar, V. Vijayaraghavan, B. Demeler, S. M. Lemon, J. C. Sacchettini, and C. C. Kao. 2008. A locking mechanism regulates RNA synthesis and host protein interaction by the hepatitis C virus polymerase. J. Biol. Chem. 283:20535-20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, K. H., J. M. Groarke, D. C. Young, R. J. Kuhn, J. L. Smith, D. C. Pevear, and M. G. Rossmann. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. U. S. A. 30:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramer, J., J. Jaeger, and T. Restle. 2006. Biochemical and pre-steady-state kinetic characterization of the hepatitis C virus RNA polymerase (NS5BΔ21, HC-J4). Biochemistry 45:3610-3619. [DOI] [PubMed] [Google Scholar]
- 13.deHaseth, P. L., M. L. Zupancic, and M. T. Record. 1998. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 180:3019-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doublie, S., M. R. Sawaya, and T. Ellenberger. 1999. An open and closed case for all polymerases. Structure 7:R31-R35. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari, E., Z. He, R. E. Palermo, and H. C. Huang. 2008. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J. Biol. Chem. 283:33893-33901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu, B., L. L. Gutshall, D. Maley, C. M. Pruss, T. T. Nguyen, C. L. Silverman, J. Lin-Goerke, S. Khandekar, C. Liu, A. E. Baker, D. J. Casper, and R. T. Sarisky. 2004. Mapping cooperative activity of the hepatitis C virus RNA-dependent RNA polymerase using genotype 1a-1b chimeras. Biochem. Biophys. Res. Commun. 313:343-350. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 18.Högbom, M., K. Jäger, I. Robel, T. Unge, and J. Rohayem. 2009. The active form of the norovirus RNA-dependent RNA polymerase is a homodimer with cooperative activity. J. Gen. Virol. 90:281-291. [DOI] [PubMed] [Google Scholar]
- 19.Honda, A., K. Mizumoto, and A. Ishihama. 1986. RNA polymerase of influenza virus. Dinucleotide-primed initiation of transcription at specific positions on viral RNA. J. Biol. Chem. 261:5987-5991. [PubMed] [Google Scholar]
- 20.Kao, C., and J. H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M.-J., and C. C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, S. J., J. H. Kim, Y. G. Kim, H. S. Lim, and J. W. Oh. 2004. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J. Biol. Chem. 279:50031-50041. [DOI] [PubMed] [Google Scholar]
- 24.Labonte, P., V. Axelrod, A. Agarwal, A. Aulabaugh, A. Amin, and P. Mak. 2002. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J. Biol. Chem. 277:38838-38846. [DOI] [PubMed] [Google Scholar]
- 25.Laurila, M. R., E. V. Makeyev, and D. H. Bamford. 2002. Bacteriophage phi 6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277:17117-17124. [DOI] [PubMed] [Google Scholar]
- 26.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Z., J. M. Robida, S. Chinnaswamy, G. Yi, J. M. Robotham, H. B. Nelson, A. Irsigler, C. C. Kao, and H. Tang. 2009. Mutations in the hepatitis C virus polymerase that increase RNA binding can confer resistance to cyclosporine A. Hepatology 50:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., H. Overton, and R. Barthenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 16:10807-10815. [DOI] [PubMed] [Google Scholar]
- 29.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 30.Ludtke, S. J., J. Jakana, J. L. Song, D. T. Chuang, and W. Chiu. 2001. A 11.5 A single particle reconstruction of GroEL using EMAN. J. Mol. Biol. 314:253-262. [DOI] [PubMed] [Google Scholar]
- 31.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 32.Ma, Y., T. Shimakami, H. Luo, N. Hayashi, and S. Murakami. 2004. Mutational analysis of the hepatitis C virus NS5B in the subgenomic replicon cell culture. J. Biol. Chem. 279:25474-25482. [DOI] [PubMed] [Google Scholar]
- 33.Monie, T. P., H. Hernandez, C. V. Robinson, P. Simpson, S. Matthews, and S. Curry. 2005. The polypyrimidine tract binding protein is a monomer. RNA 11:1803-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 35.Munakata, T., M. Nakamura, Y. Liang, K. Li, and S. M. Lemon. 2005. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102:18159-18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murali, A., X. Li, C. T. Ranjith-Kumar, K. Bhardwaj, A. Holzenburg, P. Li, and C. C. Kao. 2008. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J. Biol. Chem. 283:15825-15833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 26:279-288. [DOI] [PubMed] [Google Scholar]
- 38.Niesen, F. H., H. Berglund, and M. Vedadi. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2:2212-2221. [DOI] [PubMed] [Google Scholar]
- 39.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 41.Pathak, H. B., S. K. Ghosh, A. W. Roberts, S. D. Sharma, J. D. Yoder, J. J. Arnold, D. W. Gohara, D. J. Barton, A. V. Paul, and C. E. Cameron. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). A surface of the primary oligomerization domain functions in capsid precursor processing and VPg uridylylation. J. Biol. Chem. 277:31551-31562. [DOI] [PubMed] [Google Scholar]
- 42.Paul, A., J. H. van Boom, D. Fillippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 44.Pikaard, C. S., J. R. Haag, T. Ream, and A. T. Wierzbicki. 2008. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 13:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piccininni, S., A. Varaklioti, M. Nardelli, B. Dave, K. D. Raney, and J. E. McCarthy. 2002. Modulation of the hepatitis C virus RNA-dependent RNA polymerase activity by the non-structural (NS) 3 helicase and the NS4B membrane protein. J. Biol. Chem. 277:45670-45679. [DOI] [PubMed] [Google Scholar]
- 46.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 47.Ranjith-Kumar, C. T., Y. C. Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2002. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ranjith-Kumar, C.-T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 49.Ranjith-Kumar, C. T., and C. C. Kao. 2006. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robida, J. M., H. B. Nelson, Z. Liu, and H. Tang. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simister, P., M. Schmitt, M. Geitmann, O. Wicht, U. H. Danielson, R. Klein, S. Bressanelli, and V. Lohmann. 2009. Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J. Virol. 83:11926-11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun, J., K. E. Duffy, C. T. Ranjith-Kumar, J. Xiong, R. J. Lamb, J. Santos, H. Masarapu, M. Cunningham, A. Holzenburg, R. T. Sarisky, M. L. Mbow, and C. C. Kao. 2006. Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J. Biol. Chem. 281:11144-11151. [DOI] [PubMed] [Google Scholar]
- 53.van Dijk, A. A., E. V. Makeyev, and D. H. Bamford. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85:1077-1093. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 56.Weng, L., J. Du, J. Zhou, J. Ding, T. Wakita, M. Kohara, and T. Toyoda. 2009. Modification of hepatitis C virus 1b RNA polymerase to make a highly active JFH1-type polymerase by mutation of the thumb domain. Arch. Virol. 154:765-773. [DOI] [PubMed] [Google Scholar]
- 57.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 58.Wolk, B., B. Buchele, D. Moradpour, and C. M. Rice. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519-10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, G., E. Letteny, C.-H. Kim, and C. C. Kao. 2009. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA 15:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, C., Z. Cai, Y. C. Kim, R. Kumar, F. Yuan, P. Y. Shi, C. Kao, and G. Luo. 2005. Stimulation of hepatitis C virus (HCV) nonstructural protein 3 (NS3) helicase activity by the NS3 protease domain and by HCV RNA-dependent RNA polymerase. J. Virol. 79:8687-8697. [DOI] [PMC free article] [PubMed] [Google Scholar]